Abstract
Traditional views of the inflammasome highlight pre-existing core components being assembled under basal conditions shortly after infection or tissue damage. Recent work, however, suggests the inflammasome machinery is also subject to tunable or inducible signals that may accelerate its autocatalytic properties and dictate where inflammasome assembly takes place in the cell. Many of these immune signals operate downstream of interferon (IFN) receptors to elicit inflammasome regulators, including a new family of IFN-induced GTPases termed guanylate binding proteins (GBPs). Here, we examine the critical roles for IFN-induced GBPs in directing inflammasome subtype-specific responses and their consequences for cell-autonomous immunity against a wide variety of microbial pathogens. We discuss emerging mechanisms of action and the potential impact of these GBPs on predisposition to sepsis and other infectious or inflammatory diseases.
Introduction
Evolutionary arms races drive host-specific adaptations to microbial pathogens1. In vertebrates, they often promote expansive and increasingly complex immune repertoires that bear limited resemblance to their ancestral precursors and which can be acquired through horizontal gene transfer1,2. Two prime examples are the interferon (IFN) family of cytokines and the caspase-1 inflammasome machinery. IFNs arose in basal chordates ~500 million years ago3 while the caspase-1 inflammasome has extant functional relatives in jawed fish but not amphibians4, suggesting it originated after the teleost-tetrapod split ~450 million years ago. Both IFNs and inflammasomes co-operate in marshaling protective immunity to infection in higher species such as mammals. IFNs regulate not only the expression of many core inflammasome proteins but also direct their spatial assembly through physical and functional interactions with other interferon-induced gene products (ISGs)5-23. These interactions control both the specificity and amplitude of inflammasome activation. As such, this alliance has important consequences for mammalian host defense and the inflammatory sequelae which often accompanies infectious insult.
In this Perspective, we discuss emerging evidence on how IFNs impact inflammasome-mediated immunity and focus on a new IFN-induced GTPase family, the 65–73 kDa guanylate binding proteins (GBPs)3,24, at the interface of this relationship. IFN-induced GBPs help customize inflammasome responses to a variety of microbial signatures. They also provide a conceptual framework wherein inflammasome activation can be viewed as a dynamic process empowered by IFN-induced transcriptional signals and subject to post-translational regulation by new IFN-induced host defense proteins.
The inflammasome: A tunable molecular machine
Inflammasomes integrate environmental signals through a series of conformational switches to assemble multiprotein complexes. These signals include pathogen- and endogenous danger-associated molecular patterns (PAMPs and DAMPs) detected largely in the cytosol of macrophages, monocytes, splenic and plasmacytoid dendritic cells, T and B cells, neutrophils, keratinocytes and inflamed endothelium5,9,10,16,25-28. Complex assembly leads to caspase-1-dependent cleavage of pro-interleukin 1β (pro-IL-1β) and pro-IL-18 to their mature exported forms as part of the canonical inflammasome pathway. A second non-canonical cascade enlists caspase-11 (CASPASE-4 and -5 in humans) as an upstream intracellular lipopolysaccharide (LPS) receptor which mobilizes immunity specifically against Gram-negative bacteria11,15,29-32. Both pathways induce a lytic form of programmed cell death termed pyroptosis that eliminates infected target cells via a mechanism genetically distinct from cytokine release13,15,27-31,33,34.
Inflammasome complex formation is itself an amplifying process where large (1-2 μm) “prion-like” foci typically incorporate sensor proteins belonging to either NLR (nucleotide binding and oligomerization domain [NBD], leucine-rich repeat [LRR]) or ALR (absent in melanoma 2-like receptor) families along with multiple copies of the adaptor protein, ASC (apoptosis-associated, speck-like protein containing a CARD)35,36. ASC in turn recruits procaspase-1 which becomes autoactivated via proximity-induced nucleation to cleave its cytokine substrates as part of an “all-or-none” processing mechanism5,25,26,33. Recent crystallographic and cryo-EM studies suggest sensor and adaptor proteins exist as auto-inhibited monomers until ligand binding induces self-clustering platforms for recruiting heterotypic partners35-38.
These core components – upstream sensor, bridging adaptor and caspase effector – constitute a minimal inflammasome complex. This idea is underscored by reconstitution assays in which sensors like NLRP3 or AIM2 along with ASC and pro-caspase-1 alone are sufficient to induce foci formation and caspase-1 processing in otherwise inflammasome-deficient cells12,39. What these reconstitution systems omit, however, are important priming and activation events synonymous with inflammasome behavior under native physiological conditions5,25,26. Ectopic overexpression of the core machinery in HEK293 cells, for example, drives ASC complex assembly on its own12,39, thereby obviating the need for dsDNA-driven AIM2 or activating NLRP3 stimuli, such as K+ efflux, essential for the same process in primary macrophages40,41. Likewise, the absence of Toll-like receptor (TLR) or IFN signaling suggests these priming events are dispensible when in fact TLR and IFN signals are often obligate for inflammasome activation triggered by many microbial pathogens in vitro and in vivo5-23. TLR4 agonists such as LPS induce not only NF-κB-dependent transcription of pro-IL-1β but also type I IFN-β via TRIF-dependent phosphorylation of interferon regulatory factor 3 (IRF3)11. IFNs and ISGs are emerging as potent transcriptional and post-translational switches, respectively, for core inflammasome activities5-23. Together they move our understanding of this multiprotein complex beyond its basal, hard-wired circuitry to a more tunable molecular machine.
IFN-inducible signals for inflammasome priming & assembly
IFN-induced signals promote canonical and non-canonical inflammasome-mediated immunity to a diverse array of microbial pathogens. TLR4-TRIF-IRF3-induced IFN-β boosts canonical AIM2-dependent IL-1β secretion to Francisella tularenis or Listeria monocytogenes6,7 and helps control caspase-11-dependent pyroptosis by Gram-negative bacteria11,34. IFN-induced transcription factors STAT1 and IRF1 likewise elicit AIM2-dependent cytokine and cell death responses to Francisella novicida22,23. In addition, type II IFN-γ enhances AIM2-induced IL-1β release or NLRP3-dependent pro-IL-18 cleavage during HSV-1 and Chlamydia muridarum infections16,20. Here IFN-γ-induced upregulation of NLRP3, ASC and procaspase-1 expression may help lower the PAMP-triggered activation threshold by ~5–20 fold due to increased core protein levels12,17,18,20. This synergism is observed for the NLRP3 inflammasome across multiple settings12,13,17,18,20. By contrast, NLRP3 responses to alum, monosodium ureate (MSU) and silica crystals appear largely unaffected by IFN-γ treatment12,18,22,23,42 and inhibited by type I IFNs42. Thus sterile crystalline agents are handled differently to soluble ligands and may be sensitized by tumor necrosis factor rather than IFNs43.
