Abstract
The tools of synthetic biology allow researchers to change the ways engineered organisms respond to chemical stimuli. Decades of basic biology research and new efforts in computational protein and RNA design have led to the development of small molecule sensors that can be used to alter organism function. These new functions leap beyond the natural propensities of the engineered organisms. They can range from simple fluorescence or growth reporting to pathogen killing, and can involve metabolic coordination among multiple cells or organisms. Herein, we discuss how synthetic biology alters microorganisms’ responses to chemical stimuli resulting in the development of microbes as toxicity sensors, disease treatments, and chemical factories.
Introduction
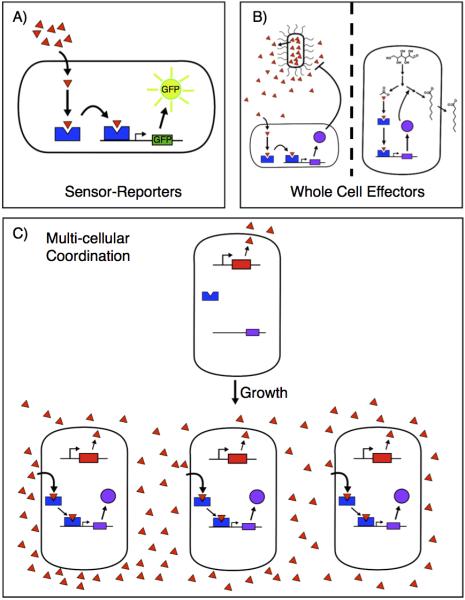
Synthetic biology allows scientists to re-program interactions between genes, proteins, and small molecules. One of the goals of synthetic biology is to produce organisms that predictably carry out desired functions and thereby perform as well-controlled so-called biological devices. Together, synthetic and chemical biology can provide increased control over biological systems by changing the ways these systems respond to and produce chemical stimuli. Sensors, which detect small molecules and direct later cellular function, provide the basis for chemical control over biological systems. The techniques of synthetic biology and metabolic engineering can link sensors to metabolic processes and proteins with many different activities. In this review we stratify the activities affected by sensors to three different levels: sensor-reporters that provide a simple read-out of small molecule levels, sensor-effectors that alter the behavior of single organisms in response to small molecules, and sensor effectors that coordinate the activities of multiple organisms in response to small molecules (Figure 1).
Figure 1.
A) Sensors (blue polygons) are the basic functional units microorganisms use to detect chemical stimuli (red triangles). Sensors act through effectors to achieve a particular function. Reporters (green sun) are simple effectors that provide an easily measured readout (growth, fluorescence) to indicate that a sensor has detected a particular compound. B) Whole cell effectors (purple circle) drastically alter cellular behavior allowing microorganisms to carry out a variety of functions. Effectors can give microorganisms the ability to incorporate signals from multiple sensors [1-4], to fight pathogens (left) [5*-11], and coordinate microbial metabolism to produce useful compounds (right) [12-17*]. C) Chemicals (red triangles), sensors (blue polygons), and effectors (purple circles) can coordinate populations of multiple cells, strains, or species with specialized abilities to detect or produce chemical stimuli resulting in the performance of tasks with labor shared among the population [18-25**,26,27]. In this example, growth leads to increased concentration of a chemical signal (red triangle) that coordinates the production of an effector (purple circle) by the entire population.
1. Sensor-Reporters
Small molecule sensors used in synthetic biology are often based on RNAs [28-34] or transcription factors [35-42] that bind to specific chemicals and influence the expression of downstream effectors. Sensors with different specificities can be mined from the literature, discovered through screens for small molecule responsive promoters [16,43], or computationally designed [36,44,45**]. Once a suitable sensor is defined, its function can be tested by placing the expression of a reporter gene under the sensor’s control (Figure 1A). Protein transcription factors are commonly used as sensors and are primarily discussed in this review. RNA based sensors that bind to a variety of ligands can be used to alter transcription, translation initiation, and ribozyme activity [46,47]. Using evolution based methods [48,49], RNAs that bind to specific ligands can be selected; however, it is often unclear how to link ligand binding function to reporter read out, and RNA based sensors for a wide array of compounds largely await development.
Simple sensor-reporter devices can be very powerful detectors of toxins or valuable chemicals. For instance, Trang and co-workers [35] show that a luciferase based bio-reporter for arsenic is capable of measuring arsenic levels in ground water from Vietnam at an accuracy better than established chemical methods. They propose to use this bio-assay in regions where expensive methods like atomic absorption spectroscopy cannot be easily performed. Similar reporters can measure levels of heavy metals, organic pollutants, and methylating compounds [37-39,42].
