Summary
Cardiovascular (CVD) risk assessment with traditional risk factors (age, sex, blood pressure, lipids, smoking and diabetes) has remained relatively invariant over the past decades despite the inaccuracies associated with this approach. However, the search for novel, robust and cost-effective risk markers of CVD risk is ongoing. A large share of the major developments in CVD risk prediction during the past five years have been made in large-scale biomarker discovery and the so called “omics” – the rapidly growing fields of genomics, transcriptomics, epigenetics and metabolomics. This review focuses on how these new technologies are helping drive primary CVD risk estimation forward in recent years, and speculates on how they could be utilized more effectively for discovering novel risk factors in the future.
Keywords: cardiovascular risk, cardiovascular disease, risk factors, biomarkers, metabolomics, genomics, epigenetics, transcriptomics
1. Introduction
“It’s tough to make predictions, especially about the future.”
-Yogi Berra
1.1 Is there a need for new cardiovascular risk factors?
The current American College of Cardiology/American Heart Association (ACC/AHA) guidelines recommend assessing traditional cardiovascular disease (CVD) risk factors every four to six years in all adults 20 to 79 years of age [1]. The concept of CVD risk factors is, therefore, an integral and central part of modern healthcare. The term “risk factor” in itself was used initially by the Framingham Heart Study investigators William Kannel and Thomas Dawber in their 1961 landmark paper “Factors of risk in the development of coronary heart disease – six year follow-up experience” [2]. In that seminal work, the authors demonstrated that high blood cholesterol levels, elevated blood pressure, smoking, and electrocardiographic abnormalities were associated with an increased risk of incident coronary heart disease over a 6-year follow-up of the Framingham cohort. After 50 years of research, their findings are still very much valid although many other modifiable (diabetes, sedentary lifestyle, and smoking) and non-modifiable (age, family history of CVD, and race) CVD risk factors have since been identified.
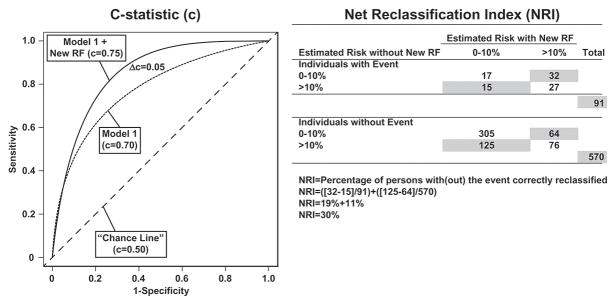
Although the discovery of the common modifiable CVD risk factors has enabled the development of population-based and personal interventions, CVD risk assessment on the individual level still lacks precision. The clinical CVD risk prediction models, i.e. risk scores, do not provide their user with confidence intervals and may not therefore reflect the within-person uncertainty in risk [3]. Even the best risk scores achieve a c-statistic (a measure of discrimination also known as the area under the ROC curve (Figure 1), of approximately only 0.80. This means that a 20% probability still exists that the risk prediction model is not able to discriminate an individual likely to develop CVD on follow-up from one less likely to do so [4]. In addition, the current ACC/AHA guidelines recommend discussing both lifestyle and pharmacological preventive interventions with individuals when their estimated 10-year risk of a CVD event of an individual exceeds 7.5% [1]. At a 10-year 7.5% CVD risk level, 92 to 93 out of 100 patients in whom treatment is initiated may not have a myocardial infarction or stroke over the next 10 years (even if the therapy would have been withheld), i.e. they may not necessarily benefit from therapy [1]. Furthermore, 67 primary prevention patients with a 5-year CVD risk of 5–10% need to be treated with statins to prevent one CVD event [5]. Because of inaccuracies like these even at the dawn of the “precision medicine” era, some scientists have argued for the redistribution of resources away from precision medicine research and efforts into public health programs [6].
Figure 1.
Concepts of two metrics for evaluating improvement in discrimination when a new risk factor (RF) is added to an existing model: 1) improvement in c-statistic (the proportion of pairs in which the person who experienced the event had a higher predicted probability of experiencing the event than the subject who did not experience the event) and 2) net reclassification index, the percentage of persons with and without the event correctly reclassified.
In this context it is important to distinguish risk prediction at the individual levels from that at the population level [7]. Of note, the primary care physician typically treats individual patients, and not populations. In order to improve risk prediction at the individual level, and thereby enhance patient care, novel risk factors and more accurate CVD risk profiling may be necessary. Eventually, improved risk prediction at the individual level may translate into improved clinical care of the individual, which in turn will eventually augment the health of the population.
1.2 Desirable features of a risk factor for the primary prevention of CVD
Epidemiological studies are usually performed in relatively asymptomatic community-dwelling adults. The results obtained from these studies about risk factors are, therefore, often best generalizable for use in primary care and primary prevention settings. There are several desirable features of a risk factor for the primary prevention of CVD (Table 1). In addition to being accurate, acceptable, reproducible, universally applicable and easily interpretable, a good risk factor should be strongly associated with the outcome (disease) and it should provide incremental value to risk prediction. Additionally, clinical use of the risk factor should translate into improvement in clinical outcomes, and it should be cost-effective when measured in primary care settings. Cost-effectiveness, however, does not only depend only on the price of the risk factor and the quality of evidence showing that screening is effective in improving outcomes, but also on the number needed to screen, price of the outcome and the benefits and harms of screening.
Table 1.
Desirable features of a risk factor for the primary prevention of cardiovascular disease.
| Characteristic |
|---|
| Easily interpretable results |
| Strong association with outcome |
| Acceptable to the population |
| Cost-effective |
| Adds to known risk prediction models (improves discrimination) |
| High sensitivity and specificity |
| Changes treatment |
| Known reference values |
| Reproducible |
| Applicable to all gender, age, and ethnic groups |
| Biologically plausible |
| Can be effectively treated |
The most important step to identifying a novel risk factor is to demonstrate an association between the risk factor and outcome with adjustment for other established and relevant risk factors. In addition to demonstration of a simple association with clinical outcome of interest, the new risk factor should also improve risk discrimination (statistically speaking) over a best available model that incorporates several known predictors of disease and that is the current standard of care [8,9]. Discrimination in this context refers to whether the risk factor adds capacity to discriminate between persons who will subsequently experience the outcome from those who will not.
The improvement in discrimination is often measured with the previously mentioned c-statistic that also has some downsides (Figure 1) [8]. Albeit being quite informative, the c-statistic only measures if the risk score is providing appropriate rank ordering of risk of non-cases and cases, not how much greater the risk is between these two groups [10]. In addition, when a new risk factor is added to a model that already includes a powerful risk factor, such as age, the increases in c-statistic are usually very small in magnitude. On the other hand, conventional risk factors such as age or sex may perform poorly when the study population consists solely of elderly, high-risk participants, especially when the age range is narrow or the gender distribution is uneven. When a novel risk factor is studied in such a population, it can easily lead to overestimation of the effect of this risk factor. Furthermore, the incremental contribution of novel risk factors can also be overestimated when the c-statistic for the baseline model is low or when all current clinically relevant variables are not included in the basic model. The results of analyses for association and improvement in discrimination must therefore always be interpreted within the context of the study population and the study methods.
Other commonly used methods for measuring improvement in risk discrimination are the net reclassification improvement and integrated discrimination improvement, which measure the new model’s ability to more accurately stratify individuals into higher or lower risk categories of clinical importance (Figure 1) [9]. In an example based on the Women’s Health Study, inclusion of HDL cholesterol with the other classical risk factors resulted in a minimal increase in c-statistic from 0.77 to 0.78. However, at the same time, over 34% of women classified at 5% to 20% 10-year risk in the model without HDL cholesterol changed risk category when HDL cholesterol was included in the model [10]. In addition, this new risk estimate was a more accurate representation of actual risk for over 99% of women reclassified. Reclassification can therefore be important for the clinician, even if the increases in c-statistic are nominal. Limitations of the reclassification metrics are that they depend on model calibration (ability to estimate the probability of an event across the whole range of prognostic estimates) and the somewhat arbitrary number of risk categories and the thresholds between categories.
1.3 Novel CVD risk factors in the 2010s
Despite the previously listed challenging requirements for new risk factors, significant advances have been made in the past 25 years in the fields of imaging, biomarker discovery and vascular assessment especially through the discovery of surrogate markers of CVD that can also be used as risk factors (Table 2). For example, assessment of subclinical atherosclerosis with ultrasound measurement of carotid intima-media thickness or with coronary calcium scoring from cardiac CT have been shown to improve risk discrimination in population studies, as compared with the traditional risk factors [11,12]. In addition to imaging, data from a few studies suggest that newer (and older) tests of vascular function such as the ankle-brachial index and aortic pulse wave velocity, a measure of large artery stiffness, may improve risk discrimination [13–15]. Out of these risk factors, coronary calcium scoring and the measurement of ankle-brachial index are already recommended by the current guidelines to inform decision making and they are consequently slowly being adopted into mainstream clinical practice [1].
Table 2.
Cardiovascular risk factors for primary prevention.
| Risk factor category | Risk factor | ||||
|---|---|---|---|---|---|
| Biomarkers | Lifestyle | Vascular testing | Genomic Research | Disease | |
| Classic (>25 yrs) | Lipids Glucose |
Sedentarism Obesity Diet Obesity Smoking Stress |
ECG-LVH | Parental history | Hypertension Atrial fibrillation Kidney failure |
| Recent (5–25 yrs) | Natriuretic peptides hs-CRP Uric acid Fibrinogen Vitamin D Apolipoproteins Interleukins Lipoprotein (a) Cystatin C Urine albumin Tissue hormones Lp-PLA2 Homocystein |
Psychosocial factors | Arterial stiffness CAC score echo-LVH Carotid IMT ABI ED |
APOE polymorphism LDLR mutations GWAS loci |
|
| Novel (<5 yrs- future) | HDL function LDL with Apo-CIII hs-Troponin Galectin-3 Soluble ST2 GDF-15 Cathepsin S NGAL Multimarker scores Metabolomics |
Sequencing loci*
Genetic risk scores Epigenetic profiling* Expression profiling* |
|||
ECG-LVH, electrocardiographic left ventricular hypertrophy; APOE, apolipoprotein E; hs-CRP, high sensitivity C-reactive protein; CAC, coronary artery calcium score; LDLR, LDL receptor; LVH, left ventricular hypertrophy; IMT, intima-media thickness; ABI, ankle-brachial index; ED, endothelial dysfunction; Lp-PLA2, lipoprotein-associated phospholipase A2; HDL, high-density lipoprotein; GWAS, genome-wide association, LDL, low-density lipoprotein; Apo-CIII, apolipoprotein C-III; LOF, loss-of-function; GDF, growth differentiation factor; NGAL, neutrophil gelatinase-associated lipocalin.
