Summary
Tumor microenvironment composition and architecture are known as a major factor in orchestrating the tumor growth and its response to various therapies. In this context, in vivo studies are necessary to evaluate the responses. However, while tumor cells can be of human origin, tumor microenvironment in the in vivo models is host-based. On the other hand, in vitro studies in a flat monoculture of tumor cells (the most frequently used in vitro tumor model) are unable to recapitulate the complexity of tumor microenvironment. Three-dimensional (3D) in vitro cell cultures of tumor cells have been proven to be an important experimental tool in understanding mechanisms of tumor growth, response to therapeutics and transport of nutrients/drugs. We have recently described a novel tool to create 3D co-cultures of tumor cells and cells in the tumor microenvironment. Our method utilizes magnetic manipulation/levitation of the specific ratios of tumor cells and cells in the tumor microenvironment (from human or animal origin) aiding in the formation of tumor spheres with defined cellular composition and density, as quickly as within 24 hours. This chapter describes the experimental protocols developed to model the 3D structure of the cancer environment using the above method.
1. Introduction
For development of cancer therapeutics in the laboratory setup or pharmaceutical industry, dose evaluation and optimization are performed in vitro prior to in vivo testing in animals. These in vitro studies are currently conducted in two-dimensional (2D) cell monocultures. However, due to the limitation of planar geometry, the model can only poorly predict in vivo behavior.
To overcome this problem, 3D in vitro cancer models are being developed lately emerging as a bridge between in vitro and in vivo models. With their spatial configuration, 3D structures are a more relevant in vitro model with better representation of the cell-to-cell and cell-to-matrix contact in the native microenvironment in vivo, compared to the standard 2D monolayer culture [1]. 3D in vitro models also enable a more realistic simulation of the transport of nutrient, gas, and signaling molecules between the cells. Growing tumor spheroids with multiple cell types represents better the native microenvironment of the tumor. The 3D spheroid approach has been studied in cancer research and has been proven to represent the complexity of tumor microenvironment in terms of cell-cell interactions and the presence of necrotic and hypoxia regions within the center of the tumors [1,2].
One of the important elements in the in vitro grown spheroids is the presence of a scaffold that mimic extracellular matrix (ECM) [3,4], an important element in the tumor stroma [5]. Tumor ECM are in general denser but much less organized than normal ECM [6], and can form physical barrier for the drug[7] as well as can cause a cell adhesion-mediated drug resistance (CAM-DR) [8] due to the change of cancer cell activity by binding to ECM [9]. In most of the developed models 3D cancer models, synthetic or naturally-derived polymers are used as a scaffold [4]. However, the addition of the scaffold may stunt the cell growth and affect the cell-cell interaction, and the static concentration of the scaffold can cause a misrepresentation in the growing in vivo environment with time. In our model, we have cancer spheroids that are grown with incorporation of fibroblasts as a part of the tumor stroma component. Fibroblasts produce fibronectin and collagen which can naturally form the fibrotic capsule in 3D [10]. In our in vitro system, fibronectin concentration can increase along with the growth of the spheroids containing fibroblasts, thus enabling a more realistic prognosis for the response of the tumor in vivo to the tested drugs. In addition to fibroblast, the tumor microenvironment in vivo is comprised of cells from various origins, including adipocytes, endothelial cells, and inflammatory cells. Together, these supporting cells and fibers may account for up to 80–90% of the total tumor volume in various malignancies [5,11]. Thus, the addition of these other cells in an in vitro model significantly changes cell-cell interactions and signaling pathways within tumors.
In this chapter, we are describing the use of magnetic levitation and 3D bioprinting [12,13] to form 3D cancer cell spheroids [10], which can be designed with various cell types. Depending on tumor types, the lesion can consist of fibroblast, adipocytes, endothelial cells, as well as immune-competent cells. This model has been utilized previously in breast cancer [10], adipose cells [14], and lung cancer [15] studies, and is currently being studied for microenvironment evaluation in co-culture with immune cells such as macrophages. This spheroid model enables the simulation of in vivo environment without using an artificial scaffold and without the need of external surface for support. Additionally, due to the ability to reach larger diameter in a short time, the 3D model is more accurate in depicting the condition in the in vivo lesions, including the presence of necrotic and hypoxia regions in the center of tumor lesion.
