Abstract
The mechanisms that coordinate and balance a complex network of opposing regulators to control Schwann cell (SC) differentiation remain elusive. Here we demonstrate that zinc-finger E-box binding-homeobox 2 (Zeb2/Sip1) transcription factor is a critical intrinsic timer that controls the onset of Schwann cell (SC) differentiation by recruiting HDAC1/2-NuRD co-repressor complexes. Zeb2 deletion arrests SCs at an undifferentiated state during peripheral nerve development and inhibits remyelination after injury. Zeb2 antagonizes inhibitory effectors including Notch and Sox2. Importantly, genome-wide transcriptome analysis reveals a Zeb2 target gene, encoding the Notch effector Hey2, as a potent inhibitor for SC differentiation. Strikingly, a genetic Zeb2 variant, which is associated with Mowat-Wilson syndrome, disrupts the interaction with HDAC1/2-NuRD and abolishes Zeb2 activity for SC differentiation. Therefore, Zeb2 controls SC maturation by recruiting HDAC1/2-NuRD complexes and inhibiting a novel Notch-Hey2 signaling axis, pointing to the critical role of HDAC1/2-NuRD activity in peripheral neuropathies caused by ZEB2 mutations.
Introduction
Schwann cells (SCs) of the peripheral nervous system (PNS) ensheath and wrap myelin membranes around axons to maximize the efficiency of rapid saltatory conduction of action potentials. Functional defects of SCs contribute to various forms of peripheral neuropathies 1–3. A recently defined autosomal dominant disorder, Mowat-Wilson syndrome (MOWS), is caused by mutations in ZEB2 (a.k.a. SIP1 or ZFHX1B), the gene that encodes a zinc finger E-box binding homeobox 2 protein. MOWS is characterized by a wide spectrum of congenital anomalies encompassing growth defects, intellectual disability, delayed motor development, and neurocristopathies with defects in neural crest derivatives 4–6. Zeb2 is critical for neurogenesis and gliogenesis in the central nervous system (CNS) 7–9. Recent studies indicate that Zeb2 can interact with the nucleosome remodeling and deacetylase complex (NuRD), suggesting a potential importance of NuRD association for Zeb2 functions 4,10. At present, whether and how Zeb2 controls SC differentiation and myelination remain elusive. Elucidation of Zeb2 functions in peripheral nerve cell development in addition to CNS neurogenesis may provide a better understanding for the etiology of neurological diseases associated with MOWS caused by ZEB2 mutations.
Development and myelination of neural crest-derived SCs are coordinately regulated by a spectrum of extracellular signals and intrinsic regulators. These comprise positive cues including neuregulin 1 (NRG1) 11,12 and laminin/integrin signaling 13,14, GPCRs such as Gpr126 15, and negative cues including Notch signaling and Sox2 16. Timely initiation of SC myelination not only demands a strict interplay between extrinsic cues and intracellular inputs, but also requires the precise balance of the activity of an intricate network of transcriptional regulators. Transcription factors such as the HMG-type protein Sox10, the POU-homeodomain proteins Oct6/Pou3f1 and Pou3f2/Brn2, and the zinc finger proteins Krox20/Egr2 and YY1 are crucial for promoting SC myelination 17–20. How these multiple signaling pathways converge and coordinate the myelination program and, more importantly, what orchestrates the fine-tuning of proper transcriptional outputs, is not fully understood.
Here, by using SC-lineage specific in vivo mutagenesis, we identified Zeb2 as a critical intrinsic timer to control the onset of SC differentiation during normal myelination and also in remyelination after peripheral nerve injury. The predominant effects of Zeb2 during SC development are channeled through directly repressing a network of inhibitory pathways including Sox2 and Notch-Hey2 signaling. We further identify that a single amino acid variant in ZEB2 seen in a MOWS patient disrupts interaction with the HDAC1/2-NuRD co-repressor complex, and abolishes the ability of ZEB2 to effect SC differentiation. Our data thus establish a critical role for HDAC1/2-NuRD-dependent Zeb2 activity in initiating SC differentiation and myelination via opposing Notch and Sox2-mediated negative regulatory circuitry.