IFN-γ has also emerged as a potent activator of caspase-11-dependent defense against cytosolic pathogens, for example, Burkholderia thailandensis in macrophages and challenged mice19. Protection can partly be ascribed to IFN-γ-induced expression of procaspase-11 during Gram-negative infection15,17,18 that is reliant on STAT1 but not IRF1 signaling44,45. Type II IFN-γ priming also heightens caspase-11-dependent responses to LPS transfected directly into the cytosol13,17,21,30. Such potentiation often exceeds that seen for autocrine IFN-β signaling, suggesting ISGs induced by IFN-γ may eventually prove more important than those governed by IFN-β for inflamasome responses to cytosolic LPS. Future studies will determine their respective contributions.
Two isolated IFN-γ-induced proteins impacting inflammasome-mediated immunity are IFI16 and protein kinase R (PKR). IFI16 encounters Kaposi sarcoma-associated herpesvirus (KHSV) in the nucleus before translocating to the cytosol for assembly with ASC and procaspase-1, showing ligand detection and inflammasome activation can be spatially segregated10. PKR engages multiple inflammasome subtypes in the cytosol (NLRP1, NLRP3, NAIP-NLRC4, AIM2)46, although this breadth of activity has been questioned47. Chemical proteomics validates its role at least for the NLRP1 inflammasome where 7-desacetoxy-6,7-dehydrogedunin (7DG), a drug targeting PKR, interferes with anthrax toxin-induced cell death48. Other PKR-related activities need to be corroborated by further analysis.
Collectively, there is mounting evidence that IFNs control inflammasome responses to bacterial and viral pathogens. Which ISGs direct these activities, however, still remains an open question for the field. Part of the answer is now furnished with the discovery of a new family of IFN-γ-induced 65–73 kDa guanylate binding proteins (GBPs)3,24. The GBPs exhibit inflammasome subtype-specific functions in both canonical and non-canonical pathways.
Discovery of GBPs as new inflammasome regulators
In 2012, we identified mammalian GBPs as new inflammasome regulators despite these proteins lacking any functional or structural similarities to NLRs, ALRs and inflammatory caspases12,24. This unexpected discovery arose from genome-wide evolutionary screens designed to uncover clues about their recently described antibacterial functions49. Previous studies using in silico identification of IFN-induced inflammasome proteins like AIM2 relied on a binary logic where the functional criteria were known, for example, DNA detection leading to IL-1β secretion. PFAM (Protein Families) databases seeded with inflammasome pyrin domain (PYD) sequences thus retrieved proteins also harboring a DNA-binding motif (HIN200)39,40. Domain accretion approaches, however, were unsuitable for mammalian GBPs because their globular N-terminal guanosine 5′-triphosphatase domains (GTPase) and C-terminal helical domains offered few insights into their potential host defense activities24,49. We thus opted for a different strategy, asking instead whether the interdomain profiles of ancestral GBPs could reveal hidden immune connections still operative in mammals, even if those domains were now distributed on seemingly unrelated proteins through evolutionary divergence12. In this way, probing the multidomain configuration of GBPs in lower organisms could suggest functional relationships in higher species that were otherwise imperceptible because they act over large protein distances24.
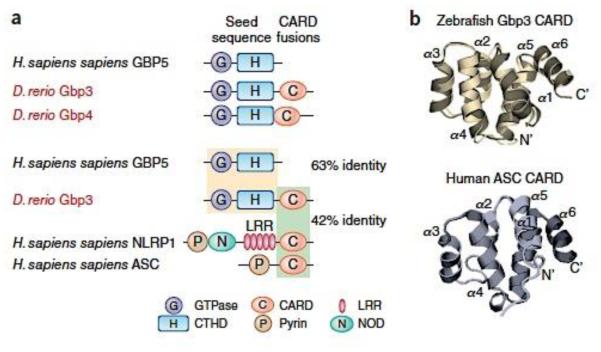
This unorthodox approach yielded striking results. Hidden Markov Modeling identified 574 GBP-related sequences (including atlastins; root hair-defective 3; GTPase, very large IFN inducible [GVIN]; and GBP families) from 91 taxa spanning vertebrates, protists, plants and algae12. Further filtering produced 193 bona fide GBPs harboring both N-terminal GTPase and C-terminal helical domains; the GTPase domains shared 60–64% identity with human GBPs12. Prominent among them were GBPs from jawed fish possessing inflammasome-related CARDs like those in ASC and NLRP1 from multiple species, including humans (Fig. 1). Recent crystallization efforts have shown these similarities extend to the atomic level; the zebrafish IGBP1 (zGbp3) CARD domain contains a 6-helix bundle fold almost identical to that in human NLRP1 or ASC (ref. 50) (Fig. 1). In addition to CARDs, we also discovered protochordates (eg. Brachiostomata) harbored GBPs fused to death effector domains (DEDs). These DEDs shared homology with human caspase-8 and FADD (Fas-associated death domain), suggesting a potential role also in inflammasome-mediated cell death. Evolutionary mining of ancient GBP-like sequences thus discovered modular links with host inflammasome and pyroptosis pathways. It opened up the possibility of a functional interplay between the GBPs and inflammasome-related NLRs, ALRs and caspases in higher species such as mammals (Fig. 1).
Fig. 1. Evolutionary links between GBPs with inflammasome machinery from fossil record studies.
(a) Example of GBP-CARD fusions in Danio rario proteins retrieved from genome-wide Hidden Markov Modeling screens of 91 taxa using human GBPs as the seed sequence12. Amino acid identities of the respective domains between human and zebrafish proteins shown. (b) Structural similarities between the CARD domains of zebrafish Gbp353 (PDB 4IRL) and human ASC (PDB 2KN6). Generated from RCSB deposits in PV Javascript viewer.
GBPs in canonical inflammasome activation
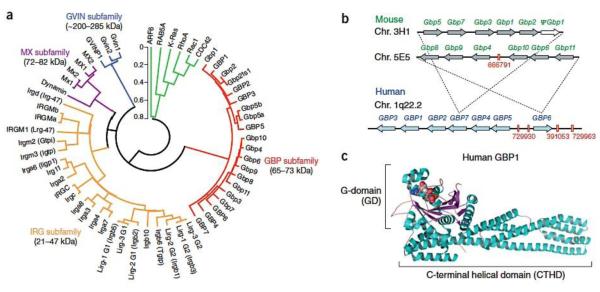
Initial experimental efforts focused on the contribution made by each human and mouse GBP to canonical inflammasome activation12. To date, 7 intact human GBP genes (GBP1-GBP7) on chromosome 1q22.2 and 11 mouse Gbp loci (Gbp1-Gbp11) distributed across two clusters on chromosomes 3H1 and 5E5 have been identified (refs. 49,51-53) (Fig. 2). GBPs form part of a 47-member IFN-inducible GTPase superfamily comprising the 21–47 kDa immunity-related GTPases (IRGs), 72–82 kDa Myxoma (MX) resistance proteins and ~200–285 kDa GVINs in both humans and mice (ref. 24) (Fig. 2). Most of these GTPases undergo nucleotide-dependent self-assembly and several target pathogen-containing vacuoles (PCVs) as first shown for the GMS (G1 motif methionine)- and GKS (G1 motif lysine)-containing IRGs54,55. These proteins are potently induced by IFN-γ with the exception of MX proteins, which preferentially respond to type I or III IFNs24. For this reason, inflammasome loss-of-function screens began by priming macrophages with IFN-γ and LPS to ensure each GBP was fully expressed for family-wide analysis.
