Abstract
Phylogeography documents the spatial distribution of genetic lineages that result from demographic processes, such as population expansion, population contraction, and gene movement, shaped by climate fluctuations and the physical landscape. Because most phylogeographic studies have used neutral markers, the role of selection may have been undervalued. In this paper, we contend that plants provide a useful evolutionary lesson about the impact of selection on spatial patterns of neutral genetic variation, when the environment affects which individuals can colonize new sites, and on adaptive genetic variation, when environmental heterogeneity creates divergence at specific loci underlying local adaptation. Specifically, we discuss five characteristics found in plants that intensify the impact of selection: sessile growth form, high reproductive output, leptokurtic dispersal, isolation by environment, and the potential to evolve longevity. Collectively, these traits exacerbate the impact of environment on movement between populations and local selection pressures—both of which influence phylogeographic structure. We illustrate how these unique traits shape these processes with case studies of the California endemic oak, Quercus lobata, and the western North American lichen, Ramalina menziesii. Obviously, the lessons we learn from plant traits are not unique to plants, but they highlight the need for future animal, plant, and microbe studies to incorporate its impact. Modern tools that generate genome-wide sequence data are now allowing us to decipher how evolutionary processes affect the spatial distribution of different kinds of genes and also to better model future spatial distribution of species in response to climate change.
Keywords: California Floristic Province, comparative phylogeography, comparative phylogeogeography, glaciation, local adaptation, migration
Phylogeographic analyses document the impact of geographic boundaries, geological changes, and climatic fluctuations on evolutionary processes that drive population divergence, speciation, and the formation of communities (1–4). Early and highly cited studies of phylogeography have emphasized the impact of recent glaciation in Europe and North America and detail population expansion and migration northward (5–7). Comparison of phylogeographic data across multiple species often reveals common patterns due to similar changes in space and time associated with specific landscapes (e.g., refs. 6 and 8–16). In regions that were glaciated during the Pleistocene, the location of refugia and regional topography can shape the postglacial migration routes in similar ways across species (3, 7, 13, 16–18). The emphasis on formerly glaciated regions where many species expanded from common refugia highlighted the congruence in patterns of historical migration and demography. However, the focus on commonality has overlooked the fact that, ultimately, phylogeography is the result of the interaction between organisms and their environment. Because species respond individualistically to climate fluctuations across the physical landscape, cooccurring species often show discordant phylogeographic patterns (e.g., ref. 19). In fact, not only do species respond individualistically, but so too can populations within species (20, 21). One way to understand these responses is to incorporate life history traits into interpretations of congruence or discordance among species in their phylogeographic patterns (22).
We propose that plant species have characteristics that amplify their sensitivity to the environment and have the potential to affect phylogeography differently than observed for most animal species. Our argument is motivated by a classic article by Bradshaw (23), who proposes that important “evolutionary consequences of being a plant” are the particularly strong impacts of natural selection on plant populations. For instance, simply the fact that plants photosynthesize means that they depend on their local environment for light, water, and nutrients to survive, which creates a strong opportunity for selection and local adaptation. In addition to their dependence on the environment, they possess certain traits that enhance the influence of selection. Sessile growth form makes plants very different from animals. Once plants are established, they must survive and reproduce in that environment whereas most animals have some ability to move if the environment is not suitable. Likewise, plants have additional traits that Bradshaw argues enhance the impact of natural selection, including high reproductive output, leptokurtic dispersal, strong influence of environment on gene flow, and potential for longevity. Obviously, these traits are not exclusive to plants. For example, algae photosynthesize, corals and many marine invertebrates are sessile, and fungi exhibit high reproductive output with leptokurtic dispersal. Nonetheless, our goal here is to explore how key characteristics found in plants illustrate an evolutionary lesson about the role of natural selection in shaping migration patterns and demographic history over space and time. We are not necessarily arguing that selection is stronger in plants than animals although Bradshaw (23) argues strongly for this case. However, we do propose that specific traits found in plants and some other taxa allow selection to shape phylogeography. In the first part of the paper, we discuss the phylogeographic impact of each of these traits. In the second part, we present two case studies that illustrate how these characteristics shape phylogeography.
Phylogeographic Consequences of Being a Plant
Sessility.
A plant is stationary once established. The obvious consequence of sessility is that it is easy to document the history of movement because the genotypes of the plants provide a clear geographic record of their distribution. More importantly, this attribute means that, once a seed is dispersed, germinates, and becomes a seedling, the plant must survive and reproduce in that same location. In contrast to animals that can move in the course of a day from one environment to another to optimize environmental conditions or can move seasonally for better foraging or mating conditions, a plant must tolerate its location and eventually reproduce, or die. Thus, spatially varying selection pressures are often conspicuous across plant populations, creating local adaptation as well as strong geographic patterns of adaptive genetic variation that will be associated with environmental gradients, sometimes even over short distances (24, 25).
An early demonstration of the importance of natural selection in plants is the work of Clausen et al. (26), which showed through a series of common garden plots that plants grew best when planted near their native environments and that plants from local environments grew better in those sites than plants derived from sites from different environments. The use of common gardens and reciprocal transplants has a long tradition in plant evolutionary biology to document genetic differences (27, 28). In forestry, the use of common gardens, also known as provenance studies, has provided a foundation for forest management practices (29, 30). Through these studies, we know that phenotypic differences among natural populations have a genetic basis. Due to the emergence of next generation sequencing, it is also possible to identify adaptive variation in natural populations by identifying outlier loci that significantly correlate with environmental gradients (31–33). These loci under selection will produce a geographic pattern of adaptive genetic variation that might differ from that demonstrated by neutral markers. Thus, the sessile nature of plants may, at a minimum, better document the spatial signature of adaptive versus neutral processes, which provides a lesson on the importance of this trait for phylogeographic studies.
Most Plant Species Exhibit a Leptokurtic Pattern of Gene Flow.
An obvious difference between plants and animals is their dispersal biology. Gene flow can occur through pollen and seeds in plants (34), which means that the geographic patterns of genetic variation are created through two separate and often asymmetric processes (35–38). Plants have chloroplasts, which are haploid and nonrecombining (like mitochondrial DNA), that prove useful for documentation of migration and demographic history. Due to their uniparental inheritance, the organellar genomes can help to separate the impact of the two processes on phylogeography, which has been demonstrated in several tree species (e.g., refs. 21 and 39–41). A unique aspect of plant dispersal is that we can see whether genes are spread through pollen or seed dispersal and colonization. This separation also allows us to determine whether environmental factors, such as phenological differences that could affect differential fertilization success between populations or biotic dispersal vectors, could create individualistic responses to climatic fluctuations and physical landscapes.
