Significance
Treatment for tuberculosis is lengthy and requires multiple-drug treatment, in part because some cells of a Mycobacterium population take longer to be killed by antibiotics. To understand the contributions of bacterial growth and cell cycle state on antibiotic tolerance, we used microfluidics-based live-cell imaging to compare growth parameters and treatment responses in single cells. We identified key growth and cell cycle classifiers that correlated with rifampicin tolerance. We used these data and modeling to define differences between cells in rifampicin-tolerant and rifampicin-susceptible mycobacterial subpopulations. This study establishes that subpopulations with simple phenotypic characteristics, such as size and cell cycle states, are associated with differences in antibiotic susceptibility, which may improve our ability to rationally design improved drug combinations.
Keywords: antibiotic susceptibility, single cell, mathematical modeling, mycobacteria, cell biology
Abstract
Mycobacteria grow and divide asymmetrically, creating variability in growth pole age, growth properties, and antibiotic susceptibilities. Here, we investigate the importance of growth pole age and other growth properties in determining the spectrum of responses of Mycobacterium smegmatis to challenge with rifampicin. We used a combination of live-cell microscopy and modeling to prospectively identify subpopulations with altered rifampicin susceptibility. We found two subpopulations that had increased susceptibility. At the initiation of treatment, susceptible cells were either small and at early stages of the cell cycle, or large and in later stages of their cell cycle. In contrast to this temporal window of susceptibility, tolerance was associated with factors inherited at division: long birth length and mature growth poles. Thus, rifampicin response is complex and due to a combination of differences established from both asymmetric division and the timing of treatment relative to cell birth.
The unusually long period of antibiotic exposure needed for treatment of tuberculosis is necessitated by the presence of Mycobacterium tuberculosis bacilli that respond slowly to drug treatment (1, 2). Although heritable drug resistance is generated by genetic mutations or acquisition of drug-resistant genes, phenotypic resistance (tolerance) enables subpopulations to resist antibiotic clearance under particular environmental conditions (3). Nongrowing, drug-tolerant cells are commonly called persister cells and are often thought of as a discrete and rare group of cells (4, 5). However, we have observed a spectrum of responses in Mycobacterium smegmatis at the single-cell level to antibiotic treatment (6). Distinct subpopulations may be generated either deterministically through a genetically encoded mechanism, which creates differences in cell physiology, or through stochastic and temporary differences in the ways individual cells interact with drugs (7). Defining these subpopulations requires methods to investigate the responses of individual cells to antibiotic stress and a quantitative framework to understand how the single-cell biology relates to the ability of a population of cells to tolerate antibiotics.
To identify the critical cell cycle parameters that define individual cells’ susceptibility to antibiotics, we have previously developed a microfluidics-based live-cell imaging system to observe the growth of individual mycobacteria (6). Using this live-cell imaging platform, we observed asymmetric division and heterogeneity in elongation rates among closely related cells, establishing that mycobacteria elongate primarily from the growth pole inherited from the parent cell at division. Corroborating this asymmetric polar growth pattern, Meniche and colleagues (8) found that the M. smegmatis cell wall synthesis machinery is localized to the subpolar region and preferentially at the old pole. Additional groups have reported asymmetric septum localization and faster growth from older poles (9, 10). Asymmetric growth has alternatively been attributed to a longer available time for the old pole to elongate between cytokinesis and division events (11, 12). Although the molecular mechanisms of asymmetric growth are not well understood, there is consensus across several experimental settings that mycobacteria divide asymmetrically such that the sister inheriting the new pole is smaller and slower growing than the sister inheriting the old pole (9–13). At division, the polarized cell generates daughter cells that are functionally distinct. One daughter cell, which we call the accelerator cell, inherits a new pole and the oldest (growing) pole and elongates faster from the old pole. Its sister cell, the alternator cell, inherits one nongrowing pole of intermediate age and one new pole. The alternator cell must switch its direction of growth by elongating from its older, previously slowly growing pole. Some accelerator cells inherit growth poles created in the immediate previous generation, whereas other accelerator cells inherit growth poles created several generations earlier. Cells with the older growth poles elongate faster and divide at a larger size than do cells with younger growth poles. This pattern of growth asymmetry quickly and deterministically creates population heterogeneity that is mirrored by differential tolerance to antibiotic treatment. Alternator cells are more tolerant to cell wall-targeting drugs (isoniazid, cycloserine, and meropenem), whereas accelerator cells are more tolerant to the transcription inhibitor rifampicin (6).
In this work, our goal was to determine the relative effect of growth pole age on rifampicin tolerance and to understand whether other cell state and growth parameters were more important. We hypothesized that characteristics of cell physiology such as the cell cycle state may be important determinants of antibiotic susceptibility. In Escherichia coli, susceptibility to antibiotics changes as cells enter different stages of the cell cycle (4, 14, 15). This may be due to differences in the state of chromosome replication or cell wall permeability. Because drug and stress tolerance is likely due to the convergence of multiple dynamic processes, we hypothesized that some combinations of parameters would be characteristic of a cell physiology that correlates with differential susceptibility to rifampicin treatment. We evaluated the growth and cell cycle states of individual M. smegmatis cells and used data-driven modeling to quantify the contribution of each factor to rifampicin response. Our study suggests that cell-to-cell variability in rifampicin response is due to a combination of differences arising from mycobacteria's asymmetric growth and division and transient changes to cell state that coordinate roughly with cell cycle and age. Thus, phenotypic rifampicin-tolerant mycobacteria do not consist of one type of persister cell, but are a dynamic and variable set of cells that arise through asymmetry and transient tolerant stages of the cell cycle.