Fig. 2. Familial and structural properties of the GBPs.
(a) Phylogenetic tree of the IFN-inducible GTPase superfamily in humans and mice49. Bracketed names depict former assignations prior to the currently adopted MGI nomenclature, except for Lirg bidomain proteins. Two IRGM and GBP5 isoforms are included, along with a GBP1 (GBP2ts1) trans-spliced isoform49. Selected H-Ras and dynamin GTPases are shown for G domain comparison (EGA package MEGA v4.0.). The scale bar indicates substitutions/site. (b) Chromosomal configuration for human and mouse GBP families. Red enumeration, NCBI gi accession for pseudogenic fragments53. Mouse chr. 3H1 is syntenic with the telomeric end of the human 1q22.2 cluster while murine 5E5 is syntenic with the human GBP6-containing centromeric end (dashed lines). (c) Crystal structure of human GBP1 (PDB 1F5N) depicting the N-terminal GTPase-domain bound to the GTP analog, GMPPNP, and C-terminal helical domain.
siRNA silencing identified two orthologs, human GBP5 and its murine Gbp5 counterpart (67% amino acid identity), needed for NLRP3 inflammasome activation. Generation of Gbp5−/− mice corroborated defects in NLRP3 inflammasome activation. Here Gbp5−/− bone marrow-derived macrophages (BMMs) showed impaired IFN-γ-induced caspase-1, IL-1β and IL-18 processing in response to ATP plus cell-wall (LPS, muramyl dipeptide [MDP], γ-D-glutamyl-meso-diaminopimelic acid [DAP])-induced NLRP3 but not flagellin-induced NLRC4 or dsDNA-mediated AIM2 activation (Ref.12)(Fig. 3). Subsequent work has verified NLRP3 dependency is more conspicuous in IFN-γ-activated20 than unactivated cells22,23, presumably because IFN-γ induces robust GBP expression49,52. Importantly, GBP5-mediated defects were independent of host species (human, mouse) or strain background (C57BL/6J, C57BL6N, BALB/c), thus ruling out contributions by congenital Casp1129, Nlrp1b26 or neighboring Gbp156 mutations to these phenotypes. Such defects also extended to stationary phase S. typhimurium, known to engage caspase-11 upstream of NLRP318,34, and Listeria monocytogenes infection, which triggers NLRP3 and AIM2 inflammasomes in the canonical pathway8,57. Use of Gbp5−/− BMMs formally excluded logrithmically-grown Salmonella-dependent cell death which strongly activates the NAIP-NLRC4 inflammasome34,58.
Fig. 3. Specific steps at which individual GBPs impact canonical vs non-canonical inflammasome cascades.
(Left) Position of individual GBPs or the Gbpchr3−/− deletion encoding 5 clustered family members (GBP1, GBP2, GBP3, GBP5, GBP7) within the canonical NLRP3 and AIM2 inflammasome pathways after type I and II IFN signaling. The other major inflammasome subtypes, NLRP1 and NLRC4-NAIP, are also depicted. (Right) Position of individual GBPs plus unassigned members of the Gbpchr3−/− deletion within the non-canonical caspase-11-dependent inflammasome pathway in response to IFN signaling. GBP2 and GBP3 are arbitrarily placed upstream of GsdmD for pyroptosis because their precise positions are currently unknown.
Remarkably, human and mouse GBP5 deficiencies failed to impact macrophage responses to sterile NLRP3 agents (eg. MSU, alum, saponin adjuvants like Q21), a demarcation subsequently seen in mutiple studies12,18,22,23,59. Because sterile agents taken up into the endolysosomal network probably engage selective scavenger receptors (such as CD36)60-61, their spatial partitioning may bypass GBP5 that assists inflammasome complex assembly primarily in the cytosol12,22,23. GBP5 activities also appear to lie outside the TAK1-JNK signaling cascade that is initiated upon lysosomal disruption to elicit NLRP3 activation62. This partitioning operates in vivo as Gbp5−/− mice exhibit pronounced defects in MDP- but not alum-induced peritonitis and impaired serum IL-1β plus IL-18 production after LPS challenge (refs. 12,22,23) (Table 1). They similarly exhibit reduced macrophage and neutrophil caspase-1 activity to microbial but not sterile triggers in situ. Gbp5−/− mice also have heightened susceptibility to bacterial infections that require caspase-1 for protection, such as Gram-positive Listeria and Gram-negative Francisella12,22,23. Administration of the caspase-1 inhibitor, z-YVAD-FMK, renders wild-type mice susceptible to Listeria but fails to further increase vulnerability in Gbp5−/− hosts12, confirming GBP5 and caspase-1 share the same pathway (Table 1). Thus, GBP5 acts as a rheostat for inflammasome responses to PAMPs but not DAMPs in vivo24.
Table 1.
Inflammasome and host defense-related phenotypes in GBP knockout mice
| GBP Knockout |
Infectious or Inflammatory Setting |
Phenotype | Reference |
|---|---|---|---|
| Gbpr1−/− | L. monocytogenes | Susceptible to to orogastric infection | 49 |
| M. bovis BCG | Susceptible to i.v. infection | 49 | |
| T. gondii | Susceptible to s.c. infection | 71 | |
| Gbp2−/− | T. gondii | Susceptible to i.p. infection | 71 |
| L.monocytogenes | Resistant to i.p. infection | 71 | |
| F. norvicida | Susceptible to i.p. infection & reduced serum IL-18 | 23 | |
| Gbp5−/− | L. monocytogenes | Susceptible to orogastric infection & insensitive to the caspase-1 inhibitor z- YVAD-FMK |
12 |
| F. norvicida | Susceptible to s.c. infection | 23 | |
| LPS i.p.challenge | Reduced serum IL-1β plus IL-18 & reduced active caspase-1 in splenic macrophages |
12 | |
| MDP i.p.challenge | Impaired peritonitis & reduced active caspase-1 in peritoneal neutrophils | 12 | |
| Alum i.p.challenge | Normal peritonitis | 12 | |
| MSU i.p.challenge | Normal peritonitis | 12 | |
| Gbpchr3−/− | T. gondii | Susceptible to i.p. infection | 65 |
| L.monocytogenes | Resistant to i.p. infection | 65 | |
| F. norvica | Susceptible to i.p. infection & reduced serum IL-18 | 23 |
i.v., intravenous; i.p., intraperitoneal; s.c. subcutaneous.