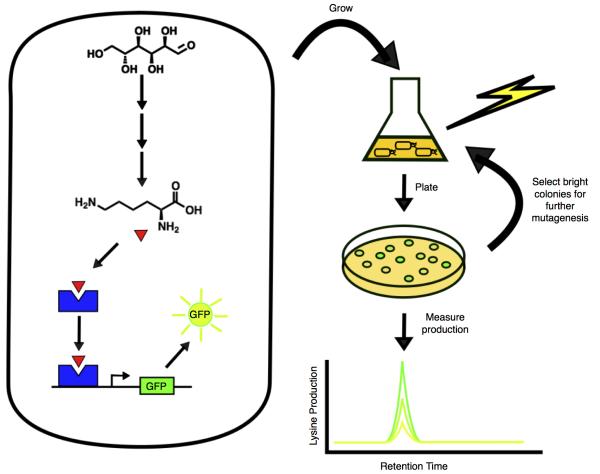
Sensor-reporter systems also allow one to screen for strains that produce a compound of interest in high yield (Figure 2). For such screening, strains need both the ability to produce a particular compound and the ability to report the amount they produce. This allows one to mutate or engineer these strains and directly measure the amount of reporter produced. High-producing strains can then be separated and used in further rounds of engineering and screening to continue enhancing yields.
Figure 2.
Using sensors to screen for high yields of a desired compound. The bacterium on the left has been engineered to produce lysine (red triangle) which is sensed by a transcription factor (blue polygon) that activates the expression of GFP. After mutagenizing this strain, lysine production by different mutants can be detected via their level of GFP expression. Mutants that produce high amounts of lysine also produce high amounts of GFP and can be picked for further mutagenesis. Directly measuring lysine quantities produced by different stains using a technique like liquid chromatography mass spectroscopy (LC-MS) confirms the correlation between GFP expression and lysine production.
The reporter-screening strategy described above has been used to enhance bacterial production of mevalonate [36], lysine [41], butanol [50], dicarboxylic acids [50], and triacetic acid lactone [51]; it is compatible with a wide variety of mutagenesis techniques. In a recent iteration of this technique, Raman et al 2014 [52**] used sensors controlling the expression of antibiotic resistance genes to select for E. coli strains with enhanced production of naringenin or glucaric acid. In their system, increased production of these compounds enhanced antibiotic resistance and provided a selective advantage over strains with lower production. Here the reporter readout was growth. The authors designed and implemented mutations enhancing production using multiplex automated genome engineering [53] and negative selection strategies to kill off any strains with mutations that simply led to high basal levels of antibiotic resistance. Iterative rounds of mutation, negative selection, and positive selection successfully improved the yields of both naringenin and glucaric acid.
2. Whole Cell Effectors
Synthetic gene circuits can lead to many more complex functions (cellular motility, memory, fuel and drug production, etc.) in response to small molecules. Here we focus on effectors with functions designed to improve health and enhance production by microbial chemical factories (Figure 1B). Some of these examples represent the first instance of transfer of synthetic circuits into real animal disease models.
Microbial Sensor-Effectors in Disease Prevention and Treatment
Pseudomonas aeruginosa, a ubiquitous bacterium capable of causing both mild and severe infections in humans, has recently been targeted using engineered E. coli. Exploiting the fact that Pseudomonas aeruginosa produce the quorum sensing molecule 3OC12HSL, researchers have generated E. coli strains that produce proteins that kill P. aeruginosa upon sensing 3OC12HSL [6,11], and other E. coli strains that respond to 3OC12HSL by migrating towards its source, releasing a protein that kills P. aeruginosa, and breaking down P. aeruginosa biofilms [5*]. The efficacy of these pathogen-killing bacteria has yet to be shown in vivo, but they provide a solid groundwork for future bacterial-based therapies.
We recently demonstrated the in vivo use of an E. coli memory device based on the lambda phage operon to record the presence of a chemical in the mouse gut [54**]. E. coli engineered with this memory circuit sustain production of beta-galactosidase after detecting anhydrotetracycline. When populating the mouse gut, these E. coli can detect anhydrotetracycline in mouse drinking water and remember its presence (i.e. continue to produce beta-galactosidase) up to 8 days after it is removed from the drinking water. This work is the first demonstration of an in vivo gut memory device and provides a platform upon which a variety of engineered gut microbiota responses to various chemicals (e.g. those indicative of infection) can be designed.
