Abstract
Intermediate phenotypes are traits positioned somewhere between genetic variation and disease. They represent a target for attempts to find disease-associated genetic variants and elucidation of mechanisms. Psychiatry has been particularly enamored with intermediate phenotypes, due to uncertainty about disease etiology, inconclusive results in early psychiatric genetic studies, and their appeal relative to traditional diagnostic categories. Here, we argue that new genetic findings are relevant to the question of the utility of these constructs. In particular, results from genome-wide association studies of psychiatric disorders now allow an assessment of the potential role of particular intermediate phenotypes. Based on such an analysis, as well as other recent results, we conclude that intermediate phenotypes are likely to be most valuable in understanding mechanism.
What’s the use of an endophenotype?
Until recently, genetic approaches in psychiatric disease had enjoyed limited success, but large-scale collaborative a genome-wide association studies (GWAS) have now begun to identify common and rare variants associated with schizophrenia, bipolar disorder and autism spectrum disorders [1–11]. An alternative to these agnostic, large-scale approaches is to focus instead on phenotypes thought to reflect disease processes. In this review we refer to these phenotypes as intermediate phenotypes, rather than endophenotypes. Definitions of ‘endophenotype’ grace every review of this area (see Box 1 for some examples), and they are not all alike. For our purposes, the critical distinction is between definitions in which an endophenotype is considered to lie on the causal pathway to disease, and those in which it is more generally an index of the likelihood that a subject has the disease (and could serve as a biomarker). This distinction, noted by Walters and Owen [12], has been treated in detail by Kendler and Neale [13]. Here we distinguish two ways in which intermediate phenotypes could be used in genetic studies, and review their utility in genetic analysis of psychiatric disease.
Box 1. Endophenotype definitions.
Cannon and Keller 2006 [87] : “Endophenotypes are intermediate phenotypes, often imperceptible to the unaided eye, that link disease-promoting sequence variations in genes (haplotypes or alleles) to lower level biological processes and link lower level biological processes to the ‘downstream’ observable syndromes that constitute diagnostic categories of disorders .”
Vrieze, Iacono and McGue 2012 [88] : “An endophenotype is a heritable, biologically based, objectively quantifiable measure that is associated with a psychiatric phenotype because both share a common genetic influence. Endophenotypes hold promise for gene finding because they are believed to (a) be more proximal to the effects of genes; (b) tap one of many facets of a disorder and thus be more etiologically homogeneous than the associated disorder; (c) be more highly heritable than the disorder, thus likely to produce larger effect sizes and boost GWAS power; or, (d) because the endophenotype deals with an etiologically homogenous facet of the disorder, be associated with fewer genes each of which may be expected to have larger individual effect sizes.”
Gottesman and Gould 2003 [89] : “1. The endophenotype is associated with illness in the population; 2. The endophenotype is heritable; 3. The endophenotype is primarily state-independent (manifests in an individual whether or not illness is active); 4. Within families, endophenotype and illness co-segregate; 5. The endophenotype found in affected family members is found in nonaffected family members at a higher rate than in the general population. … Endophenotypes represent more defined and quantifiable measures that are envisioned to involve fewer genes, fewer interacting levels and ultimately activation of a single set of neuronal circuits. The fewer the pathways that give rise to an endophenotype, the better the chances of efficiently discovering its genetic and neurobiological underpinnings.”
The first use of intermediate phenotypes is to aid gene discovery. For example, if a genetic study of major depression fails to identify a signal, then researchers may decide to focus their attention on cognitive variables instead, reasoning that individual differences in the processing of stimuli representing negative affect (such as a bias in the interpretation of sad or happy faces [14]) might be an important mechanistic pathway, better reflecting the underlying biology, and therefore more genetically tractable. In addition, they might include data indicating activity in brain areas involved in processing affective or motivational stimuli (such as the amygdala and nucleus accumbens). With results in hand, researchers can combine the additional phenotypes with the diagnostic information, searching for a subgroup in their data in which the genetic signal is enhanced. Alternatively (or additionally) they can work solely with the new phenotypes, hoping that, by solving the genetics of disease mechanism they can then later solve the genetics of disease itself (for example, a locus identified for the cognitive measures could be tested for association in the depression cohort). Of course, this approach depends on selecting the true disease model (i.e., the correct intermediate phenotypes).
The second use of intermediate phenotypes is to interpret the results of genetic discovery studies. For instance, suppose, through a genomewide association study (GWAS) or exome sequencing, a number of genes are found that are unequivocally involved in conferring risk of schizophrenia. Either the function of these genes is completely unknown, or their role in psychiatric disease was never previously suspected. Genetic analysis of intermediate phenotypes might provide clues to the role these genes play. For instance, in the case of schizophrenia studies of high-risk individuals, together with birth and cohort studies, indicate that cognitive deficits predate the onset of psychosis [15, 16]. These cognitive features are heritable and are genetically correlated with psychosis (genetic correlations have been estimated to be greater than 0.5) [17–19]. Researchers might attempt to identify loci contributing to both variation in cognitive performance and risk of psychosis, and hence begin to unravel the function of one group of genetic risk factors for schizophrenia.
Using intermediate phenotypes to find disease genes
For more than ten years intermediate phenotypes have been incorporated into genetic studies of psychiatric disease. In our opinion, their use to identify disease genes rests on two key assumptions.
The first assumption is that intermediate phenotypes are more tractable to genetic mapping than a primary phenotype, principally because they are presumed to be closer to the underlying biology. That is, we assume that genetic associations for intermediate phenotypes will be less influenced by measurement error, environmental influences, or multiple pathway effects that are likely to complicate the analysis of primary phenotypes. If this is true, and in some cases it is where we are fortunate to have a well-placed intermediate measure, then smaller sample sizes might be sufficient to identify these intermediate phenotypes. This, in turn, also implies that the genetic architecture of intermediate phenotypes will be different to that of disease phenotypes. However one must contend with the important notions that (i) these intermediate phenotypes are often not available and (ii) with biological proximity often comes a lack of clinical or real life translation (especially in the field of psychological traits).