The idea of GBPs as regulators of PAMP-sensitive inflammasome responses has grown to include other family members besides GBP5 (Fig. 3). Immunoprecipitation of endogenous GBP5 in Salmonella-infected macrophages retrieved GBP2 as a native protein partner (B.H.K., unpublished) and studies using Gbp2−/− BMMs show its requirement for IL-1β, IL-18 and cell death responses to this bacterium18. GBP2 has also recently been identified as an important cofactor for AIM2-dependent immunity to F. novicida where it cooperates with GBP5 (refs. 22,23) (Fig. 3). In this scenario, initial bacterial detection by the DNA sensor cyclic GMP-AMP synthase (cGAS) and its adaptor STING induce IRF-1-dependent GBP2 and GBP5 expression for IFN-induced intracellular killing and subsequent rounds of DNA release which then activate the AIM2 inflammasome. Cell-autonomous immunity63 thus links AIM2-mediated cytokine release for mobilizing antimicrobial defense mechanisms22,23. These mechanisms appear to manifest in vivo; Gbp2−/− mice have reduced serum IL-18 concentrations needed for IFN-γ production and host defense to F. novicida infection23,64; such mice are highly susceptible to this bacterium (Table 1).
GBPs present within a gene cluster on mouse chromosome 3H1 that contains Gbp2 and Gbp5 are likewise involved in cytokine maturation during Chlamydia trachiomatis and C. muridarum infections20. En bloc removal of this gene cluster (a 173-kilobase deletion of chr.3 [Gbpchr3−/−]65) impairs IL-1β and IL-18 release via canonical NLRP3 and AIM2 inflammasomes as well as the caspase-11-dependent noncanonical pathway (Fig. 3). GBPs facilitate faster kinetic processing of constitutive IL-18 by NLRP3 inflammasomes after either IFN-γ or LPS priming, whereas IL-1β secretion was affected only after LPS stimulation, suggesting TLR-induced NF-κB signaling is important for IL-1β expression but not IL-18 release by these bacterial species20. Multiple GBPs are therefore involved in customizing IFN-induced inflammasome responses to different intracellular bacteria.
GBPs in non-canonical inflammasome activation
A major advance in understanding bacteria-specific innate immunity has been identification of caspase-11 (caspase-4 and caspase-5 in humans) as an intracellular LPS receptor stimulating cytokine secretion via an NLRP3-ASC-caspase-1 complex and pyroptosis through a separate pathway after Gram-negative bacterial infection15,29-32. Transfection of intact LPS or its acetylated lipid A moiety directly into the cytosol binds the CARD domain of caspase-1115,30-31. Cytosolic LPS recognition bypasses TLR4 signaling and may proceed in a TRIF-independent fashion although infection by intact Gram-negative bacteria often requires the TLR4-TRIF-IRF3 circuit despite type I IFNs being dispensible for induction of procaspase-11 itself14,34. Hence other IFNs, principally IFN-γ, and other ISGs can contribute to non-canonical inflammasome-directed host defense against bacteria. Chief among these ISGs are the IFN-induced GBPs.
Proteomics-based expression analysis found GBPs were highly upregulated during Gram-negative bacterial infection of murine macrophages and genetic loss-of-function assays showed their importance for caspase-11-mediated IL-1β secretion and cell death17,18,20 (Fig. 3). GBP defects were again most evident with IFN-γ versus LPS or poly(I:C) priming, unless assays were conducted over suficiently long periods (12–18 hours) to enable sustained autocrine IFN-β signaling by the latter agents for robust GBP expression49,52. These findings also reinforce the underappreciated role for type II IFN-γ in inflammasome responses12. IFN-γ-dependent effects have been reported in macrophages infected with T3SS (ΔSPI-2), flagellin (ΔFlhD) and vacuolar (ΔSifA) mutants of S. typhimurium that preferentially engage caspase-1113,34; SdhA mutants of Legionella pneumophila; and other pathogenic species including Vibrio cholerae, Shigella flexneri, Citrobacter rodentium, Chlamydia trachiomatis and Chlamydia muridarum17,18,20. Hence GBPs are critical for non-canonical inflammasome activation by many Gram-negative bacteria.
Gbp2−/−, Gbp5−/− and Gbpchr3−/− BMMs primed with IFN-γ are also defective for pyroptosis and IL-1β release following transfection with ultrapure LPS from E.coli, Salmonella or Legionella17 (E.S.P., unpublished)(Fig. 3). Impaired responsivity in IFN-γ-activated GBP-deficient cells suggests they act downstream or independently of phagosomal disruption since LPS has already been introduced into the cytosol17. In unprimed cells, however, responses to transfected LPS appear intact, either because GBP expression is too low to affect the outcome or they operate upstream to disrupt bacterial phagosomes that release lipoglycans for detection18. Both mechanisms are plausible given GBPs assemble their antibacterial binding partners not only in the cytosol but also on PCVs12,49.
Most of the studies above used macrophages from Gbpchr3−/− mice to broadly implicate GBPs on chromosome 3H1 in the non-canonical pathway17,18. An exciting development has been the application of CRISPR/Cas9 engineering along with conventional gene targeting to individually delete all the GBPs in this cluster: Gbp1−/−, Gbp2−/−, Gbp3−/−, Gbp5−/− and Gbp7−/−12,17-18,49 (B.H.K., C.J.B., unpublished). It yielded an unexpected result: individual members such as GBP2, GBP3 and GBP5 each have distinct functions in this pathway. GBP2 binds ASC and GBP5 engages NLRP3 to facilitate partner ASC-NLRP3 assembly after heterotypic GBP2-GBP5 interactions (Fig. 3). This assembly promotes IL-1β secretion following detection of free LPS or LPS-containing microvesicles in the cytosol. In contrast, Gbp3 controls pyroptotic elimination of bacteria-infected cells independently of cytokine secretion. Such bifurcating activities invoke a new functional hierarchy, one in which individual GBPs are responsible for cytokine or cell-death decisions downstream of caspase-11 to promote cell-autonomous immunity to Gram-negative bacterial infection63.
How do the GBPs work?