Many bacteria thrive in the hypoxic and acidic conditions of the tumor-micro environment with bacterial growth resulting in many anti-tumor effects. This phenomenon has a long history dating back to the 1800s and the use of bacterial extract (Coley’s toxin) to boost the immune system in cancer treatment. There has also been extensive work using attenuated and engineered strains of bacteria to combat cancer (Reviewed in [55]). Although therapies using S. typhimurium have shown limited efficacy [56-58], an on-going phase I clinical trial using intratumoral injections of attenuated Clostridium novyi has shown promising anti-tumor activity [59**]. In addition, the natural anti-tumor activity of Salmonella typhimurium has been expanded by giving it the ability to invade cancer cells or to secrete toxins when in the vicinity of tumors [8,9,18,43]. These strategies take advantage of promoters that induce gene expression under conditions indicative of the tumor microenvironment. For example, Flentie et al 2012 [43] discovered a Salmonella typhimurium promoter responsive to low pH and placed Shiga Toxin 2 under its control in S. typhimurium. When injected into mouse xenograph tumors, this strain decreased tumor cell viability within 5 days of injection [43].
The engineered ability to sense and respond to the chemical nature of the tumor microenvironment holds promise in increasing the specificity of bacterial anti-cancer therapies, but this specificity can also be achieved through other means. For example, Salmonella selectively kill tumor cells in a mouse xenograph model when they express antibodies to cause their attachment to the tumor cells and thymidine kinase to locally convert the prodrug Ganciclovir into an apoptosis inducer [60*]. Furthermore, E. coli have been used as a means of delivering shRNA’s silencing genes important for cancer cell viability to mammalian cells and live mice [10,61,62]. Further pairing of sensors with these types of selective treatments will likely lead to therapies with exquisite specificity.
Microbial Sensor-Effectors in Microbial Chemical Factories
Often, expressing a single enzyme can be enough to get a microbe to produce a small amount of a desired product, but more complicated metabolic engineering is usually required to divert metabolism away from natural cellular processes and toward production. While sensor-reporters provide a means of detecting high producers, sensor effectors can be used to dynamically modulate metabolism in response to the production of a compound of interest or its precursors providing new means to enhance yields.
Engineered metabolic pathways require coordination to limit the production of toxic intermediates and prevent hindrances on growth. In an early example of dynamic metabolic coordination, Farmer and Liao (2000) [14] enhanced E. coli lycopene production by placing lycopene-producing genes under the control of a promoter activated in the presence of acetyl phosphate (indicative of excess flux to acetate production). E. coli produce acetyl phosphate and acetate when they have excess glucose, but acetate retards E. coli growth. Farmer and Liao [14] hypothesized that high acetyl phosphate concentrations would be indicative of a metabolic state ideal for lycopene production; instead of diverting excess glucose into toxic acetate, acetyl phosphate induction of lycopene genes could re-direct excess glucose into lycopene synthesis. This strategy successfully increased lycopene production by greater than ten-fold compared to strains with lycopene production genes under the control of a lactose inducible promoter.
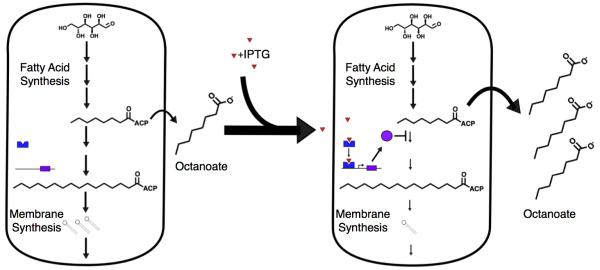
Our lab has demonstrated the use of an inducible degradation system to modulate E. coli fatty acid metabolism for the production of medium chain fatty acids [63] (Figure 3). E. coli require long chain fatty acids for normal growth, but medium chain fatty acids are potential precursors to fuel-like compounds. To divert E. coli fatty acid synthesis away from the production of long chain fatty acids, we replaced one of E. coli’s two fatty acid elongating enzymes with a mutant that can only elongate the fatty acids up to eight carbons. We then made the degradation of the second enzyme inducible by the small molecule isopropyl β-D-1-thiogalactopyranoside (IPTG). Replacing both enzymes with mutants would likely have been lethal, but the inducible degradation system allowed us to push fatty acid synthesis toward medium chain fatty acid production. These efforts, combined with further enzyme deletions designed to shunt cellular resources into fatty acid production, resulted in the production of the medium chain fatty acid octanoate at 12% theoretical yield. Future efforts could use sensors to link similar degradation systems to growth or fatty acid production instead of IPTG.
Figure 3.
Inducible inhibition of fatty acid elongation to enhance medium chain fatty acid production. In the system used in [63], fatty acid elongation was inhibited by an effector (purple circle) whose transcription was activated by exogenously added IPTG (red triangle). This slowed fatty acid synthesis and enhanced the production of the medium chain fatty acid, octanoate. Future systems could sense long chain fatty acids and avoid the need to add an exogenous inducer like IPTG.
