Abstract
The increasing amount of structural information on protein–protein interactions makes it possible to predict the structure of protein–protein complexes by comparison/alignment of the interacting proteins to the ones in cocrystallized complexes. In the predictions based on structure similarity, the template search is performed by structural alignment of the target interactors with the entire structures or with the interface only of the subunits in cocrystallized complexes. This study investigates the scope of the structural similarity that facilitates the detection of a broad range of templates significantly divergent from the targets. The analysis of the target-template similarity is based on models of protein–protein complexes in a large representative set of heterodimers. The similarity of the biological and crystal packing interfaces, dissimilar interface structural motifs in overall similar structures, interface similarity to the full structure, and local similarity away from the interface were analyzed. The structural similarity at the protein–protein interfaces only was observed in ~25% of target-template pairs with sequence identity <20% and primarily homodimeric templates. For ~50% of the target-template pairs, the similarity at the interface was accompanied by the similarity of the whole structure. However, the structural similarity at the interfaces was still stronger than that of the noninterface parts. The study provides insights into structural and functional diversity of protein–protein complexes, and relative performance of the interface and full structure alignment in docking.
Keywords: protein recognition, protein modeling, bioinformatics, structure prediction, structural templates
INTRODUCTION
Structural information on protein–protein interactions is important for characterization of biomolecular processes. Experimental approaches to structure determination of protein–protein complexes can provide the structures of only a fraction of known protein–protein interactions.1 Thus, computational structural modeling of protein complexes by protein docking procedures1–5 is a necessary complement to the experimental techniques. Modeling is also important for understanding the mechanisms of protein interactions.
Since the 80s, predictive modeling of individual protein structures has been increasingly dominated by template-based approaches. Such approaches use experimentally determined protein structures (templates) as the basis for modeling of proteins with unknown structure (targets). Extensive evaluation, including community-wide blind assessments,6 showed that the template-based predictions, in general, are significantly more accurate and reliable than the ab initio, “first principles,” models (although the latter obviously have a natural advantage when the goal is to understand the mechanisms of protein folding).
A similar paradigm shift, from the ab initio to the template-based predictions, in modeling of protein complexes has started only recently,5,7 largely due to relatively successful application of the ab initio docking, and the perceived paucity of protein–protein templates (generally, protein complexes are more difficult to crystallize than individual proteins). Despite the progress in the ab initio docking,2 modeling based on existing structures may potentially provide greater accuracy and reliability. As far as the availability of such structures is concerned, a recent study showed that current PDB already contains suitable templates for almost all known protein–protein interactions, provided the structures of the interacting proteins are determined experimentally or modeled by homology.8 Still, many such templates may be incorrect, and numerous protein encounters in a crowded cell environment may not be stable enough for crystallization (and detection by other experimental techniques). Thus, in the future, the structural modeling of protein interactions is likely to combine the ab initio and the template-based approaches.7
The template-based modeling of protein complexes involves target/template relationships based on sequence/structure similarity,9–19 with the structure similarity techniques showing a great promise in terms of the availability of templates.8 In the methods based on the structure similarity, the template search is performed by the structural alignment of the target interactors with the entire structures (full structure alignment, FSA), or the interface only (partial structure alignment, PSA) of the subunits in co-crystallized complexes. There are also multiple types of target/template relationships based on the nature of the structures involved—heterodimers versus homodimers, single-versus multi-domain structures, biological versus crystal packing complexes, and such.
In this article we analyze the spectrum of structural similarity in protein–protein complexes, as it relates to detection of suitable templates for protein docking, and the corresponding performance of the full and partial structural alignment approaches.
METHODS
We used a manually curated set of 372 two-chain bound structures (12 antibody–antigen, 66 enzyme-inhibitor, and 294 other complexes), nonredundant at 30% sequence identity, from the Dockground resource20 (http://dockground.bioinformatics.ku.edu) as our target set (Table SI in Supporting Information). The template pool consisted of 11,932 two-chain PDB complexes with sequence identity <90% between complexes, or interface fragments extracted from the full structures using 12 Å distance cut-off to define the interface (a detailed description is in our earlier publications19,21).