GBPs impact both canonical and noncanonical inflammasome pathways. How do they confer their functions? Part of the answer lies with understanding their related biochemical and cell biological properties. At the protein level, human GBPs share 40–98% amino acid identity with a bidomain architecture composed of N-terminal GTPase and C-terminal helical domains, the latter harboring 12–13 amphipathic α-helicies involved in tail-to-tail dimer contacts12,66. Both GTPase and C-terminal helical domains contribute to homotypic self-assembly, similar to the ~90 kDa dynamin-like GTPases with which they share structural similarities12,66-68. GBPs typically bind GTP, GDP or GMP with equimolar affinity and exhibit high intrinsic rates of GTPase and GDPase activity (kcat ~100–150/min) due to an internal GAP domain, oviating the need for external GTPase-activating proteins (GAPs) to accelerate GTP or GDP hydrolysis and oligomerization67,69. In addition, some family members - GBP1, GBP2 and GBP5 - harbor C-terminal CaaX motifs for isoprenylation and membrane anchorage70.
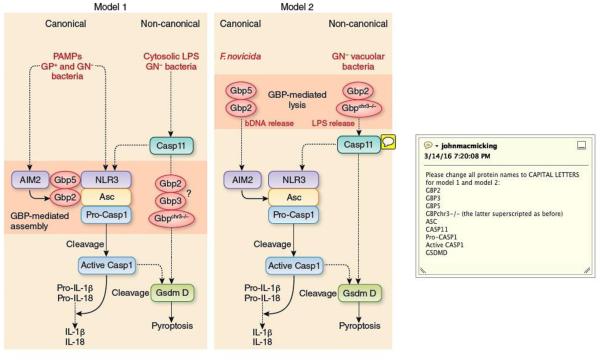
These two properties – guanine nucleotide-dependent self-assembly and membrane binding – are thought to underlie several of their antimicrobial functions. Previous studies showed their dual importance for antibacterial defense by generating nucleotide-binding and GTPase mutants of GBP1 or GBP7 unable to undergo transition-state assembly as well as GBP1 mutants lacking an intact CaaX motif for lipidation49. All mutants failed to target intracellular Listeria and Mycobacteria in IFN-γ-activated macrophages with direct loss of cell-autonomous immunity49,63. Similar functional phenotypes have been observed for Toxoplasma gondii in IFN-γ-treated murine embryonic fibroblasts and macrophages for GBP1 and GBP2 mutants71,72, while oligomerization mutants (D184) of human GBP1 interfere with control of Chlamydia trachiomatis73. Co-immunoprecipitation and direct binding studies reveal GBPs form homo- and heterotypic complexes with native partners (other GBPs and IRGs, autophagy-related proteins such as Atg4b and p62 (also known as SQSTM1), NADPH oxidase subunits, inflammasome core proteins) while membrane targeting helps deliver this antimicrobial cargo to the pathogen-containing vacuole (PVC) and autolysosomes12,49,65,70,74,75. For T. gondii, GBP-mediated delivery of a subset of GKS IRGs has been posited to disrupt the parisitophorous vacuole65 although GKS IRGs are missing from the human genome24. Thus, other mechanisms likely operate in human cells76-78, especially given the lack of biophysical evidence for human GBPs possessing direct dynamin-like “pinchase” activities that could functionally substitute for the loss of GKS IRGs24. Hence complex assembly rather than vacuolar disruption probably accounts for most of their inflammasome-related mechanisms, although the latter model could help direct where assembly takes place in the cell or predominate in specific settings (Fig. 4).
Fig. 4. Molecular mechanisms of GBPs in inflammasome activation.
(Left). Model 1 proposes some GBPs directly bind to and facilitate assembly of the core inflammasome machinery via their own multimerization in both canonical and non-canonical inflammasome pathways. Whether GBP2 and/or GBP3 assemble components of the pyroptotic arm including GsdmD remains to be tested (depicted by the question mark). GP+ (Gram-positive bacteria), GN− (Gram-negative bacteria). (Right). Model 2 proposes the GBPs lyse either pathogen-containing vacuoles (PVCs) or bacteria directly (bacteriolysis) to release ligands (eg. Gram-negative bacterial LPS or bacterial DNA [bDNA]) for downstream detection by the core inflammasome machinery.
The first inflammasome-related model proposed for the GBPs arose during their discovery as physical interactors with the core machinery12. GBP5 bound NLRP3 and facilitated inflammasome complex formation through its own self-assembly12 (Fig. 4). To date this remains the only molecular mechanism formally demonstrated and fits the emerging concept of supramolecular organizing centers (SMOCs) as important hubs for innate immunity79. GBP5 co-immunoprecipiated with native NLRP3 from IFN-γ-activated human monocytes and directly bound the PYD of NLRP3 in cell-free systems to promote ASC foci formation12. Gbp5−/− BMMs triggered by NLRP3 stimuli were correspondingly devoid of assembled ASC foci or “specks” and reconstitution with GBP5 mutants found GBP5 tetramerization was essential for generating higher oligomers of NLRP3-ASC in purified inflammasome complexes12. Thus, GBP5 acts as a SMOC cofactor for inflammasome activation. Here reconstituted assembly took place without infection; hence inflammasome activation can occur in the absence of PCV disruption although assembly may be initiated inside macrophages near PVCs or bacterial outer membrane vesicles (OMVs) harboring the requisite PAMPs.
Besides GBP5, tetrameric GBP280 also binds the core machinery, in this case ASC to assemble NLRP3 in the non-canonical pathway (B.H.K., unpublished). Because GBP2 and GBP5 heterotypically interact70, each can bring their own inflammasome partner (ASC and NLRP3, respectively) into the final complex to facilitate caspase-1 processing. This model provides an additional explanation for why GBP2 and GBP5 impacts the AIM2-ASC inflammasome22,23; GBP2 tetramerization induces ASC partner assembly while GBP5-GBP2 multimers may further promote ASC prionization (Fig. 4). Whether post-translational lipidation of GBP2 and GBP5 CaaX motifs helps initiate inflammasome oligomerization directly on endomembranes is unknown. GBP2 but not GBP5 also participates in caspase-11-dependent pyroptosis18. Here it is joined by Gasdermin D (GSDMD)21,81 and a third GBP family member, GBP3, as new components of this pathway. Notably, GBP3 does not converge on the core machinery nor Salmonella PCVs (B.H.K., unpublished) (Fig. 4). Thus individual GBPs can operate non-redundantly to confer specific defense activities, engaging different mechanistic partners like GBP1 and GBP7 that assemble p62/SQSTM1 and NADPH oxidase subunits, respectively, for autophagic and oxidative immunity to intracellular bacteria49. These alternative modes of action also fit their distinct biochemical activities, subcellular localization and pathogen susceptibility profiles24,70,82.