May not be applicable to primary prevention.
In spite of these advances in imaging and vascular assessment during the past two decades, a large share of the very recent major developments in CVD risk prediction have been made in large-scale biomarker discovery and so called “omics” the rapidly growing fields of metabolomics, genomics, epigenetics and transcriptomics (Table 2). This review will therefore focus on how these new technologies have helped push primary CVD risk estimation forward in recent years, and speculates on how they could be utilized more effectively for discovering novel risk factors in the future.
2. Biomarkers
Biomarkers are biological measures that are used as an indicator of the presence (or absence) or severity of a particular disease (or state of health). The wide definition of the term includes all imaging tests and any recordings obtained from a person, but most often biomarkers are considered to be compounds isolated from serum although urine or other biological specimens. The traditional CVD risk factors include two sets of biomarkers, namely blood glucose and lipids for diagnosing diabetes and hypercholesterolemia (Table 2). Other biomarkers, such as B-type natriuretic peptide, C-reactive protein, estimated glomerular filtration rate and urine microalbumin, have also been previously shown to be strongly associated with CVD prognosis both in patients and in the general population and used for CVD risk assessment in select settings [1,16–18].
While the traditional search for novel biomarkers has been a slow process, new methods and technologies now allow for large-scale high throughput screening of biological specimens for biomarker discovery. It seems that biomarker-based CVD risk prediction, will slowly move from the use of single biomarkers, to composite marker scores based on several biomarkers, and finally to metabolomics that allows for the precise identification of hundreds to thousands of small molecules in biological samples.
2.1 Lipoprotein and lipid-related biomarkers
High density lipoprotein (HDL) cholesterol
The reverse cholesterol transport process, which removes excess cholesterol from peripheral cells to the liver for excretion, is considered one of the most important antiatherogenic functions of HDL [19]. Numerous epidemiological studies have robustly shown that high concentrations of plasma HDL cholesterol are associated with a lower risk of CVD [17,19]. Increasing HDL cholesterol levels was therefore considered to be a promising therapeutic strategy until several randomized studies of HDL cholesterol-raising drugs, such as niacin and cholesteryl ester transfer protein inhibitors, failed to demonstrate any benefit on the clinical outcome of patients when added to statin therapy [20,21]. Furthermore, genetic mechanisms that raise plasma HDL cholesterol do not seem to lower risk of myocardial infarction in Mendelian randomization studies (a topic addresser later in this review), as they have done for low density lipoprotein (LDL)-related genetic loci [22]. In fact, a recent paper by Zanoni et al. suggested that persons with a loss-of-function variant in SCARB1, the gene encoding the major receptor for HDL cholesterol, have elevated levels of HDL cholesterol and, surprisingly, increased odds of coronary heart disease [23]. Although HDL cholesterol still remains a strong predictor of CVD outcome, due to these negative results from trials and genetic studies, epidemiological research has started to focus on the functional properties of HDL, rather than solely on its mean cholesterol concentration, and one emerging notion is that HDL may be a marker of risk, rather than a causal risk factor per se.
HDL function
One relevant functional property of HDL could be the HDL cholesterol efflux capacity, the ability of HDL to accept cholesterol macrophages, which is a key step in reverse cholesterol transport [24]. So far, two population-based studies have studied the prognostic significance of HDL cholesterol efflux capacity. In the Dallas Heart Study, baseline HDL was not predictive of CVD while a 67% reduction in CVD risk was observed in participants in the highest quartile of cholesterol efflux capacity versus the lowest quartile in regression models that also included traditional risk factors and HDL particle concentration [25]. In addition, adding cholesterol efflux capacity to traditional risk factors improved discrimination and reclassification indexes. Results from a nested case-cohort sample of a population study from United Kingdom were supportive of these findings [26]. However, higher cholesterol efflux activity has also been paradoxically associated with an increase in risk of myocardial infarction and stroke although these observations were made in a case-cohort setting and with a hospital based sample [27].
In addition to cholesterol efflux capacity, HDL particle size and number are other factors that are thought to potentially reflect HDL function but the results from studies assessing the relation between these factors and CVD outcome have been mixed [28,29]. Nonetheless, given the complexity of HDL, many potential mechanisms of HDL function will require further investigation in the future in relation to CVD risk.
Apolipoprotein C-III (Apo-CIII)-containing lipoprotein subclasses
Apo-CIII is a protein that is a component of very low density lipoprotein (VLDL), low density lipoprotein (LDL), and HDL. LDL that contains proinflammatory Apo-CIII has been shown to have a markedly altered metabolism and proatherogenic effects on vascular cells [30]. Several studies performed in patients have demonstrated an association of CVD events with blood Apo-CIII level in total plasma or in VLDL and LDL, but results from prospective cohort studies are limited [30]. In a recent report, concentrations of LDL with Apo-CIII in 1476 community-dwelling participants were associated with risk of coronary heart disease, even when adjusting for HDL cholesterol, LDL cholesterol, apoB, triglycerides, or the total cholesterol/HDL-ratio [31]. In addition to LDL with Apo-CIII, increased circulating plasma Apo-CIII also has been linked with CVD risk in the general population [30,32]. The validity of these findings are supported by the observations that humans with genetic deficiency of Apo-CIII have lower triglyceride and LDL cholesterol levels as well as reduced atherosclerosis [32]. Although more data are needed, Apo-CIII rich containing lipoproteins seem to have direct atherogenic consequences and could be potentially used as risk factors for coronary heart disease.
2.2 Cardiac biomarkers
High sensitivity troponin T/I (hs-TnT/I)
Although drastic elevations of cardiac troponins have been used as markers of acute myocardial injury in the clinic since the early 1990s, they have had no role in primary prevention of CVD. However, by the late 2000s, ultra-hs-TnT assays capable of measuring levels as low as 0.005 ng/ml had been developed that enabled researchers to assess whether minute detectable TnT levels (even in healthy individuals) would be suitable for screening the general population for cardiac dysfunction [33].
In 2010, hs-TnT was found to be associated with pathological cardiac phenotypes [34], all-cause mortality [34], heart failure [35] and CVD death [35] in two large prospective observational cohorts. These findings were later confirmed in other Northern American and European population cohorts, including when hs-TnI was used instead of hs-TnT as the biomarker [36,37], In addition, modest improvements in model discrimination were observed in all of these studies when hs-TnT was added to prediction models that included the traditional risk factors [34–37], Although hs-Tn is most likely the cardiac biomarker closest to being adopted into mainstream diagnostic and prognostic use, more research is still needed to demonstrate its cost effectiveness in different primary care settings, to determine diagnostic thresholds, and to establish parameters for hs-Tn testing for screening or other purposes.
Other cardiac biomarkers
Other biomarkers such as those of cardiac fibrosis (galectin-3 and soluble ST2) [38], apoptosis (growth differentiation factor 15 [GDF-15]) [38], inflammatory activity (cathepsin S) [39] and renal dysfunction (neutrophil gelatinase-associated lipocalin [NGAL]) [40] have also been recently used to predict CVD in population cohorts. The results of these studies are summarized in Table 3.
Table 3.
Association between biomarkers and cardiovascular outcomes in various population studies.
| Biomarker | Cohort
|
|||||
|---|---|---|---|---|---|---|
| FHS [38,41,50] | Dallas [42,48] | FINRISK [43,44] | PREVEND [39] | RB [40,46,47] | ULSAM [45,49] | |
| Galectin-3 | M, HF | M, CVDM, HF | M | CVDM* | ||
| sST2 | M, HF, CVD, S | M, CVDM | No association | |||
| GDF-15 | M, HF, CVD, S | M*, CVDM | M*, CVDM | M*, CVDM* | ||
| Cathepsin S | M, CVDM | |||||
| NGAL | M, CVDM* | |||||
FHS, Framingham Heart Study; Dallas, Dallas Heart Study; PREVEND, Prevention of REnal and Vascular End-stage Disease study; RB, Rancho Bernardo study; ULSAM, Uppsala Longitudinal Study of Adult Men; M, mortality; HF, heart failure; CVDM, cardiovascular disease mortality; CVD, cardiovascular disease; S, stroke. The biomarkers reflect cardiac fibrosis (galectin-3 and soluble ST2 [sST2]), apoptosis (growth differentiation factor 15 [GDF-15]), inflammatory activity (cathepsin S) and renal dysfunction (neutrophil gelatinase-associated lipocalin [NGAL]).
Improvement in discrimination when biomarker was added to a model that included conventional risk factors.
Evidence on the prognostic significance of most of these biomarkers is still limited or missing. Several [41–46], although not all [47], studies have found an association between CVD events and blood levels of galectin-3 or soluble ST2 (Table 3). However, although these two markers of myocardial fibrosis have been proposed as early markers of heart failure, most of these studies reported no improvement in discrimination, especially when the models included natriuretic peptides which are still one of the mainstays of a heart failure diagnosis [41,42,45,46]. Furthermore, because soluble ST2 has a low specificity, it cannot be currently considered a prime candidate for heart failure risk assessment or diagnosis [38]. The evidence of the predictive value of circulating levels of cathepsin S and NGAL are even more limited as their predictive values have been assessed in single studies, and no improvement in discrimination has been demonstrated for cathepsin S (Table 3) [48,49].
GDF-15, in contrast, seems to show some promise as a cardiac risk factor as it has been found to predict CVD events and to improve discrimination in numerous studies quite consistently (Table 3) [44,50–53]. However, no universally accepted reference values for GDF-15 currently exist and there is no evidence whether measurement of GDF-15 would improve clinical decision making over and above the currently used clinical biomarkers.
2.3 Biomarker scores
Although numerous papers on several new biomarkers have recently been published, relatively few of these biomarkers consistently associate with outcome and improve CVD risk discrimination. Instead of single biomarkers, more attention has recently been focused on composite risk scores formed of biomarkers from one or even several pathophysiological pathways – a “multimarker” approach, under the assumption that several biomarkers may outperform individual ones.
One of the first studies to take this approach was published in 2006 when the Framingham Heart Study investigators measured 10 biomarkers from distinct biologic pathways in 3209 community-dwelling participants [54]. In this study, persons with multimarker scores (based on 5 significant biomarkers) in the highest quintile had a 4-fold greater risk of dying and 2-fold greater risk of having a major CVD event as compared with those with scores in the lowest quintile. However, even with 10 biomarkers, CVD event risk discrimination was improved only marginally with an increase in c-statistic from 0.76 to 0.77. This risk score was later amended with some of the novel biomarkers for cardiac stress. This score, composed of natriuretic peptides, high sensitivity CRP, soluble ST2, GDF-15 and hs-TnI was strongly predictive of all-cause death, CVD events, and heart failure while improving the c-statistic by 0.01–0.02 in the same Framingham population [44]. Participants in the highest quartile of the multimarker score had a 3-fold risk of death, 6-fold risk of heart failure, and 2-fold risk of CVD events. Later, the same multimarker score was found to be significantly associated with incident atrial fibrillation although no improvement in risk discrimination was observed [44,55].