Complexes were modeled by spatial rearrangement of separate 3D structures of the target monomers to structurally overlap with the cocrystallized interface fragments (PSA) or full structures (FSA) of the templates. The Cα structural alignment of the monomers was performed by TM-align.22 The alignment quality was assessed by TM-score,23 which has values ranging from 0 to 1 (the average TM-score of a random structure match is 0.17). The alignment pairs had to satisfy the following criteria: (i) TM-score of at least one alignment >0.4, (ii) at least 50% of the aligned residues for both receptor and ligand (“receptor” and “ligand” are the larger and the smaller proteins in the complex, respectively) should be at the surface, and (iii) at least 40% of the interface residues in both receptor and ligand should be covered by the alignment. Transformation matrices from the alignments were applied to the target receptor and ligand to generate models. Redundant models (TM-scores of both alignments >0.9) were excluded from further consideration. The quality of the resulting models was evaluated in terms of RMSD between interface Cα atoms of the predicted and the native ligand structures, with the receptors optimally aligned (i-RMSD). The distance cutoff for the interface residues in i-RMSD calculations was 6 Å. The models were designated as higher-accuracy (i-RMSD <5 Å ) and lower-accuracy (5 Å ≤ i-RMSD <10 Å ) ones, consistent with the previous estimates for meaningful docking predictions24 and the docking funnel size.25 Sequence identities between targets and templates were calculated from alignments produced by CLUSTALW.26,27 Because models of the complexes are generated by the alignment of Cα structures, they may have a number of clashes between the side chains. The surface and the coverage requirements for the models (see above) improve the structural quality of the interface. However, significant number of clashes still remains and could be removed by minimization.
RESULTS AND DISCUSSION
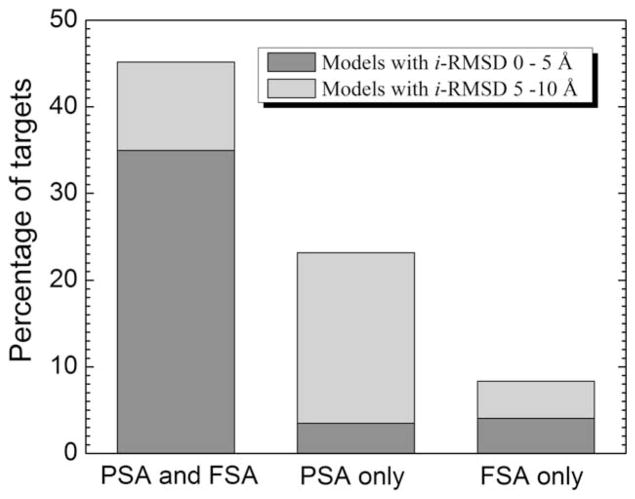
Benchmarking results of the full structure alignment (FSA) and interface/partial structure alignment (PSA) are summarized in Figure 1. Some target complexes were predicted with either higher or lower accuracy by both full and interface alignments, and some by either interface alignment or full alignment only. Accordingly, our analysis is split into these three categories. The analysis shows the structural factors responsible for either comparable or significantly better performance of each alignment protocol.
Figure 1.
Performance of full and partial structural alignments in higher and lower accuracy predictions.
Comparable performance of full and interface structure alignments
Both FSA and PSA yielded the best (smallest i-RMSD from the native structure) models with i-RMSD <5 Å—higher-accuracy best (HAB) models—for 130 targets (35% of the dataset) and 103 of those models were ranked 1. For 92 targets, the PSA HAB models have smaller i-RMSD compared to the corresponding FSA HAB models with difference in i-RMSD (Δ i-RMSD) >1 Å for 17 such models. At the same time, only in 19 cases FSA HAB model has a smaller i-RMSD compared to the corresponding PSA HAB model (in four cases Δ i-RMSD >1 Å ). PSA and FSA templates for the HAB models originated from the same X-ray structure for 125 targets.