The second assumption is that once genetic loci contributing to the intermediate phenotype have been found, this will facilitate the search for disease susceptibility loci. For instance, rather than searching the entire genome for loci associated with the primary phenotype of disease susceptibility, and suffering the penalty of applying a corrected significance threshold of P < 5×10-8 (i.e., genomewide significance), researchers will use a GWAS of the intermediate phenotype to identify putative markers for disease susceptibility. Those few found with reliable evidence in the GWAS of the intermediate phenotype can then be followed up in relation to the primary phenotype to confirm their role in disease susceptibility, requiring an appropriate but far less restrictive multiple comparison correction [20]. In other words, intermediate phenotypes can provide a reduced search space for disease susceptibility loci. We review the evidence for these assumptions in turn.
Genetic architecture
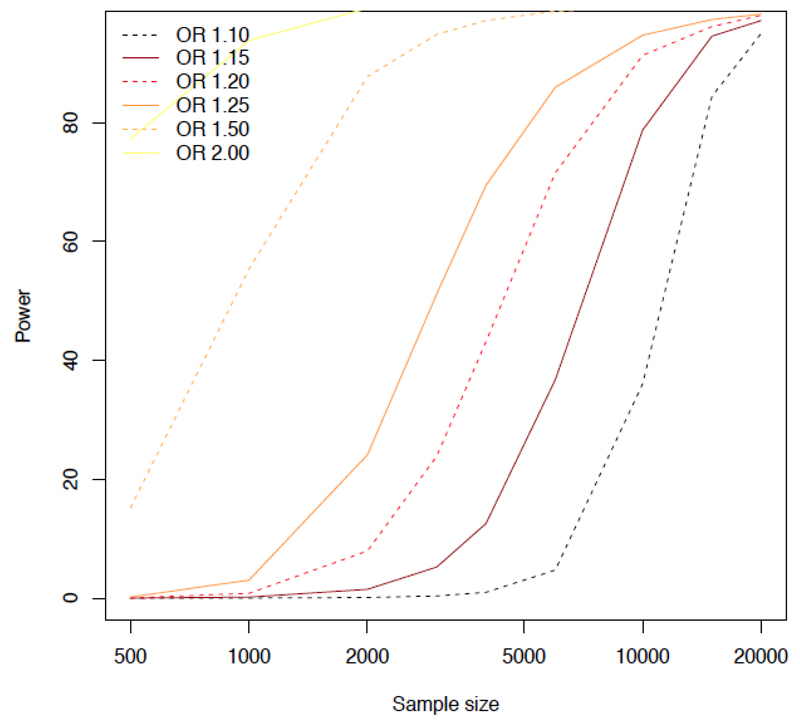
In a previous review [21] we asked whether the mean effect size of loci that contribute to psychiatric disease was different from the mean effect size of loci contributing to variation in intermediate phenotypes. We posed this question because larger effect sizes are easier to detect, requiring smaller sample sizes (in other words, the phenotypes are more genetically tractable). Since statistical power is proportional to the square of the effect size, if effect sizes are halved, then four times the sample size is required for their detection. Therefore, if genetic loci contributing to variation in intermediate phenotypes have larger effects sizes than loci contributing to psychiatric disease, it would make sense for genetic studies to focus on intermediate phenotypes (Figure 1). Our conclusion at that time was that the genetic basis of intermediate phenotypes appeared to be at least as complex as that of psychiatric disease, with no clear evidence of substantially larger individual effects [21]. However, we cautioned that we had little reliable data about the genetic architecture of complex traits in humans.
Figure 1. Statistical Power.
Simulated data are plotted to show power (vertical axis) for different sample sizes (horizontal axis) for six different effect sizes, expressed as odds ratios. The simulations use two million SNPs and are taken from [97], which contains details of the parameters used. The significance level was set at 5×10−8. Sample size is shown for the number of cases required; the simulations assume an equal number of controls.
We are now in a position to address empirically whether intermediate phenotypes are more tractable to genetic mapping than psychiatric disease primary phenotypes. The extent to which intermediate phenotypes help with gene identification depends on their genetic architecture — the number of loci, their effect sizes and the way they operate. We now know the likely genetic architecture of complex traits in general, and something of psychiatric disease in particular.
One of the most important observations to emerge is that almost all complex traits have a highly polygenic component to their variance: that is, their genetic basis consists of relatively frequent (> 5%) risk alleles at a very large number of loci, each making a small contribution to variation, or disease susceptibility. Across all diseases, the median odds ratio (OR, a measure of the extent to which a genotype, or allele, increases the risk of developing disease) is 1.25 This observation is drawn from the 1,736 studies in the National Human Genome Research Institute (NHGRI) catalog of published GWAS (564 studies of disease; accessed November 2013) [22]. Fewer than 2% of the observations have an OR greater than 2.5, and fewer than 1% have an OR greater than 4. Similarly, for quantitative traits, the amount of variance explained by a locus is much less than 0.5%. Even these estimates are likely upwardly biased due to the inclusion of smaller studies and Winner’s Curse effects [23].
What is the contribution of common genetic variation to the genetic architecture of psychiatric intermediate phenotypes? We now have answers to this question. Brain structural variation is a plausible intermediate phenotype — similar differences in brain structure have been found in unaffected individuals at increased genetic risk of psychiatric illness and affected individuals [24–26]. These phenotypes have been subject to GWAS [27–30], and the loci identified “have comparable effect sizes to those observed in other genome-wide association studies of complex traits” [27]. One marker explains just 0.58% of intracranial volume per risk allele and required 21,151 subjects (combined cases and controls in discovery and replication samples) to be identified [27]. Mapping of measures of cognitive performance [31] similarly shows genetic effects no larger than those found for psychiatric disease. Two consortia, one based in Europe (the Wellcome Trust Case Control Consortium; http://www.wtccc.org.uk/ccc2/projects/ccc2_pe.html) and one in the United States (the Consortium on the Genetics of Endophenotypes in Schizophrenia [32, 33]), have gathered cognitive, neuroanatomical and neurophysiological phenotypes that may act on the causal pathway to schizophrenia. The European consortium has so far published no genome-wide significant hits, while the United States consortium has reported linkage (not association) analyses of 12 endophenotypes, finding only one locus that exceeded genome-wide significance [34]. This result is no more successful than prior attempts to identify risk loci for schizophrenia through family-based linkage mapping [35, 36]. The authors point out: “while the biological basis of these endophenotypes may be simpler than that of schizophrenia per se, they nonetheless remain complex and appear to be highly polygenic. Conceivably, a refinement of these endophenotypes may probe a more specific physiology and thereby be sensitive to a more pure genetic signal” [34]. Is that optimistic conclusion realistic?