In 2008, Swedish investigators determined a biomarker score based on four biomarkers: TnI, N-terminal pro-B-type natriuretic peptide (nt-proBNP), cystatin C and C-reactive protein for 1135 participants of the Uppsala Longitudinal Study of Adult Men (ULSAM) [56]. As expected, all four biomarkers were significantly associated with CVD outcome. The surprising finding was the marked improvement in risk discrimination when the four biomarkers were incorporated into a model with established risk factors, with the c-statistic increasing from 0.688 to 0.784 as compared with the modest increases around 0.01–0.02 usually observed in primary prevention studies. However, as previously mentioned, all results must be interpreted within the context of their limitations. The ULSAM cohort consists solely of elderly persons of similar age and the same gender. Hence, the conventional risk factors did not perform as well as in a general adult population; therefore the lower c-statistic of the base model made it easier to improve risk prediction metrics such as the c-statistic with new biomarkers.
The initial significant findings of the Framingham and ULSAM investigators drew a great deal of attention to biomarker scores among CVD epidemiologists. In recent years, the trend in multimarker scores has been towards an ever increasing number of biomarkers as high-throughput multiplex analyses have enabled the fast and simultaneous determination of tens or even hundreds of serum biomarkers. In 2010, the number of assayed biomarkers for creating a multimarker score had already increased to 30 [57]. In this study based on a population of 8000 participants from Finland and Northern Ireland, the strongest associations between biomarker levels and CVD outcome were found for nt-proBNP, C-reactive protein, and sensitive troponin I. A biomarker score based on these three biomarkers also resulted in improved risk discrimination in internal validation. Very recently, up to 284 cardiometabolic biomarkers were analyzed in participants of a diabetes drug trial. 10 of these biomarkers (nt-proBNP, trefoil factor 3, apolipoprotein B, angiopoietin-2, osteoprotegerin, α-2-macroglobulin, glutathione S transferase α, GDF-15, hepatocyte growth factor receptor, and chromogranin A) were predictive of CVD composite outcomes, with c-statistic increasing from 0.64 for the clinical variables to 0.71 [58].
In general, the published data suggest that the simultaneous addition of several biomarkers related with several pathogenic pathways improves the risk stratification for CVD disease beyond that of a model that is based only on established risk factors and single biomarkers. One promising method of finding novel pathways may be the use of metabolomics, which allows for the screening of an even larger number of small molecules than regular laboratory testing.
2.4 Metabolomics
Metabolites are the intermediates and products of metabolism that represent a diverse group of low-molecular-weight structures such as lipids, amino acids, peptides, nucleic acids, organic acids, vitamins, thiols, and carbohydrates [59]. Metabolomics is the study of these metabolites present in cells, tissues and organs. Specifically, modern metabolomics tries to study the unique chemical fingerprints that cellular processes leave behind thereby potentially helping us understand better the mechanisms underlying health and disease. The interest in metabolomics has recently increased significantly through technological advances made in platforms commonly used to analyze metabolites, namely nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS). Modern versions of these technologies now enable the high-throughput determination of up to thousands of small molecular metabolites from peripheral blood samples [59].
Metabolite profiling has been used extensively and successfully to identify biomarkers of diabetes and the insulin-resistant state that precedes overt type 2 diabetes [60]. However, until recently, there was a paucity of evidence establishing that metabolic profiling could provide any additional value over the traditional risk factors for predicting CVD risk in epidemiological studies. Only in 2013 was it demonstrated in a case-cohort sample of the Malmö Diet and Cancer Cardiovascular Cohort that an amino acid score composed of tyrosine, phenylalanine and isoleucine that had been previously used to predict new-onset diabetes, was also associated with CVD [61]. In this study, participants in the highest quartile of the amino acid score had 2.2-fold odds of having a CVD event during follow-up. After this initial discovery with a predefined risk score, a few other studies have also demonstrated that metabolomics can also be used for the discovery of novel metabolic profiles that are associated with CVD. In 685 participants of the population-based Bruneck study, 135 lipid species from 8 different lipid classes were profiled with MS [62]. The authors observed that molecular lipid profiling results in significant improvement in CVD risk discrimination beyond the information provided by traditional risk factors, including lipid measures. In two other separate studies, Swedish investigators used MS, while a Finnish-British-American collaboration used NMR to discover metabolite profiles consisting of 4 different metabolites associated with CVD disease [63,64]. However, risk discrimination was not substantially improved in either study.
Although evidence of the benefits of non-targeted metabolite profiling in CVD risk prediction is slowly accumulating, the use of metabolomics in clinical practice is still limited by the complexity of the metabolism and its associations with other biological phenomena. Metabolomics currently remains a promising and attractive tool that is being applied predominantly in research settings; its clinical applicability remains to be demonstrated.
3. Genomic research
The first observations of genetic susceptibility to CVD were made over 50 years ago. The range of genetic variance in, for example, coronary artery disease is currently estimated to be between 40% and 60%, based on findings from twin studies [65]. However, the exact mechanisms underlying this hereditary component are still a mystery to a large extent. Even though the genetic underpinnings of some Mendelian disorders that predispose to CVD, such as familial hypercholesterolemia, were identified already 30 years ago, these rare diseases do not explain the variance in CVD prevalence on the population level.
In the 1990s and early 2000s, numerous studies tried to find an association between potential candidate genes, selected based on known disease pathways, and CVD outcomes. However, the results from these studies were often limited, could not be reproduced or explained only an insignificant part of trait variance [66]. At this time, the notion of “common disease-common variant” started to gain more popularity. This hypothesis predicts that common diseases are determined by a combination of common genetic variants with very small effects that would in aggregate result in the adverse CVD phenotype.
After the early 2000s, the development of microarrays containing ever-increasing numbers of DNA sequences has made it possible to analyze in an efficient manner more than 1 000 000 single nucleotide polymorphisms (SNPs), common variants in DNA sequence that occur at a frequency of over 1–5 %. This technology initially raised high expectations as it enabled researchers for the first time to explore the associations between complex diseases and nearly the whole genome without prior hypothesis, referred to as genome-wide association (GWA) studies. After the first cardiovascular GWA studies 10 years ago, tens of SNPs associated with coronary heart disease and stroke have since been identified including the chromosome 9p21 locus that can be considered the most robust single genetic marker of coronary artery disease today [67–69]. The initial results of these studies were slightly underwhelming, particularly considering the expenses and the high expectations that motivated these studies, as most of the significant SNPs had very small effect sizes (odds ratio <1.2) and explained together only ~10% of the estimated heritability [67]. However, an approximate joint association analysis was recently performed using genome-wide complex trait analysis software to discover 202 variants in 129 loci associated with coronary heart disease. These loci together explained 28% of the estimated heritability of coronary heart disease, a substantial proportion by any measure [70].
So where is the remaining 72% of heritability of coronary heart disease? Although a GWAS-based approach will not most likely provide definite answers to this question, several explanations may contribute to the disagreement between estimated and observed heritability. First, CVD phenotyping is difficult and most of the population-based genetic studies are large-scale collaborations by tens of cohorts from different ethnicities with their own disease definitions. For example, phenotypes of coronary artery disease have been consistently used interchangeable with that of myocardial infarction. Second, variants of low and very low allele frequencies (<1%) may not be captured by current GWA arrays even when using modern software and genotype imputation techniques. Third, current estimates of CVD heritability could be exaggerated. Fourth, it might be that most heritability is not actually missing, but instead the individual effects are too small to pass the stringent significance tests adjusted for multiple comparisons (p < 10−8). In fact, investigators have recently demonstrated that the use of SNP arrays followed by whole-genome imputation of ~17 million variants may capture nearly all of the heritable variance for human complex traits, such as height and weight [71]. However, no matter what research method is used, the notion of completely independent genetic and environmental risk factors needs reconsideration, as both are components of a larger intertwined complex molecular system that, as a whole, results in the complex phenotype.
In the next part of this review, we will focus on the post-GWA study era of CVD genomics. We will review how Mendelian randomization has helped establish if the associations of some traditional and novel CVD risk factors are truly causal, and whether genetic risk scores based on previously discovered SNPs can be used to improve CVD risk assessment. In addition, we will briefly review emerging areas of genomic research – whole genome sequencing, epigenetics, and transcriptomics – that will hopefully advance our understanding of CVD pathogenesis and can thereby also be used to improve CVD risk estimation.
3.1 Mendelian randomization
The associations observed in epidemiological studies between risk factors and CVD may be causal. However, true causality is usually challenging to demonstrate with observational study designs due to the confounding and inherent and often unavoidable biases associated with this observational approach. In the absence of an ethical, practical, and economical randomized trial, the epidemiologist must seek alternative ways to demonstrate causality.
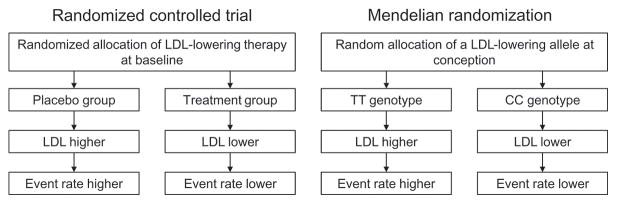
Mendelian randomization represents a novel epidemiologic study design akin to a randomized trial that can be used to address the question whether an association between a risk factor and disease is truly causal (Figure 2) [72]. The basic principle of Mendelian randomization is to use common genetic variants closely associated with a risk factor as the ‘intervention’. Random allocation of alleles at fertilization mirrors the randomization in the controlled trial setting. Because allele distribution is mainly random, Mendelian randomization should be free from any confounding or reverse causation. And because the genetic variant is closely associated with a risk factor of interest, individuals with these variants should be predisposed to the outcome of interest if the risk factor is truly causal (Figure 2).
Figure 2.
Principles of a randomized controlled trial and Mendelian randomization.
There are, however, certain assumptions and limitations to Mendelian randomization [72,73]. Most importantly, a suitable genetic variant may not be available or can have other effects beyond its effect on the specific biomarker being studied, i.e., pleiotropy. Pleiotropy can counteract any effects of the variant on the disease acting via the biomarker, or alternatively, a positive direct association between the genetic variant, and may be mistakenly be interpreted as a causal association with the biomarker. Furthermore, allocation of alleles at conception may not be perfectly random when two loci are in close proximity to each other on the same chromosome. The probability of allocation of these alleles from parents to offspring will be correlated, a phenomenon known as linkage disequilibrium. In addition, adequate statistical power is required to detect the association between the risk allele, the intermediate factor, and the outcome of interest.