Results
Measuring Cell Growth Parameters and Rifampicin Tolerance.
Our experimental setup (Fig. 1A) enabled us to provide M. smegmatis cells with a homogeneous growth environment during long-term, live-cell microscopy and control the timing and levels of rifampicin challenge (6). The initial growth period established microcolonies and allowed us to identify a predrug control population, i.e., cells that divided before the application of drug (Fig. 1B). After 10.5 h of growth, cells were challenged with rifampicin for 6 h and evaluated for antibiotic susceptibility (Fig. 1C). We defined drug-tolerant or “live” cells as those that elongated or divided with at least one elongating daughter cell during the 9.75-h recovery period. To analyze the characteristics of rifampicin-susceptible and rifampicin-tolerant cells, we calculated several cell properties during the annotation and analysis process. We used a fluorescent reporter strain expressing single-stranded DNA-binding protein fused to green fluorescent protein (SSB-GFP) (Fig. 1D) to derive cell cycle state, length, and age at the start of treatment, as well as short-term and average growth rates (Methods). In these cells, single or double foci appear at the replisome during active DNA replication (16).
Fig. 1.
Measuring drug treatment response in M. smegmatis. (A) Experiment design, cellular parameters, and microfluidics device schematic. Two syringes, controlled by microfluidic pumps, were connected to a mixing device. M. smegmatis SSB-GFP cells were seeded in a microfluidics device and then placed in a heated environmental chamber for imaging every 15 min. The media syringe pump dispensed medium for the entire duration of the experiment (26.25 h), and the drug syringe pump was activated 10 h after the start of the experiment and dispensed rifampicin for 6 h. (B) Cellular parameters measured before and during treatment. Cells that were born and divided during the predrug growth period were used as controls. Cells that were born after the time lapse began but had not divided before the drug treatment were annotated for cell length and relative growth pole age (numbers, in black: 1 = youngest pole, 4 = oldest pole) for the remainder of the experiment. Accelerator and alternator cells are denoted as “acc” or “alt,” respectively. Schematics of cells demonstrate two possible drug treatment outcomes. A cell was classified as rifampicin-tolerant if it either resumed growth during the recovery period or produced at least one daughter cell that resumed growth. (C) Microfluidic device image sequence. The brightfield image sequence depicts an M. smegmatis microcolony (Left to Right) immediately before drug treatment, immediately after drug treatment, and at the end of the recovery period. Two drug treatment outcomes are illustrated: rifampicin-tolerant (green) and rifampicin-susceptible (violet). (D) Cellular parameters measured at the start of treatment. The following cell parameters were tracked: length at birth (indicated as lengthb), length and age at division (indicated as lengthts and agets), average growth rate and elongation rate immediately before treatment start (indicated as growthtot and growthinst), and presence of SSB-foci (green dots in the schematic), which were used to determine cell cycle stage and timing (B-C-D line, where C is DNA replication stage). SSB-foci were recorded and tracked throughout the course of growth, drug treatment, and recovery.
We treated the cells in various experiments with 0.5, 2, and 3× of the minimal inhibitory concentration (MIC) of rifampicin (20, 80, and 120 µg/mL, respectively) (SI Appendix, Table S1). Our analysis focused on the cells present in the microfluidic device at the start of 3× MIC rifampicin drug treatment. This concentration corresponds roughly to the concentration of rifampicin in human pulmonary lesions after a single dose of rifampicin (17). SI Appendix, Table S1 provides a summary of the dataset, including the number of annotated cells and the number of tolerant cells for each rifampicin concentration.
To determine whether susceptible cells were nonviable or alternatively delayed in resuming growth, we performed an additional experiment, in which we evaluated the growth of 286 cells with a longer recovery period. We annotated the growth parameters of these cells when rifampicin was applied. Seventy-one percent would have been annotated as susceptible at our standard 9.75 h of recovery period. During the extended 16-h recovery period, 97% of cells classified as susceptible did not show any signs of growth in the last 6 h. To assess whether there were viable but nonculturable cells (VBNCs) among the susceptible cells, we performed an additional experiment, in which we added red-fluorescent propidium iodide (PI) stain to the chambers at the very end of the standard recovery period. PI is used to identify cells with disrupted membranes; these cells will take up the stain, whereupon the PI fluoresces upon intercalation with DNA (SI Appendix, Fig. S1) (18). Of 92 total cells, 81 did not grow after the drug treatment and were thus classified as susceptible. All 81 susceptible cells were PI+ (as in SI Appendix, Fig. S1), thus suggesting that VBNCs do not make up a significant proportion of the susceptible population.
