Abstract
Understanding how differentiation programs originate from within the gene expression landscape of hematopoietic stem cells (HSC) is crucial to develop new clinical therapies. We mapped the transcriptional dynamics underlying the first steps of commitment by tracking transcriptome changes in human HSC and eight early progenitor populations. Transcriptional programs are extensively shared, extend across lineage-potential boundaries, and are not strictly lineage-affiliated. Elements of stem, lymphoid and myeloid programs are retained in multi-lymphoid progenitors (MLP), reflecting a hybrid transcriptional state. Based on functional single cell analysis, BCL11A, SOX4 and TEAD1 governed transcriptional networks within MLPs, leading to B cell specification. Overall, we show that integrated transcriptome approaches can identify novel regulators of multipotency and uncover additional complexity in lymphoid commitment.
In homeostasis, blood production depends on a highly coordinated hierarchy of hematopoietic cells. At the apex of the hierarchy are hematopoietic stem cells (HSC), which are capable of self-renewal, have multi-lineage potential and are responsible for generating all of the lineages of hematopoietic cells in the blood. HSC self-renewal capacity and multipotentiality are gradually lost as cells progress through various multi-, oligo- and uni-lineage intermediates, eventually acquiring either erythroid, myeloid or lymphoid identity. Understanding how the genomic information present in HSC translates into such complex differentiation programs is crucial to develop new approaches in regenerative medicine and better cancer therapeutics.
At the molecular level, targeted functional studies of single or paired transcription factors have identified a relatively small number of key transcription factors that drive differentiation of progenitor cells, by directing the sequential establishment of transcriptional programs essential for terminal differentiation 1. Complex transcriptional networks integrated around the GATA1-PU.1 bimodal switch represent a paradigm for myeloid vs erythroid lineage specification 2. By contrast, differentiation into lymphoid lineages follows a more linear network architecture. Establishment of lymphoid identity requires successive and obligatory activation of E2A, Ebf1 and finally Pax5 in distinct progenitor populations 3,4. However, a clear genome-wide picture of how these master transcription factors interact with the transcriptional and epigenetic landscape in which they operate is still lacking 5,6.
To date, most mechanistic studies used murine models, but with robust sorting and functional assays, global transcriptional analyses of human hematopoietic cell types is now feasible. Initial gene expression analysis on 38 human hematopoietic cell subtypes, identified gene modules and transcription factors circuits active in stem and progenitor cell-enriched fractions and reused in terminally differentiated cells 7. One limitation of this study was the lack of highly purified immature progenitor and stem cell populations, which precluded dissection of the very first transcriptional events linked to commitment. In mouse, several studies have described the expression of lineage-affiliated transcriptional programs within multilineage progenitors 8–10. These studies support the lineage priming hypothesis, which argues that multipotent progenitors express, before lineage restriction, low levels of genes a priori known to be key determinants of distinct fates 11. However none of these studies functionally investigated whether there are additional layers of regulation upstream of the master transcription factors that affect lineage specification, or alternative molecular routes to specify any particular fate.
At the cellular level, earlier models of hematopoietic commitment described a unique binary split between myeloid and lymphoid fates, immediately downstream of a multipotent cell 12. Several recent reports challenged this view by demonstrating that lymphoid and myeloid fates remain entangled over several early cell populations. The earliest thymic progenitors (ETP) and granulocyte-monocyte progenitors (GMP), long thought to be unilineage, retained residual myeloid or lymphoid potential respectively 10,13,14. Importantly, recent studies in human cord blood and bone marrow demonstrated the existence of early lymphoid-biased progenitors that retain myeloid, but not erythroid, potential. These progenitors, termed multi-lymphoid progenitors (MLP, 13), or lymphoid-primed multipotent progenitors (LMPP, 15) by analogy with the mouse system 16, are identified as CD34+CD38−Thy1−CD45RA+ 13,15 or by high expression of L-selectin on CD34+ cells 17. In view of this flexibility in lymphoid commitment, it is likely that a large number of yet unidentified regulators orchestrate specification of lymphoid fates. The identification of MLP provides a unique opportunity to investigate the molecular mechanisms underlying lymphoid vs myeloid lineage choice in primary human hematopoietic cells.
To understand how stem, lymphoid and myeloid programs are coordinated during hematopoietic differentiation, we systematically profiled the transcriptome of MLP in the context of nine other human hematopoietic stem and progenitor cell populations, for which self-renewal and differentiation capacities are known at the single cell level 13,18. At this level of cellular resolution, we uncovered a landscape of transcriptional programs that cross population and lineage boundaries. Computational and functional mapping of transcription factors activity in the very first stages of hematopoietic differentiation revealed the molecular complexity underlying lymphoid commitment, identified new transcription factors that contribute to B cell commitment, and established that molecular regulation of B cell specification occurs at the level of MLP.
RESULTS
Transcriptional dynamics of the early stages of human hematopoiesis
To monitor global transcriptional changes during the first steps of human hematopoietic differentiation, we prospectively isolated 10 populations of cord blood (CB) HSC and early progenitors (Fig. 1a and Supplementary Table 1). This included populations of highly enriched repopulating HSC (HSC1, 1 in 10; HSC2, >1 in 100), transiently engrafting multipotent progenitors (MPP) 18 and a spectrum of early committed progenitors: common myeloid progenitors (CMP), megakaryocytic-erythroid progenitors (MEP), granulocytic-monocytic progenitors (GMP), and multi-lymphoid progenitors (MLP). MLP represents the earliest lymphoid progenitor giving rise to all lymphoid lineages (B, T and NK cells) as well as monocytes and dendritic cells 13,15. Finally, we included 3 lymphoid-restricted precursor populations: B-NK precursors 13, proB cells and the most primitive progenitors found in the thymus, ETP 19. Gene expression profiling was performed on Illumina arrays with a protocol optimized for maximal sensitivity with low cell number.
Fig. 1. Transcriptional architecture of the first steps of the human hematopoietic hierarchy.
(a) Differential expression distances overlaid on the current hierarchical model of human hematopoietic differentiation. Solid arrows represent accepted progenitor-product relationships while dashed arrows represent assumed ones. The number of genes upregulated in the precursor population is shown in bold and red and those upregulated in the product population in brown. Populations shown in orange were used for undirected pattern discovery detailed in Fig. 2. Differential expression was calculated using limma (fold change >2 and FDR<0.05) comparing the downstream population to its progenitor. (b) Principal Component Analysis of the 10 human hematopoietic stem and early progenitor cells. Only the first 2 principal components are shown here as they explain the vast majority of the variation in our dataset (Supplementary Figure 1a). All populations were purified from 3 to 5 independent pools of cord blood (CB) with the exception of ETP, which were isolated from 3 independent neonatal thymi. All analyses were performed on the DREGH genes.
To understand the general transcriptional architecture of these 10 populations, we focused on the 9898 genes (13385 probes) that change expression by at least 2-fold between any populations (hereafter termed “dynamically regulated hematopoietic”, DREGH, genes, one-way ANOVA, FDR<0.05). Principal Component Analysis (PCA, Fig. 1b and Supplementary Figure 1a) showed that the major lineage outcomes (stem, lymphoid, myelo-erythroid) are recapitulated in clusters of global transcriptional similarity, with the exception of ETP, whose transcriptome closely resembled that of myeloid progenitors. Unsupervised hierarchical clustering identified similar relationships between populations (Supplementary Figure 1b). Despite differences in repopulating HSC frequency, the two stem cell populations profiled here (HSC1 and HSC2) clustered extremely close, and differed by less than 10 genes (data not shown). We thus restricted further analyses to the HSC1 population. Finally, MLP gene expression clustered much more closely with HSC and MPP than with more differentiated lymphoid fractions.