Several Mendelian randomization studies have been performed in recent years to confirm the association between CVD and traditional risk factors such as LDL cholesterol [74], triglycerides [75], hypertension [76] and hyperglycemia [77]. Mendelian randomization has also been used to demonstrate the causal effects of lifestyle-related risk factors such as obesity [78], smoking [79] and alcohol [80] for CVD although some of the mechanisms may ultimately be through other intermediate factors such as hypertension. More interestingly, Mendelian randomization has been recently used to assess whether certain novel biomarkers that have been found to associate with CVD are truly causal and therefore also potential targets for therapy, or only ‘markers’ of disease risk when they can be used only for guiding diagnosis, prognosis and treatment.
As already briefly stated in the beginning of this review, Mendelian randomization has been utilized to demonstrate that some genetic mechanisms that raise plasma HDL cholesterol do not seem to lower risk of myocardial infarction [22]. Together with the negative results from HDL-elevating drug trials, it is now doubted whether elevated HDL is causally protective against coronary artery disease. The causalities of several other biomarkers that have been found to associate with CVD, such as uric acid [81], fibrinogen [82], adiponectin [83], lipoprotein-associated phospholipase A2 [84], secretory phospholipase A2 [84,85] and C-reactive protein [86] have also been assessed using Mendelian randomization. However, none of the genetic variants that affect the circulating levels of these biomarkers had any impact of CVD outcome and it is therefore unlikely that altering their levels would be a useful therapeutic goal for preventing CVD. In contrast, several studies have shown that genetic variants that increased lipoprotein(a) levels predispose to coronary artery disease [87], aortic valve stenosis [88] and heart failure [89] and that persons carrying the allele leading to reduced interleukin-6 receptor binding show a lower inflammatory response and reduction in coronary disease events [90].
Mendelian randomization has emerged as a valuable method for assessing the causality of a novel risk factor. However, it has several requirements and limitations that need to be taken into consideration when designing a study that uses this method. Furthermore, Mendelian randomization is an important ancillary research method that does not eliminate the need for randomized controlled studies, which still remain the golden standard for establishing causality.
3.2 Genetic risk scores
Because of the complex genetic background of traits such as CVD, no single risk allele can be used for risk prediction in the general population. Genetic risk scores (GRSs), however, can be used to incorporate the information from multiple disease-associated SNPs for more effective CVD risk prediction [91]. The simplest method for building a GRS is to sum the number of risk alleles (0, 1 or 2) across all genetic variants associated with the trait of interest. For improved accuracy, effect sizes across different variants should also be taken into account [91]. In addition to predicting CVD risk, genetic risk scores can also be used to evaluate the genetic contribution of intermediate traits and risk factors to CVD, and for Mendelian randomization.
The first epidemiological cohort studies to use a GRS to predict CVD were published in 2010. Investigators of the Women’s Genome Health Study concluded that a GRS comprising 101 SNPs associated with coronary artery disease was not significantly associated with incidence of total CVD [92]. In contrast, a more strictly selected 13-SNP GRS was strongly associated with incident coronary heart disease in a Finnish-Swedish collaboration [93]. This positive association between a GRS and coronary artery disease has since been replicated twice [94,95]. Later, several papers have demonstrated that GRSs composed of blood pressure- or intermediate trait-related SNPs can also be effectively used to predict coronary heart disease and stroke [96–99].
In spite of several studies that have demonstrated a strong association between GRSs and incident CVD, only some of these studies have demonstrated significant, albeit modest, improvements in model c-statistics [95,99]. In addition, GRS studies have not always included family history of CVD in the adjustment variables. Although the GRS approach could be relative easy to incorporate into clinical practice, it should be first robustly proven that the use of GRSs provides any additional benefit over and above the simple question “Does anyone in your family have a history of CVD at a young age?” before widespread clinical use can be recommended.
3.3 Exome and whole genome sequencing
GWA studies have been successfully used to identify common gene variants associated with CVD that have allele frequencies, but a significant share of the heritability of CVD is still unexplained. Although the common disease-common variant hypothesis has become the dogma of the day, it is also possible that some of the missing genetic component of CVD could be attributed to rare genetic variants with large effect sizes [100]. Because GWA studies are generally not effective in identifying rare and extremely rare variants, methods such as whole genome and whole exome sequencing are needed to capture these rare variants. Specifically, whole genome sequencing is used to determine the precise order of bases in the whole genome whereas whole exome sequencing determines the order of bases in a DNA molecule only in the exome, the 1% protein-coding portion of the genome.
With the recent development of next-generation sequencing methods, whole genome and exome sequencing have become considerably less expensive and feasible. In fact, even the cost of the more expensive whole-genome sequencing (without upstream costs) is now approaching three-figure numbers – a strikingly low number compared to the approximate 2.7 billion dollar price tag of the Human Genome Project [101]. This significantly reduced price along with increased availability has made DNA sequencing feasible in both clinical and epidemiological settings.
Whereas sequencing to date has mainly been limited to revealing genetic variants in a small number of affected individuals with rare forms of heart disease such as cardiomyopathies, next-generation sequencing is already being introduced at the population level [102]. Interest in rare loss-of-function gene variants significantly peaked after the discovery of several LDL-cholesterol lowering and cardioprotective PCSK9 gene mutations that led to the development of a whole new drug class, the PCSK9 inhibitors [103]. Thereafter, whole exome and candidate gene sequencing have been successfully used in epidemiological settings to find several other rare loss-of-function mutations in genes that affect both intermediate traits and CVD risk. Within the last two years, new loss-of-function mutations in genes that control LDL-cholesterol and triglyceride levels, such as APOC3, NPC1L1, LDLR, HMGCR, and APOA5, have been discovered in epidemiological cohorts [32,76,104,105]. The fact that carriers of these rare mutations also have an altered risk of CVD makes them potential targets for drug development, e.g., in the cases of NPC1L1 (cholesterol absorption inhibitors) and HMGCR (statins). In addition to rare variants associated with lipid levels, whole exome sequencing was also very recently used to discover a rare coding variant in CLCN6 that is associated with lower blood pressure and hypertension risk [106].
Although significant discoveries in the field of CVD epidemiology have been made in recent years through exome sequencing, the application of DNA sequencing for variant discovery has not always been successful. This is highlighted by the results of Peloso et al. who used an “exome array”, based on coding sequence variants discovered from previous exome-sequencing studies, to genotype over 200 000 low-frequency and rare coding sequence variants in over 56 000 individuals, and tested these variants for association with lipid levels [107]. In the end, no new genes associated with LDL cholesterol were identified and the four novel variants with large effects on HDL cholesterol and triglycerides were not associated with risk for coronary heart disease. The main challenge of the DNA sequencing approach for finding novel CVD risk factors is the notable rarity of the clinically relevant low frequency variants. To illustrate this, over 500 000 mostly rare single-nucleotide variants were identified with exome sequencing in a study by Tennessen et al. that included 2 440 community-dwelling individuals [108]. However, only 2% of the 13 595 single-nucleotide variants each person carried on average were predicted to affect protein function and 95.7% of single-nucleotide variants predicted to be functionally important were rare. In addition to the difficulty of finding rare variants with extreme effects on the phenotype, results from whole genome sequencing have suggested that in the end, common variation contributes more than rare variation to heritability of complex traits, such as HDL cholesterol [109].
These results highlight that although sequencing is an excellent method for revealing potential drug targets and the genetic underpinnings of rare hereditary diseases in family lines, variant discovery for complex disease will require the sequencing of thousands of cases and careful statistical analysis because the power to detect an association is low for most human genes. For the same reasons, non-selective sequencing in asymptomatic individuals will not most likely improve risk discrimination.
3.4 Epigenetics
Epigenetics investigates genetic variations that are caused by external or environmental factors that switch genes ‘on’ and ‘off’. These modifications are often tissue-specific and do not change the DNA sequence, but instead, affect how cells ‘read’ genes. Epigenetic markers may be heritable similar to the genetic features of DNA, but these markers differ for their potential reversibility by external factors [110]. So far, research on epigenetic phenomena related to CVD has relied mostly on cell culture and animal models that are outside the scope of this article. However, recent advances in epigenetic research methods have also enabled the epidemiologic study of epigenetic mechanisms of CVD development and progression. The results of these initial studies provide interesting new insight into the epigenetic control of gene expression in CVD.
There are several epigenetic mechanisms that control gene expression including, but not limited to, histone modification, micro-RNA (miRNA) based mechanisms and DNA methylation [110]. Of these, histone modifications are post-translational modifications that serve to allocate the genome into active regions of euchromatin, where DNA is accessible for transcription, or inactive heterochromatin regions, where DNA is more compact and less accessible for transcription [110]. Although the study of histone modification is slowly being introduced to CVD epidemiology, it has so far been mainly limited to tissue samples in small patient cohorts. We will therefore concentrate on the two other major epigenetic mechanisms – DNA methylation and miRNA-based mechanisms, although the latter will be addressed later in the context of transcriptomics.
DNA methylation is the best understood and most studied epigenetic mechanism that occurs when a methyl group is added to cytosine-guanine dinucleotides (CpGs) [111]. CpG nucleotides are statistically underrepresented in the genome but are found to be concentrated in areas such as around the transcription start sites of genes. These methylated regions, or CpG islands, are relatively stable but may vary during an individual’s lifespan [111]. Methylated regions are not actively transcribed and genes in such regions are silenced whereas nonmethylated regions remain active. DNA methylation is affected by environmental exposure and could therefore be one of the key mechanisms for the development of human disease such as cardiovascular disorders.
Earlier studies that tried to link locus-specific DNA methylation markers with CVD have produced interesting results on the underlying pathophysiological mechanisms of CVD, but their clinical potential remains unclear [112]. In addition, the majority of phenotypic variation due to DNA methylation will not be detected by this approach. Recently developed techniques that couple next-generation sequencing with simultaneous analyses for DNA methylation have now enabled performing epigenome-wide association (EWA) studies based on peripheral blood samples of population cohorts. Although no EWA study of CVD per se has yet been performed, several studies have already identified specific DNA-methylation profiles associated with traditional CVD risk factors, such as smoking [113], diabetes [114], blood lipids [115], and BMI [116,117].