Rifampicin-Tolerant Cells Are Larger.
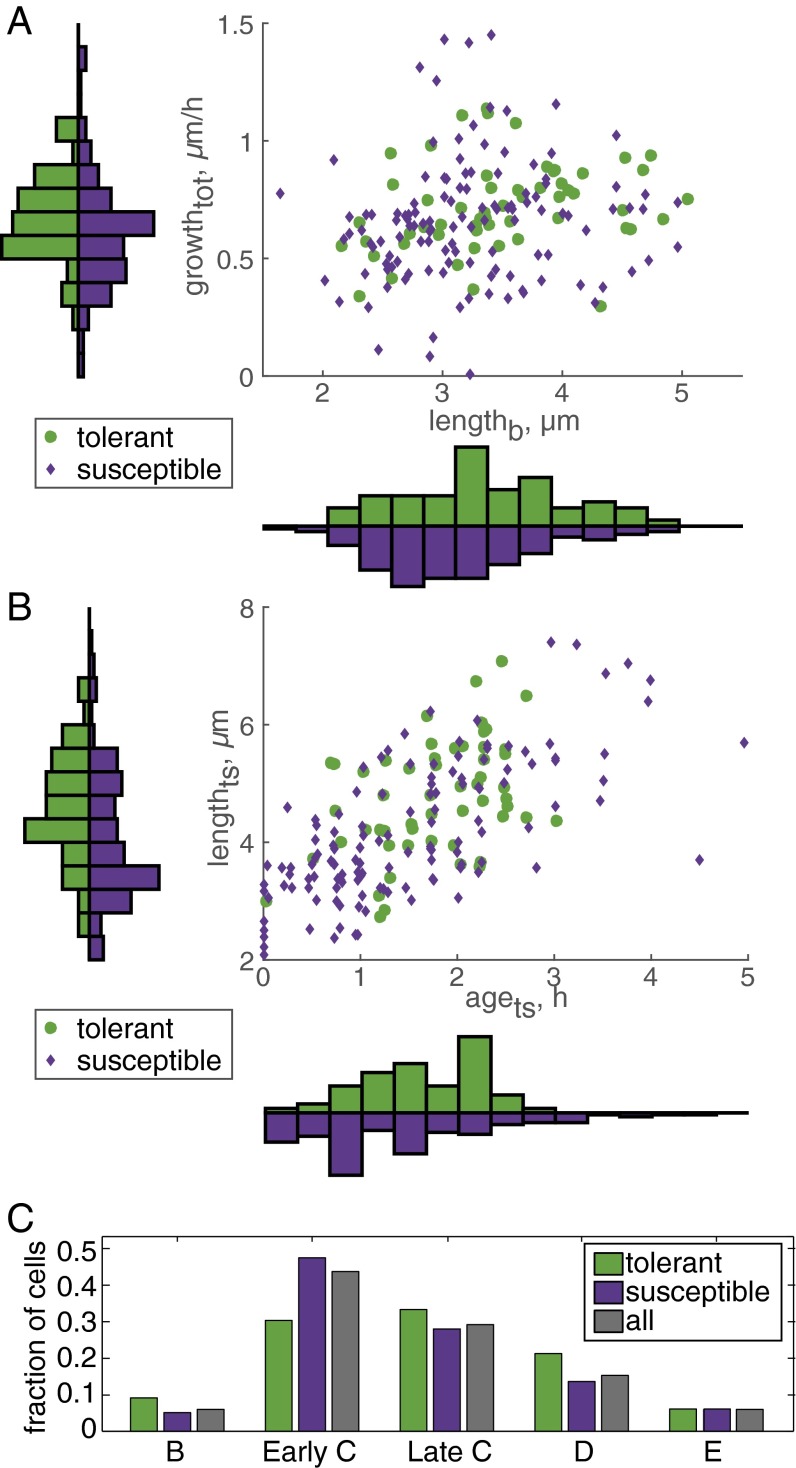
Accelerators are overrepresented in the rifampicin-tolerant subpopulations across a broad range of rifampicin concentrations (0.5, 2, and 3× MIC) (SI Appendix, Table S1). Because accelerator cells are both larger at birth (SI Appendix, Fig. S2) and faster-growing (SI Appendix, Fig. S3) than alternator cells (6), we investigated whether tolerant and susceptible subpopulations exhibit differences in size and growth rate (Fig. 2). Rifampicin-tolerant cells were significantly longer at birth (lengthb; 3.7 ± 0.7 µm) compared with susceptible cells (3.2 ± 0.8 µm) (Fig. 2A, Bottom histogram, and SI Appendix, Fig. S4). However, there were no differences in the growth rates of tolerant and susceptible subpopulations (Fig. 2A and SI Appendix, Figs. S5 and S6).
Fig. 2.