A further notable characteristic of plant dispersal biology is that gene flow is often leptokurtic, with the majority of pollen and seeds dispersing locally near where the genotype is likely to be most well adapted (23). At one extreme, plants with inbred mating systems and limited seed dispersal may have highly restricted gene flow and a high degree of genetic structure that will be enhanced by selection (42). The phylogeography of these species might be similar to animals with restricted dispersal and small population sizes. However, at the other extreme, outcrossing species with leptokurtic dispersal kernels will exhibit both local and long-distance dispersal (36, 43). In this regard, plants might show different phylogeographic patterns than animals. In plants with long-distance dispersal, selection will favor individuals who can survive in the new environment, which will then create divergence from the source population for loci under selection. However, for neutral genetic markers, long-distance dispersal will maintain high connectivity among populations (36). For example, in the Mediterranean regions of Europe, the tree and shrub species with the most genetically divergent populations were those with low seed dispersal abilities. In contrast, the genetically most diverse populations were located at intermediate latitudes, where divergent lineages from separate refugia immigrated and established (18). If animals move long distances, we do not expect strong population divergence at adaptive genetic loci because they are likely to select suitable habitats. Of course, if animals disperse and do not maintain ongoing gene exchange, population divergence will emerge. Such a scenario is illustrated in a study of 22 species of Californian amphibians and reptiles and 75 phylogeographic lineages across those species (11), which identified congruent lineage breaks for multiple species across the Central Valley, San Francisco Bay, the Sierra Nevada, and the Tehachapi and Trinity ranges of California. Such results suggest that dispersal and divergence across species are similarly affected by the landscape. In contrast, the lesson from plants is that they often have a distribution of dispersal distances and that the interaction between dispersal and selection for local adaptation can promote divergence at smaller scales for adaptive genetic variation and less divergence among neutral markers.
High Reproductive Output.
Most plant species have high fecundity and much higher offspring-to-parent ratios than animal species. This high reproductive output is reminiscent of the high volume of male gametes produced by many organisms relative to the number fertilized; in many plant species, the high reproductive output results in thousands of seeds that can survive if they land in the right environment, but few do—resulting in strong selection. For trees, high fecundity across years reduces the impact of any single reproductive episode, which buffers year-to-year variation in seed production (44). A benefit of high reproductive output is that it mitigates the uncertainty of seed survival. When combined with the leptokurtic dispersal kernel for pollen and seeds, it enhances the potential for the evolution of local adaptation within the vast majority of locally dispersed propagules and the opportunity for long distance dispersal events to transport the seeds somewhere that an individual can survive. This life history trait would enhance the ability of plants or other sessile organisms to have large species distributions. For example, it may be a contributing factor for wide ranges of invasive weedy plant species. The lesson we learn from this trait is that high reproductive output increases the potential for wide dispersal and for selection on propagule after dispersal that enhances their ability to colonize and establish in new sites.
Environmental Influence on Gene Flow.
The combination of sessility and local adaptation can create population divergence in heterogeneous landscapes due to the joint affects of isolation by distance (IBD) and isolation by environment (IBE) (45) or isolation by adaptation (IBA) (46). An implicit assumption of phylogeography is that gene flow decreases with distance and is modified by the physical landscape. Because most plant species exhibit leptokurtic dispersal that results in spatial autocorrelation of genotypes (47, 48), isolation by distance (49, 50) will almost always be involved in plant phylogeography. With IBE, the environmental differences will also shape genetic distances through a variety of processes. Such processes could include isolation caused by differences in flowering time created by environment differences (e.g., refs. 51 and 52) or nonrandom gene flow created by dissimilar pollinator communities across environments (e.g., refs. 53 and 54). IBA increases divergence at neutral loci when migrants from one locally adapted population are unable to successfully immigrate into a population in a different environment. For example, Andrew et al. (46) report that environmental variables such as soil nitrogen and vegetative cover explained more of the variation in genetic differences than IBD or landscape resistance alone. Thus, the dependence of plants on the local environment that they remain in throughout their lifespan means that the environmental heterogeneity is highly likely to shape plant phylogeography.
In case studies reported below, we highlight how species with plant characteristics are very likely to illustrate the influence of the environment on population divergence. In the literature, we also find other examples of environmental effects on the phylogeography of plants. For example, in a study of the Sonoran desert succulent Euphorbia lomelii in Baja California, temperature variables affecting phenological synchrony of flowering significantly affected geographic patterns of genetic variation after controlling for phylogeographic history (55). In another example, we learn how microhabitat preferences may create different phylogeographic outcomes in codistributed and closely related montane sedges from the Rocky Mountains (56). In that study, through the use of next generation sequence data and species distribution models, Massatti and Knowles (56) demonstrate that the ecology of the species interacts with glaciations to produce fundamental differences in the past distributions. Thus, the strong impact of the environment on patterns of geographic differentiation found in plants highlights the role of the environment on phylogeography and the opportunity for selection that needs more attention. Of course, the phylogeography of animal species is also likely to show evidence of IBE (57), but the mechanisms for those patterns are likely to differ.
Longevity, or Plants Can Be Trees.
One of the most unique differences between plants and animals is that plants can be extremely long-lived with indeterminate growth (23). The local environment experienced by a tree throughout its life span will influence both a plant’s size and its reproduction. Even a long-lived turtle grows to adult size that is more or less constant and can move to a new environment to suit its preference. In contrast, for long-lived perennial plants, selection acts strongly on the seedling stage, and it persists to act throughout their long life spans. Longevity helps maintain effective population size and genetic variation of populations (44), which means that reproductive episodes throughout a lifespan will produce genetically diverse offspring that may experience different selective pressures in the face of environmental variability. Thus, longevity can help prevent extinction of local populations (44). In addition, trees have great potential for long distance gene flow and large effective population sizes (44). The bristlecone pine (Pinus longaeva) of the White Mountains in California and Nevada provides a dramatic example of the impact of longevity and large effective population size on phylogeography (58). The bristlecone pine can live over 5,000 y and once was widespread at lower elevations throughout the Great Basin during the Last Glacial Maximum. However, its current range is restricted to isolated mountaintops at the western edge of its former distribution (59). Despite the limited and fragmented distribution of bristlecone pine, its level of genetic diversity is comparable with that of other pines. Thus, we see many tree species with widespread distributions whose longevity may have prevented population contractions from resulting in complete extinction before environmental conditions would allow expansion.
Case Studies of Plant Phylogeography
Evolutionary Lessons from a California Endemic Oak, Quercus lobata.
Study region.
California is a species-rich biogeographic region that simultaneously offers a multifaceted history of geological change, complex geographical structure, and historic climatic fluctuations (9), providing a convenient laboratory for phylogeographic study. The majority of California is within the species-rich California Floristic Province (60, 61), which contains more than 4,700 native plant species, almost half of them endemic (60, 62), and is one of the world's biodiversity hot spots (9, 63–65). Many factors could have contributed to this biogeographic pattern. First, this region is a confluence of different floras that allowed a high accumulation of species (60, 61, 66). Second, long-term environmental stability of some areas has been inferred to account for substantial paleo-endemism (61, 67). Third, topographic complexity, environmental heterogeneity, and isolation may have created high speciation rates (60). Fourth, the emergence of the Mediterranean climate enhanced the diversification of lineages since the Middle Miocene when the transition toward summer dry periods began (60). However, based on an analysis of the molecular phylogenies of 16 angiosperm clades, Lancaster and Kay (62) argue that low extinction rates, not high speciation rates, account for the high biodiversity of the California Floristic Province. Not only would the lack of glaciation foster persistence, but also the topography itself may enhance survival because plants could respond to climatic fluctuations by shifting their altitudinal distributions along steep topographic gradients with a moderating effect of montane areas on precipitation patterns. In addition, the combination of topographical gradients with only moderate temperature oscillations would allow populations to expand and contract regionally and avoid extinction (68, 69). Thus, California provides an opportunity to examine phylogeographic patterns that can be distinct from the postglacial population expansion dynamics reflected by species in Europe (18), eastern North America (19, 70), and northwestern North America (8, 10, 12).