As all of the current EWA studies are cross-sectional, the study of DNA methylation and epigenetics for CVD risk prediction is still very much in its infancy. The International Human Epigenome Consortium is in process of generating at least 1 000 reference baseline human epigenomes from different cell types, which could significantly accelerate epigenome-wide studies for many common diseases [118]. However, already at this point, the dynamic processes of epigenetics highlight the limitations associated with an approach to CVD research that only focuses on the relatively static DNA code. In fact, preliminary results suggest that combining genetic and epigenetic information could have greater utility for complex-trait prediction than what is achieved with solely genetic information [119].
3.5 Transcriptomics
Transcriptomics is very much intertwined with both genetics and epigenetics as it is the study of the transcriptome – the complete set of RNA transcripts that are produced by the genome at any time [120]. Like the epigenome, but unlike DNA sequence, which is normally fixed within an individual, the transcriptome is dynamic as considerable variability in gene expression exists in different tissues, and in response to stimuli. The main component of the transcriptome is messenger RNA (mRNA) that conveys codes from the DNA that the protein-synthesizing machinery in the ribosome translates into protein. Among several other types of RNA are non-coding RNAs, such as miRNAs, that are not translated into a protein but may have other functions, such as regulatory functions [121].
Traditional transcriptomics techniques have been slow, inaccurate, expensive or have required a significant amount of RNA which have challenged their use in epidemiology [120]. However, similar to what has happened in genetics and epigenetics, transcriptomics has recently experienced a revolution brought about by the advent of next-generation high-throughput RNA sequencing technologies. In addition to being fast and cost-effective, RNA sequencing detects more subtle changes in the transcriptome than the previous technologies and does not depend on previous knowledge of the transcriptome [120]. So far, the use of transcriptomics in the discovery of CVD risk factors has mainly been limited to patient populations. For example, gene expression data have been used to help in the discovery novel biomarkers such as soluble ST2 and GDF-15 [122,123]. Furthermore, a blood-based gene expression score for 23 genes has been effective in predicting obstructive coronary artery disease in patients referred for coronary angiography or myocardial perfusion imaging [124,125].
The connection between gene expression and CVD risk, however, has been sparsely investigated in community-based cohorts where RNA is usually limited to blood-derived sources. Three recent papers based on the Framingham Heart Study data have reported an association between platelet gene expression and several factors associated with CVD risk, such as lipids, BMI, CRP and IL-6 [126–128]. These studies are some of the first to demonstrate CVD correlates of gene expression signatures in a large community-based cohort. The results are consistent with the hypothesis that specific peripheral-blood transcripts play a role in the pathogenesis of coronary heart disease and its risk factors. However, no evidence exists yet if transcriptome profiling would improve CVD risk prediction prospectively in the population.
Another emerging field of transcriptomics besides mRNA sequencing is miRNA expression profiling. To date, more than 2500 miRNAs have been identified in the human genome [129]. MiRNAs are non-coding single stranded RNAs that typically downregulate gene expression at a posttranscriptional level [130]. MiRNAs are typically present in tissues and they, in addition to other functions, regulate cardiac growth, vascular development, and angiogenesis. In 2008, their presence was detected also in plasma and serum making them more easily accessible for wide-scale research [130]. The release of certain miRNAs has been associated to cellular damage such as cardiac injury. In addition to cross-sectional studies that have reported altered levels of miRNAs in cardiac patients, some circulating miRNAs have been effectively used to predict patients who are at high risk of developing acute coronary syndrome [131] or of heart failure after a myocardial infarction [132]. In population cohorts, the evidence of the prognostic significance of miRNAs is still very limited. In 820 participants of the Bruneck cohort, two circulating miRNAs demonstrated a negative association and one miRNA a positive association with incident myocardial infarction [133]. In addition, circulating levels of miRNA-328 were found to be associated with prevalent, but not incident atrial fibrillation in the Framingham Heart Study [134]. Although these results are very preliminary and additional research is needed, miRNAs are interesting CVD risk factors because they are also potential therapeutic targets as their function can be efficiently and specifically inhibited with chemically modified antisense oligonucleotides [135].
3.6 Challenges associated with the novel techniques of genomic research
Although next-generation sequencing provides a great opportunity for discovering novel CVD risk factors in the genome, transcriptome and epigenome, several challenges remain with these new techniques, such as the requirement for large study populations, ethical problems, imprecision of sequencing techniques [136], high upstream costs, complex data integration and complicated statistical testing (Table 4). All of these problems need to be considered and solved before these novel techniques can be used more effectively in risk factor research, and especially before they can be used in clinical medicine for diagnosing patients and for risk prediction.
Table 4.
Challenges associated with metabolomics, genomics, epigenetics and transcriptomics.
| Challenge |
|---|
| Need for large study populations |
| Complexity of an informed consent |
| DNA may also reveal properties of the relatives of the patient that have not consented for the test |
| Problems with sample preparation |
| Incomplete coverage of inherited disease genes |
| Current DNA methylation arrays target <2% of the CpG sites present in the human genome |
| Low reproducibility of detection of genetic variation with the highest potential clinical effects |
| Significant upstream costs associated with data storage and analysis |
| Need for extensive quality control, robust normalization algorithms and rigorous statistical testing |
| Challenging integration or different omics data |
| Uncertainty about clinically reportable findings |
| Need for multi-disciplinary research teams |
4. Expert commentary
There are some caveats to consider when assessing the CVD risk of a patient. A plethora of risk assessment tools that usually provide 10-year risk estimates based on the classical risk factors are currently available for the clinician. However, the treating physician must remember that these tools only provide estimates of absolute risk for the population and should not be therefore interpreted as the exact CVD risk of an individual. Second, patients may also find it difficult to understand an estimated CVD risk score. A more concrete risk presentation format such as vascular age, calculated by comparing an individual’s absolute risk to the age at which they would reach that absolute risk if they had ideal risk factors, may help motivate patients adopt healthier lifestyles even if absolute CVD risk is low [137]. Furthermore, because the risk assessment tools usually include only the classical CVD risk factors, physicians must be vigilant for other risk factors or signs of subclinical disease, such as left ventricular hypertrophy or microalbuminuria, which can markedly increase CVD risk. Finally, if a patient is expected to live for the next 50 years instead of the 10-year horizon of the risk assessment tools, even a relatively low burden of CVD risk factors is associated with significant increase in the lifetime risk of CVD [138]. Long-term risk assessment is therefore particularly relevant in younger patients.
CVD risk factor research has made significant advances during the past five years. Although the causative role of HDL cholesterol has been questioned, there are currently no considerable competitors in the market to replace the traditional CVD risk factors that have held their place for decades. However, certain novel ancillary risk factors (see Five-year view) are slowly being introduced into clinical practice. In any case, the need for novel risk factors has not disappeared, as demonstrated by the inaccuracies associated with current risk prediction models.
The search for new CVD risk factors is currently undergoing a significant revolution. In addition to the constant progress that is being made in imaging and vascular assessment, scientists are no longer looking for a single biomarker or genetic locus that would improve risk prediction. Instead, high-throughput screening of biological samples for up to thousands of biomarkers and millions of genetic variants with the use of metabolomics, genetics, epigenomics and transcriptomics is now possible. The use of these emerging techniques for finding CVD risk factors with the goal of risk prediction is still in its infancy. However, the initial findings have shown that these techniques may not only improve risk prediction, but also deepen our understanding of the pathogenesis of CVD itself. Furthermore, some of these findings, such as the discovery of rare PCSK9 mutations, have resulted in the development of novel therapeutic agents for the treatment of traditional risk factors.
While current research in CVD risk factors is showing promise, one must always keep in mind the wide gap between what we know about these risk factors and physicians’ and patients’ willingness to act upon this knowledge. Physicians’ clinical inertia, defined as “recognition of the problem, but failure to act” and patients’ non-adherence to lifestyle advice and drug treatment are among the most important obstacles in the successful prevention and treatment of chronic illnesses. These issues need to be tackled urgently because without doing so even the most accurate risk prediction tools will not be able to stop the global epidemic of CVD.
5. Five-year view
Rapid shifts in medical paradigm are extremely rare – the association between cholesterol and atherosclerosis was studied for over 50 years before any definite conclusions were being made. For this reason, CVD risk prediction will most likely be mainly based on the same old traditional risk factors for the next five years, with additional “fine tuning” of risk in a limited number of individuals with a few selected risk factors such as natriuretic peptide, urine microalbumin, hs-CRP, hs-Tn, coronary calcium scoring and measurements for arterial stiffness. After these five years, two “buzzwords” – omics and precision medicine – will most likely play increasingly central roles in CVD risk estimation. A pan-omic approach to the patient has the potential to transform cardiovascular risk prediction from CVD risk scores based on population means to truly individualized risk estimates based on a tidal wave of data spanning the spectrum of clinical, environmental, genomic and molecular information. However, the adoption of omics and precision medicine into primary, or even secondary or tertiary care, is still very far from being reality. Not even experienced researchers, let alone primary care physicians, currently know how to most effectively analyze and interpret the vast amounts of data that are produced by integrative omics.
The simple relationship between single risk factors and disease will have to be replaced by models that strive to integrate the whole field of omics into medicine. This includes the ability to disseminate, manage, and interpret the vast amounts of data in a clinical context and to ensure that omics results add value to patient care. In addition to clinical knowledge, this approach requires computational and mathematical modeling of complex biological systems, i.e., systems biology. Researchers working on CVD risk factor discovery will therefore have to transform CVD epidemiology from a science that is mainly led by physicians, public health researchers and biostatisticians to one that also includes geneticists, biochemists, nutritional scientists, and bioinformaticians as equals. Finally, to reach clinical application, even the most complex risk factor still has to pass the test of the three fundamental questions outlined by Morrow and deLemos: 1) “Can the clinician measure the risk factor?”, 2) “Does the risk factor add new information?”, and 3) “Does the risk factor help the clinician to manage patients?” [139].
6. Key issues.
CVD risk assessment with the traditional risk factors has remained relatively fixed for the past decades despite the significant inaccuracies associated with this approach
Most of the recent developments in CVD risk prediction have been made in large-scale biomarker discovery and so called “omics”
Although the causative role of HDL cholesterol is now being questioned, HDL function, Apo-CIII-containing lipoprotein subclasses, hs-Tn and GDF-15 show promise as novel biomarkers for CVD
Mendelian randomization represents a novel epidemiologic study design akin to a randomized trial that can be used to address the question whether an association between a risk factor and disease is truly causal
The so called “omics” – metabolomics, genetics, epigenomics and transcriptomics – are still in their infancy but provide a great opportunity to discover novel CVD risk factors
Initial findings have demonstrated that these emerging techniques may not only improve risk prediction, but be also useful in drug development and understanding CVD pathogenesis better
There are significant challenges associated with omics that need to be tackled before they can be effectively used in risk factor research, and especially in clinical medicine
Integrating the whole field of omics into medicine will require computational and mathematical modeling of complex biological systems, i.e., systems biology
CVD epidemiology will have to transform from a science that is mainly led by physicians, public health researchers and biostatisticians to one that also includes geneticists, biochemists, nutritional scientists, and bioinformaticians as equals.