Similar strategies have been used to improve amorphadiene [16], alpha-santalene [13], fatty acid [15,17*], and fatty acid ethyl-ester [12] production. It is important to note however, that it is unclear whether these strategies can be used at commercial scale.
3. Multi-cell Effectors
While single engineered cells are powerful tools, greater functionality can be achieved by chemically coordinating activities between multiple cells, strains, or species. The use of coordinated consortia can limit the need for researcher intervention, provide means to stabilize compound production by limiting toxicity and/or providing a selective advantage to high producers, and enhance yields. Applications of chemically coordinated cellular consortia are only just beginning to take shape, but synthetic biology provides us with the tools to chemically coordinate the activities of multiple organisms, as discussed below.
Quorum Sensing
Microorganisms use quorum sensing (QS) to measure population density through sensing the concentration of a small molecule produced by members of the population. QS allows microorganisms to restrict certain behaviors (e.g. virulence, sporulation, and light production) to particular population densities (Reviewed in [64]). Many QS molecules, production enzymes, sensors, and responsive promoters have been discovered [64]. These QS tools allow researchers to coordinate engineered activities within microorganisms. For example, researchers must measure population density when using bacteria to produce recombinant proteins or useful chemicals. If production is turned on too early or too late, yields can be limited. Quorum sensing provides a useful means through which engineered bacteria can measure their own population density and has the added advantage of preventing the use of a potentially expensive inducer molecule [22,23,65] (Figure 1C).
QS can also be used to regulate population density and composition. Mixed microbial consortia offer the possibility to partition engineered functions into optimally engineered and cooperating microbial strains or species. QS systems can be used to both activate and repress gene expression [66]. Altered gene expression due to Inter-microbe QS can be used to modify population structures as may be necessary in complicated microbial production schemes [21,24,65,67]. For example, You et al 2004 [24] used QS controlled cell lysis to regulate E. coli population density. Such a system could be used to maintain an engineered strain at a desired and/or safe level. Hong et al 2011 [21] used QS to regulate the bacterial composition of a biofilm. Such biofilm tuning could be used to effect dynamic metabolic output. QS systems can also be incorporated into logic gates [68] and have been use to drive bacterial localization to cancer cells [69]. These developments provide us with many tools, but QS systems await the realization of their full potential in synthetic systems that produce commodity chemicals or treat human disease.
Coordinating Metabolic Pathways
It is not always beneficial or possible to engineer all the components of metabolic pathways in a single organism. Different organisms have different natural metabolic propensities and sensitivities to toxic compounds. It can therefore be advantageous to use multiple organisms to carry out a complex task. For example, Minty et al 2013 [70*] co-cultured a fungus that naturally breaks down cellulose with E. coli engineered to produce isobutanol; they showed that the two can convert cellulosic biomass into isobutanol. In this example, the E. coli are essentially parasitic to the fungi because they use fungal resources and produce a compound (isobutanol) that inhibits fungal growth.
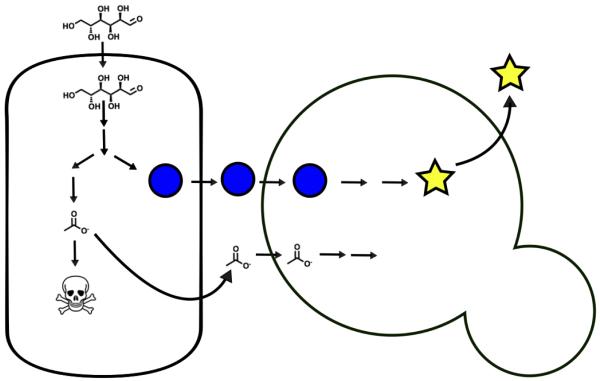
Chemicals produced by consortium members can be used to foster mutualisms, in which the members of the consortium are dependent upon one another for growth. One way to establish mutualism is to have one member of a two-component consortium consume a toxic product produced by the other member (Figure 4). For example, Zhou et al 2015 [25**] used a consortium of S. cerevisiae and E. coli to produce oxygenated taxanes (precursors to anti-cancer therapeutics). A portion of the metabolic route to the oxygenated taxanes was previously engineered in E. coli, but further processing required the use of cytochrome P450s that negatively impacted downstream steps in the pathway [71]. To avoid these negative impacts, the authors separated the pathway into components in E. coli and S. cerevisiae. The E. coli were capable of consuming provided xylose and produced oxygenated taxane precursors, but also produced toxic acetate as a byproduct. The yeast used the acetate as their carbon source thereby limiting its toxicity. The yeast then produced the cytochrome P450s and completed the pathway to oxygenated taxanes. This mutualistic consortium nearly doubled its production of oxygenated taxanes compared to a mixed culture where the E. coli and the yeast competed for glucose. A similar system was used by Bayer et al 2009 [26] to produce methyl halides from cellulosic biomass.