We can examine the idea that a specific physiology has a “purer genetic signal”, in the sense that it has a more tractable genetic architecture, by looking at what is known about the genetic basis of physiological phenotypes far simpler than psychiatric disease. It seems reasonable to propose that measures of protein concentration and metabolites levels qualify as probes for more specific physiology; in other words, they should be good intermediate phenotypes. The locus-specific effects of such phenotypes are all small: the mean effect size for 95 loci that influence blood lipids is 0.12% phenotypic variance [37], while the 17 loci so far discovered that contribute to variation in serum C-reactive protein have a mean effect size of 0.28% [38]. The 26 loci influencing serum urate levels have a mean effect size of 0.26% of phenotypic variance [39]. Effect sizes on cellular phenotypes are of a similar magnitude — 0.12% of phenotypic variance for 75 loci contributing to variation in mean red cell volume [40].
While the available GWAS data do not indicate that intermediate phenotypes will necessarily be any more genetically tractable than psychiatric disease, there is one important proviso: even a small increase in the mean locus-specific effect size has a substantial impact on power (given that power is proportional to the square of the effect size). Consistent with the notion that height and weight are more complex phenotypes (that is, less biologically coherent) than lipid levels, locus-specific effects are smaller: the mean effect size for 180 loci on height is 0.058% of phenotypic variance [41] and mean effect on weight is 0.045% of phenotypic variance for 32 loci [42]. These figures are lower than the effects found for lipids and red cell phenotypes. Thus while more than 50,000 subjects may be needed to detect a single locus contributing to major depression susceptibility [43], a comparable figure for an intermediate phenotype might be less than 10,000. Consequently, genetic analysis of the latter may be a preferable first step (cheaper and quicker) than studying disease.
One objection to this argument is that we do not know how to predict the genetic architecture of a phenotype prior to genetic mapping. Just because a phenotype qualifies as intermediate does not guarantee that it has a more tractable genetic architecture to disease. It was not clear a priori why genetic analysis of Crohn’s disease would require a few thousand cases, whereas breast, prostate and colorectal cancers required much larger samples [44]. Heritability is not a useful guide (as the example of height indicates [45]). Supposed proximity to the underlying biology (and therefore site of genetic action), while an attractive concept, has not turned out to have much predictive value. This remains an area where we must be guided by empirical findings from GWAS.
Reduced search space
One justification for finding loci that influence an intermediate phenotype is that subsequently testing the involvement of the locus in psychiatric disease will require a smaller sample size than a genome-wide analysis, since investigators will not have to impose a significance threshold that takes into account multiple testing across the entire genome. At first sight, the saving appears impressive. Ignoring correlation between markers, the required corrected 0.05 significance threshold for testing 1 million markers expressed as a Z score, is 5.40 standard deviations. Testing a single variant requires a Z score of 1.96. The reduction in sample size is then approximately the square of 1.96/5.40, or 7.6 times smaller.
We can provide numbers for the sample size savings by using the mean OR for loci recently reported for schizophrenia. The mean OR for the 27 genome-wide significant loci is 1.10[1]. Assuming a population prevalence of schizophrenia of 0.7%, and assuming both the marker and disease-causing allele have a minor allele frequency of 0.3, then to achieve 80% power to detect a single locus at a 5% significance threshold (not genome-wide) we require 4,000 cases and 4,000 controls. This appears preferable to testing 32,000 (about eight times as many for a GWAS).
However the penalty in sample size attributable to the imposition of a genome-wide significance may be much less than the penalty required to detect an even slightly smaller effect locus, which may be the case with the locus that the intermediate phenotype analysis has proposed for testing. The effect size found in the intermediate phenotype analysis is no guide to the effect size that the same locus exerts on the psychiatric disease. Assuming a slight reduction in the OR from 1.1 to 1.08, the required sample size jumps to 24,000 (i.e., 12,000 cases, 12,000 controls) [46], almost large enough to detect genome-wide significant signals for the loci with OR of 1.1 and above. In other words, the saving in sample size is not necessarily as large as first appears to be the case.
A further sample size penalty is incurred when we realize that it is unlikely that only one locus will need to be validated: a 5% uncorrected significance threshold is inappropriate. As described above, intermediate phenotypes are polygenic, and their mapping will generate a large number of loci for testing in psychiatric disease. The genetic correlation between disease and intermediate phenotype will be much less than 100%, so that many of the loci cannot both be disease risk locus and a contributor to intermediate phenotype variation; for example, the genetic correlation between cognitive phenotypes and schizophrenia is estimated to be approximately 0.5[17]. Therefore, many loci will have to be tested, and a simple 0.05 significance threshold will be inappropriate. Allowing for ten variants tested (quite probably an underestimate), the sample sizes required for the two effect sizes described above (ORs of 1.10 and 1.08) increase to 12,000 and 37,000 respectively. Once again, the savings in sample size are again not as large as first expected, and given the expense associated with the collection of many intermediate phenotype savings in sample size do not necessarily translate into greater efficiency.
Using intermediate phenotypes to find disease mechanisms
The use of intermediate phenotypes to reveal disease mechanism depends on the assumption that a complex phenotype, such as schizophrenia, addiction or depression, can be broken down into a series of simpler component units. These intermediate phenotypes are thought to be more biologically coherent than the primary disease phenotype; that is, disease classification is derived from clinical needs, primarily reproducibility, whereas intermediate phenotypes arise from neuroscience research. In this sense, the use of intermediate phenotypes reduces to the challenge to understand the biology of brain mechanisms as it relates to disease. There are two ways in which the biological coherence of an intermediate phenotype is critical for its use.