Rifampicin-tolerant cells are larger and have older growth poles than susceptible cells. (A) Scatter plot of cell birth length (lengthb), average growth rate from birth to treatment start (growthtot), and drug treatment outcome. Histograms display the distribution of tolerant and susceptible cells along each axis; distributions were normalized by setting the area to one. Each bin of the histogram of the x axis (lengthb) covers 0.29 µm of cell length, and each bin of the y axis (growth rate) covers 0.11 µm/h. Rifampicin-tolerant cells had a larger average length at birth (P < 0.005). No significant difference was detected for the average growth rate of susceptible and tolerant subpopulations. (B) Scatter plot of cell age (agets) and length (lengthts) at the start of drug treatment and drug treatment outcome. Each bin of the histogram of the x axis (agets) spans 0.36 h of cell age, and each bin of the y axis (lengthts) spans 0.43 µm of cell length at treatment start. Drug-tolerant cells had a larger average length at treatment start (P < 0.005). No significant difference was detected for the average age of susceptible and tolerant subpopulations. (C) Distribution of cell cycle stages (B prereplication, early C replication, late C replication, D postreplication, and E predivision replication) at the start of drug treatment. There was no significant difference between cell cycle distribution for tolerant and susceptible cells.
We have previously observed microcolony-to-microcolony variation in rifampicin susceptibility but also synchronized division times within individual microcolonies (6). In E. coli, susceptibility to antibiotics changes as cells enter different stages of the cell cycle (4, 14, 15). We therefore hypothesized that cell parameters that describe cell state at the start of drug treatment such as cell cycle state, cell age, and cell size may determine treatment outcome. Tolerant cells were significantly larger not only at birth but also at the start of rifampicin treatment (lengthts; 5.3 ± 1.1 µm) compared with susceptible cells (4.3 ± 1.4 µm) (Fig. 2B and SI Appendix, Fig. S7). Because older cells are longer than newly divided cells, we expected that differences in susceptible vs. tolerant subpopulations in cell age at the time of treatment (agets) would correspond with lengthts differences. Whereas lengthts was different in susceptible and tolerant subpopulations, there was no significant difference in the agets or cell cycle state of tolerant cells compared with susceptible cells (Fig. 2 B and C).
Two Rifampicin-Susceptible Subpopulations Have Distinct Growth and Cell State Properties.
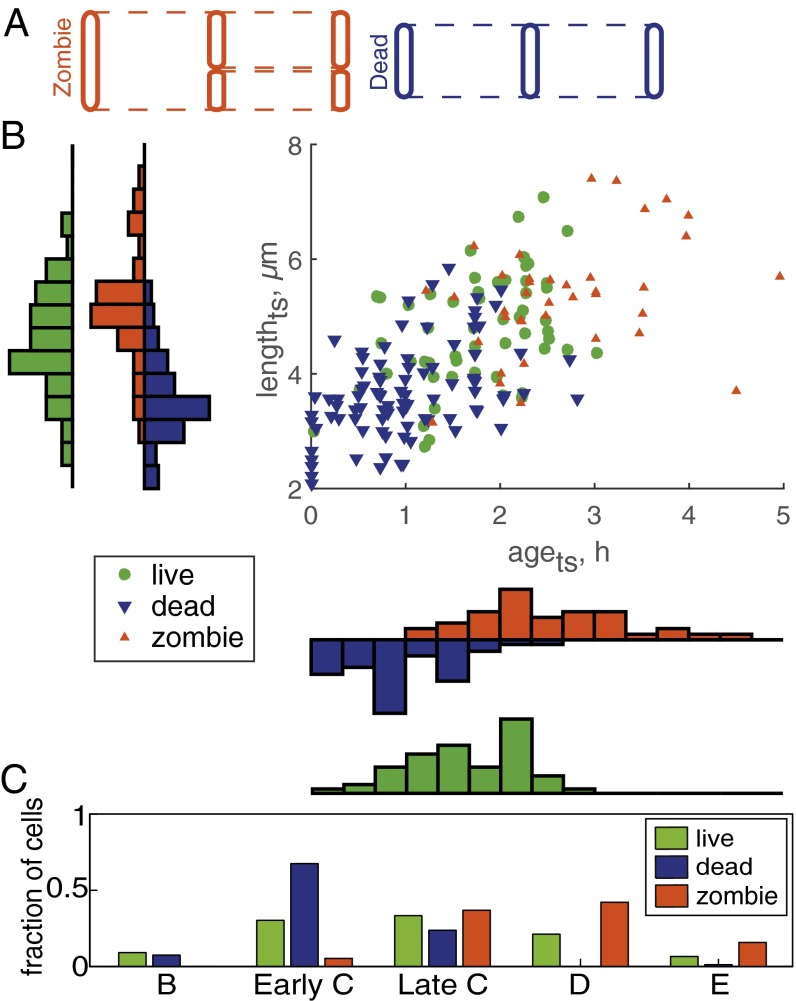
Because rifampicin-tolerant and rifampicin-susceptible cells differ significantly in size, but not in age or cell cycle state, we reasoned that other growth parameters must contribute to rifampicin susceptibility or that the drug response categorizations were too broad. Whereas most susceptible cells simply stopped growing and never resumed growth during the recovery period, around 30% of susceptible cells divided into two nongrowing daughters after the start of rifampicin treatment. We therefore created two subcategories of susceptible cells: “dead” cells that did not grow or divide during treatment or during the recovery period and “zombie” cells that were present at treatment start and divided into two nongrowing cells after the start of treatment (Fig. 3A).
