Abstract
Cellular toxicity introduced by protein misfolding threatens cell fitness and viability. Failure to eliminate these polypeptides is associated with numerous aggregation diseases. Several protein quality control mechanisms degrade non-native proteins by the ubiquitin proteasome system. Here, we use quantitative mass spectrometry to demonstrate that heat-shock triggers a large increase of ubiquitylation associated with misfolding of cytosolic proteins. We discover that the Hul5 HECT ubiquitin ligase participates in this heat-shock stress response. Hul5 is required to maintain cell fitness after heat-shock and to degrade short-lived misfolded proteins. In addition, localization of Hul5 in the cytoplasm is important for its quality control function. We identify potential Hul5 substrates in heat-shock and physiological conditions to reveal that Hul5 is required for ubiquitylation of low solubility cytosolic proteins including the Pin3 prion-like protein. These findings indicate that Hul5 is involved in a cytosolic protein quality control pathway that targets misfolded proteins for degradation.
Keywords: Ubiquitin, Protein Quality Control, HECT Ligase, Proteasome, Protein Homeostasis, Proteolysis, Misfolding, Heat-Shock, Proteotoxic Stress, Cytosol, HUL5, SSA1, PIN3, Mass Spectrometry, Proteomics
Introduction
Protein quality control degradation pathways eliminate misfolded and damaged proteins to prevent their cytotoxic accumulation and aggregation in the cell1, 2. Failure to degrade these non-native proteins is associated with numerous diseases, especially age-related neurodegenerative pathologies such as Parkinson’s and Huntington’s. Modulating proteostasis may help treat these diverse proteopathies, therefore there is an increasing need for deeper insight into protein quality control mechanisms3.
In eukaryotes, the ubiquitin proteasome system plays a major role in the degradation of misfolded proteins4, 5. Targeted proteins are specifically modified by ubiquitin ligases, which covalently attach a poly-ubiquitin chain to mediate substrate recognition and degradation by the proteasome6. Selective recognition of aberrant polypeptidesis cardinal to avoid degradation of proteins that can refold into their native state. Several compartmentalized protein quality control pathways target a variety of substrates. For instance, ER-localized misfolded proteins are targeted by the ER-associated protein degradation (ERAD) pathway7. In this pathway, non-native proteins are recognized, re-translocated to the cytoplasm, and ubiquitylated by the conserved Doa10 and Hrd1 ubiquitin ligases for proteasome degradation8, 9. Correspondingly, misfolded nuclear proteins in yeast bind the San1 ubiquitin ligase via their exposed hydrophobic regions and are subsequently directly targeted for proteolysis10–12.
It is becoming apparent that several quality control pathways target misfolded proteins in the cytoplasm. The CHIP (Carboxyl Terminus of Hsp70-interacting protein) ubiquitin ligase plays a major role in eliminating cytosolic misfolded proteins in metazoans13. Other ubiquitin ligases, such as Ubr1, target cytosolic misfolded in proteins yeast14–16. Moreover, the Ltn1/Rkr1 ligase associates with ribosomes to specifically target nascent non-stop polypeptides for degradation17. These distinct and specialized quality control pathways likely complement each other to eliminate the wide spectrum of aberrant cytosolic polypeptides.
Here, we observe that heat-shock triggers a large increase of ubiquitylation linked to misfolding of cytosolic proteins in yeast cells. We discover that Hul5 (HECT Ubiquitin Ligase 5) plays a major role in this stress response, while none of the other known ubiquitin ligases functioning in protein quality control are involved. Cytosolic Hul5 is important for cell fitness and increased ubiquitylation levels after heat-shock. We identify several potential physiological substrates to show that Hul5 is also important for the ubiquitylation and degradation of low solubility cytosolic proteins in unstressed cells. Together, these findings indicate that Hul5 is a component of a new cytosolic protein quality control pathway.
Results
Heat-induced misfolding triggers a large increase of ubiquitylation levels
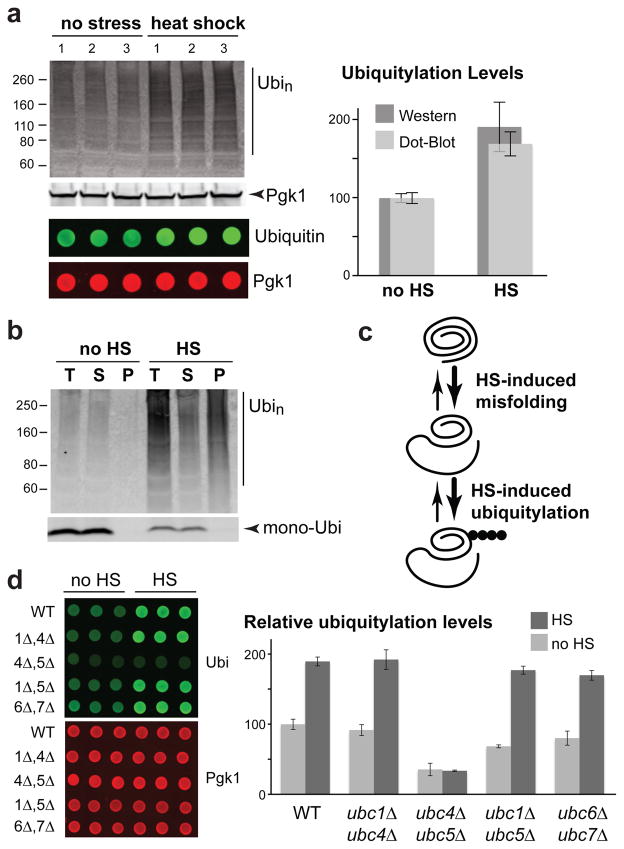
To gain holistic insight into the targeting of misfolded proteins by the ubiquitin proteasome system, we sought to identify a cellular stress that induces a strong ubiquitylation response. We found a near two-fold increase in poly-ubiquitylation in Saccharomyces cerevisiae cells after a 15 minute 45°C heat-shock treatment, as determined by both Western and dot blots (Figure 1a). The heat-shock ubiquitylation response was much stronger compared to other tested stresses (Figure S1a). We ruled out that this phenomenon was caused by a reduction in proteasome activity at high temperature, as a stronger poly-ubiquitylation increase was observed in cells treated with a proteasome inhibitor (Figure S1b, c). Notably, most of these heat-shock induced poly-ubiquitylated species were poorly soluble (Figure 1b). These data indicate that heat-induced protein misfolding causes a rapid and strong increase of poly-ubiquitylation mediated by a protein quality control pathway (Figure 1c).
Figure 1.
Heat-shock stress induces protein misfolding and poly-ubiquitylation. (a) BY4741 cells were subjected to heat-shock (HS; 15 min at 45°C) or not (noHS). Experimental triplicates were analyzed by Western (top) and dot blots (bottom) with anti-ubiquitin and anti-Pgk1 antibodies (left). The region above 70 kDa in the Western blot and the whole spotted signal in the dot blot were quantified (right). Ubiquitylation signals were normalized to Pgk1 levels and standard deviations are shown. (b) Ubiquitylation levels in total cell extract, soluble and pellet fractions after 16,000 g centrifugation from both unstressed and heat-shock treated (15 min 45°C) BY4741 cells are shown by anti-ubiquitin Western blot. (c) Schematic diagram of the proposed relationship between heat-shock and the increased ubiquitylation response. (d) Relative increase of Pgk1-normalized ubiquitylation levels after a 15 min 45°C heat-shock in the indicated E2 double deletion strains were quantified by dot blot and averaged values from three replicates are shown with standard deviations.