In the first, an intermediate phenotype includes physiology relevant to understanding disease pathogenesis. Suppose that a psychiatric disease arises in part from a disordered neuronal circuit, whose anatomical location and components are unknown. If a phenotype can be found that specifically reflects activity in that circuit, the phenotype can be described as more coherent than the disease, reflecting a process that gives rise to disease. In the extreme case, the use of intermediate phenotypes could reduce pathogenesis to a molecular change, as was possible for sickle cell anemia, an inherited blood disorder. Sickle cell patients possess a mutation that replaces the hydrophilic amino acid glutamic acid with the hydrophobic amino acid valine at the sixth position of the beta chain of the haemoglobin molecule. Losing a polar amino acid enhances the molecular aggregation of haemoglobin, altering the shape of the red blood cell so that it loses plasticity, distorts, and occludes blood vessels with life-threatening consequences. Measurements of how a normal blood cell navigates the body’s circulation, which in turn depend on information about membrane mechanics and circulatory competence, are the equivalents of the intermediate phenotypes of brain structure, memory and intelligence assayed in a study of schizophrenia.
One immediate obstacle to this application of intermediate phenotypes is that we do not know which physiology is relevant to psychiatric disease. Kendler and Neale point out that it is “difficult in humans to actually discriminate between liability-index and mediational models, especially when joint models …. are plausible alternatives” [13]. If the phenotype is a consequence of disease, or a liability-index, it will not help with attempts to understand the biological basis of psychiatric illness through genetic analysis. A key step in the use of intermediate phenotypes is obtaining evidence that the phenotype lies on the causal pathway from gene to disease. As Kendler and Neale emphasize, establishing that aetiological relationship is not easy. Indeed, we could say that a major aim of intermediate phenotype research is to establish causal pathways, rather than starting from the premise that they are on the causal pathway.
Ignorance about causal relationships is not due to a lack of theories about important pathways. For example, for major depression we can identify at least seven: (i) alterations in cAMP signaling pathway [47], (ii) impaired corticosteroid receptor signaling [48], (iii) neurotrophins [49], (iv) alterations in fibroblast growth factors [50], (v) GABAergic deficits [51], (vi) epigenetic changes at glucocorticoid receptors and brain derived neurotrophic factor [52], and (vi) alterations in glutamate [53] and (vii) alterations in serotonin signaling [54]. Each theory suggests possibilities for collecting intermediate phenotypes, at multiple levels (for instance, measuring GABA and glutamate concentrations by proton magnetic resonance spectroscopy [55]). Since we know so little about the origins of psychiatric disease, we are left in the situation of collecting a large number of intermediate phenotypes in the hope that some might aid our understanding of the disease.
The second way in which biological coherence is important is when it refers to the ease with which genetic findings can be interpreted. For some complex diseases, genes identified through GWAS of the primary phenotype (i.e., disease susceptibility) have informed us about mechanism: inflammatory bowel disease [56], multiple sclerosis [57], and type 2 diabetes [58] provide examples. However, there is a legitimate concern that genetic mapping of psychiatric disease may provide little mechanistic insight because of the complexity of the phenotype [59]. Intermediate phenotypes are expected to suffer less from this problem, since they are not defined to meet the pragmatic demands of clinical utility, but rather supposedly correspond more closely to a biological substrate. An attraction of the biological coherence assumption is that intermediate phenotypes represent more biologically meaningful units than disease, and this will make genetic analysis easier.
Genetic mapping of intermediate phenotypes is not yet sufficiently advanced to know whether in general they have greater biological coherence, at a gene level, than psychiatric disease. The few results for brain morphometry hint at relevant biology — HRK acts as a regulator of apoptosis [60], FBXW8 has a known role in promotion of dendrite growth in hippocampal [61], SBNO1 is involved in Notch signaling [62] — but these results have not be been subject to the detailed pathway and functional analyses that might indicate mechanisms [63]. Currently it is unknown whether the interpretation of mapping intermediate phenotypes will prove easier than the interpretation of mapping disease phenotypes.
A substantial literature has emerged in recent years describing attempts to take a locus identified via GWAS and attempt to show association with a relevant intermediate phenotype. This, in principle, could provide insight into the mechanistic pathway between genetic variation and disease. An example is that of six markers for schizophrenia identified using GWAS and selected for further testing for association with a number of relevant cognitive measures [64]. Five out of the six variants showed no association at a 5% significance threshold, while one (rs6904071 on chromosome 6) was associated with episodic memory in the predicted direction (the disease predisposing allele reduced performance on the cognitive task).
We reviewed the literature for intermediate phenotype analyses of markers identified in GWAS studies of psychiatric disease. Two loci have received considerable interest: one marker associated with susceptibility to bipolar disorder, rs1006737 (MAF = 0.305) in the CACNA1C gene [65] and one marker associated with schizophrenia, rs1344706 (MAF = 0.345) in the ZNF804A gene [66]. We identified 35 relevant studies (Supplemental Table). The range of phenotypes tested (brain imaging, cognitive phenotypes), the differences in subjects included (healthy, bipolar disorder, schizophrenia), and the mixture of study designs (case control or quantitative variation) precluded a meta-analysis. However, we can use the information gleaned about expected effect sizes to provide a simple estimate of power. Using the minor allele frequency of 0.305 (for rs1006737) and assuming the causative variant is in high linkage disequilibrium (r2 = 1) with this marker, then for an OR of 1.5 we need a sample size of 801 to obtain 80% power at a significance threshold of 0.05 [46]. For an OR of 1.2 (a more reasonable, but still conservative, estimate of effect size) the required sample size is 2,340, while for an OR of 1.1 it is 8,414. In fact, no study is adequately powered to detect an OR of 1.2, and only five would be able to reliably detect an OR of 1.5. The median sample size of all studies is less than 200, adequately powered only to detect an OR of 2.5 or above. In short, these studies on intermediate phenotypes are underpowered, and their results should be treated with extreme caution[23].
These examples provide a salutary lesson. A researcher might perform a genetic association study of a candidate gene or a GWAS-confirmed variant and obtain a result with a P-value less than 0.05, and therefore conclude that the hypothesized association is proven. However, recall that the genetic architecture of intermediate phenotypes resembles the genetic architecture of other complex traits. If the effect size detected is inconsistent with that expected, then the result should be treated with the same scepticism due any unexpected finding. Extraordinary claims require extraordinary evidence, and not all P-values are created equal [67]. Loci with effect size equivalent to an OR greater than 2, or explaining more than 1% of phenotypic variance, are extremely rare, while those explaining more than 5% of the variance are almost unheard of.