To capture dynamic changes in transcription upon lineage commitment, we generated precursor-product transition gene-sets, which estimate differentially expressed genes between 2 developmentally related populations (moderated t-tests, FDR < 0.05, Supplementary Table 2). We then overlaid the numbers of differentially expressed genes onto the established model of developmental relationships within the hematopoietic hierarchy 13,18 (Fig. 1a, Supplementary Table 3). Only 29 genes were differentially expressed between HSC and MPP, indicating a very high degree of transcriptional similarity between these two cell types. Eight times more genes changed upon transition from MPP to CMP (7991 genes) than from MPP to MLP (999 genes), showing a clear demarcation between the multipotent (HSC and MPP) and myeloid committed progenitors. Changes along the lymphoid branch of the hierarchy were more gradual, consistent with the idea that lymphoid specification is not a rapid binary decision point but rather an extended process characterized by progressively more committed states 20,21. Establishment of a differentiated cellular identity involves activation of lineage-specific transcriptional programs, defined as groups of genes which expression is modulated similarly during the commitment process. To identify the dominant transcriptional programs present in the early hematopoietic hierarchy, we integrated two distinct unsupervised pattern recognition methods, K-means and Short Time Series Expression Miner (STEM) 22, that we applied to the populations most relevant for studying lineage commitment: HSC1, MLP, proB, CMP, GMP, MEP. Clustering was performed independently with each algorithm on the DREGH genes to generate expression profiles (Fig. 2a, b and Supplementary Table 4). Profiles with similar kinetic and biological behavior were then grouped into clusters, which we defined as transcriptional programs. Each cluster was named based on the cell types in which the expression of the genes in that cluster is the highest, therefore relating these clusters of expression profiles to meaningful biological programs and lineage commitment routes. Both methods identified similar transcriptional programs, independently of the parameters used, and assigned similar proportions of genes to each program (Fig. 2c and Supplementary Figure 2a). The transcriptional program with the highest number of genes contained genes with high expression in HSC that were downregulated in all other populations, thereby termed stem cell program. A reciprocal progenitor program, containing genes low in HSC but upregulated, independently of lineage choice, in all other populations was the second most represented program and was enriched for cell cycle associated genes, consistent with the higher proliferative rate of progenitors. Lineage-specific programs were less abundant; among these, lymphoid-specific clusters contained more genes than myeloid or erythroid. The STEM algorithm uniquely identified a group of genes shared by MLP and GMP, termed myelo-lymphoid, which was enriched in NF-κB signaling, apoptotic and immune response genes (data not shown). K-means clustering revealed a very close association between gene-expression in CMP and MEP, which was not otherwise detected by the STEM method.
Fig. 2. Six predominant transcriptional programs are associated with commitment of human HSC.
(a, b) Heat-maps of the 14 significant gene expression profiles as derived by the STEM algorithm (a) and K-means clustering method (b) (performed specifying 14 clusters, for other values of k, see (c). Each box in the heat-map represents the mean of the expression of all the genes assigned to that profile in the indicated populations. Log transformed expression data is mean centered and hierarchically clustered by profiles. Color-coded boxes on the right of each heat-map represent the classification of each profile into a transcriptional program based on the populations in which the expression of the genes in that transcriptional program is highest. (c) Quantitative comparison of the two pattern recognition methods. The percentage of DREGH genes assigned to each transcriptional program according to the two algorithms is shown. The k-means algorithm was run independently with 3 different values of k (8, 14 and 18) chosen after applying the adjusted Figures of Merit method (FOM, Supplementary Figure 2a) to determine at which k the algorithm reaches its maximum predictive value. p = 0.89 by a two-sample Kolmogorov-Smirnov test between STEM method and k-means with k=14. An overlay of the transcriptional programs on the current model of hematopoietic differentiation is presented in Supplementary Figure 2b.
MLP participated in 4 of the 6 main transcriptional programs (Supplementary Figure 2b) and exhibited the proliferative signature of progenitors as well as elements of both myeloid and lymphoid programs, consistent with their poised developmental state. Approximately 56% of the HSC cluster genes (as measured by the STEM algorithm, Supplementary Table 5) were expressed at similar levels in MLP, despite profound differences in self-renewal capacity. Of all the DREGH genes, 38% were transcribed similarly by HSC and MLP, whereas only 16–19% showed co-expression in HSC and either MEP, GMP or ProB (Supplementary Table 5), again highlighting the transcriptional resemblance between HSC and MLP. Collectively, whereas the stem, lymphoid and myeloid transcriptional programs were interwoven, the erythroid program was more separated (Supplementary Figure 2b). Consistent with previous observations in mice 10, we observed more gradual expression changes along the lymphoid lineage, and a persistent association of lymphoid- and myeloid-associated genes over several steps of hematopoietic differentiation. Furthermore, the HSC program remained largely active in MLP, which also displayed lymphoid and myeloid elements, suggesting that these cells most likely represent a critical stage at which lineage commitment occurs.
Transcription factor architecture of early human lymphoid commitment
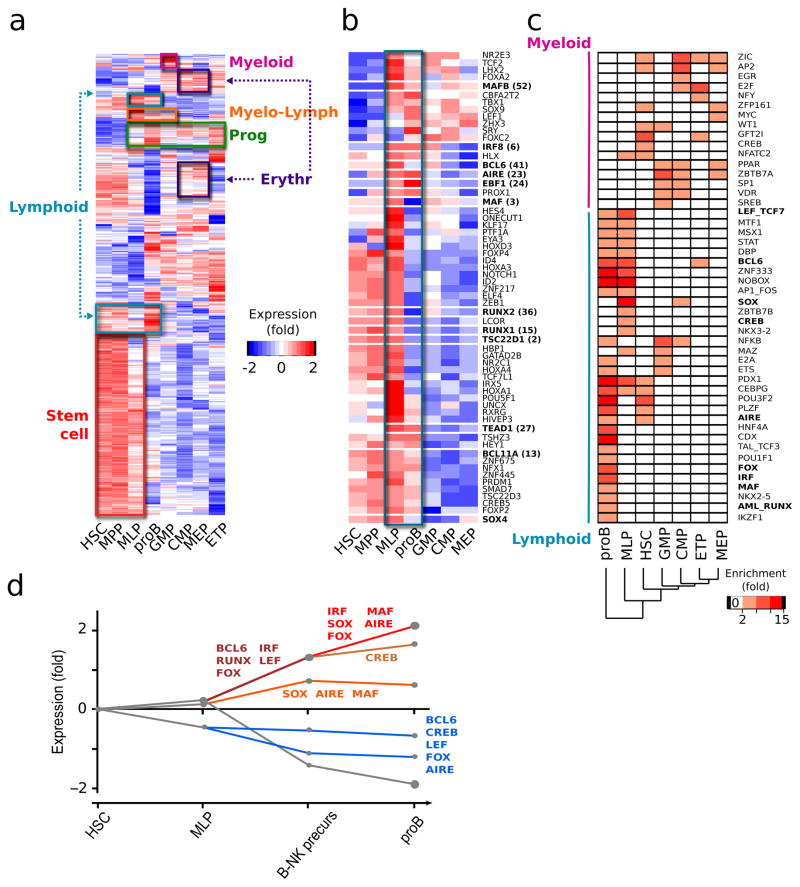
Transcription factors are master regulators of lineage commitment decisions. To capture the global complexity of transcription factors expression during commitment, we compiled a comprehensive list of differentially expressed transcription factors in hematopoiesis (see Methods), that can be mined for regulators of self-renewal and differentiation. Of 1581 putative transcriptional regulators, 477 (30%) were differentially expressed across the hierarchy (FDR<0.01, fold-change >2, Supplementary Table 6). Transcription factors expression clustered into the 6 main transcriptional programs described above (Fig. 3a), reflecting the internal structure of transcriptional changes during lineage commitment. Each transcriptional program contained transcription factors shown to be important in maintaining their respective states (EVI1, ERG and ID1 in the stem cluster; GATA1 in MEP; the myeloid-affiliated factors CEBPA and SPI1 in the GMP cluster). MLP expressed high levels of both myeloid (CEBPA) and lymphoid (IKFZ1, EBF1) determining transcription factors, again indicating that these cells are not fully committed to either lymphoid or myeloid fate.