We further characterized this stress response by determining which ubiquitin conjugating enzyme (E2) is involved, since several E2s participate in protein quality control in different cell compartments11, 18–20. We found that deletion of both UBC4 and UBC5 fully abolished the heat-shock ubiquitylation response (Figure 1d), while single deletions of these two E2s had no effect (data not shown). Deletion of UBC6 and UBC7, which are E2s involved in ERAD18, 19, had no effect either (Figure 1d). These data suggest that heat-shock induces an increase in ubiquitylation of cytosolic proteins, as Ubc4 and Ubc5 target short-lived misfolded proteins in the cytoplasm20.
Heat-shock mainly induces ubiquitylation ofcytosolic proteins
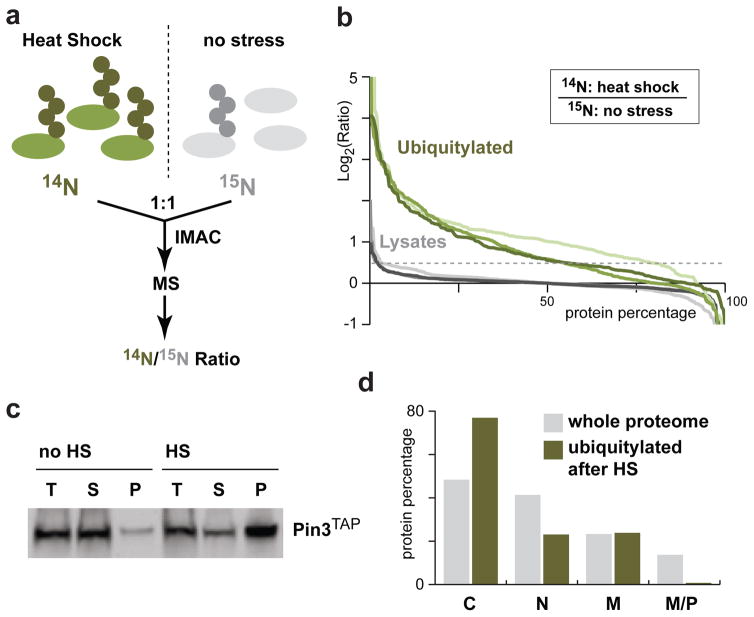
To determine whether heat-shock affects cytosolic proteins, we adapted a quantitative mass spectrometry method21 to identify proteins that were further ubiquitylated (Figure 2a). We used cells expressing an octahistidine tag fused to the N-terminus of ubiquitin (H8-Ubi) and performed immobilized metal ion affinity chromatography (IMAC) to purify tagged-ubiquitin conjugates from cells that were metabolically labeled with either 14N or 15N for quantitative analysis. We first confirmed that neither the labeling nor IMAC were introducing a bias, as most proteins were equally enriched in the two differentially labeled cell populations (Figure S2a; all MS data are reported in Table S1). To identify proteins ubiquitylated after heat-shock, we compared stressed cells (14N; 20 min at 45°C) with unstressed control cells (15N). Using this approach, we identified 155 proteins that were ubiquitylated or further ubiquitylated upon heat-shock treatment in at least two of three independent experiments (Figure 2b, S2b; Table S2). In contrast, the majority of the proteins in the total cell lysate were unaffected by heat-shock. By comparing H8-Ubi tagged (14N) and untagged (15N) cells, we verified that most of the quantified proteins were specifically enriched due to the ubiquitin tag (80%; Figure S2c) and were more ubiquitylated after heat-shock (Figure S2d). Representative quantified peptides from two ubiquitylated proteins further enriched in heat-shock treated cells are shown (Figure S2e). Rps7B is a ribosomal protein that is targeted to the proteasome in unstressed cells21. Pin3/Lsb2 is a prion-like protein that contains short poly-glutamine stretches22, 23, and is further ubiquitylated upon heat-stress24. We confirmed that endogenous TAP-tagged Pin3 was noticeably less soluble after heat-shock suggesting that the increase in ubiquitylation is due to augmentation of protein misfolding (Figure 2c). Nearly 80% of the heat-shock affected proteins are cytosolic, according to data from a yeast genome-wide GFP analysis25. The enrichment of cytosolic proteins compared to the localization profile of the proteome (Figure 2d) indicates that heat-stress causes the rapid ubiquitylation of mostly cytosolic proteins, while not predominantly affecting proteins in other compartments. This result suggests that an efficient protein quality control machinery exists that targets misfolded cytosolic proteins after heat-shock.
Figure 2.
Heat-shock mainly affects cytosolic proteins. (a) A schematic diagram of the workflow of the quantitative mass spectrometry analysis. (b) Percentage of proteins above the corresponding log2 values of the 14N heat-shock/15N no stress ratios for three independent experiments (I: light; II: medium; and III: dark). Analysis of proteins in the total cell lysate (grey: I, 486; II, 730; and III, 399) and of IMAC enriched ubiquitylated proteins (green: I, 302; II, 481; and III, 219) are shown. Proteins with a log2 ratio ≥ 0.5 are considered heat-shock affected. (c) Pin3TAP solubility was assessed prior to and after a 15 min 45°C heat-shock. An equal portion of each fraction (T: total; S: supernatant; P: pellet) was loaded on a SDS-PAGE for Western blot analysis with the anti-TAP antibody. (d) Subcellular localization of 155 proteins affected by heat-shock (green; identified as enriched in at least two of the three experiments in b) compared to the whole proteome (grey) - cytosol (C), nucleus (N), membrane (M) and mitochondria or peroxisome (M/P). Note that several proteins localize to more than one compartment.
Hul5 plays a major rolein the heat -shock ubiquitylation response
To better understand the targeting mechanism of these misfolded proteins, we next identified which ubiquitin ligase(s) may be involved in this heat-shock response. Using the dot blot heat-shock assay, we assessed 82 yeast strains that each carried a deletion of a verified or putative ubiquitin ligase gene (Table S3). Surprisingly, none of the known or suspected ubiquitin ligases involved in protein quality control8–11, 14–17, 26 showed any significant perturbation of the heat-shock ubiquitylation response (Figure S3a).
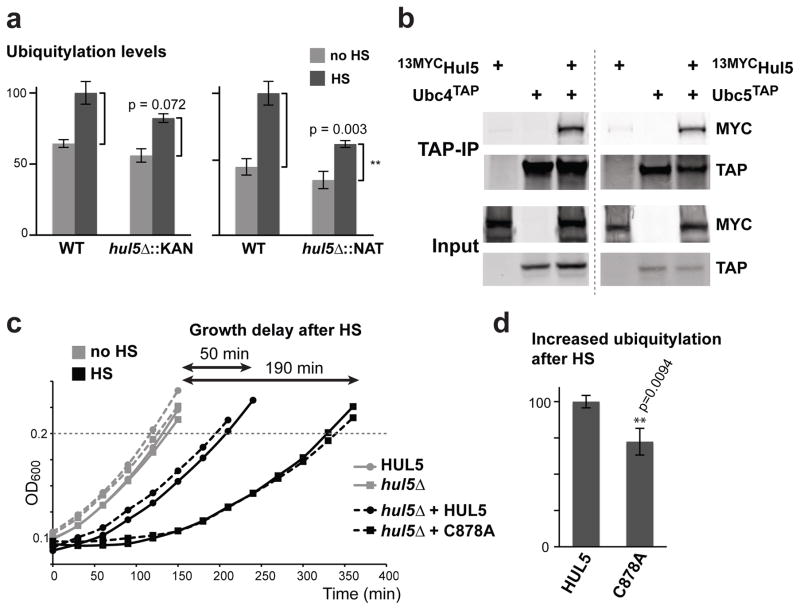
We found that deletion of HUL5, a gene encoding for a HECT (Homologous to E6AP C Terminus) ubiquitin ligase27, led to a significant reduction of the heat-shock ubiquitylation response (Figure S3a). Hul5 associates with the proteasome, and has been implicated in processing ERAD substrates and promoting proteasomal processivity28–31. We confirmed that the heat-induced ubiquitylation was reduced in two independent hul5Δ deletion strains using the dot blot assay (Figure 3a). In agreement with their role in the heat-shock ubiquitylation response, we found that Ubc4TAP and Ubc5TAP interact with 13MYCHul5 (Figure 3b, S3b). We showed that deletion of HUL5 led to a reduction of the heat-shock ubiquitylation response in a time-course between 5 to 30 minutes (Figure S3c). This data indicates that HUL5 is required at the early stage of the heat-stress response. Deletion of HUL5 resulted in a 20–50% reduction of the ubiquitylation response, suggesting that Hul5 is a major ubiquitin ligase involved in targeting misfolded proteins after heat-shock.