Conclusions
We can summarize our argument with two points. The first is that there is little evidence that the observed variation in genetic architecture for intermediate phenotypes currently available is any different to that of other complex traits. The second is that, despite this, the limited understanding of the origins of psychiatric disease makes the acquisition of intermediate phenotypes that capture the mechanisms underlying disease processes essential to the interpretation genetic findings. This process is conceptually no different from the acquisition of physiological information to interpret a molecular explanation of disease origin. In particular, the use of the term endophenotype obfuscates the traditional process of understanding pathogenesis by casting undue weight on the genetic component of the measure and by assuming that the phenotype is part of the causal pathway from genetic variant to disease [13]. We may be better served by working to improve the precision of our phenotypes, and thereby reduce measurement error (see Box 2).
Box 2. Improving measurement precision to refine gene-disease associations.
Genome-wide association studies revealed an association between several loci in the nicotinic acetylcholine receptor gene cluster CHRNA5-A3-B4 and daily cigarette consumption [90, 91]. This was of particular interest because models of nicotine dependence primarily implicated the alpha-4 and beta-2 nicotinic receptor sub-units, and not the sub-units encoded by these genes. Following the identification of this association, research using knock-out mouse models indicated that the alpha-5 sub-unit does in fact influence nicotine self-administration, via tolerance to the toxic effects of high doses of nicotine [92]. Subsequent human studies have sought to further explore this association, using refined phenotypes. These have shown that a locus within this cluster, marked primarily by two SNPs (rs1051730 and rs16969968, which are in perfect linkage disequilibrium in samples of European ancestry), is also associated with levels of cotinine, the primary metabolite of nicotine [93]. Interestingly, this association remains after adjustment for self-reported smoking, which suggests that even amongst people who smoke the same number of cigarettes there is still genetically-influenced variation in nicotine consumption. This is likely to be due to differences how a cigarette is smoked (e.g., volume of smoke inhaled per puff, number of puffs taken per cigarette), and subsequent volume of smoking inhaled. Ongoing work is investigating the potential mediation of the relationship between variation at the CHRNA5-A3-B4 gene cluster and cotinine levels by volume of smoke inhaled, using precise measures of smoking behavior – cigarettes per day is in fact a relatively noisy measure of exposure, while cotinine levels is a highly precise measure. The use of phenotypes less subject to measurement error, such as measures of cotinine levels rather than cigarettes smoked per day, has therefore refined this association, and provided an indication of the mediating mechanisms.
Stating that intermediate phenotypes are in general no more genetically tractable than other complex traits does, however, ignore the fact that there is variation in genetic architecture. As we have discussed, some phenotypes are more genetically tractable than others, and even a small increase in the mean effect size will improve the chances of gene identification. It is therefore unwise to ignore the opportunities that mapping an intermediate phenotype might provide; the danger lies in assuming that the mapping these traits will be substantially easier than using the psychiatric disease phenotype. In other words, even if one is able to access a suitable intermediate measure, larger effects should not be assumed and moreover, if use of the new phenotype only increases statistical efficiency efficiency and genetic tractability at the cost of meaningfully translated outcomes, their relevance may arguably be limited.
We expect that the best use of intermediate phenotypes will not lie in aiding gene identification but in interpreting the results of GWAS of psychiatric disease, through a variety of existing and novel study designs (see Box 3). The heterogeneous origins of many psychiatric disease, and the recognition that they may have little biological coherence (at a genetic level), suggests that genetic mapping alone may not be particularly useful for revealing how disease arises [59]. Having a phenotype that reflects a real, or even hypothesized, mechanism of how a gene acts on disease will be extremely useful. To take one example, there is considerable evidence that major depression is comorbid with anxiety disorders [68–77]. If we map depression and anxiety in the same subjects, we will be able to distinguish genetic loci that contribute specifically to one or the other disorder, and also find loci that affect both [78, 79]. This information is required for understanding the pathway by which genetic risk for depression results in disease. The expanding literature on examining the association between intermediate phenotypes and markers found to be genome-wide significant for a psychiatric disease [80–86] is testament to the perceived value of intermediate phenotypes in interpreting genetic signals.
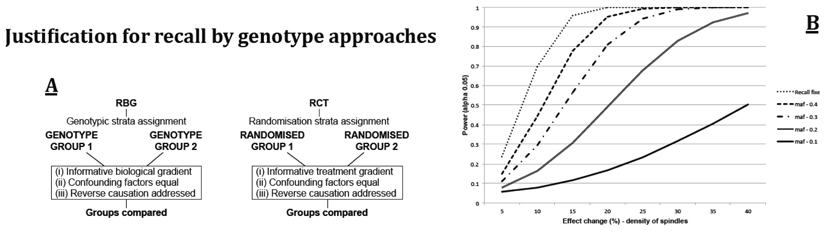
Box 3. Recall-by-genotype and intermediate phenotypes.
The exhaustive collection of intermediate phenotypes in large studies is impractical and not the most efficient approach to the allocation of finite resources. Therefore the targeted measurement of dense or precise phenotypic characteristics within selected groups is appealing. Recall-by-genotype studies provide a way of selecting subjects so as to maximize the value of collecting intermediate phenotypes. Key benefits include the ability to draw causal inferences in a manner similar to that seen in Mendelian randomisation studies [94]. Subjects in recall-by-genotype experiments are essentially “randomized” with respect to other genotypes and potential confounding factors. Analyses are less subject to bias and to the possibility that associations are due to reverse causality (Box Figure A). There are also benefits in terms of statistical efficiency. Where one wishes to measure expensive phenotypes in order to clarify the nature of a genetic association or to assess the implications of a specific risk factor, statistical power can be improved by using genotype-specific recall instead of random sampling. For example, it is far more powerful to selectively phenotype 100 rare homozygotes and 100 common homozygotes than to phenotype 200 individuals selected at random, because the latter design would select only a small number of individuals with the rare genotype. This is particularly true in the case of low frequency genetic variants (including CNVs etc.), or combinations of genetic variants that produce low frequency genotype combinations) where the increase in power is particularly dramatic (Box Figure B). The approach can also be extended to make use of polygene scores [66], where individuals at the extreme of the score distribution can be selected in a similar way. Lastly, these designs allow precise and refined phenotype measurements (otherwise unavailable in large collections) to be made within smaller targeted experiments. These are likely to further increase statistical efficiency as well as support better understanding of causal pathways.