Fig. 3. Transcription factor expression complexity during commitment.
(a) Heat-map of the expression of the 477 transcription factors differentially expressed between any 2 hematopoietic populations. Boxes highlight the transcription factors belonging to the 6 main transcriptional programs defined in Fig. 2 and Supplementary Figure 2b. (b) Heat-map of the expression of the 60 transcription factors with an early lymphoid pattern. Blue box: transcription factors expression in MLP and proB. Bold: transcription factors highlighted selected for functional validation. In both a and b, log transformed expression data is mean centered and hierarchically clustered by gene. (c) Transcription factor families whose DNA-binding motifs were found over-represented in the promoters of at least one population-specific gene set (peach to red boxes). Bold: transcription factor families of which at least one member was found among the 60 early-lymphoid specific transcription factors as defined by their expression. The gene lists used for this analysis are listed in Supplementary Table 7. (d) Dynamic regulatory map of transcription factors controlling specification to B cells generated with the DREM algorithm. y axis: log transformed expression relative to first developmental stage, HSC ; x-axis: stepwise progression along B cell commitment. Indicated here are the transcription factor families predicted to be stage-specific regulators of expression that were also found in b and c (complete output in Supplementary Figure 2). The lines represent the average expression of a group of genes and the size of the circles its standard deviation.
To better understand transcription factor activity in MLP and eventually identify novel regulators of lymphoid restriction, we integrated three computational approaches. First we assembled a list of transcription factors with an early lymphoid-specific pattern of expression (see Methods). This yielded 60 transcription factors whose expression is upregulated in MLP relative to HSC-MPP or non-lymphoid progenitors (Fig. 3b). While known regulators of B cell (EBF1, ID2, FOX and HOX family factors) and T cell (NOTCH1, HEY1, HES4) lineages were in this group, most transcription factors had no known association with lymphoid development, providing a rich resource of potential candidates whose role in lineage choices will require functional validation. Second, to identify critical transcription factors that may not exhibit differential expression, we examined overrepresentation of known binding motifs within those genes expressed across the hierarchy. We generated population-specific gene sets for all populations (genes that are most differentially upregulated in one population relative to all others, Supplementary Table 7) and looked for over-representation of 385 experimentally verified or in silico predicted cis-binding sequences of transcription factors in each gene set (see Methods). Less transcription factors family motifs were enriched in the myelo-erythroid gene sets (17 motifs in CMP, GMP, MEP), than in the MLP and proB gene sets (33 motifs, including BCL6, CEBPG, CREB, STAT and SOX family motifs, Fig. 3c). The majority of transcription factor families (11/17) controlling genes highly expressed in MLP also controlled proB specific genes, again underscoring the complexity and gradualness of establishing a differentiated lymphoid transcriptional program.
Last, we sought to predict in silico how changes in particular transcription factors expression might account for the dynamics of gene expression that lead to the establishment of B cell identity. To this end, we integrated our gene expression data with available transcription factors binding information from the Dynamic Regulatory Events Miner algorithm (DREM) 23 to deduce a global developmental map, annotated with transcription factors that are most likely to control the expression changes associated with a particular transition of early B cell commitment (Fig. 3d and Supplementary Figure 3). Of interest, SOX and MAF were predicted to first activate genes at the MLP to B-NK progenitor transition and then two distinct sets of genes in the transition to proB. A known set of B cell genes with binding sites to BCL6, AIRE and FOX were predicted to be upregulated between MLP and B-NK progenitors; similar binding sites were also in a series of apoptosis-related genes repressed in proB cells. Collectively, this integrative bioinformatics approach uncovered novel transcription factors anticipated to control B cell specification, and predicted that these factors may have complex roles by controlling distinct sets of genes at different stages of lineage specification.
Identification of early regulators of commitment in primary human MLPs
Our bioinformatic analysis predicted that lineage-specific transcriptional networks are not yet stabilized in MLP. To test this idea and identify key determinants of lymphoid vs myeloid commitment, we perturbed lineage outcomes by silencing key transcription factors in MLP. Based on the computational analysis of MLP-specific transcription factors, we studied the function of factors with DNA-binding motifs that were over-represented in the MLP-specific gene-set (Fig. 3c), or that were predicted in silico to act recurrently during B cell development (Fig. 3d), as these most likely represent key nodes in the MLP regulatory circuit (highlighted in bold in Fig. 3b). The expression of these candidates was verified across the hematopoietic hierarchy by qRT-PCR (Supplementary Figure 4a). To examine the role of these transcription factors in lymphoid development, we developed lentiviral vectors expressing shRNAs that efficiently silenced the expression for 8 of 12 candidate transcription factors (Supplementary Figure 4 b, and c) and assessed the clonal potential of sorted single MLP to differentiate into monocytic, B and NK cells in vitro. Lineage-depleted CB cells were transduced with shRNA-expressing lentiviral vectors and single MLP were sorted into plates seeded with murine stromal MS-5 cells 24 in the presence of appropriate cytokines (Supplementary Figure 4d). After 3 weeks, the number of transduced (GFP+) myeloid, B and NK cell colonies was monitored by detection of lineage-specific cell surface markers (Supplementary Figure 4e). The capacity of the MS5-MBN assay to accurately identify changes in lineage commitment was validated extensively (Supplementary Note 1, Supplementary Fig. 4f–h). In particular, shRNA-silencing of EBF1, a key master regulator of B cell specification significantly reduced the number of B cell colonies formed in our assay (p=0.049, 4 independent CB). shRNA-silencing of GATA2, not expressed in MLP, did not affect any of its developmental outcomes (Fig. 4a).
Fig. 4. Single cell shRNA silencing screen for transcription factors that determine MLP commitment to the lymphoid fate.
(a) and (b) Proportion of myeloid (CD11b+, top), lymphoid (CD19+, middle) and NK (CD56+, bottom) colonies generated from single MLP in which the indicated transcription factors were silenced. The proportions for each shRNA are normalized to single MLP from the same pool of CB transduced with control hairpins lentiviral vectors (shLacZ and-or shLUC). shBCL11A, shIRF8, shMAF, shRUNX2, shTSC22D1 and shLacZ expression are driven from the H1 promoter, while shBCL6, shEBF1, shGATA2, shSOX4, shTEAD1 and shLUC are downstream of the U6 promoter. n=3 distinct pools of CB for shBCL11A-1, shBCL6, shEBF1, n=4 for others. Non-normalized data is available in Supplementary Table 8. (c) Percentage of myeloid colonies (CD11b+) that are fully differentiated to monocytes (>95% CD14+). Dashed lines connect measurements from the same CB pool. (d) Percentage of GFP+ CD34+ CD133+ in S-G2-M after 100 MLP per well were cultured 7 days on MS5 stroma in the same conditions as for the screen described above. n≥3. (e) Methylcellulose assays from 800 GFP+ CD34+ cell sorted 3 days after transduction of Lin- CB with the indicated shRNA-expressing lentiviral vectors. Shown is the number of erythroid (top) and myeloid (bottom) colonies relative to control 12 days after plating. n≥3. For all panels: mean and SEM is shown; *: p<0.1; **:p<0.05 by paired two-tailed t-test.