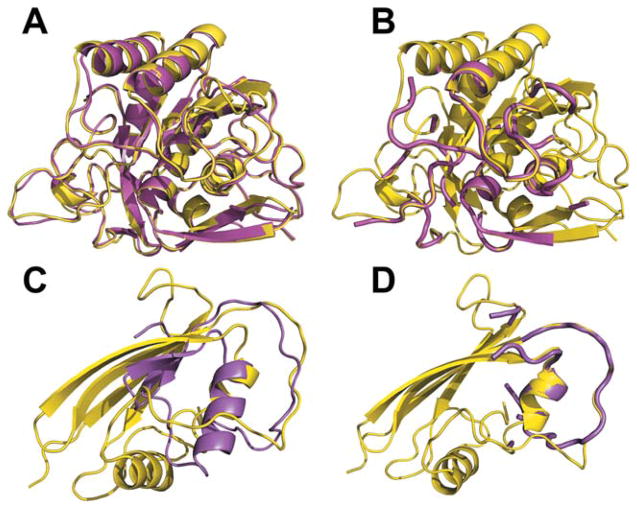
A typical reason for better PSA performance for HAB models is illustrated in Figure 2, where PSA and FSA alignments between monomers of the target (subtilisin BPN from Bacillus amyloliquefaciens, inhibited by a synthetic protein, chains L and R from 3sic) and the template (subtilisin Carlsberg from Bacillus licheniformis, inhibited by ovomucoid protein from Meleagris gallopavo, chains R and L from 1r0r) are shown. Both subtilisins have similar structures with sequence identity 70% and their FSA and PSA alignments are also similar [Fig. 2(A,B)]. The inhibitors, however, are quite dissimilar (sequence identity 12%), with similarity only in the binding loops. Thus, PSA correctly aligns the interface parts of the target and the template [Fig. 2(D)] yielding an HAB model with only 0.9 Å i-RMSD. FSA seeks to find the minimal distance between all Cα atoms of the target and the template, and the alignment of the interface loops becomes less accurate [Fig. 2(C)], with 4.9 Å i-RMSD for the resulting HAB model.
Figure 2.
Structural similarity in hetero-complexes. (A) FSA and (B) PSA alignments of the receptors (chains E of the target 3sic and template 1r0r). (C) FSA and (D) PSA alignments of the ligands (chains I).
Another reason for better HAB models by PSA is the domain structure of the target and the template monomers, as illustrated in Figure S1 (see Supporting Information). Ligands of the target (human signaling complex, chains B and A from 1ki1) and the template (another human signaling complex, chains A and B from 2nz8) have very similar structures (78% sequence identity). Receptors of the target and the template have two-domain structures, with only one of the domains participating in the binding. The structures of separate domains are very similar (albeit with 18% sequence identity), but their mutual orientation in the target and the template is different. Thus, FSA yielded an HAB model with 5.0 Å i-RMSD. PSA correctly aligned the interface parts of the target and the template [Fig. S1(B)] producing a HAB model with 0.6 Å i-RMSD. However, such extreme cases were not very common in our dataset, observed only in five targets with HAB models.
The results for lower-accuracy best (LAB) models (5 Å ≤ i-RMSD <10 Å ) do not show, however, the same advantage for the PSA algorithm. Out of 38 targets, for which both protocols yielded a LAB model, only 11 targets have PSA model with smaller i-RMSD (5 cases with Δ i-RMSD >1 Å ). For 11 targets, PSA LAB ranking was higher than FSA LAB ranking. Only for two targets, LAB models utilized the same template for both PSA and FSA.
Significantly better performance of interface structure alignment
A significant part of targets have the best model built by only one of the protocols (Fig. 1). The models were built solely by the PSA protocol for 86 targets—13 HAB and 73 LAB models, compared to 15 HAB and 16 LAB models built by FSA only. This indicates frequent occurrence of similar structural motifs at interfaces of protein–protein complexes that otherwise have dissimilar monomer structures. Significant sequence identity (>20%) between the target and the best template was found in just 11 such cases.
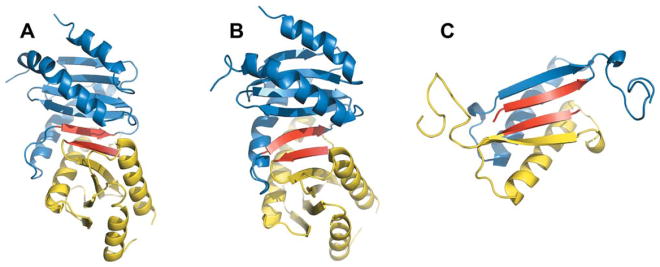
For all target/best template pairs with low sequence identity, a similar portion of the interface is quite small. An example is shown in Figure 3 for the PSA model of mice protein signaling complex (1vet) built using an interface fragment between two chains (out of 9 identical chains in the asymmetric unit) of RUVA protein from E. coli (4otc). These fragments consist of 45 and 53 residues in the template monomers, but the common structural motif (two β-strands shown in red in Fig. 3) consists of only 6 residues. The shape of these β-strands differ slightly in the target and the template X-ray structures, thus the PSA model has 6.0 Å i-RMSD due to the imprecise tilt of the ligand. The overall structures of the target and the template are so different that FSA did not find any target-template alignments with TM-score >0.4.