Figure 3.
HUL5 is required for the full ubiquitylation response and cell fitness after heat-shock. (a) Ubiquitylation levels in unstressed (no HS) and heat-shocked (HS) cells are compared between wild-type and hul5Δ strains using the dot blot assay. HUL5 was deleted by two different cassettes (KanMX6 and NatMX4). Standard deviation is shown for three replicates and p values were calculated using an unpaired Student’s t-test. (b) TAP pull-down experiments with cells expressing or not Ubc4TAP (left panel) or Ubc5TAP (right panel) at endogenous levels with or without a plasmid expressing 13MycHul5 were analyzed by Western blot using 9E10 or anti-TAP antibodies. Inputs (1%) are shown below. (c) The OD600 of the first cell doubling following a 20 min incubation at 45°C heat-shock (HS; black) or at 25°C (no HS; grey) of HUL5 (round) and hul5Δ::NAT (square) cells. hul5Δ::NAT cells carrying a centromeric plasmid (dashed lines) with HUL5 (round) or hul5-C878A (square) are also compared. Growth delay is defined by the difference of the first doubling time between the corresponding unstressed and stressed cells. Each data point is averaged from three replicates. (d) hul5Δ cells carrying either wild-type HUL5 or the catalytically inactive Hul5 (C878A) were subjected to heat-shock (15 min at 42°C). The increase in ubiquitylation levels (with standard deviations) was measured by dot blots with anti-ubiquitin and anti-Pgk1 antibodies in three replicates. An unpaired Student’s t-test was used to assess the significance of the difference between both cell populations.
While heat-shock treatment did not cause a significant loss of viability in hul5Δ cells (Figure S3d), we observed a growth delay upon recovery from the stress. To quantify this effect, we measured the growth rate of wild-type and hul5Δ cells following heat-shock treatment during the exponential growth phase. In wild-type cells, a 20-minute heat-shock at 45°C induced a 50-minute delay of the first cell doubling after incubating the cells back to 25°C (Figure 3c, S3e). In contrast, hul5Δ cells displayed a much longer growth delay of 190 minutes after heat-shock, while unstressed hul5Δ cells grew normally. Similar results were obtained with an independent hul5Δ strain (data not shown). The delay in hul5Δ cells was rescued to the normal growth rate when a wild-type copy of HUL5 expressed from its own promoter was introduced (Figure 3c, S3e). Conversely, addition of HUL5 mutated on a conserved catalytic cysteine residue of its HECT ligase domain (C878A) did not rescue the heat-shock induced growth delay of hul5Δ cells (Figure 3c, S3e). Correspondingly, cells carrying catalytically inactive Hul5 displayed a decreased heat-shock ubiquitylation response (Figure 3d). Together these results indicate that Hul5 ligase activity is required for ubiquitylation of misfolded proteins and maintenance of cell fitness after heat-shock treatment.
The cytosolic localization of Hul5 is required for its role in the heat-shock ubiquitylation response
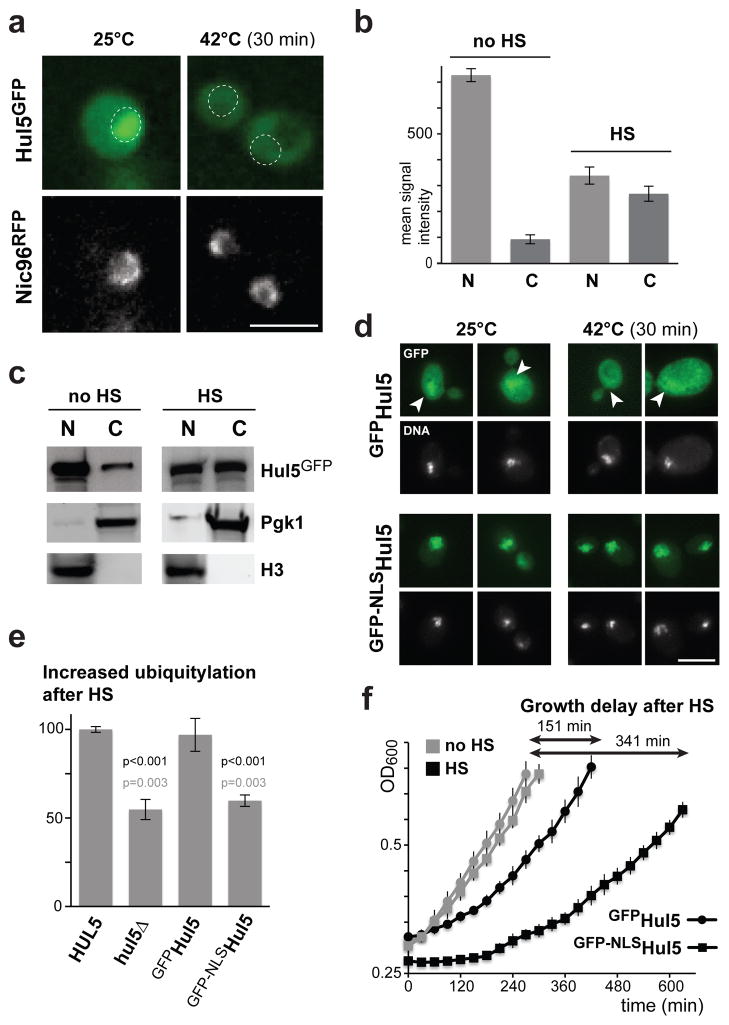
We observed by fluorescence microscopy that, while the localization of GFP-tagged Hul5 prevailed in the nucleus of unstressed cells, the pool of cytosolic Hul5 increased after heat-shock (Figure 4a, b & S4a). The change of Hul5 distribution after heat-stress was verified by subcellular fractionation of the nuclei and cytosol (Figure 4c). To determine the importance of Hul5 cytosolic localization, we added the SV40 nuclear localization signal (NLS) to a GFP-Hul5 fusion protein. We found that the NLS led to a further enrichment of Hul5 in the nucleus of unstressed cells, and blocked Hul5 relocalization to the cytoplasm after heat-shock (Figure 4d). As well, the presence of the NLS led to a reduced heat-induced ubiquitylation response and a slower growth recovery after heat-shock (Figure 4e, f, S4b). These data suggest that ubiquitylation of misfolded proteins after heat-shock requireslocalization of Hul5 tothe cytoplasm.
Figure 4.
