Abstract
Most of the efforts in elucidating the molecular relatedness and epidemiology of Staphylococcus aureus in Malaysia have been largely focused on methicillin-resistant S. aureus (MRSA). Therefore, here we report the draft genome sequence of the methicillin-susceptible Staphylococcus aureus (MSSA) with sequence type 1 (ST1), spa type t127 with Panton-Valentine Leukocidin (pvl) pathogenic determinant isolated from pus sample designated as KT/314250 strain. The size of the draft genome is 2.86 Mbp with 32.7% of G + C content consisting 2673 coding sequences. The draft genome sequence has been deposited in DDBJ/EMBL/GenBank under the accession number AOCP00000000.
Keywords: Methicillin-susceptible Staphylococcus aureus, Panton-Valentibe Leukocidin, ST1-MSSA PVL positive Malaysia isolates
Specifications
| Organism/cell line/tissue | Methicillin-susceptible Staphylococcus aureus |
| Strain | KT/314250 |
| Sequencer or array type | Illumina GA IIx |
| Data format | Assembled |
| Experimental factors | Bacterial strain |
| Experimental features | Assembled and annotated draft genome of a strain of methicillin susceptible Staphylococcus aureus PVL+ from east coast Malaysia |
| Consent | Not applicable |
| Sample source location | Pus |
1. Direct link to deposited data
2. Experimental design, materials and methods
MRSA evolved over the times from Methicillin susceptible S. aureus via acquisition of mobile genetic elements called staphylococcal cassette chromosome mec (SCCmec) [1], [2]. Thus, this makes MSSA to be a potential reservoir for the MRSA strains. S. aureus strains producing PVL have been associated with a variety of illness ranging from skin and soft tissue infections to necrotizing pneumonia as well as septicaemia that are invariably fatal [3]. The genome sequencing of KT/314,250 strain was performed using the Illumina genome analyzer IIx 100-bp paired-end reads. The paired-end reads were trimmed and assembled de novo using CLC genomics workbench 5.1 (CLC Bio, Denmark). Multi Locus Sequence Typing (MLST) was performed by using Local BLAST identification and manually aligned based on primers used to amplified seven gene fragments (arcC, aroE, glpF, gmk, pta, tpi and yqiL) [4]. Meanwhile the spa typing was assigned using DNAGear freely available Software [5]. Thus, all genotypic analysis revealed this strain as ST1, spa type t127, agr III and dru type dt10ao.
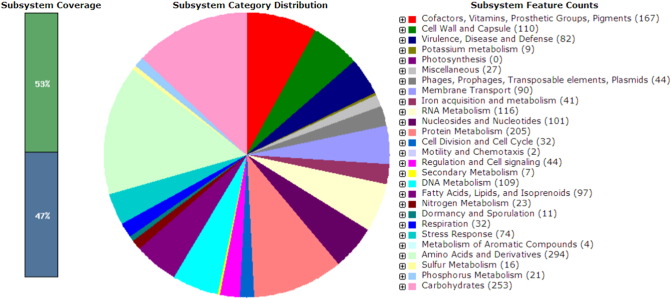
The draft genome were annotated by using free accessible bioinformatics tools Blast2GO 2.5.0 [6] and subsequently validated using Rapid Annotation Subsystem Technology (RAST) [7] and Bacterial Annotation System (BASys) [8]. Initial sequence analysis revealed a total of 69 contigs from the de novo assembly with an accumulate length of 2,846,051 bp with G + C content of 32.7%. A total of 2673 coding sequences (CDSs) and 48 RNAs regions were annotated. Of the CDS, 4.12% were associated with cell wall and capsule; 3.07% were associated with virulence, disease and defence mechanism and 2.77% were related with stress response which is contributed in host adaptation and survival (Fig. 1).
Fig. 1.
Subsystem distribution of methicillin-susceptible S. aureus KT/314,250 (based on RAST annotation server).
Nucleotide sequence accession number
The Draft genome sequence of Methicillin-susceptible Staphylococcus aureus (MSSA) KT/314,250 strain has been deposited under the accession number AOCP00000000. The version described in this paper was the first version, AOCP00000000.
Acknowledgements
This research was supported by Universiti Sultan Zainal Abidin research funds to Z.S. and C.C.Y. under grants UDM/09/BR (009) and UDM/09/BR (006), respectively.
References
- 1.Baranovich T., Zaraket H., Shabana I.I., Nevzorova V., Turcutyuicov V., Suzuki H. Molecular characterization and susceptibility of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from hospitals and the community in Vladivostok, Russia. Clin. Microbiol. Infect. 2010;16(6):575–582. doi: 10.1111/j.1469-0691.2009.02891.x. ( http://doi.org/10.1111/j.1469-0691.2009.02891.x) [DOI] [PubMed] [Google Scholar]
- 2.Chongtrakool P., Ito T., Ma X.X., Kondo Y., Trakulsomboon S., Tiensasitorn C.…Hiramatsu K. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 2006;50(3):1001–1012. doi: 10.1128/AAC.50.3.1001-1012.2006. ( http://doi.org/10.1128/AAC.50.3.1001-1012.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lina G., Piémont Y., Godail-Gamot F., Bes M., Peter M.O., Gauduchon V.…Etienne J. Involvement of Panton-valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 1999;29(5):1128–1132. doi: 10.1086/313461. ( http://doi.org/10.1086/313461) [DOI] [PubMed] [Google Scholar]
- 4.Enright M.C., Day N.P.J., Davies C.E., Peacock S.J., Spratt B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000;38(3):1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.AL-Tam F., Brunel A.-S., Bouzinbi N., Corne P., Bañuls A.-L., Shahbazkia H.R. DNAGear–a free software for spa type identification in Staphylococcus aureus. BMC Res. Notes. 2012;5:642. doi: 10.1186/1756-0500-5-642. ( http://doi.org/10.1186/1756-0500-5-642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conesa A., Götz S., García-Gómez J.M., Terol J., T. M., R. M. 2005. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. [DOI] [PubMed] [Google Scholar]
- 7.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A.…Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. ( http://doi.org/10.1186/1471-2164-9-75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Domselaar G.H., Stothard P., Shrivastava S., Cruz J.A., Guo A., Dong X.…Wishart D.S. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 2005;33(Web Server issue):W455–W459. doi: 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]