2. Materials
Nanoshuttle (n3D Biosciences, Houston, TX, USA)
Cells of interest: may consist of a combination of cancer cells, fibroblasts, myofibroblasts, immune cells or adipocytes.
Cell culture medium with and without fetal calf serum (FCS, 10%).
RPMI1640 complete medium: RPMI1640 medium, 10% human serum, 1% MEM vitamin solution and 1% penicillin/streptomycin.
Trypsin-EDTA (0.25%)
Phosphate buffered saline (PBS), pH 7.4
Vybrant DiD/DiL/DiO cell-labelling solution (Molecular Probes, Eugene, Oregon, USA)
T25 and T75 flasks
96-well low attachment plates
MagPen™ - teflon pen (n3D Biosciences, Houston, TX, USA)
Ficoll-Paque premium grade (GE Healthcare Bio-sciences AB, Uppsala, Sweden)
Ethanol
Fixation solution: 4% Paraformaldehyde in PBS
Permeablizing solution: 0.1 to 0.2% Triton X-100 in PBS
DAPI nuclear stain: 1μg/mL solution in PBS
Primary and secondary antibodies (based on the desired immunohistochemistry/immunofluorescence analysis and manufacturer’s protocol)
Histogel™ (Richard-Allan Scientific, Kalamazoo, Michigan, USA)
O.C.T. compound – frozen tissue mounting media (Sakura FInetek, Torrance, CA, USA)
Prolong Gold® - antifade mounting agent (Life technologies, Eugene, Oregon, USA)
Cytoseal™ XYL (Richard-Allan Scientific, Kalamazoo, Michigan, USA)
3. Procedures
3.1 Pre-incubation of cells with magnetic nanoparticles
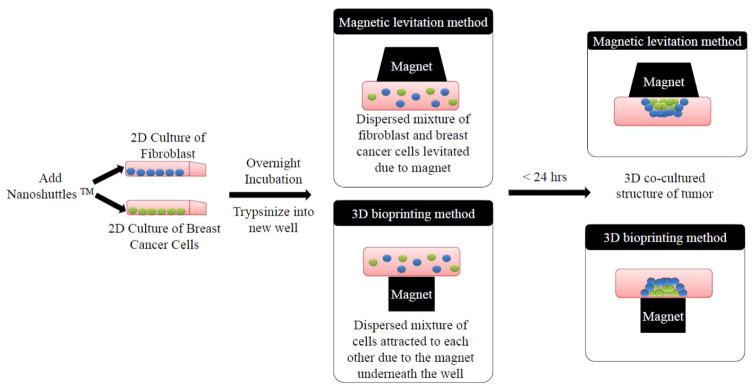
The processes for 3.1. and 3.2 is summarized schematically in Figure 1.
Fig 1.
Schematic presentation of the development of 3D in vitro breast tumor spheroid from a dispersed mixture of fibroblasts (in blue) and breast cancer cells (in green) after the addition of nanoshuttles. The magnet placed on top or on the bottom of the plate aids the attraction of the nanoshuttle internalized cells to form a 3D tumor mass composed of fibroblasts and breast cancer cells and can be grown for several days. Figure modified from ref [10].
Make sure that the cells used can grow in the same media.
Grow the cells of interest in T25 or T75 flasks (use the standard conditions).
Wash cells with PBS by removing medium and adding PBS from the side of the dish without disturbing the cell layer. Rinse the cells by spreading PBS all over the dish. Discard the PBD and repeat this step twice.
Trypsinize cells by incubation with 1 mL (T25) or 3.5 mL (T75) trypsin-EDTA for 2–5 minutes at 37°C.
After the cells are detached from the surface, neutralize trypsin by adding 3mL (T25) or 10.5 ml (T75) serum-containing medium (see Note 1). Harvest all the cells into a 15mL falcon tube and centrifuge according to the procedures for the cells.
Remove the supernatant containing trypsin and replenish with a fresh medium. Count the cells and plate them into T25 flask.
Culture cells to ~70% confluence before adding the nanoparticle assembly (see Note 2).
Prepare the nanoparticles assembly by removing them from refrigerator and allow to reach room temperature (for about 15 min). Homogenize nanoparticles before using by vigorous vortexing or pipette mixing.
Add the nanoparticles assembly to the cells with the concentration of 2–4μL/cm2. Make sure the nanoparticles were distributed evenly throughout the flask by gently tilting the flask back and forth.
Incubate the cells with nanoparticle assembly overnight to allow the uptake process.