We next tested whether silencing of 8 candidate transcription factors altered the lineage outcomes of MLP, by screening over 8500 single-cell derived colonies. Our screen identified 4 transcription factors (BCL11A, SOX4, BCL6 and TEAD1) that had significant effects on lymphoid colony formation across multiple independent experiments (Fig. 4b). Of note, the results described below were recapitulated with independent shRNAs (Fig. 4b), with the exception of BCL6 for which we could not generate a second shRNA construct that significantly reduced BCL6 mRNA levels. As such, we cannot exclude potential off-target effects in the BCL6 sliencing phenotypes described below. Silencing of two genes, SOX4 and BCL6, led to significantly fewer B and NK colonies. Also, cultures of BCL6-shRNA-silenced and SOX4-shRNA-silenced cells contained proportionally more fully differentiated monocytic colonies compared to controls, as scored by the number of CD14+ colonies (Fig. 4c). These observations suggest that BCL6 and SOX4 act in MLP or their immediate progeny to promote lymphoid differentiation while repressing myeloid fates. Silencing of two other genes, BCL11A and TEAD1, generated fewer B cell colonies (Fig. 4b), but MLP differentiated normally into NK and myeloid cells. These data suggest that BCL11A and TEAD1 act in B cell development at or downstream of B cell specification. No significant differences in the proportion of cycling (Fig. 4d) or apoptotic (data not shown) cells were observed in MLP following a week of culture on MS5 stroma, suggesting that the skewed colony distributions derived from alteration of commitment decisions. In addition, these transcription factors govern specifically the lympho-myeloid lineage choice, as we observed no skewing of myelo-erythroid output in conventional colony-forming assays upon shRNA-silencing of BCL11A, SOX4, BCL6 or TEAD1 in CD34+ cells (Fig. 4e). In summary, our functional validation of transcription factors, predicted to be important regulators based on a global computational analysis of human CB progenitors, has uncovered unique and distinct roles for BCL11A, SOX4, BCL6, and TEAD1 in human lymphoid development.
BCL11A, SOX4 and TEAD1 regulate MLP entry to B cell commitment
During B lymphopoiesis, B cell precursors transit through a continuum where alternate fates are being progressively repressed and B cell identity is gradually acquired 21. To precisely establish at which developmental stage the four transcription factors identified in the screen influence B cell differentiation, we reconstituted human hematopoiesis in xenografts, in which all steps of human B cell commitment are recapitulated 25. Equal numbers of HSC-enriched CB cells (CD34+) transduced with lentiviral vectors expressing shRNAs against BCL11A, BCL6, SOX4 or TEAD1 were injected into immune-deficient NSG mice. Efficient silencing was observed in vivo for all lentiviral vectors used (Supplementary Figure 5a). Normal percentages of total B cells (CD19+) were produced upon BCL6 silencing. By contrast, the percentage of total B cells was significantly reduced in animals transplanted with BCL11A, SOX4 and TEAD1 silenced cells (Fig. 5a, Supplementary Figures 5b–c), confirming our in vitro results.
Fig. 5. Effects of BCL11A, BCL6, SOX4 or TEAD1 silencing on B cell commitment in vivo.
Mice were transplanted with Lin− cells transduced with lentiviral vectors expressing either control shRNA or shRNAs against the candidate transcription factors that led to significant silencing in vivo. Silencing levels are shown in Supplementary Figure 5a. The composition of the human graft was analyzed 8 to 10 weeks post-transplantation. (a) Percentage of total B cells (CD19+) among human cells (CD45+ GFP+) in the injected bone. Circles represent individual animals; shLacZ, n=21; shBCL11A, n=19 (p < 0.0001); shLUC, n=23; shBCL6, n=5; shSOX4, n=12 (p < 0.0001); shTEAD1, n=14 (p = 0.0002). Raw data pertaining to this figure is available in Supplementary Table 9; results in the other bones are shown in Supplementary Figure 5b–c. (b) and (c): the quantification of each intermediate of B cell differentiation in xenografts was performed according to the flow cytometry gating strategy presented in Supplementary Figure 5d. (b) Percentage of MLP among human (GFP+) cells. shLacZ, n=4; shBCL11A, n=4; shLUC, n=18; shBCL6, n=5; shSOX4, n=12; shTEAD1, n=12. (c) Number of population doublings between the populations indicated. Population doublings were calculated as: log2 (product population/precursor population). Full quantification of each B cell progenitor population is available in Supplementary Figure 5f. Mean ± SEM is shown. *: p<0.1; **:p<0.05 by unpaired two-tailed t-test.
We then analyzed the production of the various developmental intermediates of B cell differentiation (Supplementary Figure 5d). Mice transplanted with BCL6-silenced cells had fewer MLP than controls (Fig. 5b), which were nonetheless directed along the B cell differentiation path as they gave rise to almost normal numbers of early B cells (Supplementary Figure 5e). By contrast, BCL11A and SOX4 silencing resulted in a significant increase in the proportion of MLP; a similar trend was observed upon TEAD1 silencing (Fig. 5b), suggesting a differentiation block at this stage. Quantification of population doublings at each step downstream of MLP (Fig. 5c) showed that progression from MLP to early B cells was significantly compromised upon silencing of all 3 genes, BCL11A, SOX4 and TEAD1. No changes in proliferation or apoptosis were observed in BCL11A or SOX4 silenced earlyB (Fig. 6a–b), indicating that the observed reduction in B cells is due to an early differentiation defect. Of note, upon silencing of TEAD1 transition from early B to proB was also significantly compromised (Fig. 5c), leading to a more than 4-fold decrease in proB (Supplementary Figure 5e–f), which could be explained by a trend towards increased apoptosis in the early B compartment (Fig. 6a). The few TEAD1-silenced proB cells produced, cycled significantly more than their control counterparts (Supplementary Figure 6a) generating preB cells at levels close to normal (Supplementary Figure 5e). No such changes in proliferation were observed for BCL11A shRNA-silenced or SOX4 shRNA-silenced proB and preB cells (Supplementary Figure 6a–b). These findings raised the possibility that BCL11A, SOX4 and TEAD1 might act upstream of some of the master regulators of B cell commitment. We therefore examined by qRT-PCR whether decreased levels of BCL11A, SOX4 or TEAD1 affected the expression of IKZF1, E2A, EBF1 and PAX5 in early B cells, and found that all three genes altered the expression at least one of these key lymphoid transcription factors (Fig. 6c), with SOX4 silencing having the broadest effect, decreasing expression levels of E2A, EBF1 and PAX5. Taken together, our data show that BCL11A, SOX4 and TEAD1 independently contribute to B cell commitment decisions by controlling the MLP to early B cell transition (Supplementary Figure 6c).
Fig. 6. BCL11A, SOX4 or TEAD1 KD do not affect proliferation or apoptosis of B cell progenitors but decrease expression of master regulators of B cell commitment.
(a) Percentage of AnnexinV+ cells in early B cells. (b) Percentage of cells in each phase of the cell cycle in early B cells. G0: Ki67− Hoechst−, G1: Ki67+ Hoechst−, S-G2-M: Ki67+ Hoechst+. For a and b: n=4 for shLacZ, n=11 for shLUC, n=3 for H1shBCL11A, n=6 for U6shSOX4 and n=4 for shTEAD1. Similar measurements for proB and preB populations are in Supplementary Figure 6a–b. (c) mRNA expression levels of IKZF1, E2A, EBF1 and PAX5 in Early B cells. All values were normalized to 2 housekeeping genes (GAPDH and ACTB) and are shown here relative to control. Ctrl, n=12; shBCL11A, n=2; shSOX4, n=3, shTEAD1, n=4. All measurements are from the injected femur of NSG mice 8 to 10 weeks after transplantation. Mean and SEM is shown. *: p<0.1; **:p<0.05 by unpaired two-tailed t-test.
DISCUSSION
Here, we detail the gene expression profiling of highly purified and functionally defined human HSC and progenitors, providing a resource for investigation of the earliest steps of human hematopoietic differentiation. Our bioinformatic analysis revealed a landscape of tightly interconnected transcriptional programs that contrasted with many commonly accepted predictions of a rigid demarcation of stem cell and lineage commitment circuits. Through a combined computational and functional approach, our data uncovered high molecular complexity in lymphoid commitment and identified BCL11A, SOX4 and TEAD1 as new lymphoid transcription factors acting upstream of known master regulators of B cell commitment.