Fig. 3.
Two rifampicin-susceptible subpopulations have distinct growth and cell state properties. (A) Subcategorization of susceptible cell populations. Susceptible cell classification was refined by introducing two subcategories of nonelongating susceptible cells: (i) dead cells that did not divide after the application of drug and (ii) zombie cells, which divided into two nonelongating cells after the start of rifampicin treatment. (B) Scatter plot of age (agets) and length (lengthts) at the start of drug treatment and drug treatment outcome. The histogram bin size is identical to Fig. 2B. The difference in lengthts was significant for two pairs: live–dead (P < 0.0001), and dead–zombie (P < 0.0001). The difference in birth time relative to the start of drug treatment was significant for all three pairs: live–dead (P < 0.001), live–zombie (P < 0.0005), and dead–zombie (P < 0.0001). (C) Distribution of cell cycle stages (B prereplication, early C replication, late C replication, D postreplication, and E predivision replication) at the start of drug treatment. Refined drug-susceptible cell categorization demonstrates that compared with the drug-tolerant population, the dead cell population was skewed toward early cell cycle stages (P < 0.0005), whereas the zombie population was skewed toward late cell cycle stages (P < 0.001).
We found no significant difference in the initial growth rates or birth lengths of dead and zombie cells (SI Appendix, Figs. S8–S12). However, dead and zombie cells exhibited starkly opposite length, age, and cell cycle characteristics at the start of drug treatment (Fig. 3 B and C). The median lengthts of dead cells at treatment start (3.9 ± 1.2 µm) was significantly shorter than that of the live subpopulation (5.3 ± 1.1 µm) and was similar to the birth size of newly divided cells in the predrug population (3.5 ± 0.8 µm). In contrast, zombie cells were longer than live and dead cells (5.6 ± 1.3 µm) and were closer to the cell length just before division (6.3 ± 1.2 µm). These length differences at the start of treatment were significant for both live–dead and dead–zombie cell comparisons across rifampicin concentrations (SI Appendix, Figs. S13 and S14). The division of zombie cells following exposure to rifampicin produced daughter cells (3.3 ± 0.8 µm) close to the size of newly born cells in the predrug population (3.5 ± 0.8 µm). These observations suggest that susceptible cells on the verge of division at the time of exposure to rifampicin are able to complete the cell division.
Consistent with the cell lengths among the dead, live, and zombie subpopulations, the agets of the tolerant subpopulation (1.5 ± 1.1 h) was significantly different from dead and zombie cell median ages (Fig. 3B). Dead cells were very young and were born 0.9 ± 1.0 h before rifampicin exposure, whereas zombie cells were much older, born 2.4 ± 1.0 h before the rifampicin exposure. The difference was consistent across rifampicin treatment concentrations (SI Appendix, Figs. S15 and S16). Dividing the susceptible subpopulation into dead and zombie cells revealed differences in cell cycle stages that correspond to the differences we observed in age and length (Fig. 3 B and C). Compared with live cells (and the entire population at start of treatment as control), dead cells exhibited a skewed distribution toward the B and early C cell cycle stages. The distribution in zombie cells was skewed toward late C, D, and E. Together, these data suggest that M. smegmatis cells are more susceptible to rifampicin at the early and late stages of the cell cycle.
We recategorized the susceptible cells as dead or zombie in our smaller extended recovery and PI staining data to assess whether one of these subpopulations was more likely to resume growth in the extended recovery time. Of the 204 susceptible cells, 145 were classified as dead (51% of total) and 59 were classified as zombies (20% of total). During the extended recovery period of 16 h, 96.5% of dead cells and 98.3% of zombie-descendant cells showed no signs of growth. PI staining at the end of the recovery period also failed to demonstrate noticeable differences between dead cells and zombie-descendant cells.
Partial Least-Squares Regression Enables Multivariate Description of Susceptible and Tolerant Subpopulations.
Using single parameters, we were able to describe differences among subpopulations after exposure to rifampicin. To quantitatively describe each subpopulation in terms of multiple cellular parameters, we used partial least-squares regression (PLSR). PLSR is an analytical method that links predictor variables with response variables. The following predictor (“X”) variables were analyzed: pole age, lengthb, lengthts, agets, growthinst, growthtot, stagets, start of DNA replication (C cycle) relative to the start of treatment, and concentration of rifampicin. Treatment outcome (live, dead, or zombie) was the qualitative response (“Y”) variable.