Figure 4.
Coordinating multi-microbe metabolism. Multiple organisms can be coordinated through chemical exchanges and metabolism to produce a desired compound. Here the organism on the left eats glucose and has been engineered to produce the blue circle, but also produces toxic acetate as a byproduct. The organism on the right further converts the blue circle into the end product (yellow star) and also eats the toxic acetate thereby promoting the growth of the organism on the left. Due to their linkage through acetate, the organisms are mutually dependent upon one another for optimal growth.
Further examples of metabolic coordination for the production of useful compounds are rare (Reviewed in [30]), but current research provides additional tools to link microbial metabolism. Microbes can be artificially linked through cross feeding essential metabolites [72,73], metabolites can be used for inter-strain and even inter-species signaling [27,74], and co-dependencies fostered by cross feeding can be used to select for the production of a target compound [75]. Equipped with these tools, future efforts should be able to generate many more mutualistic interactions that foster the cooperation of organisms with diverse functionalities.
Conclusion
We have come to the point in synthetic biology where there are many lab-scale or proof-of-concept examples of chemically controlled systems useful to sense small molecules, treat disease, and produce commercially useful compounds. These systems have great potential, but more attention needs to be paid to their stability, efficacy, and safety. Being that the sensor-effectors discussed above function in living, evolving organisms, it is unclear how well they will retain function when distributed in a patient or in a large-scale bioreactor. Future efforts should focus on developing these sensor-effectors for real-world application. Engineered organisms will only be useful if we can prove that their functions are reliable, predictable, and cost effective.
Acknowledgements
We thank Stephanie Hays and Jessica Polka for critical reading of the manuscript. This work was conducted with support from the National Science Foundation Graduate Research fellowship (to T.J.F.), and the Ruth L. Kirschtein National research Service Award program of Harvard Catalyst, The Harvard Clinical and Translational Science Center Award UL1 RR 025758 and financial contributions from Harvard University and its affiliated academic health care centers (to T.J.F.). The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, the National Center for Research Resources or the National Institutes of Health. This material is based upon work supported by the National Science Foundation. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
References
- 1.Callura JM, Cantor CR, Collins JJ. Genetic switchboard for synthetic biology applications. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5850–5855. doi: 10.1073/pnas.1203808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniel R, Rubens JR, Sarpeshkar R, Lu TK. Synthetic analog computation in living cells. Nature. 2013;497:619. doi: 10.1038/nature12148. + [DOI] [PubMed] [Google Scholar]
- 3.Stanton BC, Nielsen AAK, Tamsir A, Clancy K, Peterson T, Voigt CA. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nature Chemical Biology. 2014;10:99–105. doi: 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang BJ, Barahona M, Buck M. A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals. Biosensors & Bioelectronics. 2013;40:368–376. doi: 10.1016/j.bios.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang IY, Tan MH, Koh E, Ho CL, Poh CL, Chang MW. Reprogramming Microbes to Be Pathogen-Seeking Killers. Acs Synthetic Biology. 2014;3:228–237. doi: 10.1021/sb400077j. Combines functionalities of previously engineered E. coli into a single organism that effectively kills and disperses Pseudomonas aeruginosa biofilms in vitro.
- 6.Saeidi N, Wong CK, Lo TM, Nguyen HX, Ling H, Leong SSJ, Poh CL, Chang MW. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Molecular Systems Biology. 2011;7 doi: 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunstan SJ, Simmons CP, Strugnell RA. Use of in vivo-regulated promoters to deliver antigens from attenuated Salmonella enterica var. typhimurium. Infection and Immunity. 1999;67:5133–5141. doi: 10.1128/iai.67.10.5133-5141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganai S, Arenas RB, Forbes NS. Tumour-targeted delivery of TRAIL using Salmonella typhimurium enhances breast cancer survival in mice. British Journal of Cancer. 2009;101:1683–1691. doi: 10.1038/sj.bjc.6605403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryan RM, Green J, Williams PJ, Tazzyman S, Hunt S, Harmey JH, Kehoe SC, Lewis CE. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Therapy. 2009;16:329–339. doi: 10.1038/gt.2008.188. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Gao LF, Zhao LJ, Guo BF, Ji K, Tian Y, Wang JG, Yu H, Hu JD, Kalvakolanu DV, et al. Intratumoral delivery and suppression of prostate tumor growth by attenuated Salmonella enterica serovar typhimurium carrying plasmid-based small interfering RNAs. Cancer Research. 2007;67:5859–5864. doi: 10.1158/0008-5472.CAN-07-0098. [DOI] [PubMed] [Google Scholar]
- 11.Gupta S, Bram EE, Weiss R. Genetically programmable pathogen sense and destroy. ACS synthetic biology. 2013;2:715–723. doi: 10.1021/sb4000417. [DOI] [PubMed] [Google Scholar]
- 12.Zhang FZ, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nature Biotechnology. 2012;30:354–U166. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 13.Scalcinati G, Knuf C, Partow S, Chen Y, Maury J, Schalk M, Daviet L, Nielsen J, Siewers V. Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquitepene alpha-santalene in a fed-batch mode. Metabolic Engineering. 2012;14:91–103. doi: 10.1016/j.ymben.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Farmer WR, Liao JC. Improving lycopene production in Escherichia coli by engineering metabolic control. Nature Biotechnology. 2000;18:533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- 15.Xu P, Li LY, Zhang FM, Stephanopoulos G, Koffas M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11299–11304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, et al. Engineering dynamic pathway regulation using stress-response promoters. Nature Biotechnology. 2013;31:1039. doi: 10.1038/nbt.2689. + [DOI] [PubMed] [Google Scholar]
- 17.Liu D, Xiao Y, Evans BS, Zhang F. Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor-actuator. ACS synthetic biology. 2015;4:132–140. doi: 10.1021/sb400158w. Authors use a malonyl-CoA sensor to coordinate the production of free fatty acids with the avaliabilty of fatty acid precursors in E. coli. This is the first demonstration of endogenous dynamic regulation of the fatty acid synthesis process.