Box Figure. Genotype by Recall.
A – Schematic representation of how recall by genotype studies mirror randomized controlled trial designs and thus allow for the development of causal inference. B - Power to detect differences across homozygotes for a genotype “x” for a model phenotypic measurement following the recall of 200 participants either at random (given a range of minor allele frequencies, 0.1-0.4) or where balanced groups each of 100 homozygotes have been selected for assessment (“recall fixed”). Total sample size = 200, α= 0.05. For the purposes of illustration, high-cost target data are polysomngraphy-recorded neuro-oscillatory brain activity characteristics taken from [95].
We do not want to imply that intermediate phenotypes are of no value; rather we believe that they should be treated like any other complex trait, posing similar difficulties for analysis and interpretation. We expect that, if treated in this way, intermediate phenotypes will play an increasingly important role in the interpretation of GWAS results for psychiatric disease (see Box 4)
Box 4. Outstanding questions.
How can we take GWAS findings into functional insights?
Causal modelling has previously attracted attention as way to tie together gene expression and phenotypes [96], but current enthusiasm currently c s
How can we combine disease outcomes and mechanistic pathways?
Over the next few years, as sample sizes increase and genotyping and sequencing costs fall, we can expect to obtain robust association results for psychiatric diseases and the biological phenotypes that are believed to be related to them, such as assays of brain function and structure obtained from imaging. How best to combine these phenotypes remains problematic. Even with loci that have been shown to be robustly associated with psychiatric disease phenotypes, pursuing these using mechanistic intermediate phenotypes will require much larger sample sizes than is typical for studies of this kind. One possible solution is the harmonisation of data collection across multiple studies, and the formation of consortia focused on these mechanistic phenotypes. Examples already exist, such as the Enhancing Neuro Imaging Genetics through Meta-Analysis (ENIGMA) consortium (http://enigma.ini.usc.edu).
What can genetic studies tell us about environmental exposures?
It is widely assumed that once we identify loci via GWAS, this will be a starting point for greater biological insight into the mechanisms underlying the phenotype. However, GWAS will also identify variants associated not directly with the phenotype, but indirectly via an environmental modifiable exposure. In other words, these studies will be informative about environmental risk factors. Behavioural traits, which represent modifiable targets for intervention (e.g., obesity, tobacco use, alcohol use) are heritable, and GWAS are beginning to identify loci associated with these. If GWAS for a disease phenotype identifies loci also associated with a behavioural trait, this can only be explained by either a direct, pleiotropic effect on both phenotypes, or an indirect effect mediated by the intermediate modifiable or environmental exposure. For example, if loci independently shown to be associated with smoking behavior also emerge in GWAS of psychiatric disease, one possible interpretation is that this reflects a causal relationship between smoking and the disease.
How can we use genetic information to identify causal pathways?
Mendelian randomization (MR) uses genetic variants as instruments for environmental exposures. These can take the form of individual single nucleotide polymorphisms (SNPs), or polygenic risk scores, which must be robustly associated with the exposure of interest (e.g., smoking heaviness or alcohol use). The principle of MR relies on the basic (but approximate) laws of Mendelian genetics (segregation and independent assortment). If these hold then, at a population level, genetic variants will not be associated with potential confounders. The SNP or risk score is therefore a proxy for the exposure, and can be used to infer a causal relationship between the exposure and an outcome. Recall-by-genotype studies (see Box 3) retain the key characteristics of MR and can be used to explore causal pathways, focused on intermediate mechanistic phenotypes.
Supplementary Material
Acknowledgements
JF is supported by the Wellcome Trust. MRM is a member of the United Kingdom Centre for Tobacco and Alcohol Studies, a UKCRC Public Health Research: Centre of Excellence. Funding from the Wellcome Trust, British Heart Foundation, Cancer Research UK, Economic and Social Research Council, Medical Research Council, and the National Institute for Health Research, under the auspices of the UK Clinical Research Collaboration, is gratefully acknowledged. NJT and MRM work within a Unit jointly funded by the MRC (grant numbers MC_UU_12013/3 and MC_UU_12013/6) and University of Bristol.