The second model proposed for how the GBPs promote inflammasome activation is disrupting PCVs to release PAMPs for NLR/ALR detection (ref. 18) (Fig. 4). This idea has borrowed heavily from earlier work on the IFN-induced 21-47 kDa GKS IRGs which could disrupt T. gondii vacuoles or Chlamydia inclusions in mouse but not human cells55,65,72,78,83. GKS IRGs are implicated in membranolytic alterations to PCVs and an analogous role for GBPs is inferred from limited antibody access to Salmonella compartments in Gbp2−/− or Gbpchr3−/− BMMs using detergent (digitonin) sensitivity assays, along with GBP2 relocation to PCVs in wild-type cells18. This recruitment of GBP2 to Salmonella PCVs seems non-specific, however, since the same anti-GBP2 antibody (11854-AP-2) still detects stationary-phase bacilli in Gbp2−/− BMMs (B.H.K., unpublished). Moreover, impaired AIM2 inflammasome responses to F. novicida in Gbp2−/−, Gbp5−/− and Gbpchr3−/− BMMs is not accompanied by differences in digitonin permeabilization, ruling out PCV disruption by GBPs in the canonical pathway23.
PCV disruption may only be required for some vacuolar bacteria but not others. For example, GBP-dependent inflammasome activation still occurs in macrophages infected with C. muridarum, a Chlamydial species that completely blocks GBP2 recruitment to the bacterial inclusion as a host-tropic evasion strategy20. Conversely, GBP-dependent inflammasome defects are evident in Gbp2−/− BMMs transfected with purified LPS alone or co-delivered with Gram-positive Listeria that lyses the PCV via its own pore-forming toxin, listeriolysin O, to release LPS into the cytosol13,17. A similar phenotype emerges after infection with Gram-negative Shigella flexneri that bypasses host-mediated phagosomal damage altogether since it escapes via its type III secretion system (T3SS)17. These studies reveal the GBPs are still needed even after LPS has reached the cytosol. Lastly, trafficking-defective macrophages lacking the GMS IRG proteins Irgm154,83 and/or Irgm384 fail to undergo GKS IRG-mediated disruption or recruit GBP2 to Salmonella or Chlamydia trachiomatis PVCs, yet these cells have normal caspase-11 inflammasome responses (B.H.K., unpublished). So, too, does p62/SQSTM1-deficient BMMs that show impaired GBP2 recruitment to Gram-negative bacterial inclusions85,86. Hence PCV disruption is often dispensible for inflammasome activation.
Despite these exceptions to the above model, GBP-mediated lysis could still conceivably impact inflammasome mobilization through bacterial rather than vacuolar damage (bacteriolysis). This alternative mode of action is implicated against F. norvicida to release bacterial DNA (bDNA) for AIM2 inflammasome activation (Refs. 22,23) (Fig. 4). Endogenous GBP2 and GBP5 often localize nearby irregularly-shaped F. novica that are permissive for propidium iodide uptake as a measure of decreased membrane permeability or increased lysis with cytosolic exposure. The same bacterial population also appear less frequently in Gbp2−/−, Gbp5−/− and Gbpchr3−/− BMMs that contained significantly fewer ASC foci23. Notably, transfection with dA:dT or F. norvicida DNA alone activated the inflammasome in a GBP-independent manner as reported earlier12,23, underscoring the idea that GBPs must first lyse the bacterial cell envelope to release dsDNA. One important caveat to this interpretation is that ectopic expression of GBP2 and/or GBP5 do not themselves reproduce this result23, suggesting their activities must be indirect, relying on other IFN-induced proteins to provide the mechanical insult3. The identity of such IFN-induced cofactors is currently unknown but remains an important area for future studies.
GBP-mediated immunity: Implications for human disease
Predisposition of GBP-deficient mice to infection plus amelioration of LPS-induced inflammation in human monocytes and mouse macrophages have implications for sepsis, cryopyrin associated periodic syndromes (CAPS) and interferonopathies. Gram-negative septicemia remains a major problem in intensive care units (>250,000 deaths each year in the U.S. alone) following surgical, burn or infectious trauma87. Current therapies blocking the initial hyperinflammatory, cytokine-mediated phase improve survival, although patients often enter a protracted immunosuppressive stage that fails to control primary infection or renders them vulnerable to secondary hospital-acquired infections, frequently by opportunistic organisms87. Uncoupling systemic inflammation from localized host defense has long been considered one way to slow progression of this multifactorial syndrome88. Rescue of deactivated monocytes in sepsis patients with IFN-γ treatment could aid antimicrobial defense89. Conceptually, blocking IFN-induced human CASPASE-4 in monocytes would interfere with both stages32; focusing on downstream targets separating this bimodal response may therefore be more beneficial. Human GBPs selectively involved in cytokine secretion could be blocked early on while GBP-mediated pyroptotic clearance of infection or bystander cell death may be manipulated at later times.
CAPS and interferonopathies are two other clinical areas where GBP-based therapies could be applicable in the future90,91. CAPS span familial cold autoinflammatory [FCAS] and Muckle-Wells syndromes as well as neonatal-onset multisystem inflammatory disorders [NOMIDs]90. They often result from autosomal dominant gain-of-function NLRP3 mutations that can lead to increased inflammasome assembly and activity. GBP5-specific inhibitors thus could limit CAPS-associated inflammatory sequelae while congenital GBP5 C-terminal truncations interferring with self-assembly66 may act as polygenic contributors to these inflammatory syndromes.
Lastly, interferonopathies are type I IFN-dependent inflammatory disorders including Aircardi-Goutières syndrome that arise from mutations in nucleic acid-sensing genes (eg. TREX1, SAMHD1, RNASEH2A, ADAR, IFIH1)91. These, together with recent SLE-associated gain-of-function TMEM173 (STING) mutations92, may enlist GBP2 and GBP5 for regulating AIM2 inflammasome responses to cytosolic DNA22,23. Again, therapeutic agents targeting these GBPs could serve as possible treatment avenues in the future.
Concluding remarks
The past 5 years have witnessed a cavalcade of discovery on the IFN-inducible GBPs during antibacterial host defense and inflammasome activation3,24. GBPs operate against at least 10 bacterial species triggering canonical or non-canonical inflammasome responses to control IL-1β plus IL-18 secretion and pyroptosis12,17,18,22,23 . Allelic deletions in Gbp1−/−, Gbp2−/−, Gbp3−/−, Gbp5−/−, Gbp7−/− and Gbpchr3−/− mice or macrophages often yield profound susceptibility or inflammatory phenotypes, as do GBP-defective human cells12,17,18,22,23,49,59,65,71-73,79. The latter remains an understudied area with important consequences for disease. Because clues to their inflammasome-related activities first arose from broad evolutionary analysis12, mining the fossil record should continue to yield insights about how IFN-induced GBPs mobilize protective immunity across the animal kingdom.
Acknowledgements
We apologize to colleagues whose work has not cited due to reference limits and acknowledge support for discoveries described in this review from the following sources: HHMI Investigator Program, NIH NIAID (R01 AI068041-09, AI108834-01A1), American Asthma Foundation Research Program (Scholar Award 14-0073), and Rainin Foundation (Innovator Award 14H7).