The case.
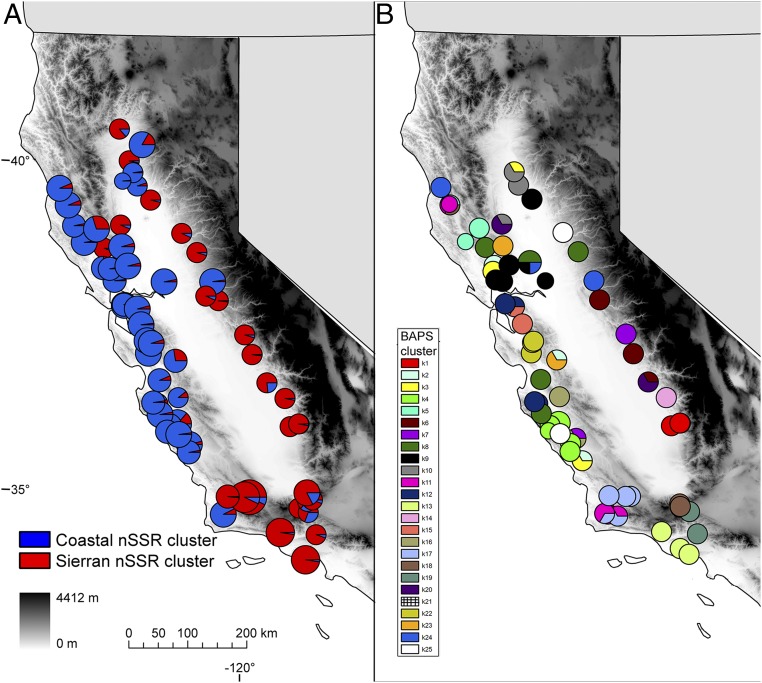
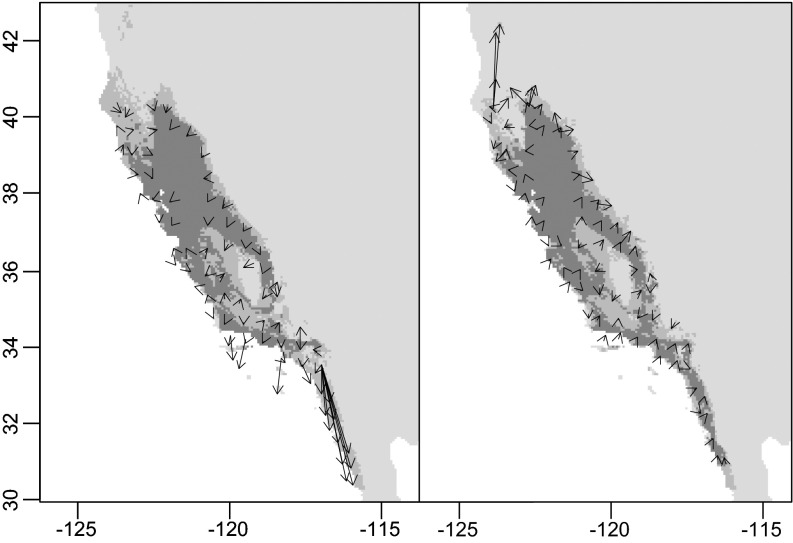
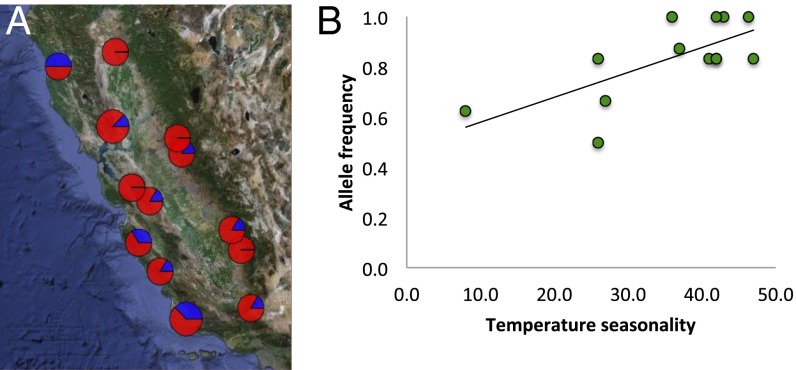
Q. lobata Née (valley oak) offers an excellent system to illustrate some of the phylogeographic lessons to be learned from plants, particularly in a region that has experienced limited glaciation. Valley oak is one of the four major endemic tree oaks of California. Its distribution along the eastern foothills of the Coast Ranges and western foothills of the Sierra Nevada surrounding the Central Valley covers a large portion of California. This distribution has remained relatively stable within California through global climate cycles (69), yet valley oak experiences diverse environments and geography across its distribution. We have analyzed the evolutionary history using several kinds of evidence (67): (i) nuclear microsatellite markers to assess the overall genetic structure of the species using STRUCTURE (71) (Fig. 1A) and environmental associations with constrained ordinations; (ii) chloroplast genetic markers, using BAPS (72) to assess patterns of colonization by seed dispersal over time (Fig. 1B); (iii) species distribution modeling to assess the potential shifts in distribution during glacial and interglacial periods (Fig. 2). In addition, we have evidence from next generation sequencing data to investigate the role of natural selection on divergence (73) (Fig. 3).
Fig. 1.
Maps of genetic structure of Q. lobata inferred from the nuclear and chloroplast microsatellite genotypes. (A) STRUCTURE analysis of nuclear microsatellites. (B) Genetic clusters using BAPS. Pie charts represent the proportion of genetic assignment to each inferred genetic cluster. Background gradient (gray scale) represents elevation (figures modified from ref. 69).
Fig. 2.
Vectors represent the local migration direction of Q. lobata during recent interglacial and glacial periods predicted by species distribution models. (A) Predicted movement from the last interglacial period (∼120 ka) to the Last Glacial Maximum (21 ka). (B) Movement from the Last Glacial Maximum to the present. Dark gray background represents areas where Q. lobata was inferred to be present in both time periods, medium gray indicates present in one time period, and background light gray is present in no time period (modified from ref. 69).
Fig. 3.
Association of an outlier SNP from a growth gene with geography and climate gradient detected through environmental analysis. (A) Map of allele frequency of a gene putatively involved in local growth adaptation. Pie charts represent proportion of each allele in each sample site. (B) Regression of temperature seasonality with allele frequency of the same growth gene (data taken from ref. 73).