References
- 1.Ripke S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013 doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sklar P, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ripke S, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 5.Xu B, et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 6.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malhotra D, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72:951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vissers LE, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 9.Xu B, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girard SL, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 11.O'Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walters JT, Owen MJ. Endophenotypes in psychiatric genetics. Mol Psychiatry. 2007;12:886–890. doi: 10.1038/sj.mp.4002068. [DOI] [PubMed] [Google Scholar]
- 13.Kendler KS, Neale MC. Endophenotype: a conceptual analysis. Mol Psychiatry. 2010;15:789–797. doi: 10.1038/mp.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surguladze SA, et al. Recognition accuracy and response bias to happy and sad facial expressions in patients with major depression. Neuropsychology. 2004;18:212–218. doi: 10.1037/0894-4105.18.2.212. [DOI] [PubMed] [Google Scholar]
- 15.Brewer WJ, et al. Memory impairments identified in people at ultra-high risk for psychosis who later develop first-episode psychosis. Am J Psychiatry. 2005;162:71–78. doi: 10.1176/appi.ajp.162.1.71. [DOI] [PubMed] [Google Scholar]
- 16.Fuller R, et al. Longitudinal assessment of premorbid cognitive functioning in patients with schizophrenia through examination of standardized scholastic test performance. Am J Psychiatry. 2002;159:1183–1189. doi: 10.1176/appi.ajp.159.7.1183. [DOI] [PubMed] [Google Scholar]
- 17.Toulopoulou T, et al. Impaired intellect and memory: a missing link between genetic risk and schizophrenia? Arch Gen Psychiatry. 2010;67:905–913. doi: 10.1001/archgenpsychiatry.2010.99. [DOI] [PubMed] [Google Scholar]
- 18.Owens SF, et al. Genetic overlap between episodic memory deficits and schizophrenia: results from the Maudsley Twin Study. Psychol Med. 2011;41:521–532. doi: 10.1017/S0033291710000942. [DOI] [PubMed] [Google Scholar]
- 19.Toulopoulou T, et al. Substantial genetic overlap between neurocognition and schizophrenia: genetic modeling in twin samples. Arch Gen Psychiatry. 2007;64:1348–1355. doi: 10.1001/archpsyc.64.12.1348. [DOI] [PubMed] [Google Scholar]
- 20.Broer L, et al. Distinguishing true from false positives in genomic studies: p values. European journal of epidemiology. 2013;28:131–138. doi: 10.1007/s10654-012-9755-x. [DOI] [PubMed] [Google Scholar]
- 21.Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychol Med. 2007;37:163–180. doi: 10.1017/S0033291706008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Button KS, et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci. 2013;14:365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- 24.Hulshoff Pol HE, et al. Overlapping and segregating structural brain abnormalities in twins with schizophrenia or bipolar disorder. Arch Gen Psychiatry. 2012;69:349–359. doi: 10.1001/archgenpsychiatry.2011.1615. [DOI] [PubMed] [Google Scholar]
- 25.Harms MP, et al. Structural abnormalities in gyri of the prefrontal cortex in individuals with schizophrenia and their unaffected siblings. Br J Psychiatry. 2010;196:150–157. doi: 10.1192/bjp.bp.109.067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brans RG, et al. Heritability of changes in brain volume over time in twin pairs discordant for schizophrenia. Arch Gen Psychiatry. 2008;65:1259–1268. doi: 10.1001/archpsyc.65.11.1259. [DOI] [PubMed] [Google Scholar]
- 27.Stein JL, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taal HR, et al. Common variants at 12q15 and 12q24 are associated with infant head circumference. Nat Genet. 2012;44:532–538. doi: 10.1038/ng.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikram MA, et al. Common variants at 6q22 and 17q21 are associated with intracranial volume. Nat Genet. 2012;44:539–544. doi: 10.1038/ng.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bis JC, et al. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet. 2012;44:545–551. doi: 10.1038/ng.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Need AC, et al. A genome-wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Hum Mol Genet. 2009;18:4650–4661. doi: 10.1093/hmg/ddp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calkins ME, et al. The Consortium on the Genetics of Endophenotypes in Schizophrenia: model recruitment, assessment, and endophenotyping methods for a multisite collaboration. Schizophr Bull. 2007;33:33–48. doi: 10.1093/schbul/sbl044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenwood TA, et al. Initial heritability analyses of endophenotypic measures for schizophrenia: the consortium on the genetics of schizophrenia. Arch Gen Psychiatry. 2007;64:1242–1250. doi: 10.1001/archpsyc.64.11.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenwood TA, et al. Genome-wide linkage analyses of 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry. 2013;170:521–532. doi: 10.1176/appi.ajp.2012.12020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mowry BJ, et al. The molecular genetics of schizophrenia: an update. Aust N Z J Psychiatry. 1997;31:704–713. doi: 10.3109/00048679709062684. [DOI] [PubMed] [Google Scholar]
- 36.Lewis CM, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dehghan A, et al. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–738. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kottgen A, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45:145–154. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Harst P, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wray NR, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry. 2012;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park JH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purcell S, et al. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 47.Duman RS, et al. A molecular and cellular theory of depression. Arch Gen Psychiatry. 1997;54:597–606. doi: 10.1001/archpsyc.1997.01830190015002. [DOI] [PubMed] [Google Scholar]
- 48.Holsboer F. The corticosteroid receptor hypothesis of depression. Neuropsychopharmacology. 2000;23:477–501. doi: 10.1016/S0893-133X(00)00159-7. [DOI] [PubMed] [Google Scholar]
- 49.Samuels BA, Hen R. Neurogenesis and affective disorders. Eur J Neurosci. 2011;33:1152–1159. doi: 10.1111/j.1460-9568.2011.07614.x. [DOI] [PubMed] [Google Scholar]
- 50.Turner CA, et al. The fibroblast growth factor family: neuromodulation of affective behavior. Neuron. 2012;76:160–174. doi: 10.1016/j.neuron.2012.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luscher B, et al. The GABAergic deficit hypothesis of major depressive disorder. Mol Psychiatry. 2011;16:383–406. doi: 10.1038/mp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paul IA, Skolnick P. Glutamate and depression: clinical and preclinical studies. Ann N Y Acad Sci. 2003;1003:250–272. doi: 10.1196/annals.1300.016. [DOI] [PubMed] [Google Scholar]
- 54.Mann JJ. Role of the serotonergic system in the pathogenesis of major depression and suicidal behavior. Neuropsychopharmacology. 1999;21:99S–105S. doi: 10.1016/S0893-133X(99)00040-8. [DOI] [PubMed] [Google Scholar]