To quantify the relative contributions of each predictor variable to a particular outcome, we evaluated the regression coefficients of the PLSR analysis. For live cells, the strongest contributing variables were a negative association with alternator cells and a positive association with accelerator cells that had older growth poles and larger birth length (Fig. 4A). Thus, cells with a live outcome are more likely to be accelerator cells that had a large birth size. For dead cells, the strongest contributing factors were negative associations with length at treatment start and age at treatment start, negative associations with late cell cycle stages such as D and E (DNA replication following a D period but before division), and positive associations with early cell cycle stages B and early C (Fig. 4A). We note that that the relatively short B cycle was not detected in all of the cells because some cells may have begun replication before birth (E period), whereas in others, the B period may have been too short to detect in the 15-min imaging intervals. This imaging interval could not be shortened without phototoxicity effects. We conclude that dead cells are characterized as young, small, and in the early stages of the cell cycle. For zombie cells, the strongest contributing factors were positive associations with length at treatment start and age at treatment start, positive associations with late cell cycle stages such as D and E, and negative associations with early cell cycle stages B and early C (Fig. 4A). Zombie cells are therefore characterized by being very large, old, and in the late stages of the cell cycle at the time of rifampicin application.
Fig. 4.
Using phenotypic characteristics to predict rifampicin (rif) treatment outcome. (A) Partial least-squares regression coefficients (coef) of cellular parameters. The correlations are shown between rifampicin treatment outcome and the phenotypic M. smegmatis characteristics that served as outcome predictor variables. The y axis represents relative correlation strength (maximum 1.0). Positive values represent covariable relationships between the input–output variable pair, whereas negative values represent negative correlation. SEs displayed as error bars at the end of each column were calculated with jack-knifing. Arrows at the bottom represent the most important contributions to each of the treatment outcomes; arrow direction represents positive (up) and negative (down) correlations. (B) PLSR loading plot of cellular parameters. The loading plot demonstrates the relative influence of predictor values on drug treatment outcome, i.e., how well x variables correlate with y, and how responses vary in relation to each other. The low contribution values are 1, B cycle state; 2, elongation rate immediately before treatment start (growthinst, µm/h); 3, pole age 2 (young accelerator); 4, C cycle duration (h); 5, drug concentration (µg/mL); 6, average growth rate (growthtot, µm/h); 7, late C cycle state; and 8, E cycle state. (C) Flowchart of phenotype selection rules for predicting rifampicin treatment outcome. The categorization process consisted of splitting the bulk population into accelerators and alternators and then defining zombie and dead cells by using cell length and age thresholds derived from the main dataset. At each sequential step, cells that met the predefined selection criteria were assigned to a particular drug treatment classification: zombie, dead, and, finally, live. Drug tolerance response was then determined for cells in a separate test set. Sixty-percent of the cells in a test set were correctly identified as either live, dead, or zombie, compared to 37% when using random selection.
Many of the cellular parameters are interrelated. To quantify the relationships among variables in each principal component, we evaluated the loadings (Fig. 4B). In loading plots, the projection of predictor variables and a response variable to a new space creates a linear combination of loading coefficients that approximate the original variables, allowing examination of the relationships among them. The variables situated near each other tend to be correlated. The graph may be additionally interpreted by identifying a Y variable of interest and constructing a line through that variable and through the origin. Other X and Y variables should be orthogonally projected onto that line. Variables on opposite sides of the projected line are negatively correlated, whereas variables on the same side of the line are positively correlated to variables situated near them. We observed a close proximity of growth pole age above 3 and cell length at birth in the loadings (Fig. 4B), suggesting that the previously observed link between pole age and tolerance of accelerator cells to rifampicin may be adequately described strictly in terms of a cell's size at birth. Additionally, the substantial second-component variation for positions of agets and lengthts on the loading plot suggests that these variables do not always travel together, and therefore each has a unique biological significance in relation to rifampicin susceptibility.
Parameter Sorting Enables Moderate Prediction of Tolerant, Dead, and Zombie Subpopulations.
We have used analysis of single (Figs. 2 and 3) and multiple cellular parameters (Fig. 4 A and B) to characterize the relationships of cell growth parameters to treatment outcome. We next attempted to predict the live–dead–zombie outcome of a treatment by codifying these descriptors. We sorted cells in a test set population according to their phenotypic parameters. Both high-contribution parameters (lengthts and agets) (Fig. 4B and SI Appendix, Table S2) were chosen for the filter. In addition, we included two medium-contribution parameters (accelerator and alternator status, lengthb) (Fig. 4B and SI Appendix, Table S2) because they captured live-cell population variability particularly well (Fig. 4A). Other medium-contribution parameters (early C and D cell cycle stage) were not chosen because dead and zombie cell populations were described by high-contribution parameters. Two separate filter sets were created for accelerator and alternator cells, due to their varying functional characteristics. The length and age filtering parameters were determined using accelerator/alternator box plot graphs derived from our main dataset (SI Appendix, Figs. S17–S19). The filters were then tested against a separate test set consisting of 190 annotated cells.
The filtering process for both the accelerators and alternator cells in the test set consisted of two similar steps (Fig. 4C). First, a cell length and age-based filter was run against the test set to select cells that were deemed similar to zombie cells. The remaining cells were then subjected to a second round of filtering to select the dead subpopulation. Cells that remained after the two filtering steps were considered to be the live subpopulation. In total, our sorting algorithm improved the total prediction accuracy (60%) by 23 percentage points (SI Appendix, Table S2) compared with a purely random baseline (37%) (refer to SI Appendix for calculations of the random baseline). At 60% accuracy, our data and model suggest that simple growth parameters beyond accelerator and alternator cell types are associated with cell physiologies that are important in rifampicin tolerance at a single-cell level.