- 18.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. Journal of Molecular Biology. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 19.Bacchus W, Fussenegger M. Engineering of synthetic intercellular communication systems. Metabolic Engineering. 2013;16:33–41. doi: 10.1016/j.ymben.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Chen AY, Deng ZT, Billings AN, Seker UOS, Lu MY, Citorik RJ, Zakeri B, Lu TK. Synthesis and patterning of tunable multiscale materials with engineered cells. Nature Materials. 2014;13:515–523. doi: 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong SH, Hegde M, Kim J, Wang XX, Jayaraman A, Wood TK. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device. Nature Communications. 2012;3 doi: 10.1038/ncomms1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nocadello S, Swennen EF. The new pLAI (lux regulon based auto-inducible) expression system for recombinant protein production in Escherichia coli. Microbial Cell Factories. 2012;11 doi: 10.1186/1475-2859-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsao CY, Hooshangi S, Wu HC, Valdes JJ, Bentley WE. Autonomous induction of recombinant proteins by minimally rewiring native quorum sensing regulon of E. coli. Metabolic Engineering. 2010;12:291–297. doi: 10.1016/j.ymben.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 24.You LC, Cox RS, Weiss R, Arnold FH. Programmed population control by cell-cell communication and regulated killing. Nature. 2004;428:868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 25.Zhou K, Qiao K, Edgar S, Stephanopoulos G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nature Biotechnology. 2015 doi: 10.1038/nbt.3095. Develop a mutualistic consortium of E. coli and S. cerevisiae to produce a precursor to the cancer therapeutic paclitaxel. One of a small number of examples of mutalism used to increase production of a useful product.
- 26.Bayer TS, Widmaier DM, Temme K, Mirsky EA, Santi DV, Voigt CA. Synthesis of Methyl Halides from Biomass Using Engineered Microbes. Journal of the American Chemical Society. 2009;131:6508–6515. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- 27.Weber W, Baba M, Fussenegger M. Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10435–10440. doi: 10.1073/pnas.0701382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Seo SW, Jang S, Shin SI, Lim CH, Roh TY, Jung GY. Synthetic RNA devices to expedite the evolution of metabolite-producing microbes. Nature Communications. 2013;4 doi: 10.1038/ncomms2404. [DOI] [PubMed] [Google Scholar]
- 29.Buskirk AR, Landrigan A, Liu DR. Engineering a ligand-dependent RNA transcriptional activator. Chemistry & Biology. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 30.Jagmann N, Philipp B. Reprint of Design of synthetic microbial communities for biotechnological production processes. Journal of Biotechnology. 2014;192:293–301. doi: 10.1016/j.jbiotec.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Ceres P, Trausch JJ, Batey RT. Engineering modular 'ON' RNA switches using biological components. Nucleic Acids Research. 2013;41:10449–10461. doi: 10.1093/nar/gkt787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muranaka N, Sharma V, Nomura Y, Yokobayashi Y. An efficient platform for genetic selection and screening of gene switches in Escherichia coli. Nucleic Acids Research. 2009;37:e39. doi: 10.1093/nar/gkp039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. Journal of the American Chemical Society. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 34.Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014;159:925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trang PT, Berg M, Viet PH, Van Mui N, Van Der Meer JR. Bacterial bioassay for rapid and accurate analysis of arsenic in highly variable groundwater samples. Environmental science & technology. 2005;39:7625–7630. doi: 10.1021/es050992e. [DOI] [PubMed] [Google Scholar]
- 36.Tang SY, Cirino PC. Design and application of a mevalonate-responsive regulatory protein. Angewandte Chemie. 2011;50:1084–1086. doi: 10.1002/anie.201006083. [DOI] [PubMed] [Google Scholar]