Nuclear DNA phylogeography supports a role for high genetic connectivity among populations, most likely through regionally widespread pollen dispersal (Fig. 1A). In one study, a clear east–west split was observed (69), and, in others, there was a north–south subdivision (74–76). A key insight from this work is that patterns of genetic variation on the landscape are not just a product of geographic features but are also a product of climatic influences on migration patterns and local selection. Multivariate analyses suggest that climate explains at least as much genetic variation as spatial/geographic forces do (69, 76), consistent with a growing body of work in other oak species (77). In valley oak, a signature of climate impacts from both the present and the Last Glacial Maximum implies a stability of these populations in this topographically complex region that has lasting impacts. Furthermore the east–west split is associated with both geographic separation and niche differences detected through species distribution models (69), a finding corroborated in other oak species in the region (78, 79). The strong associations of overall genetic structure with climate are most likely due to isolation by environment (45).
The phylogeographic analyses of chloroplast DNA variation support the inference that valley oak’s distribution was stable and that migration was local (Fig. 1A). We observed many more haplotypes than observed in Europe, and those haplotypes were only locally distributed rather than dispersed widely across the species range as observed in regions with large impacts of glaciation (69, 80). Species distribution models based on climate data indicate that valley oak habitat has been largely stable throughout its current distribution since at least the last interglacial period over 100 ka (69). The limited distribution changes that did occur were likely local elevational shifts, which are suggested by the short distances of vectors between interglacial, glacial, and contemporary periods (Fig. 2). This finding is in stark contrast to similar niche models in an eastern North American white oak, Quercus alba, which expanded and contracted dramatically in response to climate change (69). Habitat stability and the restricted movement of valley oak in a diverse climate landscape may have promoted local adaptation.
Phylogeographic studies use neutral genetic markers to study migration and demographic history, but a growing body of research investigates geographic patterns of adaptive genetic variation. In valley oak, landscape genomic analyses to test for natural selection identify a number of candidate genes for local adaptation to climate (73, 75). What is most striking is that allele frequencies in these genes do not typically align with neutral genetic structure and instead follow other climate gradients (Fig. 3). This pattern demonstrates that adaptive alleles may readily disperse on the landscape despite population structure, such as shown for introgressed genes from invasive salamanders (81). Another interesting initial finding is that significant evidence of natural selection along climate gradients could be detected with relatively small samples sizes, suggestive of strong local adaptation to environments (75). Sork et al. (73) found that several functional genes have allele frequencies that correlate with climate gradients in a pattern different from neutral genetic structure. In a landscape genomic study of balsam fir, a gene associated with circadian rhythm showed evidence of local adaptation to temperature (82). A step forward for phylogeography will be to incorporate adaptive genetic variation into a historical demographic context to infer when adaptations arose in response to particular environmental factors (83). Nonetheless, these studies demonstrate how dispersal patterns and environmental forces come together to generate local adaptation and current patterns of phylogeography.
Returning to the five characteristics of plants, valley oak illustrates several lessons about features common to plants. The sessile nature of valley oaks leaves them vulnerable to the local environment around them. As a result, the climate environment has particularly strong influence on genetic variation at the landscape level, leading to isolation by environment and local adaptation. Having two modes of dispersal leads to a dual pattern of local genetic structure with effective regional gene flow. These modes, along with their leptokurtic distribution, permit adaptive genetic variation that enhances fitness to spread relatively freely (presumably through pollen dispersal) despite regional or local genetic structure. In the case of this tree, longevity has led to a lasting signature of past climate on present patterns of genetic variation. Long generation times may hamper the ability of trees to respond to future climate change. On the other hand, high reproductive output, effective gene flow, and large effective population sizes may enhance adaptive responses.
Evolutionary Lessons from California’s State Lichen, Ramalina menziesii.
Study region.
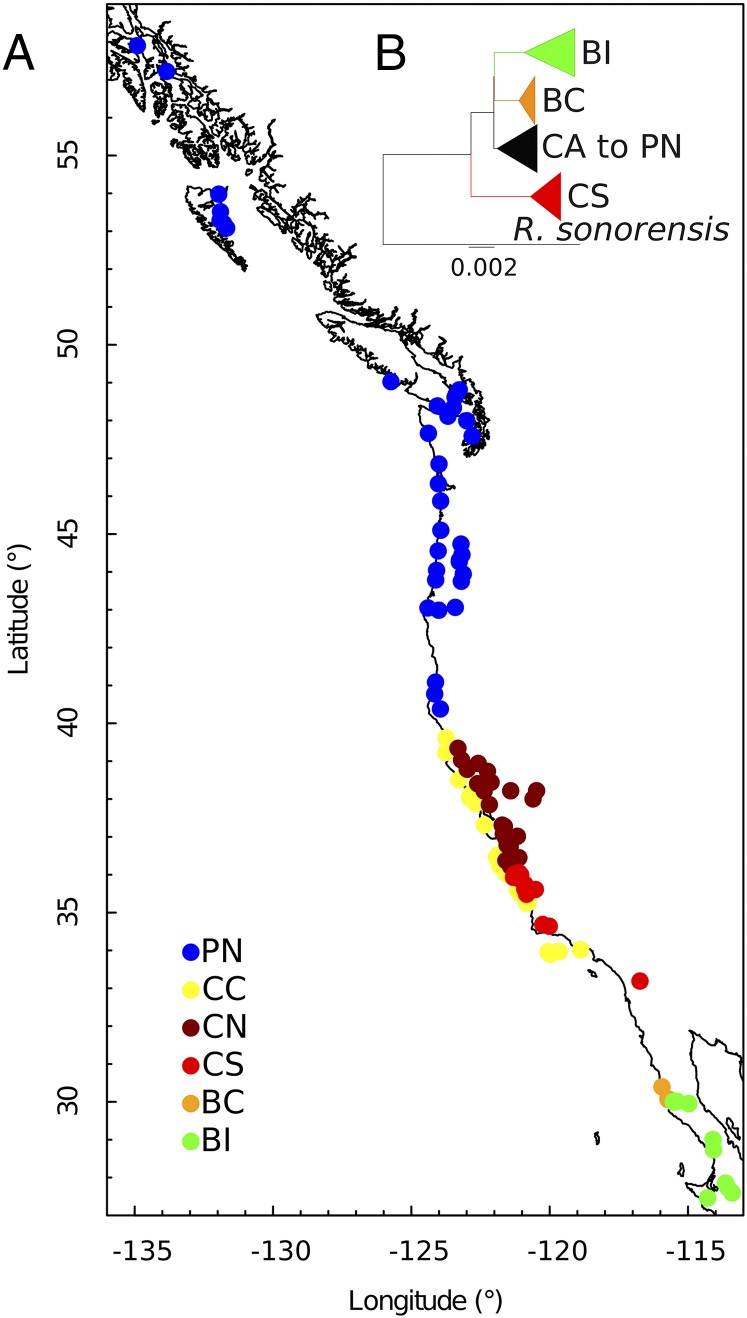
The widely distributed lace lichen R. menziesii Tayl. (Fig. 4.) covers a dramatic latitudinal range centered in California but extends south to Baja California of Mexico and north along the western coast of North America to Alaska. The lichen is found in six ecoregions, from fog desert in south ranges to the coniferous forest of the Pacific Northwest and Alaska. This unusual species distribution provides an opportunity to study phylogeographic processes across glaciated and unglaciated regions, to determine whether diversity is higher in the ecoregions within the California Floristic Province than other ecoregions (Fig. 5A). Within each ecoregion, the lichen will be found on a subset of phorophyte species (i.e., the plant on which the lichen grows). Through an analysis of the four low-copy nuclear genes (Fig. 5B), we found evidence that the center of genetic diversity and probably the origin of this lichen is the California Floristic Province (84).