Discussion
To understand whether growth pole age and other growth and cell cycle factors distinguish rifampicin-tolerant and rifampicin-susceptible mycobacteria, we have constructed a compendium of cellular growth and cell cycle parameters and their relation to rifampicin treatment response. Reconciliation of how the single growth and cell cycle parameters correlated with drug response required separation of rifampicin-susceptible cells into two distinct categories. We found that age and cell length at the start of drug treatment strongly correlated with drug susceptibility, with susceptible cells frequently either in early or late, but not intermediate, stages of the cell cycle. In contrast, drug tolerance was closely correlated with long cell length and advanced growth pole age at birth. By creating a rule-based filter, we were able to capture a high degree of variability in treatment outcome with these very simple functional parameters. Thus, moderate cell-to-cell differences in growth state reflect important differences in cell physiology that affect the bacterium’s ability to experience or respond to rifampicin. These growth parameters are generated through a combination of differences in cell cycle state at the time of drug treatment and in cell size arising from asymmetric division. The bacterial drug response field has concentrated on identifying single factors of tolerance (19); however, our findings illustrate that it may be critical to evaluate the roles of several cell state factors simultaneously in single cells.
Although many of the growth factors are connected to each other, some factors were not overrepresented in either susceptible or tolerant subpopulations. For example, cell size correlates with growth rate at the population level, but cell size and growth rate are not strictly correlated in single cells. It is widely reported that rapidly growing E. coli cells are more susceptible in bulk to antibiotic treatment, but that single-cell growth rate is not associated with isoniazid susceptibility in M. smegmatis (20–22). Using multivariate PLSR modeling, we found that cell size, but not growth rate, was significantly associated with rifampicin susceptibility. Thus, our study emphasizes that factors that alter drug susceptibility in bulk may not translate to differential antibiotic susceptibilities at the single-cell level.
We initially hypothesized that dead and zombie cells may be different in their response to rifampicin and that one of these populations may contain VBNCs. However, all of the dead and zombie-descendant cells stained with PI, indicating that their membranes were extremely disrupted. Instead, our data suggest that susceptible cells are at the early or late (but not intermediate) stages of the cell cycle, and therefore cells are more susceptible to rifampicin just before or after division. It remains to be discovered how death is mechanistically similar or dissimilar in these two subpopulations. It may be that dead and zombie cells are in the same cellular state, but are only separated by a division event that is not dependent on new transcription.
Our data and modeling suggest that mycobacterial growth and cell cycle state reflect important multifactorial differences in cell physiology that together influence the ability of individual bacteria to tolerate rifampicin treatment. Whereas other studies have sought to identify growth outliers associated with persister cells, we find here that even moderate variation in growth and cell cycle parameters is associated with differential antibiotic susceptibility (19). These states may arise through a combination of inherent size differences from asymmetric growth and division and the timing of treatment in relation to the cell cycle state of individual cells. How these temporal and inherent growth parameters reflect cell physiologies with various abilities to tolerate rifampicin stress will be an important question for future studies. Cells born large may be more tolerant if they have more copies of rifampicin’s target, the beta subunit of RNA polymerase (RpoB), per chromosome copy because of their larger cell volume; more copies of RpoB may enable cells to maintain a sufficient pool of functional protein to survive and recover. Another possibility is that cells near division or born small may be more drug-permeable or have decreased efflux capabilities, thus exposing these vulnerable cells to more drug.
A time window of susceptibility may not be unique to mycobacteria. Other studies have found that E. coli tightly control protein induction at stationary exit and synchronize the timing of a lag before regrowth following antibiotic treatment (14, 15). Mycobacteria may use similar mechanisms to adapt the timing of their stress response. Alternatively, this time window may arise from changes to cell state due to cell cycle-related patterns of transcription and translation as in other microbes (23, 24). Our findings that mycobacterial drug tolerance has variable components via the timing of birth and cell size at birth open a new conceptual framework of rapid and cyclic changes to drug susceptibility that may be anticipated and quantified. The suggested presence of phenotypic subpopulations of cells in pathogenic mycobacteria that may be preferentially targeted by certain antibiotics could lead to a more rational method of designing drug combination therapies.
Methods
Cell Culturing and Microscopy.
We used M. smegmatis strain mc2155 transformed with a hygromycin-resistant replicating plasmid expressing single-stranded binding protein fused to green fluorescent protein (SSB-GFP), as described previously (16). Time-lapse images were acquired every 15 min for a duration of 26.25 h using a widefield DeltaVision PersonalDV (Applied Precision) with a hardware-based autofocus. See also SI Appendix, SI Methods.
Data Analysis.