Hul5 redistribution to the cytoplasm is important for its role in the heat-shock response. (a) Localization of Hul5GFP was assessed in a strain that carries Nic96mRFP as a nuclear periphery marker, in both unstressed and heat-shocked (30 min at 42°C) cells. Images were taken with a 40X objective and in focus z-stacks were flattened with the Wavelet extended function. The nuclear positions are marked by circles and the scale bar represents 5 μM. (b) Histograms of the mean Hul5GFP signal intensities (per pixel; with standard error) measured in a 0.7 μm2 area of the nuclei (N) and cytoplasm (C) of unstressed (25°C) and heat-shocked (30 min at 42°C) cells (n=100), after subtraction of the average background signal from untagged cells. (c) Levels of Hul5GFP in both nuclear (N) and cytosolic (C) fractions were analyzed by Western blot using anti-GFP, anti-Pgk1 and anti-Histone H3 antibodies after sub-fractionation of both unstressed (no HS) and heat-shocked (HS; 30 min at 42°C) cells. (d) The localization of GFPHul5 (upper panels) and GFP-NLSHul5 (lower panels) expressed from a plasmid was assessed in hul5Δ cells grown at 25°C and subjected to heat-shock (30 min at 42°C). DNA was stained with Hoechst (shown below GFP images), single-stack images were taken with a 63X objective, arrowheads point to the nucleus and scale bar is 5 μM. (e) The increase of ubiquitylation levels after heat-shock (15 min at 45°C) is compared using the dot blot assay between HUL5 and hul5Δ cells with an empty vector, and hul5Δ cells expressing GFPHul5 or GFP-NLSHul5 from a plasmid. Standard deviation is shown for three replicates and p values were calculated using an unpaired Student’s t-test (in black or in grey when comparing to HUL5 or GFPHul5 expressing cells, respectively). (f) OD600 following a 20 min heat-shock at 45°C (HS; black) or 20 min at 25°C (no HS; grey) of hul5Δ cells carrying a plasmid with GFPHul5 (round) or GFP-NLSHul5 (square) are compared. Each data point is averaged (with standard deviations) from three replicates.
Hul5 is required for the increased ubiquitylation of misfolded proteins in the absence of SSA-chaperone activity
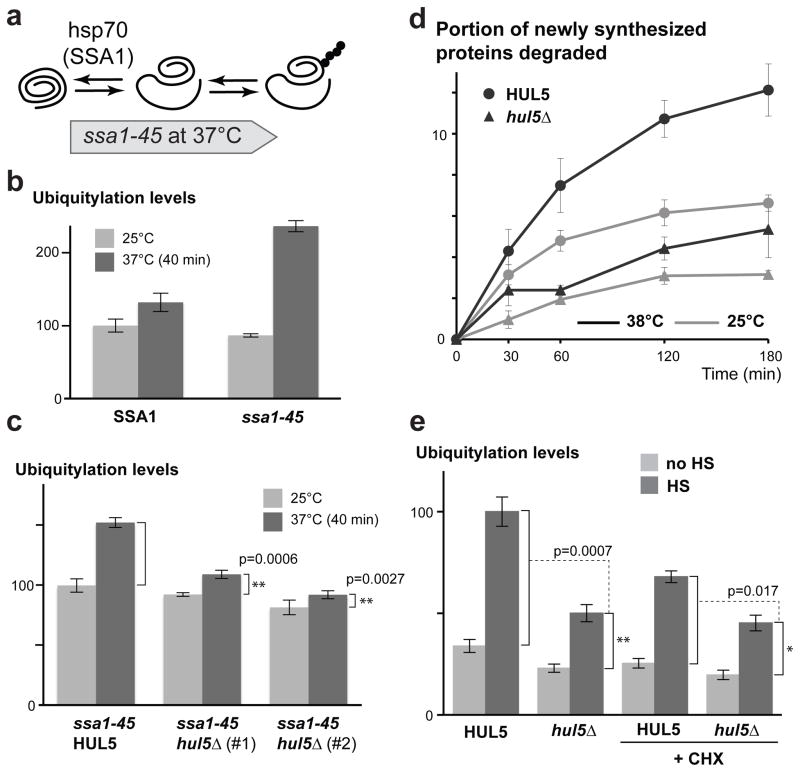
To further characterize HUL5 function in protein homeostasis, we assessed its role in the ubiquitylation of proteins misfolded in absence of SSA (Stress Seventy sub-family A) chaperone activity. The Hsp70 sub-family, which is composed of four members (Ssa1-4), is a major folding system in the cytosol32, 33. We reasoned that inactivation of SSA-chaperone activity should be followed by increased ubiquitylation of misfolded polypeptides (Figure 5a). Indeed, we observed a two-fold increase of ubiquitylation levels in thermo-sensitive cells carrying the ssa1-45 mutant allele, in which the three SSA2, 3 and 4 genes are deleted, after a shift to the non-permissive temperature (37°C) for 40 minutes (Figure 5b). In contrast, there was only a minor ubiquitylation increase in the control cells (ssa2Δ, 3Δ, 4Δ) expressing wild-type SSA1. We next assessed whether HUL5 is important for the ubiquitylation response caused by SSA-chaperone inactivation. Remarkably, we saw a dramatic reduction of ubiquitylation in two independent hul5Δ strains carrying the ssa1-45 allele (Figure 5c). These data indicate that HUL5 is a major player in targeting misfolded cytosolic proteins in an SSA-chaperone independent pathway (since the ubiquitylation increase occurs in the absence of SSA activity).
Figure 5.
HUL5 is essential for the ubiquitylation of proteins misfolded in absence of SSA chaperone activity and for the degradation of pulse-labeled misfolded polypeptides. (a) Schematic diagram of the proposed model for the SSA hsp70 chaperone inactivation. (b) The bar graph shows Cdc28-normalized ubiquitylation levels in SSA1 and ssa1-45 cells (ssa2-4Δ) measured before and after shifting cells from 25°C to 37°C for 40 min. The ubiquitylation levels were quantified by dot blot assay from four replicates and shown with standard errors. (c) The increased ubiquitylation levels in ssa1-45 HUL5 or ssa1-45 hul5Δ cells (two independent strains) were compared using five replicates for each assessed strain as described for (b). The p values were determined by Student’s t-test. (d) Wild-type (BY4741) and hul5Δ cells were subjected to 35S-pulse labeling (5 min) followed by a 3 h chase. The graph shows the averaged percentage (with standard errors) of pulse-labeled proteins that were degraded at each time point from cells incubated at 25°C or 38°C from three replicates. (e) Histograms of averaged ubiquitylation signals measured by dot blots in unstressed (no HS; light grey) and heat-shocked (HS; dark grey) cells in three replicates are shown with standard deviations. Cells were grown at 25°C and pre-treated or not for 15 min with 100 μg/ml cycloheximide (CHX) prior to heat-shock (15 min at 45°C). Student’s t-test was used to assess the significance of the ubiquitylation level differences between the indicated strains.
Hul5 targets misfolded proteinsfor degradation
We next determined whether HUL5 is important for the degradation of misfolded ubiquitylated polypeptides. It was previously shown that newly synthesized proteins have a higher turnover rate at high temperature due to increased misfolding34. We therefore performed pulse-chase labeling to determine whether HUL5 is important for the short half-life of these misfolded proteins. As reported, shifting cells from 25°C to 38°C led to a higher proteolysis rate of 35S pulse-labeled proteins in wild-type cells (Figure 5d). Conversely, this increase of proteolysis was strongly reduced in hul5Δ cells, indicating that HUL5 plays a major role in the degradation of short-lived misfolded proteins. To further examine whether Hul5 solely targets newly synthesized proteins, we repeated the heat-shock induced ubiquitylation assay in the absence of protein synthesis. Pre-treatment with the translational elongation inhibitor cycloheximide led to a lower heat-induced ubiquitylation response that was further reduced in the absence of HUL5 (Figure 5e). These results indicate that Hul5 likely targets both newly synthesized and long-lived misfolded proteins. Remarkably, degradation of short-lived proteins was also affected at ambient temperature in hul5Δ cells (Figure 5d). A substantial portion of short-lived proteins is likely degraded due to misfolding in physiological conditions and Hul5 may play an important role in their proteolysis andin protein homeostasis in general.