Fig. 4.
Photographs of lace lichen (R. menziesii). (A) Close-up photography of lace lichen thallus (photograph taken by S.W.). (B) Deciduous valley oak (Q. lobata) heavily colonized by lace lichen.
Fig. 5.
Spatial distribution of sampling sites and clades of the lichen-forming fungus R. menziesii along western North America. (A) Sampling sites, color-coded by ecoregion. (B) Bayesian Markov Chain Monte Carlo tree generated in BEAST on the basis of four nuclear genes, with ecoregion-specific clades color-coded as indicated in the figure. AK, Alaska; BCC, Baja California coastal; BCI, Baja California interior; CA, California; CAI, California interior (based on data reported in ref. 84)
The case.
R. menziesii, which shares many features with plants, offers a unique opportunity to examine the impact of those features. One key difference among most vascular plants and lichens is that lichens do not tap a root system into a water reservoir, but depend on ambient precipitation and fog to satisfy their demands for water. Nonetheless, like plants, their needs for humidity, nutrients, and light for photosynthesis and their sessile lifestyle make them very dependent on the local environment. Lichens are created by the symbiotic relationship between a mycobiont and a photobiont. In this lichen, the haploid lichen-forming ascomycete R. menziesii associates with the haploid photosynthetic green alga Trebouxia decolorans (predominantly) (84–87) and lives for 15–20 y or longer in its lace-like growth form (Fig. 4). This symbiosis also provides an opportunity to use comparative phylogeography to understand the extent to which environmental factors have contributed to the evolutionary history of a species with similar features to many plant species, including the absence of longevity.
To understand the phylogeographic history of the lichen, it is most illustrative to investigate the fungal genome because it is the lichenized fungus that creates the intricate thallus structure of the lichen R. menziesii. Below, we discuss how dispersal and high reproductive output shape the complex history of the fungal genome and its green-algal photobiont, and we compare the phylogeographic history of the two symbiotic taxa that have experienced the same environment during evolution because of their association. The dispersal of both the lichen mycobiont and photobiont are important to the phylogeography of R. menziesii. The fungus reproduces mainly sexually with high reproductive output and is able to disperse broadly (86). To form a new lichen thallus, a germinating fungal ascospore has to associate with a compatible local algal strain through a process termed “relichenization” that involves horizontal transmission of the photobiont. The lace lichen has high dispersal capability combined with high reproductive output. In fact, it has been hypothesized for taxa with microscopic dispersal stages, such as lichens that disperse with spores, that dispersal does not limit their geographic distributions: In these cases, everything is everywhere, and “the environment selects” (88). In the case of the lace lichen, spores must land where T. decolorans (or in a small region, an alga similar to Trebouxia jamesii) is present or lichen will not establish (87). The mycobiont must be sufficiently specialized to be compatible with the photobiont’s physiology but not so specialized that it cannot relichenize with a range of green algal genotypes. For this particular lichen, high dispersal capability and high reproductive output of the microscopic spores are essential, given that the photobiont is not codispersed in these propagules.
An interesting feature of the lace lichen is that it can persist in a broad range of habitats and ecoregions, most likely due to local adaptation of the photosynthetic green alga, rather than the mycobiont. For example, in a local-scale study, the green algal populations (but not the mycobiont) showed genetic differentiation across their oak phorophytes (85, 86). In our species-wide genetic analysis of T. decolorans, we found that the abundance of strains differs among phorophyte species and that green-algal populations are differentiated according to phorophyte (87). Through a multivariate analysis of the genetic association of T. decolorans across the landscape with climate and geographic location, we found a significant influence of climate, as well as a significant effect of the species of the phorophyte where the green-alga genotype was found. These findings highlight the potential role of the environment as a selecting force shaping the phylogeographic structure of this symbiotic association by acting on the photobiont. An advantage of this system could be that the fungus associates with locally adapted algal partners (89), which may allow the lichen to have a broad distribution range.
The lace lichen study system also offers a noteworthy opportunity to study comparative phylogeography. First, we compared the genealogical trees from a subset of samples that had mycobiont and photobiont genotypes. We discovered some similarities in their clustering with certain ecoregions, but overall the phylogenetic trees were largely incongruent among symbionts, suggesting little coevolution between fungal and algal genotypes (90). Next, we compared recent migration patterns for different types of genetic markers to see whether the symbionts were showing similar patterns of movements, which one might expect given that they coexist within the same individual and may respond to climatic fluctuations in a similar way (90). The analysis revealed that the mycobiont and photobiont of lace lichen showed a general tendency to move south, with different patterns of movement across ecoregions for each of the markers (Fig. 6). The slightly similar clustering of haplotypes in the genealogical trees probably reflects the barriers imposed by differences across ecoregions. However, the overall lack of concordance between the phylogeographic patterns of these two symbiotic taxa might be explained better by dissimilar migration (90). In short, the differences in the dispersal abilities of the two taxa, and possibly the differences in their microhabitat preferences, illustrate that plant life history traits can disrupt cophylogeography of mutualistic cooccurring taxa. Overall, our comparative analyses illustrate that the contribution of both migration and local adaptation is shaping the phylogeography of the involved taxa.
Fig. 6.
Comparison of recent migration patterns of R. menziesii and T. decolorans across ecoregions as inferred from coalescent analysis for three kinds of markers: (A) fungal nuclear genes, (B) T. decolorans nuclear gene, and (C) T. decolorans chloroplast gene (maps modified from ref. 90).
The lace lichen, which shares many traits found in plants, also illustrates the evolutionary lesson that selection matters in phylogeography. Their sessile nature and their physiology make them sensitive to the local environment around them with a particularly large opportunity for selection to create locally adapted genotypes. The impact of selection is reflected in fungal and algal clades restricted to regions with unique climatic conditions (84, 87) and in photobiont strains differentiated among phorophyte species (85). Lace lichen’s widespread dispersal capability, along with its high reproductive output, and its ability to associate with locally adapted algal strains have allowed it to occupy an exceptionally wide geographic range across multiple climate zones, encompassing tropical/subtropical dry-to-humid temperate conditions. Climatic signatures are apparent in its phylogeographic pattern, with several fungal and algal clades being restricted to regions with unique climatic conditions. A unique feature of this study system is the opportunity to conduct a comparative phylogeographic analysis of the lace lichen for two organisms experiencing the exact same environments and physical barriers to their migration. The fact that they show incongruent phylogeographic structure within ecoregions and lack of comigration among ecoregions is a dramatic illustration of how selection by habitat, generation time, and dispersal abilities can create different histories and different evolutionary trajectories, even for taxa sharing the same environment.