Figure 3.
Interface structural similarity of hetero- and homo-dimers. (A) PSA model and (B) X-ray structure of the target complex (chains A and B of 1vet). (C) X-ray structure of the template complex (chains B and C of 4otc). Similar parts of the interfaces are in red for both receptors and ligands.
Hetero-complexes with local interface similarity and dissimilar global folds of interactors were previously studied in detail by Keskin and Nussinov.28,29 Our results indicate that in many cases (58 targets with LAB PSA-only models) interface is also similar between het-ero- and homo-dimeric complexes (e.g., the complex shown in Fig. 3). These targets and templates are primarily from different organisms (only 7 homo-dimeric templates were from the same species as the hetero-dimeric targets for at least one of the monomers). In eight cases, the interfaces of the homo-dimeric templates exist only in biological units. Figure S2 (Supporting Information) shows the complex of colicin E3 with its immunity protein, where PSA yielded the LAB model (7.3 Å i-RMSD) based on the X-ray structure of colicin E3 homodimer. The biological function of colicin is to kill excess E. coli cells by binding and cleaving the enemy cell DNA. To prevent the host cell suicide, the colicins form complexes with their immunity proteins inhibiting DNA binding site.30 In either case, colicins do not exist in vivo as homodimers. Thus, the case is an example of a biological complex modeled on a crystal packing interface. Because of structural dissimilarity of the colicin E3 and its immunity protein (19% sequence identity and TM-score <0.2) FSA failed to produce models for this template (FSA also did not find any other template with TM-score >0.4). Because of the absence of clear and unambiguous criteria for distinguishing biological and crystallographic interfaces, we were not able to provide the exact number of such cases.
Significantly better performance of full structure alignment
Best templates for 31 targets (8.3% of the dataset) have structurally dissimilar interfaces, but the non-interface structure is similar enough to produce near-native FSA models (FSA only in Fig. 1). Our analysis revealed three categories of such complexes.
In the first group (seven cases), the interface loop(s) connecting the otherwise similar interface β-strands in the target and the template have different lengths. Figure S3 (Supporting Information) shows an example of target 1itb (ligand complex with human interleukin-1beta) and template 1cvs (ligand complex with human fibroblast growth factor 2). Ligands of both the target and the template belong to the cytokine superfamily in SCOP classification and their overall structures are quite similar, albeit with only 15% sequence identity. FSA protocol correctly superimposes ligand structures [Fig. S3(D)] yielding an HAB model with 4.8 Å i-RMSD. The main structural difference between the ligands is the length of the interface loop connecting two β-strands that are partially at the interface. This loop is longer, while the interface part of the β-strand is shorter in the target structure. Thus PSA aligns wrong loop and strands parts [Fig. S3(C)], generating a significantly less accurate LAB model (7.3 Å i-RMSD).
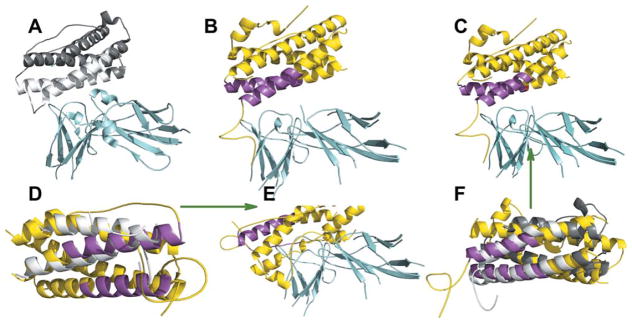
The second group (eight targets) consists of the complexes with a four-helix bundle motif, where only parts of the helices participate in binding. Figure 4 illustrates such a case for target 1f6f (ligand complex with Ovis aries placental lactogen) and template 1pvh (ligand complex with human leukemia inhibitor factor). Both monomers with α-helical structures belong to the same long-chain cyto-kines SCOP family with the sequence identity between them only 7%. The overall structure of these monomers is very similar, resulting in an HAB FSA model (4.5 Å i-RMSD). PSA, however, aligns the interface parts of the template helices to non-interface parts of the target helices producing an incorrect model (15.0 Å i-RMSD).
Figure 4.