Hul5 is required for the ubiquitylation of low solubility cytosolic proteins in physiological conditions
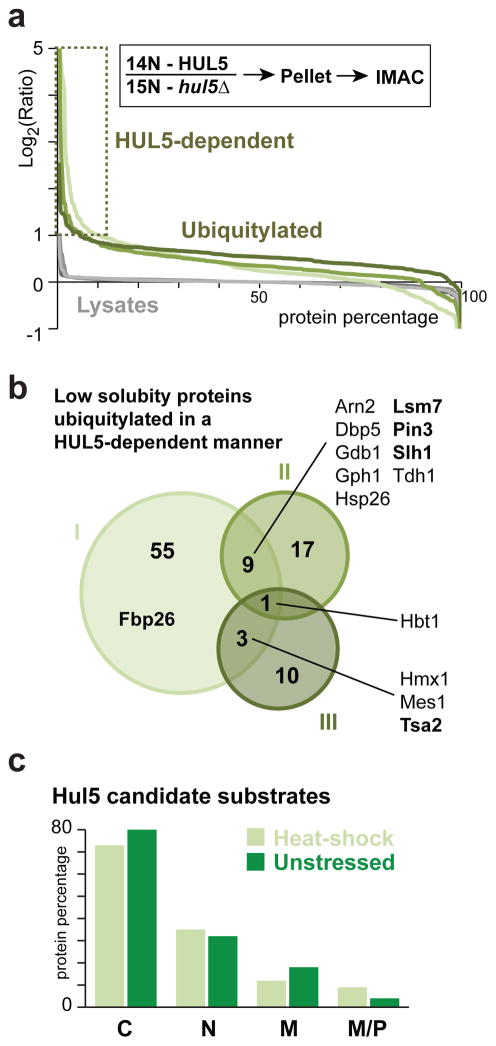
We next identified misfolded proteins that are ubiquitylated by Hul5 in physiological conditions. We reasoned that Hul5 substrates should be more ubiquitylated in wild-type cells compared to hul5Δ cells. We also supposed that misfolded or aggregated proteins should be enriched in the cell pellet due to their lower solubility. We therefore compared metabolically labeled wild-type (14N) and hul5Δ (15N) cells expressing H8-Ubi grown at 25°C and not subjected to heat-shock stress, then lysed in a native buffer prior to fractionation by centrifugation. Proteins in the cell pellet were next resolubilized in a denaturing buffer for IMAC followed by mass spectrometry analysis (Figure S5a). We identified several HUL5-dependent ubiquitylated proteins (Figure 6a). In contrast, we found that deletion of HUL5 caused no significant perturbation of the ubiquitylation profile when isolating conjugated proteins from the whole cell lysate (Figure S5b). These results indicate that, in physiological conditions, Hul5 specifically affects ubiquitylation of low solubility proteins, but not the overall ubiquitylation levels in the cell. In three independent experiments, we identified a total of 95 candidate proteins that were less ubiquitylated in hul5Δ cells, including thirteen proteins that were identified in at least two analyses (Figure 6b; Table S2). Noticeably, a small pool of Hul5 candidate physiological substrates, including Pin3, was also identified in the original heat-shock analysis (Figure S5c). We confirmed that HUL5 plays an important role in targeting misfolded proteins after heat-shock (20 min, 45°C), as approximately 20% of the ubiquitylated proteins quantified by mass spectrometry were less abundant in hul5Δ cells (Figure S5d). As well, only a small portion of Hul5 candidate substrates was identified in both stressed and unstressed conditions (Figure S5e). This data suggests that predominantly different proteins are ubiquitylated due to misfolding by Hul5 in physiological and heat-shock stress conditions. Notably, the majority of proteins requiring HUL5 for ubiquitylation in unstressed or stressed cells were cytosolic (Figure 6c), further emphasizing the importance of Hul5 in targeting cytosolic misfolded proteins.
Figure 6.
HUL5 is required for ubiquitylation of low solubility cytosolic proteins. (a) 14N and 15N metabolic labeling was performed for wild-type and hul5Δ cells, respectively. Percentage of proteins above the corresponding log2 values of the 14N/15N ratios in three independent experiments (I: light; II: medium; and III: dark). Analysis of proteins in the total cell lysate (grey: I, 345; II, 346; and III, 240) and of IMAC enriched ubiquitylated proteins (green: I, 661; II, 430; and III, 267) are shown. Proteins with a IMAC-log2 ratio ≥ 1 are considered Hul5 candidate substrates. (b) Venn diagram representing all 95 proteins identified as more ubiquitylated in experiments I to III in a. Bold protein names are confirmed Hul5 substrates in Figure 7, S6. (c) Histogram showing the subcellular localization of the 99 (light green) and 95 (dark green) Hul5 candidate substrate proteins identified in heat-shock stressed and unstressed cells, respectively: cytosol (C), nucleus (N), membrane (M) and mitochondria or peroxisome (M/P).
Ubiquitylated Hul5 substrates accumulatewith less soluble polypeptides
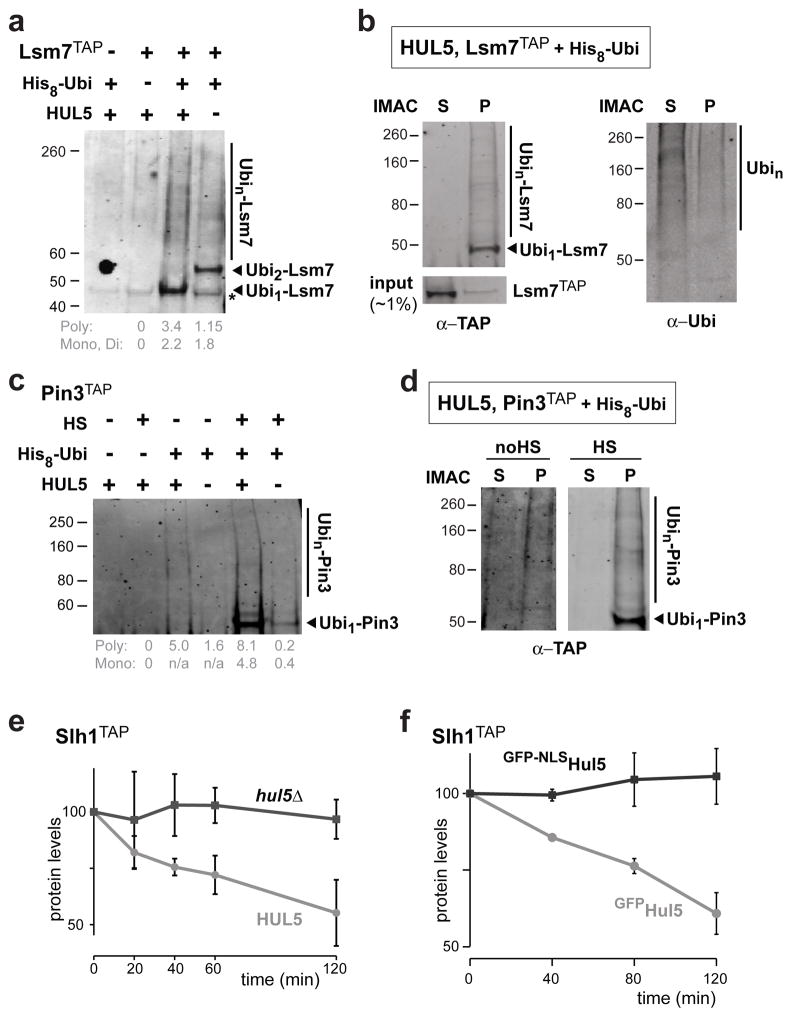
To verify the data, we selected a few identified proteins for additional analysis. We first focused on Lsm7, a small protein involved in cytosolic RNA decay35, which was identified as a Hul5 candidate substrate in unstressed cells. For validation, we subjected cells that expressed H8-Ubi and endogenous C-terminally TAP-tagged Lsm7 to IMAC, followed by an anti-TAP Western blot. High-molecular weight species corresponding to poly-ubiquitylated Lsm7TAP (Ubin-Lsm7) were detected at greater levels in wild-type cells compared to hul5Δ cells, confirming the data obtained by mass spectrometry (Figure 7a). Remarkably, ubiquitylated Lsm7TAP species (after IMAC) were only found in the low solubility fraction, while the majority of unmodified Lsm7TAP and most ubiquitylated proteins were soluble in the cell (Figure 7b). This result indicates that ubiquitylation of Lsm7 is associated to less soluble polypeptides. We also found that poly-ubiquitylation of TAP-tagged Pin3 required HUL5 (Figure 7c). We confirmed that these ubiquitylated species were only present in the pellet fraction (Figure 7d). Since Pin3 was less soluble and further ubiquitylated in HUL5-dependent manner after heat-shock (Figure 2c, S5d), we repeated the IMAC after shifting the cells to 45°C for 20 minutes. Indeed, Pin3TAP was further ubiquitylated after heat-shock and this increase was mostly abrogated in hul5Δ cells (Figure 7c). We also verified that HUL5 is required for the ubiquitylation of two other Hul5 substrate candidates: Tsa2 (thioredoxin peroxidase) and Fbp6 (fructose-2,6-bisphosphatase) (Figure S6a, b). These four TAP-tagged proteins have long half-lives in our growth conditions (data not shown), suggesting that only a small portion of these proteins are misfolded and degraded. In contrast, levels of Slh1TAP, another Hul5 candidate substrate both in physiological and heat-shock conditions, were found to decrease after a two-hour cycloheximide treatment (Figure 7e) in a proteasome dependent manner (data not shown). Slh1 is a putative RNA helicase that inhibits translation of non-poly(A) mRNA in the cytoplasm36. We confirmed that ubiquitylation levels of Slh1TAP were reduced by HUL5 deletion (Figure S6c) and only low solubility Slh1TAP was ubiquitylated (Figure S6d). As Slh1TAP ubiquitylation levels increase after heat-shock (Figure S5d) and its protein half-life decreases at higher temperature (Figure S6e), we reasoned that Slh1 turnover could be associated with misfolding. In agreement, we observed that deletion of HUL5 stabilized Slh1TAP (Figure 7e). As well, Slh1 was stable when Hul5 localization was constrained to the nucleus upon addition of the SV40 NLS (Figure 7f), further indicating that Hul5 targets substrates in the cytoplasm. All together, these results confirm that Hul5 is important for the targeting of low solubility cytosolic proteins.