Conclusions
The overarching evolutionary lesson from plants and these two case studies that occupy regions with high environmental heterogeneity and high species diversity is the contribution of natural selection in the response of species to climate fluctuations and the presence of major physical barriers. The geographic location of early phylogeographic studies in glaciated regions where populations have expanded and contracted from unglaciated refugia has taught us about the impact of neutral demographic patterns on species distributions. Common physical barriers have often led to concordance among multiple species that highlights shared impacts of neutral demographic processes. Increasingly, however, we are finding that cooccurring species are responding individualistically (22, 91). As studies have been expanded to regions where populations have persisted over longer time frames than the last glaciation and to regions with high environmental heterogeneity promoting ecological impacts of gene movement, we observe the effect of nonneutral processes. Selection pressures across diverse environments will enhance population divergence and often lead to discordance among species in the same region. Selection against migrants from different environments will result in divergent genome-wide genetic signatures across populations. In addition, the movement of pollen, seeds, or individuals across a landscape allows the introduction of variants at certain loci that might enhance local adaptation of the populations. The phylogeography of adaptive variation may differ from that of neutral markers. Plants may have traits that can exacerbate or simply highlight the impact of natural selection, but the phylogeographic history of all organisms will be affected by both neutral and adaptive processes. The increased availability of datasets with large numbers of loci derived from next generation sequencing will create the next phase of phylogeography that allows us to examine the history of movement of not just species but the processes that led to the shifts in species distributions. Future studies that use the new genomic tools that include neutral and adaptive genetic markers, incorporate information about adaptive and demographic responses, and include the contribution of life history traits will more accurately model species-specific shifts in species distributions due to climate change.
Acknowledgments
We thank the organizers of this symposium, Francisco Ayala and John Avise, its participants, and two anonymous referees. V.L.S. thanks Ryan Harrigan, Brad Shaffer, and Steve Weller for valuable discussion and Krista Beckley for help with the figures. We thank our collaborators on valley oak phylogeography: Frank Davis, Delphine Grivet, Maki Ikegami, Robert Westfall, and Jianli Zhao. We acknowledge the following funding sources: the National Geographic Society (V.L.S. and S.W.), the Swiss National Science Foundation (S.W.), European Commission Marie Curie Fellowship “Lichenomics” 302589 (to S.W.), the University of California, Los Angeles (P.F.G. and V.L.S.), and the Chinese Academy of Sciences (J.-M.C.).
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution X: Comparative Phylogeography,” held January 8–9, 2016, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/ILE_X_Comparative_Phylogeography.
This article is a PNAS Direct Submission.
References
- 1.Avise JC. Phylogeography: The History and Formation of Species. Harvard University Press; Cambridge, MA: 2000. [Google Scholar]
- 2.Hickerson MJ, et al. Phylogeography’s past, present, and future: 10 years after Avise, 2000. Mol Phylogenet Evol. 2010;54(1):291–301. doi: 10.1016/j.ympev.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 3.Comes HP, Kadereit JW. The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci. 1998;3(11):432–438. [Google Scholar]
- 4.Hewitt GM. Speciation, hybrid zones and phylogeography: Or seeing genes in space and time. Mol Ecol. 2001;10(3):537–549. doi: 10.1046/j.1365-294x.2001.01202.x. [DOI] [PubMed] [Google Scholar]
- 5.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405(6789):907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 6.Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc Lond. 1996;58(3):247–276. [Google Scholar]
- 7.Avise JC, et al. Intraspecific phylogeography: The mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Evol Syst. 1987;18:489–522. [Google Scholar]
- 8.Brunsfeld S, Sullivan J, Soltis D, Soltis P. Comparative phylogeography of northwestern North America: A synthesis. Special Pub-Brit Ecol Soc. 2001;14:319–340. [Google Scholar]
- 9.Calsbeek R, Thompson JN, Richardson JE. Patterns of molecular evolution and diversification in a biodiversity hotspot: The California Floristic Province. Mol Ecol. 2003;12(4):1021–1029. doi: 10.1046/j.1365-294x.2003.01794.x. [DOI] [PubMed] [Google Scholar]
- 10.Carstens BC, Brunsfeld SJ, Demboski JR, Good JM, Sullivan J. Investigating the evolutionary history of the Pacific Northwest mesic forest ecosystem: Hypothesis testing within a comparative phylogeographic framework. Evolution. 2005;59(8):1639–1652. [PubMed] [Google Scholar]
- 11.Rissler LJ, Hijmans RJ, Graham CH, Moritz C, Wake DB. Phylogeographic lineages and species comparisons in conservation analyses: A case study of california herpetofauna. Am Nat. 2006;167(5):655–666. doi: 10.1086/503332. [DOI] [PubMed] [Google Scholar]
- 12.Soltis DE, Gitzendanner MA, Strenge DD, Solits PA. Chloroplast DNA intraspecific phylogeography of plants from the Pacific Northwest of North America. Plant Syst Evol. 1997;206(1-4):353–373. [Google Scholar]
- 13.Hewitt GM. Post-glacial re-colonization of European biota. Biol J Linn Soc Lond. 1999;68(1-2):87–112. [Google Scholar]
- 14.Ralston J, Kirchman JJ. Continent-scale genetic structure in a boreal forest migrant, the Blackpoll Warbler (Setophaga striata) Auk. 2012;129(3):467–478. [Google Scholar]
- 15.Ruegg KC, Smith TB. Not as the crow flies: A historical explanation for circuitous migration in Swainson’s thrush (Catharus ustulatus) Proc Biol Sci. 2002;269(1498):1375–1381. doi: 10.1098/rspb.2002.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shafer AB, Cullingham CI, Côté SD, Coltman DW. Of glaciers and refugia: A decade of study sheds new light on the phylogeography of northwestern North America. Mol Ecol. 2010;19(21):4589–4621. doi: 10.1111/j.1365-294X.2010.04828.x. [DOI] [PubMed] [Google Scholar]
- 17.Sproul JS, et al. Climate oscillations, glacial refugia, and dispersal ability: Factors influencing the genetic structure of the least salmonfly, Pteronarcella badia (Plecoptera), in Western North America. BMC Evol Biol. 2015;15(1):279. doi: 10.1186/s12862-015-0553-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petit R, et al. Glacial refugia: Hotspots but not melting pots of genetic diversity. Science. 2003;300(5625):1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- 19.Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS. Comparative phylogeography of unglaciated eastern North America. Mol Ecol. 2006;15(14):4261–4293. doi: 10.1111/j.1365-294X.2006.03061.x. [DOI] [PubMed] [Google Scholar]