Indistinguishable interface and global structures. (A) X-ray structures of template (chains A and B from 1pvh) and (B) target (chains A and C from 1f6f). (C) FSA and (E) PSA models of the target, and (F) FSA and (D) PSA alignments, for the target and template ligands. The receptors in are in cyan, ligands in the target are in gray and those in the template are in yellow. The interface of ligand helixes is in magenta for the target and white for the template.
In the third group (16 targets), there is a local structural similarity between the target and the template away from the interface. The sequence identities between the target and the template monomers in all such cases were <10%. An example is shown in Figure S4 (Supporting Information) for the target 1v74 (colicin D with its immunity protein) and template 2vhz (colicin E5 with its immunity protein). Despite the biological function similarity of the target and the template, their overall structures, including interfaces, are quite dissimilar. However, the same mutual orientations of noninterface helices and part of a β-strand (arrows in Supporting Information Fig. S4) in the target and the template yielded a 5.8 Å i-RMSD FSA model.
CONCLUDING REMARKS
Performance of full and interface structural alignment approaches for modeling protein–protein complexes was analyzed in terms of structural similarity between targets and templates. Templates for higher-accuracy models often shared global structural similarity with the targets. Both methods perform similarly well in placing the best higher-accuracy models to the top of the predictions pool. However, most of the PSA models are more accurate than the corresponding FSA models—a structural match at the interface has a greater influence on i-RMSD than a match away from the interface.
The sequence identity between at least one target-template pair was <20% for most complexes in the dataset, indicating that the conservation of protein-protein interfaces, detectable by structural alignment, extends beyond simple amino acid similarity,28,29 with significant level of conservation observed among remote structural neighbors.31 Similar structures of one pair of the target and the template monomers and dissimilar structures of the other pair is a common feature in all higher-accuracy PSA models. Thus, if it is known that a protein binds different proteins at a single binding site (e.g., enzyme–inhibitor complexes), the partial structural alignment is a better choice. However, dissimilar interface loops or predominantly helical structure of the target and detected templates make full structural alignment a better modeling alternative. Full alignment also works better if structural similarity is observed for noninter-face regions of the target and the template.
The templates for lower-accuracy models typically share only local structural similarity with the targets. Thus, for remote structure homologs, PSA is the only approach. Similar fragment may consist of only a few interface residues, and cannot be detected by the full structure comparison. Similar protein–protein interfaces often exist in structurally and functionally diverse proteins from distant organisms. Interface structural similarity was observed between hetero- and homo-complexes, including crystal packing complexes. This suggests that conservation of protein-protein interfaces to a significant extent is determined by their geometry rather than simple amino acid conservation. Partial and full structural alignment methods are complementary to each other and their combined use significantly improves the pool of putative templates for protein-protein docking.
Supplementary Material
Acknowledgments
Grant sponsor: NIH; Grant number: R01 GM074255.
Rohita Sinha contributed to the docking by structure alignment approach at earlier stages of the development.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Wass MN, David A, Sternberg MJE. Challenges for the prediction of macromolecular interactions. Curr Opin Struct Biol. 2011;21:382–390. doi: 10.1016/j.sbi.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Lensink MF, Wodak SJ. Docking and scoring protein interactions: CAPRI 2009. Proteins. 2010;78:3073–3084. doi: 10.1002/prot.22818. [DOI] [PubMed] [Google Scholar]
- 3.Vajda S, Camacho CJ. Protein–protein docking: is the glass half-full or half-empty? Trends Biotechnol. 2004;22:110–116. doi: 10.1016/j.tibtech.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Vakser IA, Kundrotas P. Predicting 3D structures of protein–protein complexes. Curr Pharm Biotech. 2008;9:57–66. doi: 10.2174/138920108783955209. [DOI] [PubMed] [Google Scholar]