- 37.Moser F, Horwitz A, Chen J, Lim W, Voigt CA. Genetic sensor for strong methylating compounds. ACS synthetic biology. 2013;2:614–624. doi: 10.1021/sb400086p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LT B-M, G M, AE F. Environmental Sensing of Heavy Metals Through Whole Cell Microbial Biosensors: A Synthetic Biology Approach. ACS Synth Biol. 2014 doi: 10.1021/sb500286r. [DOI] [PubMed] [Google Scholar]
- 39.Gu MB, Mitchell RJ, Kim BC. Whole-cell-based biosensors for environmental biomonitoring and application. Advances in biochemical engineering/biotechnology. 2004;87:269–305. doi: 10.1007/b13533. [DOI] [PubMed] [Google Scholar]
- 40.Duan F, March JC. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11260–11264. doi: 10.1073/pnas.1001294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Binder S, Schendzielorz G, Stabler N, Krumbach K, Hoffmann K, Bott M, Eggeling L. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biology. 2012;13 doi: 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Behzadian F, Barjeste H, Hosseinkhani S, Zarei AR. Construction and Characterization of Escherichia coli Whole-Cell Biosensors for Toluene and Related Compounds. Current Microbiology. 2011;62:690–696. doi: 10.1007/s00284-010-9764-5. [DOI] [PubMed] [Google Scholar]
- 43.Flentie K, Kocher B, Gammon ST, Novack DV, McKinney JS, Piwnica-Worms D. A Bioluminescent Transposon Reporter-Trap Identifies Tumor-Specific Microenvironment-Induced Promoters in Salmonella for Conditional Bacterial-Based Tumor Therapy. Cancer Discovery. 2012;2:624–637. doi: 10.1158/2159-8290.CD-11-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Looger LL, Dwyer MA, Smith JJ, Hellinga HW. Computational design of receptor and sensor proteins with novel functions. Nature. 2003;423:185–190. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- 45.Tinberg CE, Khare SD, Dou JY, Doyle L, Nelson JW, Schena A, Jankowski W, Kalodimos CG, Johnsson K, Stoddard BL, et al. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501:212. doi: 10.1038/nature12443. + Computationally design, screen, and crystallize ligand binding proteins with high affinity for their target small molecules. Provides a platform to computationally design new small molecule biosensors.
- 46.Wittmann A, Suess B. Engineered riboswitches: Expanding researchers' toolbox with synthetic RNA regulators. FEBS letters. 2012;586:2076–2083. doi: 10.1016/j.febslet.2012.02.038. [DOI] [PubMed] [Google Scholar]
- 47.Michener JK, Thodey K, Liang JC, Smolke CD. Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways. Metabolic engineering. 2012;14:212–222. doi: 10.1016/j.ymben.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 49.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 50.Dietrich JA, Shis DL, Alikhani A, Keasling JD. Transcription Factor-Based Screens and Synthetic Selections for Microbial Small-Molecule Biosynthesis. Acs Synthetic Biology. 2013;2:47–58. doi: 10.1021/sb300091d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang SY, Qian S, Akinterinwa O, Frei CS, Gredell JA, Cirino PC. Screening for enhanced triacetic acid lactone production by recombinant Escherichia coli expressing a designed triacetic acid lactone reporter. Journal of the American Chemical Society. 2013;135:10099–10103. doi: 10.1021/ja402654z. [DOI] [PubMed] [Google Scholar]
- 52.Raman S, Rogers JK, Taylor ND, Church GM. Evolution-guided optimization of biosynthetic pathways. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:17803–17808. doi: 10.1073/pnas.1409523111. Develop sensor-selectors for the production of naringenin and glucaric acid that enable positive and negative selection schemes as well as iterative rounds of mutagenesis. Method should be applicable to the production of a wide variety of compounds provided reponsive transcription factors are avaliable or can be engineered.
- 53.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kotula JW, Kerns SJ, Shaket LA, Siraj L, Collins JJ, Way JC, Silver PA. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4838–4843. doi: 10.1073/pnas.1321321111. Develop a bacterial memory device that functions in the mouse gut and reports on the presence of chemicals in mouse drinking water. First in vivo demonstration of a bacterial memory circuit.