Figure 7.
Hul5 targets proteins that are specifically ubiquitylated in the low solubility cellular fraction. (a–d) Validation of the Hul5 substrate candidates Lsm7 and Pin3 using TAP-tagged strains expressing H8-Ubi. IMAC was performed in denaturating conditions to pull down ubiquitylated proteins, and anti-TAP or anti-ubiquitin antibody was used for Western blot analysis. The asterisk denotes unspecific signal. Corresponding signal intensities for poly- and mono-ubiquitin were measured by subtracting the background signal in control cells (a, c). Solubility of ubiquitylated Lsm7TAP (b) and Pin3TAP (d) were assessed by comparing both soluble and pellet fractions subjected to IMAC and analyzed by Western blots with anti-TAP (b, d) and anti-ubiquitin (b). A 20 min 45°C heat-shock (HS) was also applied to Pin3TAP expressing cells (c, d). (e–f) Turnover of Slh1 is dependent on cytosolic Hul5. Protein levels of Slh1TAP were monitored by Western blot after the addition of 100 μg/ml cycloheximide to both HUL5 and hul5Δ cells grown at 25°C (e), and to hul5Δ cells expressing either GFPHul5 or GFP-NLSHul5 and shifted to 38°C (f). Relative averaged signal intensities (with standard deviations) were quantified and normalized to Pgk1 levels in three independent experiments.
Discussion
Previous studies on yeast cytosolic protein quality control were mainly based on “model” or “bait” substrates14, 16, 17. Our approach, instead, primarily focused on global misfolding responses in the cell. We identified HUL5 as major player in the ubiquitylation of cytosolic misfolded proteins under physiological conditions and in response to stresses that induce misfolding (heat-shock, SSA-inactivation). Moreover, the Hul5 catalytic HECT domain and its cytosolic localization are important for cell fitness following heat-shock stress. The absence of Hul5 also strongly decreases the degradation rate of short-lived misfolded proteins. We found that Hul5 substrates are mainly cytosolic and these ubiquitylated species are specifically associated with the low solubility cellular fraction. Overall, our data show that Hul5 participates in a quality control degradation pathway targeting misfolded cytosolic proteins.
Deletion of other ubiquitin ligases involved in cytosolic quality control pathways, such as Ubr1 and Ltn1, show no significant defect in the heat-shock ubiquitylation response, suggesting that these ligases are not involved in this pathway. The Hul5 quality control machinery is also possibly distinct to the previously identified pathway responsible for the degradation of the misfolded VHL (Von Hippel-Lindau) model substrate. Turnover of VHL requires SSA-chaperone activity as well as the Sse1 and Sti1 co-factors37. Similarly, deletion of SSE1 or STI1 did not affect the increase of ubiquitylation levels after heat-shock (data not shown). All together, these results further indicate that there are several distinct cytosolic protein quality control pathways in the cell.
Hul5 binds to the proteasome and has been proposed to antagonize the Ubp6 de-ubiquitylating enzyme29, 31. Interestingly, chemical inhibition of Usp14, the mammalian Ubp6 orthologue, was recently found to enhance proteasome function and to accelerate the degradation of proteins involved in proteotoxic stress such as ATXN338. Hence, Hul5’s possible novel role in the ubiquitylation of misfolded proteins and its Ubp6-antagonistic activity are consistent. It will be interesting to determine whether Hul5 remains associated to the proteasome during the ubiquitylation of low solubility proteins.
In unstressed cells, the majority of Hul5 localizes to the nucleus. Strikingly, only about 5% of Hul5 candidate substrates are solely nuclear (most nuclear proteins affected by HUL5 deletion are also cytosolic)25. Why then does Hul5 not target more nuclear proteins? One possibility is that Hul5 requires another cofactor in the cytoplasm. The low levels of cytosolic Hul5 may be sufficient for targeting a basal level of misfolded proteins in the cytoplasm of unstressed cells. In agreement, turnover of Slh1 is abrogated when GFP-NLSHul5 is sequestered to the nucleus, and blocking Hul5 cytosolic localization mimics the hul5Δ phenotype after heat-shock. Our results indicate that Hul5 ubiquitylates low solubility cytosolic substrates within the cytoplasm.
Intriguingly, most Hul5-candidate substrates were mono-ubiquitylated and the mono-ubiquitylation levels were not affected by HUL5 deletion to the same extent than the poly-ubiquitylation signal (Figure 7a–d, S6a, b). These results suggest that Hul5 may, in agreement with previous work29, act as an E4 ubiquitin ligase to further ubiquitylate misfolded proteins targeted by another E3 ligase. Noticeably, deletion of Ufd2, another E4 ligase, did not affect the heat-induced ubiquitylation response (Figure S3a), suggesting that Hul5 is specific for this quality control pathway. One possibility is that misfolded proteins are targeted by this quality control machinery in a two-step ubiquitylation process that precludes the “en masse” targeting of transiently misfolded polypeptides that can refold (Figure S7f). In addition, the redistribution of Hul5 to the cytoplasm during heat-shock may permit the rapid targeting of a larger number of misfolded proteins. The discovery of cofactors that work in conjunction with Hul5 will provide further insight into the recognition and targeting mechanisms of these misfolded proteins in cells.