Several principles underlying loss of stemness and lineage commitment emerge from our analysis of global patterns of gene expression. First, transcriptional programs are shared among cell types with similar lineage potential (MLP and proB; CMP and MEP; MLP and GMP, both monocyte precursors). Second, transcriptional programs can cross physiological lineage boundaries. For example, GMP, which give rise to myeloid cells, and ETP, which generate T cells in the thymus, also display very similar transcription profiles. This similarity is not surprising in view of the myeloid potential of ETP 14 and the capacity of GMP to produce T cells when appropriately stimulated 10,13. A third important principle is that HSC programs do not terminate abruptly but persist for many stages. HSC and transiently repopulating MPP differ only by a handful of genes raising the possibility that the predominant regulation of self-renewal does not occur at the transcriptional level. This conclusion remains cautious as limitations in microarray technology may not capture subtle, but important transcriptional differences. The stem cell program is also partially carried over into MLP, which continue to express many but not all HSC genes. This phenomenon can be interpreted in view of the lineage priming hypothesis, which postulates that genes important for differentiation into a particular fate are already expressed at low levels in HSC. However, the expression of many genes, including some with established stem cell functions (i.e. EVI1, ERG, ID1), is maintained at similar or slightly lower levels in MLP, indicating that MLP retain components of the stem cell circuitry. A fourth conclusion is that MLP do not possess a unique transcriptional program but rather exhibit partially established stem, myeloid and lymphoid transcriptional programs. Thus, we propose that the molecular regulation of myeloid or lymphoid fate acquisition occurs physiologically at the level of MLP.
The computational description of the transcriptional programs and transcription factors architecture presented here suggests a model in which lymphoid specification proceeds more gradually and involves more molecular players than does commitment to myeloid fate. Taking into account potential functions of epigenetic regulators and miRNAs not analyzed here, our results, together with those of other groups 7,26 indicate that the molecular circuitry underlying entry into B cell specification is very likely more complex than previously assumed. Based on the current view of hematopoiesis 20, in which there is no early obligatory separation between myeloid and lymphoid fates, it seems likely that myeloid differentiation is a default commitment program that needs to be shut down for other lineages to be specified 27. Accordingly, in the thymus, T cell specification requires down-regulation of pan-progenitor genes, which is achieved through multiple distinct repressor functions 28. Likewise we propose the existence, in B cell commitment, of an additional layer of transcription factors regulation, composed of molecules such as BCL11A, SOX4, TEAD1 and IKZF1 10 that sets the stage for activation of the self-sustaining EBF1-PAX5 axis 29, itself required and sufficient for the establishment of the full B cell differentiation program. We speculate that this molecular organization makes entry into B cell specification more adaptable to shifting demands.
Three of the transcription factors for which we describe a role in the very early stages of lymphoid commitment have previously been implicated in later steps of lymphopoiesis. BCL6 plays key roles in germinal center B cells 30 and is required for formation of a diverse B cell repertoire 31. Our results obtained with a single shRNA for BCL6 should be interpreted with caution, but they suggest that this transcription factor is also active during much earlier stages of hematopoiesis by regulating MLP formation or differentiation. Our data on BCL11A and SOX4 KD are consistent with the phenotype of BCL11A and SOX4-deficient mouse models, in which there is no B cell development 32–34, but we further show that they direct MLP commitment to the B cell lineage rather than limiting later differentiation steps. The fourth transcription factor identified, TEAD1, has not been previously associated with hematopoiesis. TEAD1 functions with the transcriptional coactivator YAP downstream of the Hippo tumor suppressor pathway 35. YAP overexpression in the mouse does not alter HSC self-renewal or differentiation 36. Thus, by showing that TEAD1 first activates the MLP to early B progression, and also favors transition to the proB cell stage, we provide evidence that the Hippo pathway fucntions in lymphopoiesis.
The dataset presented here represents a resource to identify cell type-specific gene regulatory networks, which when integrated with future RNA-seq, genome-wide chromatin occupancy and epigenetic modification analyses will shed further light on how hematopoietic cells are driven to commitment. We make available stem and progenitor specific gene expression sets as well as transcriptional program signatures that will, among other uses, facilitate classification of tumor subtypes based on their transcriptional homology to normal progenitors 37,38 and inform on their cell of origin. In addition, our data will contribute to improved methods for hematopoietic differentiation of pluripotent stem cells, by serving as a molecular roadmap with which to compare engineered cell types to their normal counterparts. Finally, the principle of obligatory sharing of transcriptional programs in the first steps of differentiation uncovered here could be a general design principle conserved in other stem cell driven tissues.
ONLINE METHODS
Primary samples
All CB samples were obtained with informed consent according to the procedures approved by the institutional review boards of the University Health Network and Trillium Hospital. Lineage depletion of CB samples was achieved by negative selection with the StemSep Human Progenitor Cell Enrichment Kit (Stem Cell Technologies) according to the manufacturer’s protocol.
Isolation of cell populations for gene expression profiling
Lin- cells were thawed and stained at 1×106 cells per 100uL with the following antibodies (all from BD, 1:100, unless otherwise stated): CD45RA FITC (1:25), CD90 PE (1:50), CD135 PE (1:10), CD49f PECy5, CD7 PECy5 (Beckman Coulter), CD38 PECy7, CD10 APC (1:25), CD34 APCCy7. Cells were sorted into low-binding 1.5mL tubes (Axygen) with a FACSAria (BD) instrument. Purity was >95%. Freshly sorted populations of progenitor cells were pelleted and resuspended in TRIzol (Invitrogen).
Microarray mRNA profiling and data pre-processing
RNA extraction, cDNA synthesis, Pre-Amplification were carried out as described in 39. Whole-genome gene expression analysis was performed using the Human HT-12 WG DASL v4.0 R2 assay, which interrogates ≈ 29K targets corresponding to ≈ 21K genes 40.
Bioinformatic analyses
If not specified, all bioinformatics analyses were performed with R (version 2.12.1) and Bioconductor (version 2.10). Pearson’s correlations and hierarchical clustering were performed by using the “cor” and “hclust” R functions. For PCA, we first compared eigenvalues from real data PCA to randomized data PCA to evaluate which components are the most relevant (Supplementary Figure 1a), and then used the “dudi.pca” function of the Ade4 package (version 1.4–16). Pearson correlation coefficient based hierarchical clustering and PCA of all samples, as well as the percentage of presence calls (p-detection values <0.05) for each sample, were used to assess quality control. Data were quantile normalized (“normalizeQuantiles” command from the limma package; version 3.6.9), then log2-transformed. All subsequent analyses were carried out with this dataset. The DREGH list was generated using GeneSpringGX software (Agilent), running one-way analysis of variation (ANOVA) analysis on all 10 populations profiled in this study, with Tukey HSD post-hoc test and Benjamini-Hochberg multiple testing correction. We considered genes with a multiple test adjusted p-value <0.05 and an absolute fold-change >2. This analysis resulted in 13385 probes, corresponding to 9898 genes. All other differential expression tests were performed with the limma package (version 3.6.9), which calculates the moderated t-test statistic for a particular contrast. All t-tests scores were controlled for multiple hypothesis testing using the Benjamini-Hochberg method. Unless otherwise stated, genes were considered differentially expressed for adjusted p-values <0.05. Three main group of contrasts were generated: (1) population-specific gene sets in which the mean expression of a gene in a particular population (e.g MLP) is compared to the mean expression in all other populations; (2) transcriptional programs gene sets, in which the samples contrasts were chosen to best reflect the transcriptional programs identified by undirected pattern discovery; (3) precursor-product transitions gene sets, where the expression of each gene in a particular population (eg: B-NKprec) is compared to its expression in its closest known progenitor population (eg: MLP) independently of all other samples. A summary of all differential expression lists generated is presented in Supplementary Table 2.