References
- 1.Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annu. Rev. Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 2.Aravind L, Dixit VM, Koonin EV. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291:1279–1284. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- 3.MacMicking JD. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 2012;12:367–382. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Angosto D, et al. Evolution of inflammasome functions in vertebrates: Inflammasome and caspase-1 trigger fish macrophage cell death but are dispensable for the processing of IL-1β. Innate Immun. 2012;18:815–824. doi: 10.1177/1753425912441956. [DOI] [PubMed] [Google Scholar]
- 6.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J. Exp. Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones JW, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl Acad. Sci. USA. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rathinam VA, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dombrowski Y, et al. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci. Transl. Med. 2011;3:82ra38. doi: 10.1126/scitranslmed.3002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerur N, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rathinam VA, et al. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell. 2012;150:606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shenoy AR, et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–485. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- 13.Aachoui Y, et al. Caspase-11 protects against bacteria that escape the vacuole. Science. 2013;339:975–978. doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Case CL, et al. Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proc. Natl Acad. Sci. USA. 2013;110:1851–1856. doi: 10.1073/pnas.1211521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strittmatter GE, et al. IFN-γ primes keratinocytes for HSV-1-induced inflammasome activation. J. Invest. Dermatol. 2016;136:610–620. doi: 10.1016/j.jid.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 17.Pilla DM, et al. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc. Natl Acad. Sci. USA. 2014;111:6046–6051. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meunier E, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 19.Aachoui Y, et al. Canonical Inflammasomes drive IFN-© to prime Caspase-11 in defense against a cytosol-Invasive bacterium. Cell Host Microbe. 2015;18:320–332. doi: 10.1016/j.chom.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finethy R, et al. Guanylate Binding Proteins enable rapid activation of canonical and noncanonical inflammasomes in Chlamydia-infected macrophages. Infect Immun. 2015;83:4740–4749. doi: 10.1128/IAI.00856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kayagaki N, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 22.Man SM, et al. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 2015;16:467–475. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meunier E, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 2015;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim BH, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. IFN-inducible GTPases in host cell defense. Cell Host Microbe. 2012;12:432–444. doi: 10.1016/j.chom.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Moltke J, Ayres JS, Kofoed EM, Chavarría-Smith J, Vance RE. Recognition of bacteria by inflammasomes. Annu. Rev. Immunol. 2013;31:73–106. doi: 10.1146/annurev-immunol-032712-095944. [DOI] [PubMed] [Google Scholar]
- 27.Chen KW, et al. The neutrophil NLRC4 inflammasome selectively promotes IL-1β maturation without pyroptosis during acute Salmonella challenge. Cell Rep. 2014;8:570–582. doi: 10.1016/j.celrep.2014.06.028. [DOI] [PubMed] [Google Scholar]
- 28.Knodler LA, et al. Noncanonical inflammasome activation of caspase-4/caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microbe. 2014;16:249–256. doi: 10.1016/j.chom.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kayagaki N, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 30.Kayagaki N, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 31.Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 32.Casson CN, et al. S. Human caspase-4 mediates non-canonical inflammasome activation against gram-negative bacterial pathogens. Proc. Natl Acad. Sci. USA. 2015;112:6688–6693. doi: 10.1073/pnas.1421699112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 2010;8:471–483. doi: 10.1016/j.chom.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broz P, et al. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Z, et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 2015;350:399–404. doi: 10.1126/science.aac5489. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 2015;350:404–409. doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muñoz-Planillo R, et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guarda G, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Franchi L, Eigenbrod T, Núñez G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 2009;183:792–796. doi: 10.4049/jimmunol.0900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee CK, Smith E, Gimeno R, Gertner R, Levy DE. STAT1 affects lymphocyte survival and proliferation partially independent of its role downstream of IFN-gamma. J. Immunol. 2000;164:1286–1292. doi: 10.4049/jimmunol.164.3.1286. [DOI] [PubMed] [Google Scholar]
- 45.Schauvliege R, Vanrobaeys J, Schotte P, Beyaert R. Caspase-11 gene expression in response to lipopolysaccharide and interferon-gamma requires nuclear factor-kappa B and signal transducer and activator of transcription (STAT) 1. J. Biol Chem. 2002;277:41624–41630. doi: 10.1074/jbc.M207852200. [DOI] [PubMed] [Google Scholar]
- 46.Lu B, et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488:670–674. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Y, Franchi L, Núñez G. The protein kinase PKR is critical for LPS-induced iNOS production but dispensable for inflammasome activation in macrophages. Eur. J. Immunol. 2013;43:1147–1152. doi: 10.1002/eji.201243187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hett EC, et al. Chemical genetics reveals a kinase-independent role for protein kinase R in pyroptosis. Nat. Chem. Biol. 2013;9:398–405. doi: 10.1038/nchembio.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim BH, et al. A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 50.Jin T, Huang M, Smith P, Jiang J, Xiao TS. Structure of the caspase-recruitment domain from a zebrafish guanylate-binding protein. Acta. Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013;69:855–860. doi: 10.1107/S1744309113015558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olszewski MA, Gray J, Vestal DJ. In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J. Interferon Cytokine Res. 2006;26:328–352. doi: 10.1089/jir.2006.26.328. [DOI] [PubMed] [Google Scholar]
- 52.Degrandi D, et al. Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J. Immunol. 2007;179:7729–7740. doi: 10.4049/jimmunol.179.11.7729. [DOI] [PubMed] [Google Scholar]