Materials & Methods
Plasmids
All plasmids are further described in Table S3. HUL5 (BPM309) and hul5-C878A (BPM310) were subcloned into pRS316 with endogenous promoter and termination regions from pJH84 and pJH85 (D. Finley), respectively. For Myc13Hul5 (BPM325), HUL5 was inserted into a pRS316 plasmid that contains the high expression GPD promoter, the N-terminal Myc13 tag and the terminator sequence from PGK1. For GFPHul5 (BPM341) and GFP-NLSHul5 (BPM345), the Hul5 promoter region, the GFP+39 with or without a 3′ NLS (SPKKKRKVEAS), and the HUL5 coding sequence with 3′UTR containing an ARS sequence were inserted into pRS306.
Cell & Heat-shock Assays
All yeast strains are listed in Tables S3. Hul5GFP and Nic96RFP were generated by a genomic insertion of GFP+39 and RFP25 at the 3′ end of the coding sequences by homologous recombination. For heat-shock assays, overnight saturated cultures were diluted and grown to exponential phase in YPD at 25°C to an OD600 of 1–1.5 before heat-shock at 45°C for the indicated times (typically 15 min). For western and dot blots, cells were washed twice and snap frozen in liquid nitrogen. Lysis was performed with glass beads in pre-warmed 1x SDS-PAGE Laemmli sample buffer without reducing agent and dye. All samples were normalized after Bradford prior to multiplex analysis with mouse anti-ubiquitin (1:2,500; MAB1510) and rabbit anti-Pgk1 or anti-PSTAIR (Cdc28; 1:1,000) antibodies. Fluorescent secondary antibodies (1:10,000; LI-COR) were used for quantification analysis by Odyssey Infrared Imaging System. For dot blot assay, 3 μl of the normalized samples (5–10 μg proteins) were spotted and dried overnight on a nitrocellulose membrane. Membranes were re-hydrated with 1 × TBS and processed as other Western blots. For the proteasome inhibition experiment, 20 μM MG132 was added to pdr5Δ cells as indicated. Cells for the SSA-experiment were similarly processed and incubated at the indicated temperatures. To assess solubility, cells were lysed (100 mM HEPES, 1% Triton-100X, 300 mM NaCl and protease inhibitors) with glass beads, pre-cleared at 2,000 g, and fractionated at 16,000 g for 10 min. For the cell fitness assay exponentially growing cells in SD-URA media were incubated for 20 min at 25°C or 45°C and transferred to a 96 well plate placed on the Infinite 200 PRO plate reader system (Tecan; Figure 3c) or in a incubator (Figure 4f) with constant shaking at 25°C.
IMAC - Purification of Ubiquitylated Proteins
In all experiments, about 1 μl of MagneHis (Promega) was used per 100 μg of protein extract. For mass spectrometry analysis, cell metabolic labeling (each in 500 ml) was done in YNB media at 25°C as previously described21. Equal amounts of cells were mixed and lysed in HU buffer (8 M urea, 100 mM HEPES pH 8, 0.05% SDS, 10 mM chloroacetemide, 1 mM PMSF, 10 mM imidazole, and protease inhibitors cocktail) by glass beads. Following 90 min incubation at ambient temperature, nickel beads were washed three times in HU buffer with 1% SDS, followed by three washes in HU buffer with 0.5% Triton-X100 and three washes in HU buffer. For pellet pre-enrichment, cells were first lysed in 100 mM HEPES pH 8, 1% Triton-100X, 300 mM NaCl, 1 mM PMSF, 1 mM phenanthroline, 10 mM chloroacetemide and protease inhibitors by glass beads and centrifuged at 16,000 g. Cell pellets were then resolubilized in HU buffer with 0.05% SDS. For TAP validation, 150 ml cells carrying H8-Ubi plasmid (RDB1851/BPM30)21 or empty vector were grown in SD-URA prior to lysis in HU buffer with 0.05% SDS, and elution from the beads was performed after adding one volume of 2 M imidazole, one volume of HU buffer and one volume of 3x SDS-PAGE buffer. IMAC from both the insoluble and soluble fractions was performed with a 1:1 mixture of the HU and lysis buffers with 0.05% SDS. Samples were also analyzed with rabbit anti-TAP antibody (1:1,000; CAB1001).
Mass Spectrometry Analysis
Samples were prepared as before21. The IMAC samples of Hul5 substrate identification experiments (Fig. 6a, S5d) were purified and fractionated on SCX stage tips to generate five fractions as described previously40, 41. All the total cell lysate samples (Fig. 2, S2, 6, S5) and the other IMAC samples (Fig. 2, S2, S5b) were purified with C18stage tips without fractionation. Purified peptides were analyzed using a linear-trapping quadrupole Orbitrap mass spectrometer (LTQ-Orbitrap; ThermoFisher Scientific) or a LTQ-Obitrap Velos (Fig. 6a) on-line coupled to an Agilent 1100 Series nanoflow HPLC using a nanospray ionization source (ThermoFisher Scientific). SCX fractions were run with a 90 min gradient, and other unfractionated samples were run for a 240 min gradient or 120 min with the Velos Instrument. The LTQ-Orbitrap was set to acquire a full-range scan at 60,000 resolution from 300 to 1600 Th and to fragment the top five peptide ions in each cycle in the Orbitrap, and the top 15 ions (90 min gradient) or the top 10 ions (120 min gradient) in the Velos instrument. Parent ions were then excluded from MS/MS for the next 30 sec, as well as singly charged ions. The Orbitrap was continuously recalibrated using lock-mass function42.
Analysis of mass spectrometry data
Centroided fragment peak lists were processed to Mascot generic format using DTA Supercharge (http://msquant.sourceforge.net) with LTQ-Orbitrap data or Proteome Discover with the Velos instrument data. Fragment spectra were searched using the Mascot algorithm against Saccharomyces Genome Database (SGD_051107) using the following parameters: peptide mass accuracy 10 ppm; fragment mass accuracy 0.6 Da; trypsin; 2 13C, fixed modification (carbamidomethyl), variable modifications (deamidation and oxidation; oxidation only for Velos data), ESI-TRAP fragment characteristics. Typical false positive rate for peptide identification (ion score ≥ 15, peptide length ≥ 6 a.a.) was estimated between 0.5 and 1.3% by using a Decoy database with Mascot. Mascot results were modified by a 15N labeling script (http://msquant.sourceforge.net/#N15 support) and 14N/15N ratios of identified peptides were quantified by MSQuant (v1.5b7; minimum of two unique peptides per protein to be selected). All peptide quantifications were manually validated and the averaged log2 (14N/15N) values of the quantified proteins were set to a maximum of 5. As none of the perturbations affected the ratio distributions of the proteins in the total cell lysates, we used these ratio distributions as reference to estimate the confidence of 14N/15N enrichment in the IMAC for a given threshold43. We reported the portion of ratios in the total cell lysate of each sample that were above the established threshold (e.g., log2 (14N/15N) ≥ 0.5) as an estimate of the false positive rates for the enrichment (all between 3–5%; Table S1).
Pull down and Subcellular Fractionation
For co-immunoprecipitation, cells with BPM325 (Myc13Hul5) or pRS316, after a 20 min incubation at 45°C, were lysed in IP buffer (100 mM HEPES pH 8, 20 mM MgAc, 300 mM NaAC, 10% glycerol, 1% NP-40, 10 mM EGTA, 0.1 mM EDTA, 1x protease inhibitor cocktail and 1 mM PMSF), and TAP-tagged proteins were pulled-down with IgG coupled Dynal magnetic beads (Invitrogen) incubated for 3 hours at 4°C followed by three washes with IP buffer. The nuclei and cytosolic fractions were prepared as previously described44 from Hul5GFP cells collected at exponential phase with additional procedures as follows. The spheroblasts were lysed (20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% Tween 20, 1 mM PMSF, 1x protease inhibitor cocktail) with 8 strokes in a chilled tight-fitting pestle dounce homogenizer, after 15 min incubation on ice. Unbroken cells and debris were removed by centrifugation for 5 min at 300 g at 4°C. The cytoplasmic fraction (supernatant) was collected after a spin at 13,000 g for 20 min at 4°C and the nuclear enriched fraction (pellet) was washed once with lysis buffer before collection. Normalized volumes of cytoplasmic fraction and nuclei fraction were then subjected to Western blotting analysis and also probed with mouse anti-MYC (9E10; 1:5000), anti-GFP (1:3,000; Roche) with anti-mouse ECL-coupled IgGs (1:3,000; BioRad) and rabbit anti-H3 (1:3,000) antibodies.