Transcriptional programs derivation
The STEM (Short Time Series Expression Miner) algorithm 22 was downloaded from http://gene.ml.cmu.edu/stem/. This clustering method first defines a set of representative model profiles, which correspond to possible patterns of gene expression across the conditions analyzed in the experiment. Based on correlation coefficients each gene is assigned to the closest profile in terms of expression. The number of genes expected randomly for each profile is also computed (random permutation of original values for each gene, renormalization then assignment to profiles, repeated over 500 permutations) and serves as a basis to calculate statistical significance of each profile. Statistical significant profiles represent the dominant expression profiles present in the dataset. By the STEM method, the number of profiles is thus unbiased as determined by the algorithm and not by the user. The parameters used for STEM clustering were set at a maximum of 50 model profiles, a maximum unit change between time points of 1, and a minimum correlation for clustering similar profiles >0.5. For GO enrichment within this program, the p-values were corrected with 500 randomizations and were considered significant for FDR<0.05. As the STEM algorithm was first implemented to analyze temporal expression profiles, the analysis was performed with 3 different population orders which all yielded similar results. K-means clustering was performed in R with the function “kmeans” setting number of clusters k to 8, 10 or 14. To determine these values of k, objectives values, we used the Figures of Merit method (FOM 41, implemented in MeV software).
Both algorithms output a number of “profiles” (predefined for K-means, but automatically (unbiased) calculated by STEM based on the dataset). As pattern recognition methods will find profiles that, even though distinct in intensity of expression, represent the same kinetic or biological behavior, similar “profiles” (based on correlation coefficient) were then grouped into clusters, which are effectively our “transcriptional programs”. This was performed independently for STEM or k-means run with different parameters. Each cluster was named based on the cell types in which the expression of the genes in that cluster is the highest, therefore relating these clusters of expression profiles to meaningful biological programs.
To quantify the degree of similarity between HSC and progenitors populations of distinct lineages, we first calculated the median standard deviation of all DREGH genes among HSC biological replicates. We then considered that a particular gene was similarly expressed in HSC and a more differentiated population (MLP, GMP, MEP or ProB) if its expression in the latter was within a standard deviation from its value in HSC.
Pathway enrichment analyses
The likelihood of over-representation of Gene Ontology categories in particular gene-lists was estimated using the Explain software suite from Biobase, which is derived from a hypergeometric distribution.
Transcription factor architecture analyses
The list of annotated transcription factor or regulators of transcription was annotated by aggregating GO and KEGG categories containing “transcription factor” or “transcription regulator activity”, which yielded 1581 genes. 477 of them were differentially expressed at FDR<0.01 at fold-change >2 by one-way ANOVA analysis with Tukey HSD post-hoc test and Benjamini-Hochberg multiple testing correction (GeneSpringGX software; Agilent).
Assembly of a list of transcription factor with an early lymphoid-specific pattern of expression
Five differential expression lists from Supplementary Table 2 were selected to generate a scoring method for transcription factors with an early lymphoid pattern of expression: the MLP-specific gene set (#2), HSC_MLP co-expressed genes (#8), the HSC to MLP transition gene set (#14), the MLP to Pro-B transition gene set (#19) and the MLP and GMP comparison (#24). Each differential expression list was restricted to transcription factors which had FDR<0.05 and absolute fold-change >2, and was then ranked by fold-change. Rank positions scores (RPS) were assigned with the most upregulated transcription factor as +1 and the most downregulated as −1. For every list the number of significantly upregulated and downregulated transcription factors were respectively designated nUP and nDOWN. There were 452 unique transcription factors across these 5 lists. For each of these, an overall lymphoid score (λ) was calculated as the harmonic sum of renormalized ranks, as follows:
This strategy gives more weight to the most differentially expressed genes in any list. Transcription factors with negative overall lymphoid scores as well as transcription factors with higher expression in HSC relative to MLP were eliminated. As our initial transcription factor list contained many transcriptional regulators that are not transcription factor per se, we manually discarded the latter ones, which resulted in the 60 transcription factor listed in Fig. 3b. In this list, we found known regulators of B cell (EBF1, ID2, FOX and HOX family factors), T cell (NOTCH1, HEY1, HES4) and myeloid development (RUNX1). Factors highlighted in bold in Fig. 3b were selected for further functional validation, based on the fact that they were also predicted to bind to the promoters of genes either highly expressed in MLP (Fig. 3c) or dynamically regulated during B cell development (Fig. 3d).
Transcription factor binding motifs enrichment
Transcription factor binding motifs enrichment analysis was run independently on each of the 8 population-specific gene sets (Supplementary Table 2). Each of these was restricted to the differentially upregulated genes in a certain population (e.g. MLP) with FDR<0.05, log2 average expression >7 and a fold-change cut-off resulting sin gene lists ranging from 300 to 1000 genes (Supplementary Table 7). Explain3.0 software from Biobase was used to determine which transcription factors most likely control the genes in each dataset. The software makes use of 2 databases: TRANSFAC (experimentally-validated transcription factor binding sites and their target genes) and TRANSPRO (vertebrate promoter sequences annotated with their characteristics). Two independent algorithms, F-MATCH 42 and P-MATCH 43 were used to find transcription factor binding sites (TFBS) and compare the number of sites found in these query sequences against a background gene set. We chose a restricted version of the DREGH list (5658 genes) as a background set, as using an annotated list of housekeeping genes which promoters are GC rich leads to false positive over-representation of AT-rich matrices in all our population specific gene sets. Furthermore, we hypothesized that using a set of pan-hematopoietic genes as background would enhance detection of population-specific transcription factor families. Promoters were scanned for the presence of motifs in a window spanning 500 bp upstream and 100bp downstream of the transcription-start site (TSS). After search, matrices cut-offs and window positions were optimized. Only best supported promoters of the TRANSPro database were used. p-value cut-offs of 0.01 and 0.05 were used respectively for the F-MATCH and P-MATCH algorithms. Only transcription factor families found significantly enriched by both algorithms were retained. An enrichment score (ES) was calculated for each of these based on the geometric mean of the negative logarithms of the F-MATCH and P-MATCH values as follows:
Transcription factor-annotated regulatory event map along B cell commitment
An annotated pan-hematopoietic transcription factor-target interaction database was generated by searching for all known predicted and known transcription factor binding sites in the promoters of the DREGH gene list. Explain3.0 software was used to run the F-MATCH algorithm without any background set, using the whole collection (13957) of experimentally verified and predicted TFBS and positional weight matrices (PWM) present in the TRANSPRO database. Of the 9898 genes present in the DREGH list, 9523 had annotated promoters in the TRANSPRO database. We also incorporated transcription factor-target interactions derived from ChIP-on-chip datasets 44,45. This information was collapsed into a matrix containing 466453 transcription factor-target interactions. To simplify model building with DREM software (see below), the transcription factor-target interactions database was restricted to 255843 entries corresponding to the transcription factor families that were found to be enriched in the promoters of at least one of our population-specific gene sets.
Dynamic Regulatory Events Miner (DREM) software 23 was downloaded from http://gene.ml.cmu.edu/drem/ and was used to build a regulatory event map. The expression data input contained log2 transformed signal data from the DREGH list, restricted to HSC, MLP, Early B, ProB datasets in this order, which reflects progression along B cell commitment. All signals were normalized to the first time point, HSC. Genes were kept in the analysis even in the absence of transcription factor input data. The minimum absolute expression change between time points was set to 0.5. The model was built using the transcription factor-gene interaction data jointly with the expression data to produce a more biologically coherent model. A maximum of three paths out of a split event was enforced and no path merging was allowed. Transcription factors associated to nodes or splits with a score <0.01 based on the hypergeometric distribution were considered in the analysis. Of note, as the transcription factor data was used to learn the model, the score does not represent a true p-value but the lower the score the more significant the association. Gene Ontology annotation was run within DREM software with FDR<0.01 (with Bonferroni correction).