- 53.Shenoy AR, et al. Emerging themes in IFN-γ-induced macrophage immunity by the p47 and p65 GTPase families. Immunobiology. 2007;212:771–784. doi: 10.1016/j.imbio.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 55.Martens S, et al. Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47-resistance GTPases. PLoS Pathog. 2005;1:e24. doi: 10.1371/journal.ppat.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Staeheli P, Prochazka M, Steigmeier PA, Haller O. Genetic control of interferon action: mouse strain distribution and inheritance of an induced protein with guanylate-binding property. Virology. 1984;137:135–142. doi: 10.1016/0042-6822(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 57.Kim S, et al. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 2010;40:1545–1551. doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rupper AC, Cardelli JA. Induction of guanylate binding protein 5 by gamma interferon increases susceptibility to Salmonella enterica serovar Typhimurium-induced pyroptosis in RAW264.7 cells. Infect. Immun. 2008;76:2304–2315. doi: 10.1128/IAI.01437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marty-Roix R, et al. Identification of QS-21 as an inflammasome-activating molecular component of saponin adjuvants. J Biol Chem. 2016;291:1123–1136. doi: 10.1074/jbc.M115.683011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheedy FJ, et al. CD36 coordinates NLRP3 inflammasome activation by facilitating intracellular nucleation of soluble ligands into particulate ligands in sterile inflammation. Nat. Immunol. 2013;14:812–820. doi: 10.1038/ni.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okada M, Matsuzawa A, Yoshimura A, Ichijo H. The lysosome rupture-activated TAK1-JNK pathway regulates NLRP3 inflammasome activation. J. Biol. Chem. 2014;289:32926–32936. doi: 10.1074/jbc.M114.579961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Randow F, MacMicking JD, James LC. Cellular self-defense: How cell-autonomous immunity protects against pathogens. Science. 2013;340:701–706. doi: 10.1126/science.1233028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pierini R, Perret M, Djebali S, Juruj C, Michallet MC, Förster I, Marvel J, Walzer T, Henry T. ASC controls IFN-γ levels in an IL-18-dependent manner in caspase-1-deficient mice infected with Francisella novicida. J. Immunol. 2013;191:3847–3857. doi: 10.4049/jimmunol.1203326. [DOI] [PubMed] [Google Scholar]
- 65.Yamamoto M, et al. A cluster of interferon-γ-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 2012;37:302–313. doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 66.Wehner M, Herrmann C. Biochemical properties of the human guanylate binding protein 5 and a tumor-specific truncated splice variant. FEBS J. 2010;277:1597–605. doi: 10.1111/j.1742-4658.2010.07586.x. [DOI] [PubMed] [Google Scholar]
- 67.Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature. 2006;440:101–104. doi: 10.1038/nature04510. [DOI] [PubMed] [Google Scholar]
- 68.Syguda A, et al. Tetramerization of human guanylate-binding protein 1 is mediated by coiled-coil formation of the C-terminal α-helices. FEBS J. 2012;279:2544–2554. doi: 10.1111/j.1742-4658.2012.08637.x. [DOI] [PubMed] [Google Scholar]
- 69.Abdullah N, Srinivasan B, Modiano N, Cresswell P, Sau AK. Role of individual domains and identification of internal gap in human guanylate binding protein-1. J. Mol. Biol. 2009;386:690–703. doi: 10.1016/j.jmb.2008.12.060. [DOI] [PubMed] [Google Scholar]
- 70.Britzen-Laurent N, et al. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS One. 2010;5:e14246. doi: 10.1371/journal.pone.0014246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Degrandi D, et al. Murine guanylate binding protein 2 (mGBP2) controls Toxoplasma gondii replication. Proc. Natl Acad. Sci. USA. 2013;110:294–299. doi: 10.1073/pnas.1205635110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Selleck EM, et al. Guanylate-binding protein 1 (Gbp1) contributes to cell-autonomous immunity against Toxoplasma gondii. PLoS Pathog. 2013;9:e1003320. doi: 10.1371/journal.ppat.1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tietzel I, El-Haibi C, Carabeo RA. Human guanylate binding proteins potentiate the anti-chlamydia effects of interferon-gamma. PLoS One. 2009;4:e6499. doi: 10.1371/journal.pone.0006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Virreira Winter S, et al. Determinants of GBP recruitment to Toxoplasma gondii vacuoles and the parasitic factors that control it. PLoS One. 2011;6:e24434. doi: 10.1371/journal.pone.0024434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee Y, et al. p62 Plays a Specific Role in Interferon-©-Induced Presentation of a Toxoplasma Vacuolar Antigen. Cell Rep. 2015;13:223–233. doi: 10.1016/j.celrep.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 76.Niedelman W, Sprokholt JK, Clough B, Frickel EM, Saeij JP. Cell death of gamma interferon-stimulated human fibroblasts upon Toxoplasma gondii infection induces early parasite egress and limits parasite replication. Infect Immun. 2013;81:4341–4349. doi: 10.1128/IAI.00416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ohshima J, et al. Role of mouse and human autophagy proteins in IFN-γ-induced cell-autonomous responses against Toxoplasma gondii. J. Immunol. 2014;192:3328–3335. doi: 10.4049/jimmunol.1302822. [DOI] [PubMed] [Google Scholar]
- 78.Selleck EM, et al. A noncanonical autophagy pathway restricts Toxoplasma gondii growth in a strain-specific manner in IFN-γ-activated human cells. MBio. 2015;6:e01157–15. doi: 10.1128/mBio.01157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kagan JC, Magupalli VG, Wu H. SMOCs: supramolecular organizing centres that control innate immunity. Nat. Rev. Immunol. 2014;14:821–826. doi: 10.1038/nri3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kravets E, et al. The GTPase activity of murine guanylate-binding protein 2 (mGBP2) controls the intracellular localization and recruitment to the parasitophorous vacuole of Toxoplasma gondii. J. Biol Chem. 2012;287:27452–27466. doi: 10.1074/jbc.M112.379636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 82.MacMicking JD. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 2004;25:601–609. doi: 10.1016/j.it.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 83.Tiwari S, Choi HP, Matsuzawa T, Pypaert M, MacMicking JD. Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P(2) and PtdIns(3,4,5)P(3) promotes immunity to mycobacteria. Nat. Immunol. 2009;10:907–917. doi: 10.1038/ni.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haldar AK, et al. IRG and GBP host resistance factors target aberrant, "non-self" vacuoles characterized by the missing of "self" IRGM proteins. PLoS Pathog. 2013;9:e1003414. doi: 10.1371/journal.ppat.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ohtsuka S, et al. SQSTM1/p62/A170 regulates the severity of Legionella pneumophila pneumonia by modulating inflammasome activity. Eur. J. Immunol. 2014;44:1084–1092. doi: 10.1002/eji.201344091. [DOI] [PubMed] [Google Scholar]
- 86.Haldar AK, et al. Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc. Natl Acad. Sci. USA. 2015;112:E5628–5637. doi: 10.1073/pnas.1515966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 2013;13:862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.MacMicking JD, et al. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell. 1995;81:641–650. doi: 10.1016/0092-8674(95)90085-3. [DOI] [PubMed] [Google Scholar]
- 89.Döcke WD, et al. Monocyte deactivation in septic patients: restoration by IFN-gamma treatment. Nat. Med. 1997;3:678–681. doi: 10.1038/nm0697-678. [DOI] [PubMed] [Google Scholar]
- 90.Broderick L, De Nardo D, Franklin BS, Hoffman HM, Latz E. The inflammasomes and autoinflammatory syndromes. Annu. Rev. Pathol. 2015;10:395–424. doi: 10.1146/annurev-pathol-012414-040431. [DOI] [PubMed] [Google Scholar]
- 91.Crow YJ, Manel N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 92.Jeremiah N, et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin Invest. 2014;124:5516–5520. doi: 10.1172/JCI79100. [DOI] [PMC free article] [PubMed] [Google Scholar]