Microscopy
Hul5 GFP Nic96mRFPcells were grown to log phase at 25°C and either imaged immediately or subjected to heat shock at 42°C for 30 min. GFP+/−NLSHul5 expressing cells were similarly treated but incubated with Hoechst 33342 (2.5 μg/ml) for 30 min prior to imaging. Cells were imaged with a Zeiss Axio Observer Inverted Microscope equipped with a Zeiss Colibri LED illuminator and a Zeiss Axiocam Ultra High Resolution Monochrome Digital Camera Rev 3.0. Image stacks were acquired with a 40X or 63X objective, at a step of 0.3 μm or in single stack, and analyzed with Zeiss Axiovision software.
35S labeling and measurement of protein degradation
The procedure was adapted with minor modifications from a previous study34. Yeast cells were grown in SD medium supplemented with 6 essential amino acids, uracil and 20 g/ml tyrosine45 to OD600 of 0.8. Cells were washed twice and incubated in SD-Met media with tyrosine for 50 min. 50 μCi/ml of EXPRE35S35S protein labeling mix (1175 Ci/mmol, PerkinElmer) was added for 5 min to label newly synthesized proteins before two washes with ice-cold chase media containing Met (6 mg/ml), Cys (0.5 mg/ml) and cycloheximide (0.5 mg/ml). Cells were then incubated at 25°C or 38°C. Cell aliquots were mixed to a final 10% TCA and incubated overnight at 4°C. Radioactivity in both TCA-soluble and -insoluble fractions was measured in a MicroBeta2 Radiometric Detector (PerkinElmer). Percentage (pi) of short-lived protein degradation was calculated by subtracting the background TCA-soluble signal in t0 from the TCA-soluble signal (ti), and then divided by the t0 signal of the TCA-insoluble fraction.
Statistical analysis
Data were statistically analyzed as indicated (mean, s.d. and s.e.m.) and statistical significance for the ubiquitylation response was assessed by two-tailed unpaired student’s t-test.
Supplementary Material
Acknowledgments
The authors express gratitude to the following people for providing reagents: D. Finley (HUL5 plasmids); J. Thorner (anti-Pgk1 antibody); L. Howe (anti-H3 antibody); A. Chruscicki (Dynal-IgG); S. Jentsch (E2 double deletion strains); E. Craig (ssa1-45 strains); L. Conibear (TAP strains); R. Deshaies, C. Boone & M. Roberge (deletion strains). We also thank P. Kaiser for comments on the manuscript, J. Gsponer for discussion, T.M lab members for their encouragement and discussion and the following people for their support: L. Foster and N. Stoynov (mass spectrometry analysis); A. Chang (microscopy); P. Hieter and J. Stoepel (plate reader analysis); E. Jan (35S labeling and Odyssey). T.M. is supported by a grant from the Canada Institutes of Health Research (CIHR) and is a CIHR New Investigator. V.M. is supported by a grant from the CIHR.
Abbreviations
- ERAD
endoplasmic reticulum associated protein degradation
- H8-Ubi
octahistidine tagged ubiquitin
- IMAC
immobilized metal ion affinity chromatography
- NLS
nuclear localization signal
Footnotes
Contributions
N.N.F. and T.M. conceived the project and designed experiments; N.N.F. carried out experiments; A.H.M.N. designed and carried out the SSA1 analysis and developed reagents; V.M. designed and carried out part of the localization analysis; N.N.F., A.H.M.N., V.M and T.M. analyzed the data; N.N.F., V.M. and T.M. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interest
References
- 1.McClellan AJ, Tam S, Kaganovich D, Frydman J. Protein quality control: chaperones culling corrupt conformations. Nat Cell Biol. 2005;7:736–741. doi: 10.1038/ncb0805-736. [DOI] [PubMed] [Google Scholar]
- 2.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 3.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 4.Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rock KL, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 6.Chau V, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 7.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 10.Rosenbaum JC, et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 12.Fredrickson EK, Rosenbaum JC, Locke MN, Milac TI, Gardner RG. Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol Biol Cell. 2011;22:2384–2395. doi: 10.1091/mbc.E11-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connell P, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 14.Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci U S A. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasad R, Kawaguchi S, Ng DT. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21:2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nillegoda NB, et al. Ubr1 and ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell. 2010;21:2102–2116. doi: 10.1091/mbc.E10-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biederer T, Volkwein C, Sommer T. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J. 1996;15:2069–2076. [PMC free article] [PubMed] [Google Scholar]
- 19.Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 20.Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayor T, Graumann J, Bryan J, MacCoss MJ, Deshaies RJ. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol Cell Proteomics. 2007;6:1885–1895. doi: 10.1074/mcp.M700264-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 23.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chernova TA, et al. Prion induction by the short-lived, stress-induced protein lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol Cell. 2011;43:242–252. doi: 10.1016/j.molcel.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 26.Xie Y, Varshavsky A. UFD4 lacking the proteasome-binding region catalyses ubiquitination but is impaired in proteolysis. Nat Cell Biol. 2002;4:1003–1007. doi: 10.1038/ncb889. [DOI] [PubMed] [Google Scholar]
- 27.Wang G, Yang J, Huibregtse JM. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol Cell Biol. 1999;19:342–352. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aviram S, Kornitzer D. The ubiquitin ligase Hul5 promotes proteasomal processivity. Mol Cell Biol. 2010;30:985–994. doi: 10.1128/MCB.00909-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crosas B, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 30.Kohlmann S, Schafer A, Wolf DH. Ubiquitin ligase Hul5 is required for fragment-specific substrate degradation in endoplasmic reticulum-associated degradation. J Biol Chem. 2008;283:16374–16383. doi: 10.1074/jbc.M801702200. [DOI] [PubMed] [Google Scholar]
- 31.Leggett DS, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 32.Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 33.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 34.Medicherla B, Goldberg AL. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol. 2008;182:663–673. doi: 10.1083/jcb.200803022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tharun S, et al. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 36.Searfoss A, Dever TE, Wickner R. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p-Slh1p. Mol Cell Biol. 2001;21:4900–4908. doi: 10.1128/MCB.21.15.4900-4908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 38.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tam YY, Fagarasanu A, Fagarasanu M, Rachubinski RA. Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J Biol Chem. 2005;280:34933–34939. doi: 10.1074/jbc.M506208200. [DOI] [PubMed] [Google Scholar]
- 40.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 41.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 42.Olsen JV, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Wilde IB, Brack M, Winget JM, Mayor T. Proteomic characterization of aggregating proteins after the inhibition of the ubiquitin proteasome system. J Proteome Res. 2011;10:1062–1072. doi: 10.1021/pr1008543. [DOI] [PubMed] [Google Scholar]
- 44.Ducker CE, Simpson RT. The organized chromatin domain of the repressed yeast a cell-specific gene STE6 contains two molecules of the corepressor Tup1p per nucleosome. EMBO J. 2000;19:400–409. doi: 10.1093/emboj/19.3.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller MJ, Xuong NH, Geiduschek EP. A response of protein synthesis to temperature shift in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979;76:5222–5225. doi: 10.1073/pnas.76.10.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.