Lentiviral vector constructs and transduction
All shRNA sequences used in this study were derived from the TRC library (http://www.broadinstitute.org/rnai/public/), synthesized as 5′-P oligonucleotides and cloned into pLKO vectors 46 in which the puromycin resistance cassette was replaced by GFP. Hairpins were placed under control of the H1 or U6 promoter. Viral particles were produced as described 47 and titrated on 293T cells. For transduction Lin- CB cells were thawed, incubated for 3–5 hours in X-VIVO 10 medium (BioWhittaker, Waldersville, MD) supplemented with 1% BSA and the following cytokines (all from R&D Systems): SCF (100 ng/ml), FLT3L (100 ng/ml), TPO (50 ng/ml) and IL-7 (10 ng/ml). Cells were then incubated in the same medium supplemented with virus at a multiplicity of infection of 50–130 Transforming Units/mL for 16 (MS5-MBN assay) to 24 hours (in vivo assays).
MS5-MBN assay
4–5×105 MS5 stromal cells 24 were seeded in 96-well plates (Nunc) coated with 0.2% gelatin (SIGMA), in H5100 medium (Stem Cell Technologies) supplemented with Pen/Strep, L-Glutamine, and cytokines (all from R&D Systems): SCF (100 ng/ml), FLT3L (10 ng/ml), TPO (50 ng/ml), IL-2 (10 ng/ml), IL-7 (20 ng/ml), IL-6 (20 ng/ml), G-CSF (20 ng/ml) and GM-CSF(20 ng/ml). 48 hours later, and 12 hours after transduction, transduced Lin- cells were washed and 1 or 2 MLP cells were sorted onto stroma. Half of the medium was changed weekly and 3 weeks after seeding, colonies were harvested, resuspended by physical dissociation, filtered through 96-well filter plates (Pall life sciences, Ann Harbor, IL) and the whole content of each well was screened by flow cytometry. Myeloid cell colonies were defined as CD56− CD11b+ whether CD14+ (monocytic) or CD14− (non-monocytic). B cell colonies were identified as CD11b− CD19+, while NK cell colonies were CD11b− CD56+. As MLP were sorted at single cell level prior to GFP appearance, each plate contained both GFP+ and GFP− colonies, which can serve as an internal negative control. Colonies were retained for analysis if containing > 10 GFP+ (GFPpos colony) or >10 GFP− (GFPneg colony, untransduced) cells. For plates seeded with MLP incubated with control lentiviral vectors, the number of colonies containing >5 Myeloid, B and/or NK cells were counted independently for GFPpos and GFPneg colonies. For plates derived from MLP exposed to candidate lentiviral vectors, only GFPpos colonies were considered and the proportion of Myeloid, B or NK colonies formed in these plates was compared to that of control plates seeded with MLP derived from the same pool of CB (paired two-tailed t-test). A minimum of 20 GFPpos colonies per CB pool, and a minimum of 3 independent transductions and CB pools were screened.
In vivo experiments
All animal experiments were done in accordance to institutional guidelines approved by University Health Network Animal care committee. NSG mice (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ; Jackson Laboratory) were sublethally irradiated (250rad) 24 hours prior to intra-femoral injection. Lin- CB transduced with control or KD lentiviral constructs were harvested 72 hours post-transduction and stained for CD34 (CD34-APC, BD, 1:100). Live (SytoxBlue-, 1:104, Life Technologies) GFP+ CD34+ cells were sorted on a FACSAria flow-cytometer and 3×104–5×104 cells/mouse were injected intrafemorally. Mice were sacrificed 8–10 weeks post-transplantation, the injected femur and other bones were flushed separately in Iscove’s modified Dulbecco’s medium (IMDM) and cells were stained with the following antibodies (all from BD and dilution 1:100 unless otherwise specified): CD19 PE, GlyA PE(Beckman Coulter), CD45 PECy5 (Beckman Coulter), CD14 PECy7 (1:200, Beckman Coulter), CD33 APC, CD15 V450; or for assessment of all the steps of B cell differentiation with: CD19 PE, CD33 PECy5 (Beckman Coulter), CD34 PerCPE710 (eBiosciences), CD38 PECy7, CD10 APC, CD20 APCCy7, CD45RA BV (1:50, Biolegend).
Cell cycle and apoptosis assays
For the assessment of proliferation and survival in the MS5-MBN assay, 100 CD34+ CD38− or 50–100 CD34+ CD38− CD45RA+ cells were sorted into 96 well plates, coated with MS5 cells as described above, 12 hours after lentiviral vector transduction. After 7 days of culture, the cells were harvested and stained for the following cell surface markers (all antibodies from BD at 1:100 unless otherwise specified): CD45 PECy5 (Beckman Coulter), CD34 PECy7, CD133 APC (1:50), CD11b APCCy7. Cells were then fixed in Cytofix-Cytoperm (BD) for 15mn on ice, and stained with Ki67 PE (1:30, BD) and cPARP Alexa700(BD, 1:50), washed and then incubated with Hoechst 33342 (1:104, Life Technologies). For ex-vivo cell cycle analyses, cells were stained with CD10 PECy5 (1:50), CD19 PECy7, CD34 APC, CD33 Al700 (1:50), CD20 APCCy7 (all from BD and 1:100 unless otherwise specified). Cells were fixed as above and stained for Ki67 PE and Hoechst. For apoptosis detection ex vivo, Annexin V Apoptosis Detection Kit (BD PharMingen) and SYTOX Blue Dead Cell Stain (Life Technologies) were used according to manufacturer’s protocols, after cell surface staining with: CD33 PECy5 (Beckman Coulter), CD34 PECy5.5 (eBiosciences), CD19PECy7 (BD), CD10 APC (BD), CD20APCCy7 (BD) (all 1:100).
Quantitative RT-PCR
RNA was extracted from 2×104–5×105 cells in TRIzol (Life Technologies) supplemented with 25 ug linear polyacrilamyde (LPA, Life Technologies) according to the manufacturer’s protocol. cDNA was reverse-transcribed with the SuperScript VILO cDNA synthesis kit (Life Technologies) and purified with QIAquick PCR purification Kit (QIAGEN). Real-time PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) and 200 nM primers (Qiagen) on an Applied Biosystems 7900HT instrument. All primers used were Quantitect Primer Assays (QIAGEN) with the exception of ACTB (For: CCTGGCACCCAGCACAAT, Rev: GGGCCGGACTCGTCATAC). SDS software (Applied Biosystems) was used for absolute gene expression quantification using the standard curve method. Two house-keeping genes were used (ACTB and GAPDH) and data presented is relative to the geometric mean of expression of these 2 genes.
shRNAs used in this study
shBCL11A-a:CCGGTCGCACAGAACACTCATGGATTCTCGAGAATCCATGAGT GTTCTGTGCGTTTTTG, TRCN0000033449; shBCL6-a: CCGGTCCACAGTGACA AACCCTACAACTCGAGTTGTAGGGTTTGTCACTGTGGTTTTTG, TRCN0000013606; shEBF1-b: CCGGTCCCTCAGATCCAGTGATAATTCTCGAGAA TTATCACTGGATCTGAGGGTTTTTG, TRCN0000013829; shGATA2-b: CCGGTGT GCAAATTGTCAGACGACAACTCGAGTTGTCGTCTGACAATTTGCACTTTTTG, TRCN0000019264; shIRF8-a: CCGGTGCCTCACACCAGAGATCATTTCTCGAGAA ATGATCTCTGGTGTGAGGCTTTTTG, TRCN0000020988; shMAF-b: CCGGTATT TGCAGTCATGGAGAACCACTCGAGTGGTTCTCCATGACTGCAAATTTTTTG, TRCN000000254; shRUNX2-a: CCGGTCAGCACTCCATATCTCTACTACTCGAGT AGTAGAGATATGGAGTGCTGTTTTTG, TRCN0000013655; shSOX4-a: CCGGTC CTTTCTACTTGTCGCTAAATCTCGAGATTTAGCGACAAGTAGAAAGGTTTTTG, TRCN0000018213; shTEAD1-b: CCGGTCCAGAAGGAAATCTCGTGATTCTCG AGAATCACGAGATTTCCTTCTGGTTTTTGG, TRCN0000277979; shTSC22D1-a: CCGGTGCCTCTTTCTTCTCAAACAATCTCGAGATTGTTTGAGAAGAAAGAGGC TTTTTG, TRCN0000013288; shBCL11A-c: CCGGTGCTCAAGATGTGTGGC AGTTTCTCGAGAAACTGCCACACATCTTGAGCTTTTTG, TRCN0000033450; shTEAD1-a: CCGGTCCAGAAGGAAATCTCGTGATTCTCGAGAATCACGAGATT TCCTTCTGGTTTTTG, TRCN0000015799.