Footnotes
Declaration of interest
This work was partially supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study (contracts N01-HC-25195 and HHSN268201500001I). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Reference annotations
* Of interest
** Of considerable interest
- 1.Goff DC, Lloyd-Jones DM, Bennett G, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;129:S49–73. doi: 10.1161/01.cir.0000437741.48606.98. [DOI] [PubMed] [Google Scholar]
- 2.Kannel WB, Dawber TR, Kagan A, et al. Factors of risk in the development of coronary heart disease--six year follow-up experience. The Framingham Study. Ann Intern Med. 1961;55:33–50. doi: 10.7326/0003-4819-55-1-33. [DOI] [PubMed] [Google Scholar]
- 3.Ladapo JA, Goldfeld KS. Statistical uncertainty in 10-year Framingham risk of coronary heart disease and cardiovascular disease. J Am Coll Cardiol. 2014;63:377–8. doi: 10.1016/j.jacc.2013.07.108. [DOI] [PubMed] [Google Scholar]
- 4.Bitton A, Gaziano TA. The Framingham Heart Study's impact on global risk assessment. Prog Cardiovasc Dis. 2010;53:68–78. doi: 10.1016/j.pcad.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor F, Huffman MD, Macedo AF, et al. Statins for the primary prevention of cardiovascular disease. Cochrane Database Syst Rev. 2013;1:CD004816. doi: 10.1002/14651858.CD004816.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayer R, Galea S. Public Health in the Precision-Medicine Era. N Engl J Med. 2015;373:499–501. doi: 10.1056/NEJMp1506241. [DOI] [PubMed] [Google Scholar]
- 7.Rockhill B, Kawachi I, Colditz GA. Individual risk prediction and population-wide disease prevention. Epidemiol Rev. 2000;22:176–80. doi: 10.1093/oxfordjournals.epirev.a018017. [DOI] [PubMed] [Google Scholar]
- 8.Pencina MJ, D'Agostino RB, D'Agostino RB, Jr, et al. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–72. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 9.Moons KG, de Groot JA, Linnet K, et al. Quantifying the added value of a diagnostic test or marker. Clin Chem. 2012;58:1408–17. doi: 10.1373/clinchem.2012.182550. [DOI] [PubMed] [Google Scholar]
- 10.Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115:928–35. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- 11.Folsom AR, Kronmal RA, Detrano RC, et al. Coronary artery calcification compared with carotid intima-media thickness in the prediction of cardiovascular disease incidence: the Multi-Ethnic Study of Atherosclerosis (MESA) Arch Intern Med. 2008;168:1333–9. doi: 10.1001/archinte.168.12.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elias-Smale SE, Proenca RV, Koller MT, et al. Coronary calcium score improves classification of coronary heart disease risk in the elderly: the Rotterdam study. J Am Coll Cardiol. 2010;56:1407–14. doi: 10.1016/j.jacc.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell GF, Hwang SJ, Vasan RS, et al. Arterial stiffness and cardiovascular events: the Framingham Heart Study. Circulation. 2010;121:505–11. doi: 10.1161/CIRCULATIONAHA.109.886655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattace-Raso FU, van der Cammen TJ, Hofman A, et al. Arterial stiffness and risk of coronary heart disease and stroke: the Rotterdam Study. Circulation. 2006;113:657–63. doi: 10.1161/CIRCULATIONAHA.105.555235. [DOI] [PubMed] [Google Scholar]
- 15.Ankle Brachial Index Collaboration. Fowkes FG, Murray GD, et al. Ankle brachial index combined with Framingham Risk Score to predict cardiovascular events and mortality: a meta-analysis. JAMA. 2008;300:197–208. doi: 10.1001/jama.300.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weir MR. Microalbuminuria and cardiovascular disease. Clin J Am Soc Nephrol. 2007;2:581–90. doi: 10.2215/CJN.03190906. [DOI] [PubMed] [Google Scholar]
- 17.Di Angelantonio E, Chowdhury R, Sarwar N, et al. B-type natriuretic peptides and cardiovascular risk: systematic review and meta-analysis of 40 prospective studies. Circulation. 2009;120:2177–87. doi: 10.1161/CIRCULATIONAHA.109.884866. [DOI] [PubMed] [Google Scholar]
- 18.Sarnak MJ, Levey AS, Schoolwerth AC, et al. Kidney disease as a risk factor for development of cardiovascular disease: a statement from the American Heart Association Councils on Kidney in Cardiovascular Disease, High Blood Pressure Research, Clinical Cardiology, and Epidemiology and Prevention. Hypertension. 2003;42:1050–65. doi: 10.1161/01.HYP.0000102971.85504.7c. [DOI] [PubMed] [Google Scholar]
- 19.Navab M, Reddy ST, Van Lenten BJ, et al. HDL and cardiovascular disease: atherogenic and atheroprotective mechanisms. Nat Rev Cardiol. 2011;8:222–32. doi: 10.1038/nrcardio.2010.222. [DOI] [PubMed] [Google Scholar]
- 20.AIM-HIGH Investigators. Boden WE, Probstfield JL, et al. Niacin in patients with low HDL cholesterol levels receiving intensive statin therapy. N Engl J Med. 2011;365:2255–67. doi: 10.1056/NEJMoa1107579. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz GG, Olsson AG, Abt M, et al. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N Engl J Med. 2012;367:2089–99. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 22**.Voight BF, Peloso GM, Orho-Melander M, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–80. doi: 10.1016/S0140-6736(12)60312-2. Mendelian randomization was used to demonstrate that genetic mechanisms that raise plasma HDL concentration do not seem to lower risk of myocardial infarction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zanoni P, Khetarpal SA, Larach DB, et al. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science. 2016;351:1166–71. doi: 10.1126/science.aad3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenson RS, Brewer HB, Jr, Davidson WS, et al. Cholesterol efflux and atheroprotection: advancing the concept of reverse cholesterol transport. Circulation. 2012;125:1905–19. doi: 10.1161/CIRCULATIONAHA.111.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohatgi A, Khera A, Berry JD, et al. HDL cholesterol efflux capacity and incident cardiovascular events. N Engl J Med. 2014;371:2383–93. doi: 10.1056/NEJMoa1409065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saleheen D, Scott R, Javad S, et al. Association of HDL cholesterol efflux capacity with incident coronary heart disease events: a prospective case-control study. Lancet Diabetes Endocrinol. 2015;3:507–13. doi: 10.1016/S2213-8587(15)00126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li XM, Tang WH, Mosior MK, et al. Paradoxical association of enhanced cholesterol efflux with increased incident cardiovascular risks. Arterioscler Thromb Vasc Biol. 2013;33:1696–705. doi: 10.1161/ATVBAHA.113.301373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackey RH, Greenland P, Goff DC, Jr, et al. High-density lipoprotein cholesterol and particle concentrations, carotid atherosclerosis, and coronary events: MESA (multi-ethnic study of atherosclerosis) J Am Coll Cardiol. 2012;60:508–16. doi: 10.1016/j.jacc.2012.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parish S, Offer A, Clarke R, et al. Lipids and lipoproteins and risk of different vascular events in the MRC/BHF Heart Protection Study. Circulation. 2012;125:2469–78. doi: 10.1161/CIRCULATIONAHA.111.073684. [DOI] [PubMed] [Google Scholar]
- 30.Wyler von Ballmoos MC, Haring B, Sacks FM. The risk of cardiovascular events with increased apolipoprotein CIII: A systematic review and meta-analysis. J Clin Lipidol. 2015;9:498–510. doi: 10.1016/j.jacl.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Mendivil CO, Rimm EB, Furtado J, et al. Low-density lipoproteins containing apolipoprotein C-III and the risk of coronary heart disease. Circulation. 2011;124:2065–72. doi: 10.1161/CIRCULATIONAHA.111.056986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute. Crosby J, Peloso GM, et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. Rare mutations that disrupt APOC3 function and lead to a reduced risk of coronary heart disease were discovered with whole exome sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewandrowski KB. Cardiac markers of myocardial necrosis: a history and discussion of milestones and emerging new trends. Clin Lab Med. 2014;34:31–41. doi: 10.1016/j.cll.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 34.de Lemos JA, Drazner MH, Omland T, et al. Association of troponin T detected with a highly sensitive assay and cardiac structure and mortality risk in the general population. JAMA. 2010;304:2503–12. doi: 10.1001/jama.2010.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.deFilippi CR, de Lemos JA, Christenson RH, et al. Association of serial measures of cardiac troponin T using a sensitive assay with incident heart failure and cardiovascular mortality in older adults. JAMA. 2010;304:2494–502. doi: 10.1001/jama.2010.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saunders JT, Nambi V, de Lemos JA, et al. Cardiac troponin T measured by a highly sensitive assay predicts coronary heart disease, heart failure, and mortality in the Atherosclerosis Risk in Communities Study. Circulation. 2011;123:1367–76. doi: 10.1161/CIRCULATIONAHA.110.005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeller T, Tunstall-Pedoe H, Saarela O, et al. High population prevalence of cardiac troponin I measured by a high-sensitivity assay and cardiovascular risk estimation: the MORGAM Biomarker Project Scottish Cohort. Eur Heart J. 2014;35:271–81. doi: 10.1093/eurheartj/eht406. [DOI] [PubMed] [Google Scholar]
- 38.Gaggin HK, Januzzi JL., Jr Biomarkers and diagnostics in heart failure. Biochim Biophys Acta. 2013;1832:2442–50. doi: 10.1016/j.bbadis.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Cheng XW, Shi GP, Kuzuya M, et al. Role for cysteine protease cathepsins in heart disease: focus on biology and mechanisms with clinical implication. Circulation. 2012;125:1551–62. doi: 10.1161/CIRCULATIONAHA.111.066712. [DOI] [PubMed] [Google Scholar]
- 40.Helanova K, Spinar J, Parenica J. Diagnostic and prognostic utility of neutrophil gelatinase-associated lipocalin (NGAL) in patients with cardiovascular diseases--review. Kidney Blood Press Res. 2014;39:623–9. doi: 10.1159/000368474. [DOI] [PubMed] [Google Scholar]
- 41.Ho JE, Liu C, Lyass A, et al. Galectin-3, a marker of cardiac fibrosis, predicts incident heart failure in the community. J Am Coll Cardiol. 2012;60:1249–56. doi: 10.1016/j.jacc.2012.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Boer RA, van Veldhuisen DJ, Gansevoort RT, et al. The fibrosis marker galectin-3 and outcome in the general population. J Intern Med. 2012;272:55–64. doi: 10.1111/j.1365-2796.2011.02476.x. [DOI] [PubMed] [Google Scholar]