- 5.Zacharias M. Accounting for conformational changes during protein–protein docking. Curr Opin Struct Biol. 2010;20:180–186. doi: 10.1016/j.sbi.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Moult J, Fidelis K, Kryshtafovych A, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)—round IX. Proteins. 2011;79(Suppl 10):1–5. doi: 10.1002/prot.23200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vakser IA. Low-resolution structural modeling of protein interac-tome. Curr Opin Struct Biol. 2013;23:198–205. doi: 10.1016/j.sbi.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kundrotas PJ, Zhu Z, Janin J, Vakser IA. Templates are available to model nearly all complexes of structurally characterized proteins. Proc Natl Acad Sci USA. 2012;109:9438–9441. doi: 10.1073/pnas.1200678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell RB, Alber F, Aloy P, Davis FP, Korkin D, Pichaud M, Topf M, Sali A. A structural perspective on protein–protein interactions. Curr Opin Struct Biol. 2004;14:313–324. doi: 10.1016/j.sbi.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Lu L, Lu H, Skolnick J. MULTIPROSPECTOR: an algorithm for the prediction of protein–protein interactions by multimeric threading. Proteins. 2002;49:350–364. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- 11.Korkin D, Davis FP, Alber F, Luong T, Shen M, Lucic V, Kennedy MB, Sali A. Structural modeling of protein interactions by analogy: application to PSD-95. PLoS Comp Biol. 2006;2:1365–1376. doi: 10.1371/journal.pcbi.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukherjee S, Zhang Y. Protein–protein complex structure predictions by multimeric threading and template recombination. Structure. 2011;13:955–966. doi: 10.1016/j.str.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan RA, EL-Manzalawy Y, Dobbs D, Honavar V. Predicting protein–protein interface residues using local surface structural similarity. BMC Bioinform. 2012;13:41. doi: 10.1186/1471-2105-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konc J, Janezic D. ProBiS algorithm for detection of structurally similar protein binding sites by local structural alignment. Bioinfor-matics. 2010;26:1160–1168. doi: 10.1093/bioinformatics/btq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunther S, May P, Hoppe A, Frommel C, Preissner R. Docking without docking: ISEARCH—prediction of interactions using known interfaces. Proteins. 2007;69:839–844. doi: 10.1002/prot.21746. [DOI] [PubMed] [Google Scholar]
- 16.Keskin O, Nussinov R, Gursoy A. PRISM: protein–protein interaction prediction by structural matching. Methods Mol Biol. 2008;484:505–521. doi: 10.1007/978-1-59745-398-1_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, Maniatis T, Califano A, Honig B. Structure-based prediction of protein–protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kundrotas PJ, Lensink MF, Alexov E. Homology-based modeling of 3D structures of protein–protein complexes using alignments of modified sequence profiles. Int J Biol Macromol. 2008;43:198–208. doi: 10.1016/j.ijbiomac.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Sinha R, Kundrotas PJ, Vakser IA. Docking by structural similarity at protein–protein interfaces. Proteins. 2010;78:3235–3241. doi: 10.1002/prot.22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douguet D, Chen HC, Tovchigrechko A, Vakser IA. DOCK-GROUND resource for studying protein–protein interfaces. Bioin-formatics. 2006;22:2612–2618. doi: 10.1093/bioinformatics/btl447. [DOI] [PubMed] [Google Scholar]
- 21.Sinha R, Kundrotas PJ, Vakser IA. Protein docking by the interface structure similarity: how much structure is needed? PloS One. 2012;7:e31349. doi: 10.1371/journal.pone.0031349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucl Acid Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 24.Tovchigrechko A, Wells CA, Vakser IA. Docking of protein models. Protein Sci. 2002;11:1888–1896. doi: 10.1110/ps.4730102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunjan J, Tovchigrechko A, Gao Y, Vakser IA. The size of the intermolecular energy funnel in protein–protein interactions. Proteins. 2008;72:344–352. doi: 10.1002/prot.21930. [DOI] [PubMed] [Google Scholar]
- 26.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2. 0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acid Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keskin O, Nussinov R. Favorable scaffolds: proteins with different sequence, structure and function may associate in similar ways. Protein Eng. 2005;18:11–24. doi: 10.1093/protein/gzh095. [DOI] [PubMed] [Google Scholar]
- 29.Keskin O, Nussinov R. Similar binding sites and different partners: implications to shared proteins in cellular pathways. Structure. 2007;15:341–354. doi: 10.1016/j.str.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Keeble AH, Joachimiak LA, Mate MJ, Meenan N, Kirkpatrick N, Baker D, Kleanthous C. Experimental and computational analyses of the energetic basis for dual recognition of immunity proteins by colicin endonucleases. J Mol Biol. 2008;379:745–759. doi: 10.1016/j.jmb.2008.03.055. [DOI] [PubMed] [Google Scholar]
- 31.Zhang QC, Petrey D, Norel R, Honig BH. Protein interface conservation across structure space. Proc Natl Acad Sci USA. 2010;107:10896–10901. doi: 10.1073/pnas.1005894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.