- 55.Sanacora G, et al. Subtype-specific alterations of gamma-aminobutyric acid and glutamate in patients with major depression. Arch Gen Psychiatry. 2004;61:705–713. doi: 10.1001/archpsyc.61.7.705. [DOI] [PubMed] [Google Scholar]
- 56.Lees CW, et al. New IBD genetics: common pathways with other diseases. Gut. 2011;60:1739–1753. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- 57.Beecham AH, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med. 2010;363:2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 59.Kendler KS. What psychiatric genetics has taught us about the nature of psychiatric illness and what is left to learn. Mol Psychiatry. 2013;18:1058–1066. doi: 10.1038/mp.2013.50. [DOI] [PubMed] [Google Scholar]
- 60.Sborgi L, et al. Characterization of a novel interaction between Bcl-2 members Diva and Harakiri. PLoS One. 2010;5:e15575. doi: 10.1371/journal.pone.0015575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Litterman N, et al. An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning. PLoS Biol. 2011;9:e1001060. doi: 10.1371/journal.pbio.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coyle-Thompson CA, Banerjee U. The strawberry notch gene functions with Notch in common developmental pathways. Development. 1993;119:377–395. doi: 10.1242/dev.119.2.377. [DOI] [PubMed] [Google Scholar]
- 63.Wang K, et al. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 64.Walters JT, et al. The role of the major histocompatibility complex region in cognition and brain structure: a schizophrenia GWAS follow-up. Am J Psychiatry. 2013;170:877–885. doi: 10.1176/appi.ajp.2013.12020226. [DOI] [PubMed] [Google Scholar]
- 65.Ferreira MA, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008 doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Browner WS, Newman TB. Are all significant P values created equal? The analogy between diagnostic tests and clinical research. Jama. 1987;257:2459–2463. [PubMed] [Google Scholar]
- 68.Zimmerman M, et al. Diagnostic co-morbidity in 2300 psychiatric out-patients presenting for treatment evaluated with a semi-structured diagnostic interview. Psychol Med. 2008;38:199–210. doi: 10.1017/S0033291707001717. [DOI] [PubMed] [Google Scholar]
- 69.Angst J. Comorbidity of anxiety, phobia, compulsion and depression. Int Clin Psychopharmacol. 1993;8(Suppl 1):21–25. doi: 10.1097/00004850-199309001-00003. [DOI] [PubMed] [Google Scholar]
- 70.Blazer DG, et al. The prevalence and distribution of major depression in a national community sample: the National Comorbidity Survey. Am J Psychiatry. 1994;151:979–986. doi: 10.1176/ajp.151.7.979. [DOI] [PubMed] [Google Scholar]
- 71.Kessler RC, et al. Comorbidity of DSM-III-R major depressive disorder in the general population: results from the US National Comorbidity Survey. British Journal of Psychiatry. 1996;168:17–30. [PubMed] [Google Scholar]
- 72.Merikangas KR, et al. Comorbidity and boundaries of affective disorders with anxiety disorders and substance misuse: results of an international task force. Br J Psychiatry Suppl. 1996:58–67. [PubMed] [Google Scholar]
- 73.Pini S, et al. Prevalence of anxiety disorders comorbidity in bipolar depression, unipolar depression and dysthymia. J Affect Disord. 1997;42:145–153. doi: 10.1016/s0165-0327(96)01405-x. [DOI] [PubMed] [Google Scholar]
- 74.Mineka S, et al. Comorbidity of anxiety and unipolar mood disorders. Annual Review of Psychology. 1998;49:377–412. doi: 10.1146/annurev.psych.49.1.377. [DOI] [PubMed] [Google Scholar]
- 75.Kessler RC, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) Jama. 2003;289:3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 76.Alonso J, et al. 12-Month comorbidity patterns and associated factors in Europe: results from the European Study of the Epidemiology of Mental Disorders (ESEMeD) project. Acta Psychiatr Scand Suppl. 2004:28–37. doi: 10.1111/j.1600-0047.2004.00328.x. [DOI] [PubMed] [Google Scholar]
- 77.Hunt C, et al. Generalized Anxiety Disorder and major depressive disorder comorbidity in the National Survey of Mental Health and Well-Being. Depress Anxiety. 2004;20:23–31. doi: 10.1002/da.20019. [DOI] [PubMed] [Google Scholar]
- 78.Consortium C-DGotPG. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee SH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013 doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krug A, et al. Effect of CACNA1C rs1006737 on neural correlates of verbal fluency in healthy individuals. Neuroimage. 2010;49:1831–1836. doi: 10.1016/j.neuroimage.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 81.Krug A, et al. A genome-wide supported variant in CACNA1C influences hippocampal activation during episodic memory encoding and retrieval. Eur Arch Psychiatry Clin Neurosci. 2013 doi: 10.1007/s00406-013-0428-x. [DOI] [PubMed] [Google Scholar]
- 82.Thimm M, et al. Effects of a CACNA1C genotype on attention networks in healthy individuals. Psychol Med. 2011;41:1551–1561. doi: 10.1017/S0033291710002217. [DOI] [PubMed] [Google Scholar]
- 83.Wessa M, et al. The CACNA1C risk variant for bipolar disorder influences limbic activity. Mol Psychiatry. 2010;15:1126–1127. doi: 10.1038/mp.2009.103. [DOI] [PubMed] [Google Scholar]
- 84.Jogia J, et al. The impact of the CACNA1C gene polymorphism on frontolimbic function in bipolar disorder. Mol Psychiatry. 2011;16:1070–1071. doi: 10.1038/mp.2011.49. [DOI] [PubMed] [Google Scholar]
- 85.Bigos KL, et al. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch Gen Psychiatry. 2010;67:939–945. doi: 10.1001/archgenpsychiatry.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Erk S, et al. Brain function in carriers of a genome-wide supported bipolar disorder variant. Arch Gen Psychiatry. 2010;67:803–811. doi: 10.1001/archgenpsychiatry.2010.94. [DOI] [PubMed] [Google Scholar]
- 87.Cannon TD, Keller MC. Endophenotypes in the genetic analyses of mental disorders. Annu Rev Clin Psychol. 2006;2:267–290. doi: 10.1146/annurev.clinpsy.2.022305.095232. [DOI] [PubMed] [Google Scholar]
- 88.Vrieze SI, et al. Confluence of genes, environment, development, and behavior in a post Genome-Wide Association Study world. Dev Psychopathol. 2012;24:1195–1214. doi: 10.1017/S0954579412000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 90.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ware JJ, et al. Association of the CHRNA5-A3-B4 gene cluster with heaviness of smoking: a meta-analysis. Nicotine Tob Res. 2011;13:1167–1175. doi: 10.1093/ntr/ntr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fowler CD, et al. Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature. 2011;471:597–601. doi: 10.1038/nature09797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Munafo MR, et al. Association between genetic variants on chromosome 15q25 locus and objective measures of tobacco exposure. J Natl Cancer Inst. 2012;104:740–748. doi: 10.1093/jnci/djs191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 95.Wamsley EJ, et al. Reduced sleep spindles and spindle coherence in schizophrenia: mechanisms of impaired memory consolidation? Biol Psychiatry. 2012;71:154–161. doi: 10.1016/j.biopsych.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spencer CC, et al. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5:e1000477. doi: 10.1371/journal.pgen.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.