- 43.Daniels LB, Clopton P, Laughlin GA, et al. Galectin-3 is independently associated with cardiovascular mortality in community-dwelling older adults without known cardiovascular disease: The Rancho Bernardo Study. Am Heart J. 2014;167:674–82. doi: 10.1016/j.ahj.2013.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang TJ, Wollert KC, Larson MG, et al. Prognostic utility of novel biomarkers of cardiovascular stress: the Framingham Heart Study. Circulation. 2012;126:1596–604. doi: 10.1161/CIRCULATIONAHA.112.129437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen LQ, de Lemos JA, Das SR, et al. Soluble ST2 is associated with all-cause and cardiovascular mortality in a population-based cohort: the Dallas Heart Study. Clin Chem. 2013;59:536–46. doi: 10.1373/clinchem.2012.191106. [DOI] [PubMed] [Google Scholar]
- 46.Jagodzinski A, Havulinna AS, Appelbaum S, et al. Predictive value of galectin-3 for incident cardiovascular disease and heart failure in the population-based FINRISK 1997 cohort. Int J Cardiol. 2015;192:33–9. doi: 10.1016/j.ijcard.2015.05.040. [DOI] [PubMed] [Google Scholar]
- 47.Hughes MF, Appelbaum S, Havulinna AS, et al. ST2 may not be a useful predictor for incident cardiovascular events, heart failure and mortality. Heart. 2014;100:1715–21. doi: 10.1136/heartjnl-2014-305968. [DOI] [PubMed] [Google Scholar]
- 48.Jobs E, Ingelsson E, Riserus U, et al. Association between serum cathepsin S and mortality in older adults. JAMA. 2011;306:1113–21. doi: 10.1001/jama.2011.1246. [DOI] [PubMed] [Google Scholar]
- 49.Daniels LB, Barrett-Connor E, Clopton P, et al. Plasma neutrophil gelatinase-associated lipocalin is independently associated with cardiovascular disease and mortality in community-dwelling older adults: The Rancho Bernardo Study. J Am Coll Cardiol. 2012;59:1101–9. doi: 10.1016/j.jacc.2011.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daniels LB, Clopton P, Laughlin GA, et al. Growth-differentiation factor-15 is a robust, independent predictor of 11-year mortality risk in community-dwelling older adults: the Rancho Bernardo Study. Circulation. 2011;123:2101–10. doi: 10.1161/CIRCULATIONAHA.110.979740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohatgi A, Patel P, Das SR, et al. Association of growth differentiation factor-15 with coronary atherosclerosis and mortality in a young, multiethnic population: observations from the Dallas Heart Study. Clin Chem. 2012;58:172–82. doi: 10.1373/clinchem.2011.171926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallentin L, Zethelius B, Berglund L, et al. GDF-15 for prognostication of cardiovascular and cancer morbidity and mortality in men. PLoS One. 2013;8:e78797. doi: 10.1371/journal.pone.0078797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andersson C, Preis SR, Beiser A, et al. Associations of Circulating Growth Differentiation Factor-15 and ST2 Concentrations With Subclinical Vascular Brain Injury and Incident Stroke. Stroke. 2015;46:2568–75. doi: 10.1161/STROKEAHA.115.009026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang TJ, Gona P, Larson MG, et al. Multiple biomarkers for the prediction of first major cardiovascular events and death. N Engl J Med. 2006;355:2631–9. doi: 10.1056/NEJMoa055373. [DOI] [PubMed] [Google Scholar]
- 55.Rienstra M, Yin X, Larson MG, et al. Relation between soluble ST2, growth differentiation factor-15, and high-sensitivity troponin I and incident atrial fibrillation. Am Heart J. 2014;167:109–115. doi: 10.1016/j.ahj.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zethelius B, Berglund L, Sundström J, et al. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N Engl J Med. 2008;358:2107–16. doi: 10.1056/NEJMoa0707064. [DOI] [PubMed] [Google Scholar]
- 57.Blankenberg S, Zeller T, Saarela O, et al. Contribution of 30 biomarkers to 10-year cardiovascular risk estimation in 2 population cohorts: the MONICA, risk, genetics, archiving, and monograph (MORGAM) biomarker project. Circulation. 2010;121:2388–97. doi: 10.1161/CIRCULATIONAHA.109.901413. [DOI] [PubMed] [Google Scholar]
- 58**.Gerstein HC, Pare G, McQueen MJ, et al. Identifying Novel Biomarkers for Cardiovascular Events or Death in People With Dysglycemia. Circulation. 2015;132:2297–304. doi: 10.1161/CIRCULATIONAHA.115.015744. 284 biomarkers were assayed in a large sample to identify up to 15 cardiometabolic biomarkers as independent determinants of CVD outcomes or death. [DOI] [PubMed] [Google Scholar]
- 59.Zhang A, Sun H, Wang P, et al. Recent and potential developments of biofluid analyses in metabolomics. J Proteomics. 2012;75:1079–88. doi: 10.1016/j.jprot.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 60.Roberts LD, Koulman A, Griffin JL. Towards metabolic biomarkers of insulin resistance and type 2 diabetes: progress from the metabolome. Lancet Diabetes Endocrinol. 2014;2:65–75. doi: 10.1016/S2213-8587(13)70143-8. [DOI] [PubMed] [Google Scholar]
- 61.Magnusson M, Lewis GD, Ericson U, et al. A diabetes-predictive amino acid score and future cardiovascular disease. Eur Heart J. 2013;34:1982–9. doi: 10.1093/eurheartj/ehs424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62*.Stegemann C, Pechlaner R, Willeit P, et al. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation. 2014;129:1821–31. doi: 10.1161/CIRCULATIONAHA.113.002500. Metabolomic profiling of lipids was used to discover molecular lipid signatures for cardiovascular disease in a population-based cohort. [DOI] [PubMed] [Google Scholar]
- 63.Ganna A, Salihovic S, Sundström J, et al. Large-scale metabolomic profiling identifies novel biomarkers for incident coronary heart disease. PLoS Genet. 2014;10:e1004801. doi: 10.1371/journal.pgen.1004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64*.Würtz P, Havulinna AS, Soininen P, et al. Metabolite profiling and cardiovascular event risk: a prospective study of 3 population-based cohorts. Circulation. 2015;131:774–85. doi: 10.1161/CIRCULATIONAHA.114.013116. Metabolite profiling in large population-based cohorts identified phenylalanine, monounsaturated fatty acids, and polyunsaturated fatty acids as biomarkers for cardiovascular risk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marenberg ME, Risch N, Berkman LF, et al. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–6. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 66.Casas JP, Cooper J, Miller GJ, et al. Investigating the genetic determinants of cardiovascular disease using candidate genes and meta-analysis of association studies. Ann Hum Genet. 2006;70:145–69. doi: 10.1111/j.1469-1809.2005.00241.x. [DOI] [PubMed] [Google Scholar]
- 67.CARDIoGRAMplusC4D Consortium. Deloukas P, Kanoni S, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Traylor M, Farrall M, Holliday EG, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–62. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holdt LM, Teupser D. Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler Thromb Vasc Biol. 2012;32:196–206. doi: 10.1161/ATVBAHA.111.232678. [DOI] [PubMed] [Google Scholar]
- 70.Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–30. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang J, Bakshi A, Zhu Z, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47:1114–20. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 73.Swerdlow DI, Hingorani AD, Humphries SE. Genetic risk factors and Mendelian randomization in cardiovascular disease. Curr Cardiol Rep. 2015;17 doi: 10.1007/s11886-015-0584-x. 33,015-0584-x. [DOI] [PubMed] [Google Scholar]
- 74.Ference BA, Yoo W, Alesh I, et al. Effect of long-term exposure to lower low-density lipoprotein cholesterol beginning early in life on the risk of coronary heart disease: a Mendelian randomization analysis. J Am Coll Cardiol. 2012;60:2631–9. doi: 10.1016/j.jacc.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 75.Triglyceride Coronary Disease Genetics Consortium and Emerging Risk Factors Collaboration. Sarwar N, Sandhu MS, et al. Triglyceride-mediated pathways and coronary disease: collaborative analysis of 101 studies. Lancet. 2010;375:1634–9. doi: 10.1016/S0140-6736(10)60545-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ference BA, Julius S, Mahajan N, et al. Clinical effect of naturally random allocation to lower systolic blood pressure beginning before the development of hypertension. Hypertension. 2014;63:1182–8. doi: 10.1161/HYPERTENSIONAHA.113.02734. [DOI] [PubMed] [Google Scholar]
- 77.Benn M, Tybjaerg-Hansen A, McCarthy MI, et al. Nonfasting glucose, ischemic heart disease, and myocardial infarction: a Mendelian randomization study. J Am Coll Cardiol. 2012;59:2356–65. doi: 10.1016/j.jacc.2012.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nordestgaard BG, Palmer TM, Benn M, et al. The effect of elevated body mass index on ischemic heart disease risk: causal estimates from a Mendelian randomisation approach. PLoS Med. 2012;9:e1001212. doi: 10.1371/journal.pmed.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rode L, Bojesen SE, Weischer M, et al. High tobacco consumption is causally associated with increased all-cause mortality in a general population sample of 55,568 individuals, but not with short telomeres: a Mendelian randomization study. Int J Epidemiol. 2014;43:1473–83. doi: 10.1093/ije/dyu119. [DOI] [PubMed] [Google Scholar]
- 80.Holmes MV, Dale CE, Zuccolo L, et al. Association between alcohol and cardiovascular disease: Mendelian randomisation analysis based on individual participant data. BMJ. 2014;349:g4164. doi: 10.1136/bmj.g4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Palmer TM, Nordestgaard BG, Benn M, et al. Association of plasma uric acid with ischaemic heart disease and blood pressure: mendelian randomisation analysis of two large cohorts. BMJ. 2013;347:f4262. doi: 10.1136/bmj.f4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ken-Dror G, Humphries SE, Kumari M, et al. A genetic instrument for Mendelian randomization of fibrinogen. Eur J Epidemiol. 2012;27:267–79. doi: 10.1007/s10654-012-9666-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yaghootkar H, Lamina C, Scott RA, et al. Mendelian randomization studies do not support a causal role for reduced circulating adiponectin levels in insulin resistance and type 2 diabetes. Diabetes. 2013;62:3589–98. doi: 10.2337/db13-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Talmud PJ, Holmes MV. Deciphering the Causal Role of sPLA2s and Lp-PLA2 in Coronary Heart Disease. Arterioscler Thromb Vasc Biol. 2015;35:2281–9. doi: 10.1161/ATVBAHA.115.305234. [DOI] [PubMed] [Google Scholar]
- 85.Holmes MV, Simon T, Exeter HJ, et al. Secretory phospholipase A(2)-IIA and cardiovascular disease: a mendelian randomization study. J Am Coll Cardiol. 2013;62:1966–76. doi: 10.1016/j.jacc.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elliott P, Chambers JC, Zhang W, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA. 2009;302:37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kamstrup PR, Tybjaerg-Hansen A, Nordestgaard BG. Genetic evidence that lipoprotein(a) associates with atherosclerotic stenosis rather than venous thrombosis. Arterioscler Thromb Vasc Biol. 2012;32:1732–41. doi: 10.1161/ATVBAHA.112.248765. [DOI] [PubMed] [Google Scholar]