(optional) Cell staining for live cell tracking purposes: add 5μL Vybrant DiD/Dil/DiO cell-labeling solution for each mL culture medium into the flask containing cells 20 minutes before cells are to be trypsinized.
3.2 Spheroid formation
Trypsinize the cells incubated overnight with nanoshuttle ( as described in 3.1 step 2–4)
After centrifugation, remove the supernatant and re-suspend cells with fresh medium. (see Note 3)
(optional) For co-culture of different cell types (Figures 2, 3): mix the cell types needed in the model at the appropriate ratio. (see Note 4)
Count the cell number using hemacytometer or any cell counter instrument.
Seed the number of cells needed to grow the spheroids in the appropriate plate (see Notes 6–7).
Place the lid inserts, magnet drive, and well-plate lid (when using magnetic levitation drive), or place the plate directly on top of the 96 well spheroid drive (when using bioprinting kit).
Place the cells at 37°C, 5% CO2, cell spheres will start forming after a couple of hours of incubation. Depending on the size of spheres needed, incubate the cells for 1–5 days.
Replace the medium every 1–3 days depending on the cell types. This can be done by tilting the plate while keeping the drive magnet underneath the plate. The spheres will be retained by the magnet on the bottom. Aspire the medium carefully without damaging the spheres and replenish the well with fresh medium (see Notes 8–9).
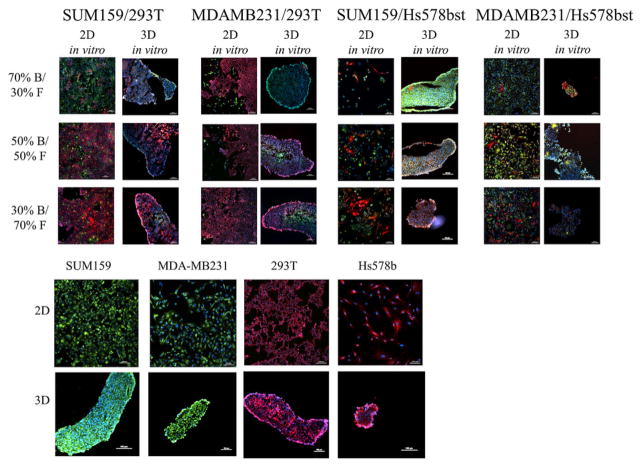
Figure 2.
Comparison of breast tumor model in 2D and 3D culture with different ratios and types of breast cancer cells (in green) and different types of fibroblasts (in red), grown for 3 days.
Figure 3.
3D in vitro breast tumor model with different ratios and types of breast cancer cells (in green) and primary fibroblasts (in red) co-cultured for 3 days using magnetic levitation system. All cells were counterstained with DAPI (blue) for nucleus. (A) Confocal microscopy images of the spheres. (B–D) Fluorescent images of the spheres taken after cryo-sectioning (4 micron slices) (B) MDA-MB-231 and Hs578bst (C) MDA-MB-231 and HPF (D) MDA-MB-231 and CAF. Images were taken with 10 × objective magnification, scale bar = 100 μm.
3.3 (Optional) Harvesting immune cells from human/ mouse blood using gradient centrifugation method
Obtain buffy coat from blood donation service (human) or harvest blood from mice.
Mix buffy coat 1:1 with PBS
Prepare 20mL of Ficoll on 50 mL falcon tubes.
Carefully overlay the buffy coat on top of the Ficoll layer. Make sure the layer is undisturbed.
Centrifuge at 1200g and 4°C for 30 min.
After centrifugation, the cells will form several layers. Mononuclear cells will form a ring between the ficoll and the plasma layer. Remove most of the plasma to gain access to the mononuclear cells.
Harvest mononuclear cells and combine the cells from two tubes into one 50 mL tube. Fill the tube with fresh PBS.
Centrifuge the cells at 300g for 7 minutes. Remove PBS and replenish with the fresh PBS. Repeat the washing step two times.
After the last centrifugation, resuspend cells with RPMI1640 complete medium.
Plate the cells at 0.5–1.5*106 cells/cm2 density on the low attachment flasks. Incubate for 1 hour at 37°C and 5% CO2.
After 1 hour incubation, the monocytes and macrophages are attached to the bottom of the flask, while lymphocytes are still in suspension. Remove lymphocytes for further experiments if needed. Wash monocytes twice with PBS, and replenish with fresh medium and place the cells back into the incubator.