- 20.Hu FS, Hampe A, Petit RJ. Paleoecology meets genetics: Deciphering past vegetational dynamics. Front Ecol Environ. 2009;7(7):371–379. [Google Scholar]
- 21.Gugger PF, Sugita S, Cavender-Bares J. Phylogeography of Douglas-fir based on mitochondrial and chloroplast DNA sequences: Testing hypotheses from the fossil record. Mol Ecol. 2010;19(9):1877–1897. doi: 10.1111/j.1365-294X.2010.04622.x. [DOI] [PubMed] [Google Scholar]
- 22.Papadopoulou A, Knowles LL. Toward a paradigm shift in comparative phylogeography driven by trait-based hypotheses. Proc Natl Acad Sci USA. 2016;113:8018–8024. doi: 10.1073/pnas.1601069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradshaw AD. Some of the evolutionary consequences of being a plant. Evol Biol. 1972;5:25–47. [Google Scholar]
- 24.Wright JW, Stanton ML, Scherson R. Local adaptation to serpentine and non-serpentine soils in Collinsia sparsiflora. Evol Ecol Res. 2006;8(1):1–21. [Google Scholar]
- 25.Antonovics J. The effects of a heterogeneous environment on the genetics of natural populations. Am Sci. 1971;59(5):593–599. [PubMed] [Google Scholar]
- 26.Clausen J, Keck DD, Hiesey WM. Heredity of geographically and ecologically isolated races. Am Nat. 1947;81(797):114–133. doi: 10.1086/281507. [DOI] [PubMed] [Google Scholar]
- 27.Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol Lett. 2004;7(12):1225–1241. [Google Scholar]
- 28.Linhart YB, Grant MC. Evolutionary significance of local genetic differentiation in plants. Annu Rev Ecol Syst. 1996;27:237–277. [Google Scholar]
- 29.Mátyás C. Climatic adaptation of trees: Rediscovering provenance tests. Euphytica. 1996;92(1-2):45–54. [Google Scholar]
- 30.Langlet O. Two hundred years genecology. Taxon. 1971;20(5-6):653–721. [Google Scholar]
- 31.Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185(4):1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hancock AM, Rienzo AD. Detecting the genetic signature of natural selection in human populations: Models, methods, and data. Annu Rev Anthropol. 2008;37:197–217. doi: 10.1146/annurev.anthro.37.081407.085141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sork VL, et al. Putting the landscape into the genomics of trees: approaches for understanding local adaptation and population responses to changing climate. Tree Genet Genomes. 2013;9(4):901–911. [Google Scholar]
- 34.Levin DA. Dispersal versus gene flow in plants. Ann Mo Bot Gard. 1981;68(2):233–253. [Google Scholar]
- 35.Oddou-Muratorio S, Petit RJ, Le Guerroue B, Guesnet D, Demesure B. Pollen- versus seed-mediated gene flow in a scattered forest tree species. Evolution. 2001;55(6):1123–1135. doi: 10.1111/j.0014-3820.2001.tb00632.x. [DOI] [PubMed] [Google Scholar]
- 36.Sork VL, Smouse PE. Genetic analysis of landscape connectivity in tree populations. Landsc Ecol. 2006;21(6):821–836. [Google Scholar]
- 37.Sork VL, Smouse PE, Grivet D, Scofield DG. Impact of asymmetric male and female gamete dispersal on allelic diversity and spatial genetic structure in valley oak (Quercus lobata Née) Evol Ecol. 2015;29(6):927–945. [Google Scholar]
- 38.Hamilton MB, Miller JR. Comparing relative rates of pollen and seed gene flow in the island model using nuclear and organelle measures of population structure. Genetics. 2002;162(4):1897–1909. doi: 10.1093/genetics/162.4.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burban C, Petit RJ. Phylogeography of maritime pine inferred with organelle markers having contrasted inheritance. Mol Ecol. 2003;12(6):1487–1495. doi: 10.1046/j.1365-294x.2003.01817.x. [DOI] [PubMed] [Google Scholar]
- 40.Latta RG, Mitton JB. A comparison of population differentiation across four classes of gene marker in limber pine (Pinus flexilis James) Genetics. 1997;146(3):1153–1163. doi: 10.1093/genetics/146.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liepelt S, Bialozyt R, Ziegenhagen B. Wind-dispersed pollen mediates postglacial gene flow among refugia. Proc Natl Acad Sci USA. 2002;99(22):14590–14594. doi: 10.1073/pnas.212285399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loveless MD, Hamrick JL. Ecological determinants of genetic structure in plant populations. Annu Rev Ecol Syst. 1984;15:65–95. [Google Scholar]
- 43.Hardy OJ. How fat is the tail? Heredity (Edinb) 2009;103(6):437–438. doi: 10.1038/hdy.2009.120. [DOI] [PubMed] [Google Scholar]
- 44.Petit RJ, Hampe A. Some evolutionary consequences of being a tree. Annu Rev Ecol Evol Syst. 2006;37:187–214. [Google Scholar]
- 45.Wang IJ, Bradburd GS. Isolation by environment. Mol Ecol. 2014;23(23):5649–5662. doi: 10.1111/mec.12938. [DOI] [PubMed] [Google Scholar]
- 46.Andrew RL, Ostevik KL, Ebert DP, Rieseberg LH. Adaptation with gene flow across the landscape in a dune sunflower. Mol Ecol. 2012;21(9):2078–2091. doi: 10.1111/j.1365-294X.2012.05454.x. [DOI] [PubMed] [Google Scholar]
- 47.Smouse PE, Peakall R. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity (Edinb) 1999;82(5):561–573. doi: 10.1038/sj.hdy.6885180. [DOI] [PubMed] [Google Scholar]
- 48.Vekemans X, Hardy OJ. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol. 2004;13(4):921–935. doi: 10.1046/j.1365-294x.2004.02076.x. [DOI] [PubMed] [Google Scholar]
- 49.Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236(4803):787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- 50.Wright S. Isolation by distance. Genetics. 1943;28(2):114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanton ML, Galen C, Shore J. Population structure along a steep environmental gradient: Consequences of flowering time and habitat variation in the snow buttercup, Ranunculus adoneus. Evolution. 1997;51(1):79–94. doi: 10.1111/j.1558-5646.1997.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 52.Yamagishi H, Allison TD, Ohara M. Effect of snowmelt timing on the genetic structure of an Erythronium grandiflorum population in an alpine environment. Ecol Res. 2005;20(2):199–204. [Google Scholar]
- 53.Bradshaw HD, Schemske DW. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature. 2003;426(6963):176–178. doi: 10.1038/nature02106. [DOI] [PubMed] [Google Scholar]
- 54.Vickery RK. Pollinator preferences for yellow, orange, and red flowers of Mimulus verbenaceus and M. cardinalis. Great Basin Nat. 1992;52(2):145–148. [Google Scholar]
- 55.Dyer RJ, Nason JD, Garrick RC. Landscape modelling of gene flow: Improved power using conditional genetic distance derived from the topology of population networks. Mol Ecol. 2010;19(17):3746–3759. doi: 10.1111/j.1365-294X.2010.04748.x. [DOI] [PubMed] [Google Scholar]
- 56.Massatti R, Knowles LL. Microhabitat differences impact phylogeographic concordance of codistributed species: Genomic evidence in montane sedges (Carex L.) from the Rocky Mountains. Evolution. 2014;68(10):2833–2846. doi: 10.1111/evo.12491. [DOI] [PubMed] [Google Scholar]