- 88.Smith JG, Luk K, Schulz CA, et al. Association of low-density lipoprotein cholesterol-related genetic variants with aortic valve calcium and incident aortic stenosis. JAMA. 2014;312:1764–71. doi: 10.1001/jama.2014.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamstrup PR, Nordestgaard BG. Elevated Lipoprotein(a) Levels, LPA Risk Genotypes, and Increased Risk of Heart Failure in the General Population. JACC Heart Fail. 2016;4:78–87. doi: 10.1016/j.jchf.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 90.IL6R Genetics Consortium Emerging Risk Factors Collaboration. Sarwar N, Butterworth AS, et al. Interleukin-6 receptor pathways in coronary heart disease: a collaborative meta-analysis of 82 studies. Lancet. 2012;379:1205–13. doi: 10.1016/S0140-6736(11)61931-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol. 2013;42:1134–44. doi: 10.1093/ije/dyt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Paynter NP, Chasman DI, Pare G, et al. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA. 2010;303:631–7. doi: 10.1001/jama.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ripatti S, Tikkanen E, Orho-Melander M, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vaarhorst AA, Lu Y, Heijmans BT, et al. Literature-based genetic risk scores for coronary heart disease: the Cardiovascular Registry Maastricht (CAREMA) prospective cohort study. Circ Cardiovasc Genet. 2012;5:202–9. doi: 10.1161/CIRCGENETICS.111.960708. [DOI] [PubMed] [Google Scholar]
- 95.Tikkanen E, Havulinna AS, Palotie A, et al. Genetic risk prediction and a 2-stage risk screening strategy for coronary heart disease. Arterioscler Thromb Vasc Biol. 2013;33:2261–6. doi: 10.1161/ATVBAHA.112.301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.International Consortium for Blood Pressure Genome-Wide Association Studies. Ehret GB, Munroe PB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fava C, Sjögren M, Olsson S, et al. A genetic risk score for hypertension associates with the risk of ischemic stroke in a Swedish case-control study. Eur J Hum Genet. 2015;23:969–74. doi: 10.1038/ejhg.2014.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Havulinna AS, Kettunen J, Ukkola O, et al. A blood pressure genetic risk score is a significant predictor of incident cardiovascular events in 32,669 individuals. Hypertension. 2013;61:987–94. doi: 10.1161/HYPERTENSIONAHA.111.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99*.Ibrahim-Verbaas CA, Fornage M, Bis JC, et al. Predicting stroke through genetic risk functions: the CHARGE Risk Score Project. Stroke. 2014;45:403–12. doi: 10.1161/STROKEAHA.113.003044. Adding a genetic risk score based on 324 single-nucleotide polymorphisms to Framingham Stroke Risk Score model resulted in a significant, albeit modest, improvement in discrimination of stroke prediction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schork NJ, Murray SS, Frazer KA, et al. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–9. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) Bethesda, MD: NIH/National Human Genome Research Institute; 2016. [last accessed 22 February 2016]. NIH Available at: / www.genome.gov/sequencingcosts/ [Google Scholar]
- 102.Biswas A, Rao VR, Seth S, et al. Next generation sequencing in cardiomyopathy: towards personalized genomics and medicine. Mol Biol Rep. 2014;41:4881–8. doi: 10.1007/s11033-014-3418-9. [DOI] [PubMed] [Google Scholar]
- 103.Cohen JC, Boerwinkle E, Mosley TH, Jr, et al. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–72. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 104.Do R, Stitziel NO, Won HH, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–6. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Myocardial Infarction Genetics Consortium Investigators. Stitziel NO, Won HH, et al. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med. 2014;371:2072–82. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu B, Pulit SL, Hwang SJ, et al. Rare Exome Sequence Variants in CLCN6 Reduce Blood Pressure Levels and Hypertension Risk. Circ Cardiovasc Genet. 2016;9:64–70. doi: 10.1161/CIRCGENETICS.115.001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peloso GM, Auer PL, Bis JC, et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94:223–32. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tennessen JA, Bigham AW, O'Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morrison AC, Voorman A, Johnson AD, et al. Whole-genome sequence-based analysis of high-density lipoprotein cholesterol. Nat Genet. 2013;45:899–901. doi: 10.1038/ng.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–90. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 112.Aslibekyan S, Claas SA, Arnett DK. Clinical applications of epigenetics in cardiovascular disease: the long road ahead. Transl Res. 2015;165:143–53. doi: 10.1016/j.trsl.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reynolds LM, Wan M, Ding J, et al. DNA Methylation of the Aryl Hydrocarbon Receptor Repressor Associations With Cigarette Smoking and Subclinical Atherosclerosis. Circ Cardiovasc Genet. 2015;8:707–16. doi: 10.1161/CIRCGENETICS.115.001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chambers JC, Loh M, Lehne B, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3:526–34. doi: 10.1016/S2213-8587(15)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Irvin MR, Zhi D, Joehanes R, et al. Epigenome-wide association study of fasting blood lipids in the Genetics of Lipid-lowering Drugs and Diet Network study. Circulation. 2014;130:565–72. doi: 10.1161/CIRCULATIONAHA.114.009158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116*.Demerath EW, Guan W, Grove ML, et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet. 2015;24:4464–79. doi: 10.1093/hmg/ddv161. An epigenome-wide association study revealed adiposity traits associated with DNA methylation at numerous CpG sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dick KJ, Nelson CP, Tsaprouni L, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383:1990–8. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 118.Bae JB. Perspectives of international human epigenome consortium. Genomics Inform. 2013;11:7–14. doi: 10.5808/GI.2013.11.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shah S, Bonder MJ, Marioni RE, et al. Improving Phenotypic Prediction by Combining Genetic and Epigenetic Associations. Am J Hum Genet. 2015;97:75–85. doi: 10.1016/j.ajhg.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Weinberg EO, Shimpo M, De Keulenaer GW, et al. Expression and regulation of ST2, an interleukin-1 receptor family member, in cardiomyocytes and myocardial infarction. Circulation. 2002;106:2961–6. doi: 10.1161/01.CIR.0000038705.69871.D9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kempf T, Eden M, Strelau J, et al. The transforming growth factor-beta superfamily member growth-differentiation factor-15 protects the heart from ischemia/reperfusion injury. Circ Res. 2006;98:351–60. doi: 10.1161/01.RES.0000202805.73038.48. [DOI] [PubMed] [Google Scholar]
- 124.Rosenberg S, Elashoff MR, Beineke P, et al. Multicenter validation of the diagnostic accuracy of a blood-based gene expression test for assessing obstructive coronary artery disease in nondiabetic patients. Ann Intern Med. 2010;153:425–34. doi: 10.7326/0003-4819-153-7-201010050-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Thomas GS, Voros S, McPherson JA, et al. A blood-based gene expression test for obstructive coronary artery disease tested in symptomatic nondiabetic patients referred for myocardial perfusion imaging the COMPASS study. Circ Cardiovasc Genet. 2013;6:154–62. doi: 10.1161/CIRCGENETICS.112.964015. [DOI] [PubMed] [Google Scholar]
- 126.Freedman JE, Larson MG, Tanriverdi K, et al. Relation of platelet and leukocyte inflammatory transcripts to body mass index in the Framingham heart study. Circulation. 2010;122:119–29. doi: 10.1161/CIRCULATIONAHA.109.928192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McManus DD, Beaulieu LM, Mick E, et al. Relationship among circulating inflammatory proteins, platelet gene expression, and cardiovascular risk. Arterioscler Thromb Vasc Biol. 2013;33:2666–73. doi: 10.1161/ATVBAHA.112.301112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Koupenova M, Mick E, Mikhalev E, et al. Sex differences in platelet toll-like receptors and their association with cardiovascular risk factors. Arterioscler Thromb Vasc Biol. 2015;35:1030–7. doi: 10.1161/ATVBAHA.114.304954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.miRBase. Manchester, United Kingdom: University of Manchester; 2014. [Last accessed 22 February 2016]. Available at: www.mirbase.org/index.shtml. [Google Scholar]
- 130.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood- based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oerlemans MI, Mosterd A, Dekker MS, et al. Early assessment of acute coronary syndromes in the emergency department: the potential diagnostic value of circulating microRNAs. EMBO Mol Med. 2012;4:1176–85. doi: 10.1002/emmm.201201749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Matsumoto S, Sakata Y, Suna S, et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res. 2013;113:322–6. doi: 10.1161/CIRCRESAHA.113.301209. [DOI] [PubMed] [Google Scholar]
- 133.Zampetaki A, Willeit P, Tilling L, et al. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol. 2012;60:290–9. doi: 10.1016/j.jacc.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 134.McManus DD, Lin H, Tanriverdi K, et al. Relations between circulating microRNAs and atrial fibrillation: data from the Framingham Offspring Study. Heart Rhythm. 2014;11:663–9. doi: 10.1016/j.hrthm.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–38. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 136**.Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole-genome sequencing. JAMA. 2014;311:1035–45. doi: 10.1001/jama.2014.1717. This article highlighted the imprecisions of modern sequencing techniques that should be considered when determining their role in clinical medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lopez-Gonzalez AA, Aguilo A, Frontera M, et al. Effectiveness of the Heart Age tool for improving modifiable cardiovascular risk factors in a Southern European population: a randomized trial. Eur J Prev Cardiol. 2015;22:389–96. doi: 10.1177/2047487313518479. [DOI] [PubMed] [Google Scholar]
- 138.Berry JD, Dyer A, Cai X, et al. Lifetime risks of cardiovascular disease. N Engl J Med. 2012;366:321–9. doi: 10.1056/NEJMoa1012848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Morrow DA, de Lemos JA. Benchmarks for the assessment of novel cardiovascular biomarkers. Circulation. 2007;115:949–52. doi: 10.1161/CIRCULATIONAHA.106.683110. [DOI] [PubMed] [Google Scholar]