After 7 days of incubation, monocytes will be differentiated to macrophages and ready to be used for further experiments.
3.4 Experimental design for drug toxicity with and without macrophages
See Note 10 for sphere handling
Count immune cells needed for the experiment. The ratio of number of macrophages to cancer cells can range from 1:10 to 1:100, depending on the model. Carefully remove the medium on the wells containing spheroids with the magnet drive latching underneath the plate. Add the macrophages to the spheres in a lower volume than needed to allow addition of drugs onto the wells.
Prepare the drug in the concentration to be tested for the appropriate volume. Working volume in 96-well plates is between 100 to 200 μL. Take into consideration the volume already added in step 3.3.1.
Incubate spheres with the drugs for the duration of the experiment in the incubator at 37°C and 5% CO2.
While in incubation, the growth and changes in spheroids can be followed by inverted microscopy imaging. This method can also be utilized to follow the permeation of drugs into the spheroids (figure 4). Hold the plate flat while transferring from incubator to microscope stage (see Note 11).
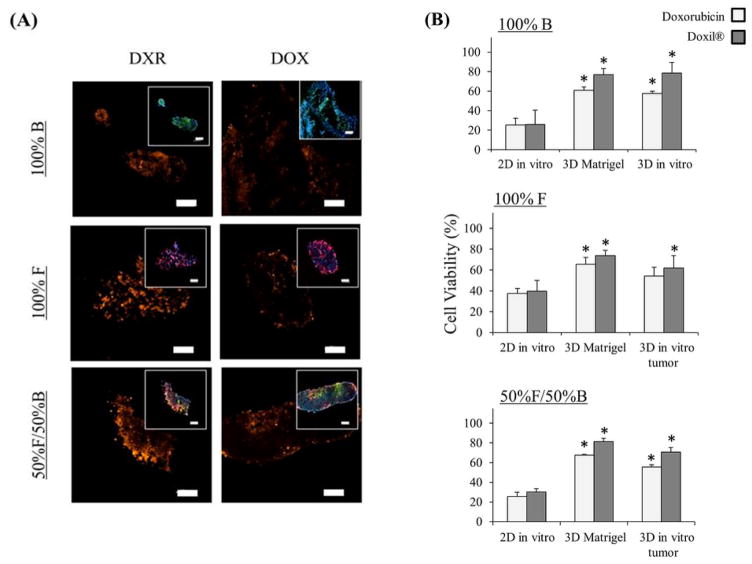
Figure 4.
Distribution and therapeutic efficacy of doxorubicin HCl and Doxil® (doxorubicin liposomes) in 3D in vitro tumors: (A) Fluorescent images of 3D in vitro tumors composed of mono- or co-culture of fibroblast (in red) and breast cancer cells (in green) –inset images, comparing 72 h treatment with doxorubicin and Doxil ®, blue – nucleus and orange – fluorescent emission from doxorubicin, Scale bar = 100 μm, (B) Viability assay treated with either doxorubicin or Doxil ® (100 nM) for 72 h, comparing on three different in vitro systems: (1) 2D in vitro (grown for 1 day), (2) 3D in vitro (grown for 1 day), and (3) 3D Matrigel™ (grown for 7 days) * = statistically significant difference to 2D in vitro with the same treatment, n = 4, p < 0.05.
3.5 Additional handling with Teflon pen for transferring spheroids to new containers
All the tools involved for transferring spheroids need to be sterile or otherwise sterilized by treatment with 70% (v/v) ethanol. The Teflon part can also be autoclaved. Caution: the rod magnet cannot be autoclaved.
Insert the rod magnet to the Teflon pen, handle the assembly either with forceps or by hand. Caution: use plastic forceps, metal forceps will be attracted to the magnet and harder to handle.
Remove the magnetic drive from the plate before removing the spheroids.
Prepare the new container with fresh medium or fixing agent where the spheroids will be transferred to before starting the process.
Place the Teflon pen facing downwards in the well, reach for the surface where the spheroids are located. The spheres will be attracted to the magnet rod and attached to the Teflon pen. Lid the Teflon pen and remove the magnet rod from the Teflon pen, the spheres will remain on the pen.