Statistical analysis was performed using MATLAB 2015b (The Mathworks). Significance of association between drug-tolerant and drug-susceptible accelerator and alternator cells was examined with a Fisher test. All other distributions were compared with one another using a Wilcoxon rank sum test. The significance threshold was set at P = 0.01. All provided subpopulation values are medians, except for agets, which was a mean value. SD (S-value) for sample averages was calculated in MATLAB. PLSR analysis was performed in SIMCA (v14). Refer to SI Appendix, SI Methods, for additional methods.
Supplementary Material
Acknowledgments
We thank members of the B.B.A. laboratory, A. Sonenshein, S. Fortune, and E. Rubin for feedback and helpful discussions. This work was supported by an Alfred P. Sloan Foundation Research Fellowship and NIH Director’s New Innovator Award 1DP2LM011952-01 (to B.B.A.); the Harvard Laboratory of Systems Pharmacology Center Grant P50GM107618 (to B.B.A. and M.C.); an NIH K99AI114952 Grant (to S.T.); and Turkish Academy of Sciences GEBIP Fellowship and TUBITAK 115S934 Grant (to M.C.). The work for the making of the microfluidic devices at the BioMEMS Resource Center was supported by NIH 5P41EB002503-12.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600372113/-/DCSupplemental.
References
- 1.Connolly LE, Edelstein PH, Ramakrishnan L. Why is long-term therapy required to cure tuberculosis? PLoS Med. 2007;4(3):e120. doi: 10.1371/journal.pmed.0040120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry CE, 3rd, et al. The spectrum of latent tuberculosis: Rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009;7(12):845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sacchettini JC, Rubin EJ, Freundlich JS. Drugs versus bugs: In pursuit of the persistent predator Mycobacterium tuberculosis. Nat Rev Microbiol. 2008;6(1):41–52. doi: 10.1038/nrmicro1816. [DOI] [PubMed] [Google Scholar]
- 4.Allison KR, Brynildsen MP, Collins JJ. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr Opin Microbiol. 2011;14(5):593–598. doi: 10.1016/j.mib.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5(1):48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 6.Aldridge BB, et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science. 2012;335(6064):100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kester JC, Fortune SM. Persisters and beyond: Mechanisms of phenotypic drug resistance and drug tolerance in bacteria. Crit Rev Biochem Mol Biol. 2014;49(2):91–101. doi: 10.3109/10409238.2013.869543. [DOI] [PubMed] [Google Scholar]
- 8.Meniche X, et al. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci USA. 2014;111(31):E3243–E3251. doi: 10.1073/pnas.1402158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joyce G, et al. Cell division site placement and asymmetric growth in mycobacteria. PLoS One. 2012;7(9):e44582. doi: 10.1371/journal.pone.0044582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh B, et al. Asymmetric growth and division in Mycobacterium spp.: Compensatory mechanisms for non-medial septa. Mol Microbiol. 2013;88(1):64–76. doi: 10.1111/mmi.12169. [DOI] [PubMed] [Google Scholar]
- 11.Santi I, Dhar N, Bousbaine D, Wakamoto Y, McKinney JD. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat Commun. 2013;4(May):2470. doi: 10.1038/ncomms3470. [DOI] [PubMed] [Google Scholar]
- 12.Kieser KJ, Rubin EJ. How sisters grow apart: Mycobacterial growth and division. Nat Rev Microbiol. 2014;12(8):550–562. doi: 10.1038/nrmicro3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manina G, Dhar N, McKinney JD. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe. 2015;17(1):32–46. doi: 10.1016/j.chom.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Gefen O, Gabay C, Mumcuoglu M, Engel G, Balaban NQ. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc Natl Acad Sci USA. 2008;105(16):6145–6149. doi: 10.1073/pnas.0711712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature. 2014;513(7518):418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- 16.Sukumar N, Tan S, Aldridge BB, Russell DG. Exploitation of Mycobacterium tuberculosis reporter strains to probe the impact of vaccination at sites of infection. PLoS Pathog. 2014;10(9):e1004394. doi: 10.1371/journal.ppat.1004394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prideaux B, et al. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat Med. 2015;21(10):1223–1227. doi: 10.1038/nm.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stiefel P, Schmidt-Emrich S, Maniura-Weber K, Ren Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015;15(1):36. doi: 10.1186/s12866-015-0376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levin BR, Rozen DE. Non-inherited antibiotic resistance. Nat Rev Microbiol. 2006;4(7):556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert P, Collier PJ, Brown MR. Influence of growth rate on susceptibility to antimicrobial agents: Biofilms, cell cycle, dormancy, and stringent response. Antimicrob Agents Chemother. 1990;34(10):1865–1868. doi: 10.1128/aac.34.10.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 22.Shah D, et al. Persisters: A distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grünenfelder B, et al. Proteomic analysis of the bacterial cell cycle. Proc Natl Acad Sci USA. 2001;98(8):4681–4686. doi: 10.1073/pnas.071538098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker N, Nghe P, Tans SJ. Generation and filtering of gene expression noise by the bacterial cell cycle. BMC Biol. 2016;14(1):11. doi: 10.1186/s12915-016-0231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.