- 57.Sexton JP, Hangartner SB, Hoffmann AA. Genetic isolation by environment or distance: Which pattern of gene flow is most common? Evolution. 2014;68(1):1–15. doi: 10.1111/evo.12258. [DOI] [PubMed] [Google Scholar]
- 58.Schierenbeck KA. Phylogeography of California: An Introduction. Univ of California Press; Oakland, CA: 2014. [Google Scholar]
- 59.Lee SW, Ledig FT, Johnson DR. Genetic variation at allozyme and RAPD markers in Pinus longaeva (Pinaceae) of the White Mountains, California. Am J Bot. 2002;89(4):566–577. doi: 10.3732/ajb.89.4.566. [DOI] [PubMed] [Google Scholar]
- 60.Baldwin BG. Origins of plant diversity in the California floristic province. Annu Rev Ecol Evol Syst. 2014;45:347–369. [Google Scholar]
- 61.Raven PH, Axelrod DI. Origin and Relationships of the California Flora. Univ of California Press; Berkeley, CA: 1978. [Google Scholar]
- 62.Lancaster LT, Kay KM. Origin and diversification of the California flora: Re-examining classic hypotheses with molecular phylogenies. Evolution. 2013;67(4):1041–1054. doi: 10.1111/evo.12016. [DOI] [PubMed] [Google Scholar]
- 63.Loarie SR, et al. Climate change and the future of California’s endemic flora. PLoS One. 2008;3(6):e2502. doi: 10.1371/journal.pone.0002502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mittermeier RA, et al. Hotspots Revisited: Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions. Conservation International; Washington, DC: 2005. [Google Scholar]
- 65.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403(6772):853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 66.Swenson NG, Howard DJ. Clustering of contact zones, hybrid zones, and phylogeographic breaks in North America. Am Nat. 2005;166(5):581–591. doi: 10.1086/491688. [DOI] [PubMed] [Google Scholar]
- 67.Stebbins GL, Major J. Endemism and speciation in the California flora. Ecol Monogr. 1965;35(1):2–35. [Google Scholar]
- 68.Grivet D, Deguilloux MF, Petit RJ, Sork VL. Contrasting patterns of historical colonization in white oaks (Quercus spp.) in California and Europe. Mol Ecol. 2006;15(13):4085–4093. doi: 10.1111/j.1365-294X.2006.03083.x. [DOI] [PubMed] [Google Scholar]
- 69.Gugger PF, Ikegami M, Sork VL. Influence of late Quaternary climate change on present patterns of genetic variation in valley oak, Quercus lobata Née. Mol Ecol. 2013;22(13):3598–3612. doi: 10.1111/mec.12317. [DOI] [PubMed] [Google Scholar]
- 70.McLachlan JS, Clark JS, Manos PS. Molecular indicators of tree migration capacity under rapid climate change. Ecology. 2005;86(8):2088–2098. [Google Scholar]
- 71.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corander J, Marttinen P, Sirén J, Tang J. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics. 2008;9(1):539. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sork VL, et al. Landscape genomic analysis of candidate genes for climate adaptation in a California endemic oak, Quercus lobata. Am J Bot. 2016;103(1):33–46. doi: 10.3732/ajb.1500162. [DOI] [PubMed] [Google Scholar]
- 74.Ashley MV, Abraham ST, Backs JR, Koenig WD. Landscape genetics and population structure in Valley Oak (Quercus lobata Née) Am J Bot. 2015;102(12):2124–2131. doi: 10.3732/ajb.1500182. [DOI] [PubMed] [Google Scholar]
- 75.Gugger PF, Cokus SJ, Pellegrini M, Sork VL. Association of transcriptome-wide sequence variation with climate gradients in valley oak (Quercus lobata) Tree Genet Genomes. 2016;12(2):1–14. [Google Scholar]
- 76.Sork VL, et al. Gene movement and genetic association with regional climate gradients in California valley oak (Quercus lobata Née) in the face of climate change. Mol Ecol. 2010;19(17):3806–3823. doi: 10.1111/j.1365-294X.2010.04726.x. [DOI] [PubMed] [Google Scholar]
- 77.Riordan EC, et al. Association of genetic and phenotypic variability with geography and climate in three southern California oaks. Am J Bot. 2016;103(1):73–85. doi: 10.3732/ajb.1500135. [DOI] [PubMed] [Google Scholar]
- 78.Ortego J, Gugger PF, Sork VL. Climatically stable landscapes predict patterns of genetic structure and admixture in the Californian canyon live oak. J Biogeogr. 2015;42(2):328–338. [Google Scholar]
- 79.Ortego J, Noguerales V, Gugger PF, Sork VL. Evolutionary and demographic history of the Californian scrub white oak species complex: An integrative approach. Mol Ecol. 2015;24(24):6188–6208. doi: 10.1111/mec.13457. [DOI] [PubMed] [Google Scholar]
- 80.Grivet D, Sork VL, Westfall RD, Davis FW. Conserving the evolutionary potential of California valley oak (Quercus lobata Née): A multivariate genetic approach to conservation planning. Mol Ecol. 2008;17(1):139–156. doi: 10.1111/j.1365-294X.2007.03498.x. [DOI] [PubMed] [Google Scholar]
- 81.Fitzpatrick BM, et al. Rapid spread of invasive genes into a threatened native species. Proc Natl Acad Sci USA. 2010;107(8):3606–3610. doi: 10.1073/pnas.0911802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fitzpatrick MC, Keller SR. Ecological genomics meets community-level modelling of biodiversity: Mapping the genomic landscape of current and future environmental adaptation. Ecol Lett. 2015;18(1):1–16. doi: 10.1111/ele.12376. [DOI] [PubMed] [Google Scholar]
- 83.Gavin DG, et al. Climate refugia: Joint inference from fossil records, species distribution models and phylogeography. New Phytol. 2014;204(1):37–54. doi: 10.1111/nph.12929. [DOI] [PubMed] [Google Scholar]
- 84.Sork VL, Werth S. Phylogeography of Ramalina menziesii, a widely distributed lichen-forming fungus in western North America. Mol Ecol. 2014;23(9):2326–2339. doi: 10.1111/mec.12735. [DOI] [PubMed] [Google Scholar]
- 85.Werth S, Sork VL. Identity and genetic structure of the photobiont of the epiphytic lichen Ramalina menziesii on three oak species in southern California. Am J Bot. 2010;97(5):821–830. doi: 10.3732/ajb.0900276. [DOI] [PubMed] [Google Scholar]
- 86.Werth S, Sork VL. Local genetic structure in a North American epiphytic lichen, Ramalina menziesii (Ramalinaceae) Am J Bot. 2008;95(5):568–576. doi: 10.3732/ajb.2007024. [DOI] [PubMed] [Google Scholar]
- 87.Werth S, Sork VL. Ecological specialization in Trebouxia (Trebouxiophyceae) photobionts of Ramalina menziesii (Ramalinaceae) across six range-covering ecoregions of western North America. Am J Bot. 2014;101(7):1127–1140. doi: 10.3732/ajb.1400025. [DOI] [PubMed] [Google Scholar]
- 88.Baas Becking LGM. Geobiologie: Of Inleiding tot de Milieukunde. Van Stockum and Zoon; The Hague, The Netherlands: 1934. [Google Scholar]
- 89.Usher KM, Bergman B, Raven JA. Exploring cyanobacterial mutualisms. Annu Rev Ecol Evol Syst. 2007;38:255–273. [Google Scholar]
- 90.Chen J-M, Werth S, Sork VL. Comparison of phylogeographical structures of a lichen-forming fungus and its green algal photobiont in western North America. J Biogeogr. 2016;43(5):932–943. [Google Scholar]
- 91.Prates I, et al. 2016. Inferring responses to climate dynamics from historical demography in neotropical forest lizards. Proc Natl Acad Sci USA 113:7978–7985.