Place the Teflon pen downwards to the new contained or well where the spheroids are to be transferred to. The spheroids will be transferred to the new container as soon as it touches the medium. Additionally, the magnetic drive plate can be placed underneath the wells to increase the attraction to the bottom of the well and ease the transfer. In case of transferring the spheres to OCT fluid, do not let all the fluid freeze before the transfer. Let a fluid area where the spheroids are to be placed, as the spheres would not attach to the frozen area.
Close the plate.
3.6 Spheroid fixation process for immunohistochemistry or fluorescence imaging
Follow step 3.1.3 to remove medium and wash with PBS, repeat two times.
Add the preferred fixation agents, such as 4% (w/v) paraformaldehyde, fix the spheres for 10 minutes at room temperature. (see Note 12)
Wash the culture three times with PBS (repeat step 3.1.3).
The culture can be frozen on the OCT compound for further processing by transferring them to the suitable cassettes (see chapter 3.4), or paraffin embedded for sectioning (figure 5). For paraffin embedding, it is recommended to fix the spheroids on Histogel™ before placing them on the cassettes to avoid the loss of sample due to the size. This can be done by heating up the Histogel™ to 60°C and place them on the mold for OCT compound. Transfer the spheroids to the Histogel™ (see Note 7 or chapter 3.4), and overlay the culture with additional thin layer of Histogel™. After the Histogel™ solidified, the sample can be removed from the mold and placed to the cassettes and be further processed for sectioning and staining.
For immunostaining of intracellular proteins, the cell membrane will need to be permeabilized by adding 0.1% (v/v) Triton X-100 as fixation solution for 10 minutes. Then remove the fixing solution and wash with PBS three times (repeat step 3.1.3). (see Note 12)
Add 1% BSA as blocking solution and incubate for 5 minutes at room temperature. Increase the incubation time to 1 hour if you are working with the whole spheroid instead of the cell sections.
Remove blocking solution, add primary antibody in appropriate concentration and incubate for 30 min at 37°C, 4 hours at room temperature, or overnight at 4°C (see Note 13). Protect the cells from light starting from this step if fluorescent antibody is used.
Wash the spheroids three times with PBS, repeat step 3.5.7 for secondary antibody if needed. Protect the cells from light.
After washing three times with PBS, add nuclear stain e.g. DAPI (1μg/mL) for counterstaining. Incubate for 15 minutes at 37°C (see Note 14).
Aspirate the DAPI solution and wash culture three times with PBS. The culture can be stored protected from light in PBS for several months at 4°C.
If needed, the spheroids can be transferred to the slides using the teflon pen or pipet tip (see Chapter 3.4)
Fix the spheroids with an antifade reagent (e.g. Prolong Gold®), cover the slide with coverslip and seal the sample with mounting media (e.g. CytoSeal™ XYL).
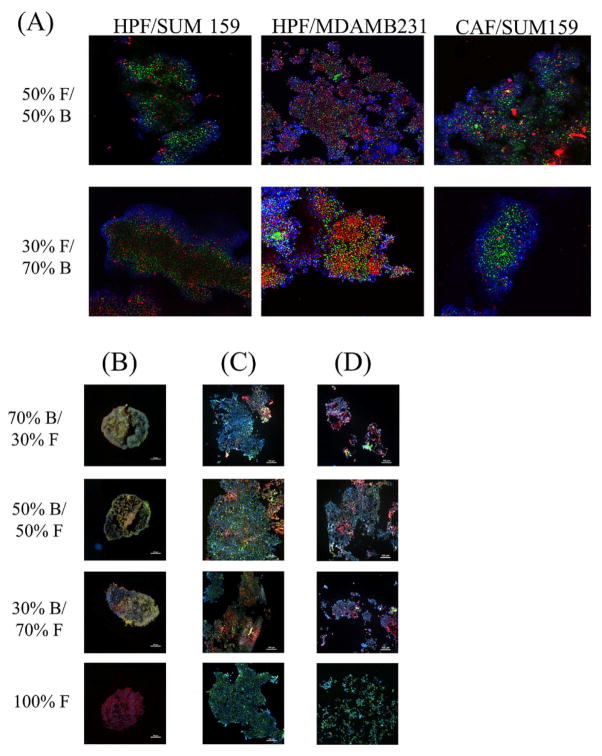
Figure 5.