- 55.Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nature reviews. Cancer. 2010;10:785–794. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thamm DH, Kurzman ID, King I, Li Z, Sznol M, Dubielzig RR, Vail DM, MacEwen EG. Systemic administration of an attenuated, tumor-targeting Salmonella typhimurium to dogs with spontaneous neoplasia: phase I evaluation. Clinical cancer research. 2005;11:4827–4834. doi: 10.1158/1078-0432.CCR-04-2510. [DOI] [PubMed] [Google Scholar]
- 57.Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ, Sherry RM, Topalian SL, Yang JC, Stock F. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. Journal of Clinical Oncology. 2002;20:142–152. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nemunaitis J, Cunningham C, Senzer N, Kuhn J, Cramm J, Litz C, Cavagnolo R, Cahill A, Clairmont C, Sznol M. Pilot trial of genetically modified, attenuated Salmonella expressing the E. coli cytosine deaminase gene in refractory cancer patients. Cancer gene therapy. 2003;10:737–744. doi: 10.1038/sj.cgt.7700634. [DOI] [PubMed] [Google Scholar]
- 59.Roberts NJ, Zhang L, Janku F, Collins A, Bai RY, Staedtke V, Rusk AW, Tung D, Miller M, Roix J, et al. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Science translational medicine. 2014;6:249ra111. doi: 10.1126/scitranslmed.3008982. Demonstrate effective tumor treatment using attenuated bacteria in a phase I clinical trial.
- 60.Massa PE, Paniccia A, Monegal A, de Marco A, Rescigno M. Salmonella engineered to express CD20-targeting antibodies and a drug-converting enzyme can eradicate human lymphomas. Blood. 2013;122:705–714. doi: 10.1182/blood-2012-12-474098. Demonstrate selective targeting of tumor cells using an attenuated strain of S. typhimurium expressing antibodies that direct it to tumors and thymidine kinase to convert the prodrug Ganciclovir into an apoptosis inducer at the site of the tumor. This is a direct demonstration of a bacterial means to target a specific tumor subtype.
- 61.Xiang S, Fruehauf J, Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nature biotechnology. 2006;24:697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]
- 62.Li Z, Yin PH, Yang SS, Li QY, Chang T, Fang L, Shi LX, Fang GE. Recombinant attenuated Salmonella typhimurium carrying a plasmid co-expressing ENDO-VEGI151 and survivin siRNA inhibits the growth of breast cancer in vivo. Molecular medicine reports. 2013;7:1215–1222. doi: 10.3892/mmr.2013.1308. [DOI] [PubMed] [Google Scholar]
- 63.Torella JP, Ford TJ, Kim SN, Chen AM, Way JC, Silver PA. Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11290–11295. doi: 10.1073/pnas.1307129110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annual Review of Cell and Developmental Biology. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 65.Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: Interfacing natural and engineered gene networks. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adams BL, Carter KK, Guo M, Wu HC, Tsao CY, Sintim HO, Valdes JJ, Bentley WE. Evolved Quorum sensing regulator, LsrR, for altered switching functions. ACS synthetic biology. 2014;3:210–219. doi: 10.1021/sb400068z. [DOI] [PubMed] [Google Scholar]
- 67.Balagadde FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You LC. A synthetic Escherichia coli predator-prey ecosystem. Molecular Systems Biology. 2008;4 doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shong J, Collins CH. Quorum sensing-modulated AND-gate promoters control gene expression in response to a combination of endogenous and exogenous signals. ACS synthetic biology. 2014;3:238–246. doi: 10.1021/sb4000965. [DOI] [PubMed] [Google Scholar]
- 69.Wu HC, Tsao CY, Quan DN, Cheng Y, Servinsky MD, Carter KK, Jee KJ, Terrell JL, Zargar A, Rubloff GW, et al. Autonomous bacterial localization and gene expression based on nearby cell receptor density. Molecular systems biology. 2013;9:636. doi: 10.1038/msb.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Minty JJ, Singer ME, Scholz SA, Bae CH, Ahn JH, Foster CE, Liao JC, Lin XN. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14592–14597. doi: 10.1073/pnas.1218447110. Demonstrate that a consortium of E. coli and fungi can convert cellulosic biomass into isobutanol. This represents an important step in the move away from using simple sugars as carbon sources for biofuels production.
- 71.Ajikumar PK, Xiao WH, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wintermute EH, Silver PA. Emergent cooperation in microbial metabolism. Molecular Systems Biology. 2010;6 doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2149–2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Silva-Rocha R, de Lorenzo V. Engineering multicellular logic in bacteria with metabolic wires. ACS synthetic biology. 2014;3:204–209. doi: 10.1021/sb400064y. [DOI] [PubMed] [Google Scholar]
- 75.Harcombe W. Novel Cooperation Experimentally Evolved between Species. Evolution. 2010;64:2166–2172. doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]