Supplementary Material
Acknowledgments
We thank the Trillium Hospital obstetrics unit for providing CB samples; P.A. Penttilä, A. Khandi, L. Jamieson and S. Zhao at the SickKids Flow Cytometry Facility for sorting; V. Voisin and G. Bader for bioinformatic advice and critical review of the manuscript; S. Bashir for statistical advice; A. Neumann for help with cloning and plasmids; M. Doedens for help with intrafemoral injections and O. Gan for critical review of the manuscript. E.L. was supported by the Swiss National Science Foundation, Roche and the FSBMB (Fondation Suisse pour les Bourses en Médecine et Biologie), S.Z. by VR (Swedish Research Council). This work was funded by Genome Canada through the Ontario Genomics Institute; Ontario Institute for Cancer Research and a Summit Award with funds from the province of Ontario; the Canadian Institutes for Health Research; a Canada Research Chair; and the Princess Margaret Hospital Foundation, the Terry Fox Research Institute, Canadian Cancer Society Research Institute and in part by the Ontario Ministry of Health and Long Term Care (OMOHLTC). The views expressed do not necessarily reflect those of the OMOHLTC.
Footnotes
AUTHORS CONTRIBUTIONS
E.L. designed and performed experiments, analyzed data and wrote the manuscript; S.D. designed and performed experiments, analyzed data and edited the manuscript; S.Z. designed and performed experiments; I.P. cloned lentiviral vectors plasmids and performed RT-PCR; J.C. and C.A. performed gene expression profiling experiments; J.B.F. supervised gene expression profile experiments, J.E.D. supervised the study and wrote the manuscript.
DATABASE ACCESSION NUMBERS
Raw data is available at GEO database (GSE42414), processed data on a dedicated website (http://jdstemcellresearch.ca/index_files/ResearchData.htm).
References
- 1.Orkin SH, Zon LI. Hematopoiesis: An Evolving Paradigm for Stem Cell Biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enver T, Pera M, Peterson C, Andrews PW. Stem Cell States, Fates, and the Rules of Attraction. Cell Stem Cell. 2009;4:387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Zandi S, Bryder D, Sigvardsson M. Load and lock: the molecular mechanisms of B-lymphocyte commitment. Immunol Rev. 2010;238:47–62. doi: 10.1111/j.1600-065X.2010.00950.x. [DOI] [PubMed] [Google Scholar]
- 4.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- 6.Ji H, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novershtern N, et al. Densely Interconnected Transcriptional Circuits Control Cell States in Human Hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers SM, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Månsson R, et al. Molecular Evidence for Hierarchical Transcriptional Lineage Priming in Fetal and Adult Stem Cells and Multipotent Progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Ng SYM, Yoshida T, Zhang J, Georgopoulos K. Genome-wide Lineage-Specific Transcriptional Networks Underscore Ikaros-Dependent Lymphoid Priming in Hematopoietic Stem Cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu M, et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 12.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 13.Doulatov S, et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 14.Luc S, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012 doi: 10.1038/ni.2255. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goardon N, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 16.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Kohn LA, et al. Lymphoid priming in human bone marrow begins before expression of CD10 with upregulation of L-selectin. Nat Immunol. 2012 doi: 10.1038/ni.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notta F, et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 19.Hao QL, et al. Human intrathymic lineage commitment is marked by differential CD7 expression: identification of CD7– lympho-myeloid thymic progenitors. Blood. 2008;111:1318–1326. doi: 10.1182/blood-2007-08-106294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10:120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, Iida R, Shimazu T, Kincade PW. Replenishing B lymphocytes in health and disease. Current Opinion in Immunology. 2012;24:196–203. doi: 10.1016/j.coi.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ernst J, Nau GJ, Bar-Joseph Z. Clustering short time series gene expression data. Bioinformatics. 2005;21(Suppl 1):i159–168. doi: 10.1093/bioinformatics/bti1022. [DOI] [PubMed] [Google Scholar]
- 23.Ernst J, Vainas O, Harbison CT, Simon I, Bar-Joseph Z. Reconstructing dynamic regulatory maps. Molecular Systems Biology. 2007;3 doi: 10.1038/msb4100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh K, et al. Reproducible establishment of hemopoietic supportive stromal cell lines from murine bone marrow. Exp Hematol. 1989;17:145–153. [PubMed] [Google Scholar]
- 25.Rossi MI, et al. Relatively normal human lymphopoiesis but rapid turnover of newly formed B cells in transplanted nonobese diabetic/SCID mice. J Immunol. 2001;167:3033–3042. doi: 10.4049/jimmunol.167.6.3033. [DOI] [PubMed] [Google Scholar]
- 26.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawamoto H, Ikawa T, Masuda K, Wada H, Katsura Y. A map for lineage restriction of progenitors during hematopoiesis: the essence of the myeloid-based model. Immunol Rev. 2010;238:23–36. doi: 10.1111/j.1600-065X.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic Transformations of Genome-wide Epigenetic Marking and Transcriptional Control Establish T Cell Identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos PM, Borghesi L. Molecular resolution of the B cell landscape. Current Opinion in Immunology. 2011;23:163–170. doi: 10.1016/j.coi.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 2012;247:172–183. doi: 10.1111/j.1600-065X.2012.01112.x. [DOI] [PubMed] [Google Scholar]
- 31.Duy C, et al. BCL6 is critical for the development of a diverse primary B cell repertoire. The Journal of Experimental Medicine. 2010;207:1209–1221. doi: 10.1084/jem.20091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schilham MW, et al. Defects in cardiac outranscription factorlow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 33.Liu P, et al. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 34.Yu Y, et al. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med. 2012 doi: 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nature Cell Biology. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansson L, Larsson J. Normal hematopoietic stem cell function in mice with enforced expression of the Hippo signaling effector YAP1. PLoS ONE. 2012;7:e32013. doi: 10.1371/journal.pone.0032013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eppert K, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nature Medicine. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan JB, et al. Highly Parallel Genome-Wide Expression Analysis of Single Mammalian Cells. PLoS ONE. 2012;7:e30794. doi: 10.1371/journal.pone.0030794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.April C, et al. Whole-genome gene expression profiling of formalin-fixed, paraffin-embedded tissue samples. PLoS ONE. 2009;4:e8162. doi: 10.1371/journal.pone.0008162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeung KY, Haynor DR, Ruzzo WL. Validating clustering for gene expression data. Bioinformatics. 2001;17:309–318. doi: 10.1093/bioinformatics/17.4.309. [DOI] [PubMed] [Google Scholar]
- 42.Kel AE, et al. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chekmenev DS, Haid C, Kel AE. P-Match: transcription factor binding site search by combining patterns and weight matrices. Nucleic Acids Res. 2005;33:W432–437. doi: 10.1093/nar/gki441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ci W, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113:5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basso K, et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115:975–984. doi: 10.1182/blood-2009-06-227017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 47.Mazurier F, Gan OI, McKenzie JL, Doedens M, Dick JE. Lentivector-mediated clonal tracking reveals intrinsic heterogeneity in the human hematopoietic stem cell compartment and culture-induced stem cell impairment. Blood. 2004;103:545–552. doi: 10.1182/blood-2003-05-1558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.