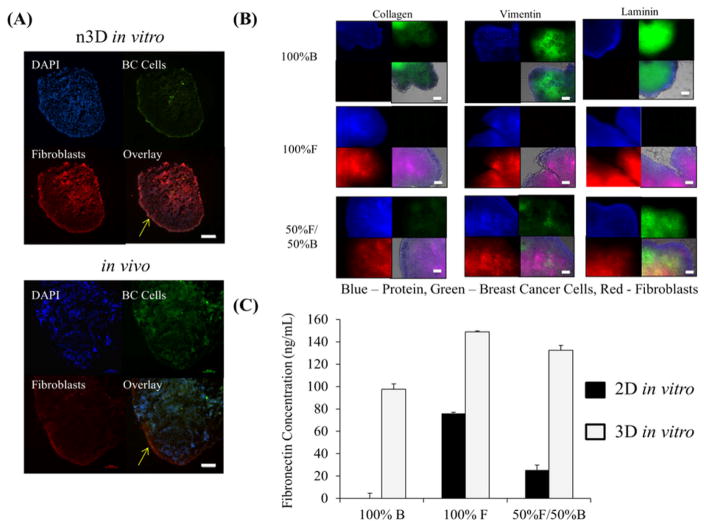
Characterization of in vitro 3D co-cultures: (A) Fluorescent images comparing the phenotype between 3D in vitro co-culture grown in 6 well plates and in vivo tumors composed of breast cancer cells (green signal) and fibroblasts (red signal) after 7 days growth. Blue signal is from DAPI, staining the nucleus. Yellow arrows indicate the fibrotic capsule formed in both 3D in vitro and in vivo tumors. Scale bar = 100 μm. (B) Collagen, fibronectin, and vimentin (in blue) immuno-fluorescent whole mount staining on 7 days old 3D in vitro mono- and co-cultures for breast cancer (in green) to fibroblast cells (in red) grown in 24 well plate, overlayed on brightfield image of tumor, Scale bar = 100 μm (C) Fibronectin concentration detected in 7 days grown 2D and 3D mono- and co-cultures of breast cancer and fibroblasts cells. F = fibroblasts (293T) and B = breast cancer cells (SUM159).
Acknowledgments
The authors would like to acknowledge Nano 3D Biosciences Inc for technical support and Susan G. Komen PDF12229449 Award, NIH U54CA143837 and NIH 1U54CA151668-01 for the financial support.
Footnotes
For serum-sensitive cells, use trypsin neutralizing solution with medium 3.1.4.
Cells can also be plated together with nanoshuttle. Count the cells to reach at least 70% confluency to optimize the nanoshuttle uptake and avoid overcrowding.
The cell pellet will appear brown due to the nanoparticles taken up by the cells. If the brown color is not homogenized within the cells, the incubation time should be increased to ensure the optimum nanoshuttle uptake.
If the 3D culture consists of different cell types, maintain the spheroids in the cell medium of the most demanding cell type, check the growth curves of other cell types in this growth medium.
It is recommended to plate 500–5000 cells in 50–100 μL for each well of 96-well plates or 100000–300000 cells in 350 μL for each well of 24-well plates.
Only use low-attachment plates to avoid cell attachment to the bottom of the well.
For plates with magnetic levitation system with magnets on the lid, use less medium than usual. This step is to ensure that the medium do not reach the lid, which can cause the attachment of the cells to the lid.
Optionally, sterile gel-loading fine pipet tips can be used to avoid having the spheroids suctioned when removing the medium.
Remove medium delicately without disrupting the spheroids. If the cells are easily disrupted, then the spheroids may need longer time to form. Incubate the spheroid further before removing the medium.
The spheres can be transferred to other plates by using pipette with larger diameter (1ml tips). In case the size of the spheres are bigger than the pipette diameter or the spheroids fall apart easily, the transfer can be done by using Teflon pen (see chapter 3.4).
For the magnetic levitation magnets, the lid insert is translucent and the spheroids should be easily imaged with the lid insert on. However, if the lid insert disturb the imaging, the lid and magnetic drive magnet can be removed in sterile environment, leaving the plate only to be covered by the transparent plate lid.
Optionally, cells can be fixed with 100% methanol, ethanol or isopropyl alcohol, or 100% acetone for 30 min at 4°C. No permeabilization is needed for fixation with methanol and acetone, but methanol may compromise the epitope. Permeabilizing agent such as Triton X-100 is harsh to the cells and optionally, milder agents such as Tween 20, Saponin, Digitonin and Leucoperm can be used in concentration of 0.2–0.5% for 30 mins. Therefore, fixation and permeabilization protocol will need to be optimized.
For immunofluorescence staining: make sure that no dyes in similar excitation and emission wavelength to the dye used in immunofluorescence were added for live cell imaging (step 9 of chapter 3.1).
DAPI is only recommended for histological slices, since the dye will not penetrate through the whole spheroids for most cell types.
References
- 1.Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Court W, Lomas C, Mendiola M, Hardisson D, Eccles SA. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012:10. doi: 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamada KM, Cukierman E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell. 2007;130(4):601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Lukashev ME, Werb Z. ECM signalling: Orchestrating cell behaviour and misbehaviour. Trends Cell Biol. 1998;8(11):437–441. doi: 10.1016/s0962-8924(98)01362-2. [DOI] [PubMed] [Google Scholar]
- 4.Talukdar S, Mandal M, Hutmacher DW, Russell PJ, Soekmadji C, Kundu SC. Engineered silk fibroin protein 3D matrices for in vitro tumor model. Biomaterials. 2011;32(8):2149–2159. doi: 10.1016/j.biomaterials.2010.11.052. [DOI] [PubMed] [Google Scholar]
- 5.De Kruijf EM, Van Nes JGH, Van De Velde CJH, Putter H, Smit VTHBM, Liefers GJ, Kuppen PJK, Tollenaar RAEM, Mesker WE. Tumor-stroma ratio in the primary tumor is a prognostic factor in early breast cancer patients, especially in triple-negative carcinoma patients. Breast Cancer Res Treat. 2011;125(3):687–696. doi: 10.1007/s10549-010-0855-6. [DOI] [PubMed] [Google Scholar]
- 6.Lu P, Weaver VM, Werb Z. The extracellular matrix: A dynamic niche in cancer progression. Journal of Cell Biology. 2012;196(4):395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dittmer J, Leyh B. The impact of tumor stroma on drug response in breast cancer. Seminars in Cancer Biology. 2015;31:3–15. doi: 10.1016/j.semcancer.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 8.McMillin DW, Negri JM, Mitsiades CS. The role of tumour-stromal interactions in modifying drug response: Challenges and opportunities. Nature Reviews Drug Discovery. 2013;12(3):217–228. doi: 10.1038/nrd3870. [DOI] [PubMed] [Google Scholar]
- 9.Watt FM, Huck WTS. Role of the extracellular matrix in regulating stem cell fate. Nature Reviews Molecular Cell Biology. 2013;14(8):467–473. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 10.Jaganathan H, Gage J, Leonard F, Srinivasan S, Souza GR, Dave B, Godin B. Three-dimensional in vitro co-culture model of breast tumor using magnetic levitation. Sci Rep. 2014:4. doi: 10.1038/srep06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downey CL, Simpkins SA, White J, Holliday DL, Jones JL, Jordan LB, Kulka J, Pollock S, Rajan SS, Thygesen HH, Hanby AM, Speirs V. The prognostic significance of tumour-stroma ratio in oestrogen receptor-positive breast cancer. Br J Cancer. 2014;110(7):1744–1747. doi: 10.1038/bjc.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haisler WL, Timm DM, Gage JA, Tseng H, Killian TC, Souza GR. Three-dimensional cell culturing by magnetic levitation. Nat Protoc. 2013;8(10):1940–1949. doi: 10.1038/nprot.2013.125. [DOI] [PubMed] [Google Scholar]
- 13.Souza GR, Molina JR, Raphael RM, Ozawa MG, Stark DJ, Levin CS, Bronk LF, Ananta JS, Mandelin J, Georgescu MM, Bankson JA, Gelovani JG, Killian TC, Arap W, Pasqualini R. Three-dimensional tissue culture based on magnetic cell levitation. Nat Nanotechnol. 2010;5(4):291–296. doi: 10.1038/nnano.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daquinag AC, Souza GR, Kolonin MG. Adipose tissue engineering in three-dimensional levitation tissue culture system based on magnetic nanoparticles. Tissue Eng Part C Methods. 2013;19(5):336–344. doi: 10.1089/ten.tec.2012.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tseng H, Gage JA, Raphael RM, Moore RH, Killian TC, Grande-Allen KJ, Souza GR. Assembly of a three-dimensional multitype bronchiole coculture model using magnetic levitation. Tissue Eng Part C Methods. 2013;19(9):665–675. doi: 10.1089/ten.tec.2012.0157. [DOI] [PubMed] [Google Scholar]