Results
Dynamic Zeb2 expression during SC lineage development
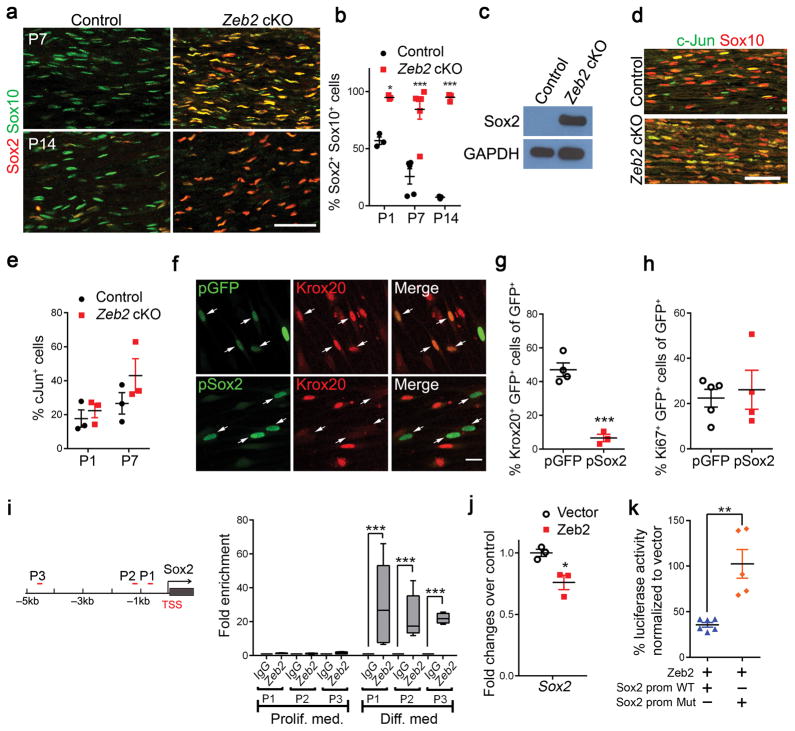
To examine Zeb2 expression in SCs, we co-immunolabeled for Zeb2 and a SC lineage marker (Sox10) in developing sciatic nerves (Fig. 1a). The vast majority of Zeb2-positive cells were Sox10+ at postnatal day 7 (P7). In keeping with its function as transcription regulator, Zeb2 was detected in the nuclei of Sox10+ SCs. Concurrent with the onset of myelination in the developing nerves, Zeb2 was highly expressed in SCs during the first 2 postnatal weeks, and the signal gradually declined as the animal reaches adulthood at 10 weeks (Fig. 1a).
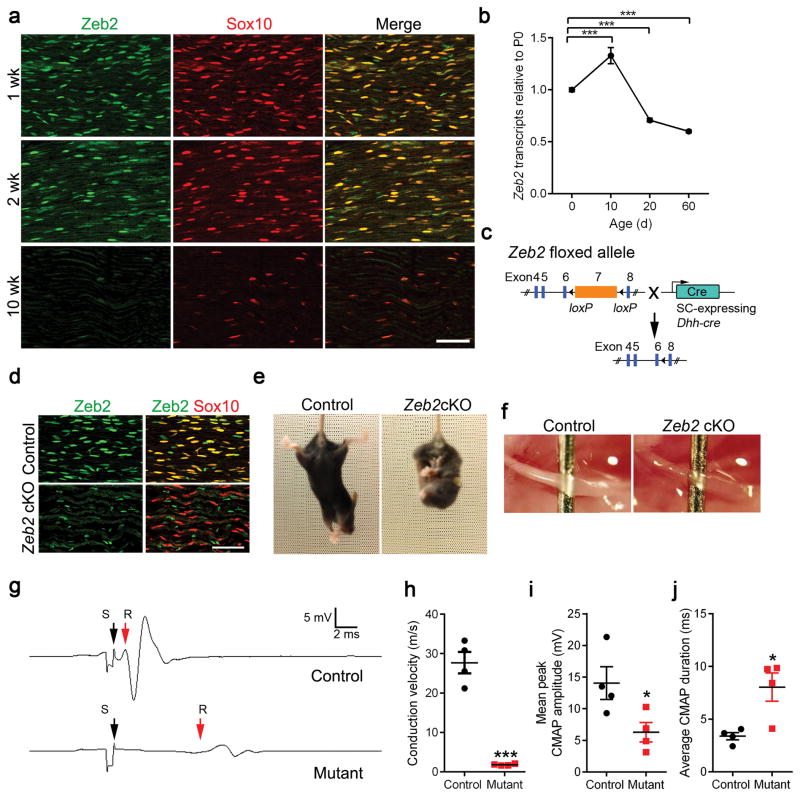
Figure 1. SC-specific deletion of Zeb2 results in peripheral nerve hypomyelination.
(a) Co-localization of Zeb2 (green) with Sox10 (red) in SC nuclei from 1 to 10 weeks by immunofluorescence labeling. Scale bar: 100 μm. The experiment was repeated 3 times on 3 control and mutant tissues.
(b) qRT-PCR showing Zeb2 transcript levels in mouse sciatic nerves at various developmental age (n = 3 control tissues at each time point; mean ± S.E.M***P < 0.001; one-way ANOVA with multiple comparisons test; P(P10) = 0.0003, P(P20) = 0.0009, P(P60) < 0.0001; F (3,12) = 61.91).
(c) A diagram showing that exon 7 of the floxed Zeb2 allele is excised by Dhh-cre.
(d) Immunostaining showing co-labeling of Zeb2 (green) with Sox10+ (red) SC nuclei in mutant sciatic nerves at P7. Scale bars: 50 μm. n = 3 control and mutant tissues.
(e) Tail suspension test showing abnormal hindlimb reflex in Zeb2 cKO (Zeb2loxP/loxP;Dhh-cre+/−) but not in control littermate (Zeb2loxP/+;Dhh-cre+/−) at P21.
(f) Appearance of sciatic nerves from control and Zeb2 cKO mice at P7.
(g) Recordings of CMAPs from P42 control and Zeb2 cKO. S: Stimulus (black arrow); R: Initiation of CMAP response (red arrow).
(h) Nerve conduction velocities in Zeb2 cKO and control littermates at P42. Data are presented as mean ± S.E.M (n = 4 control and mutant tissues; ***P < 0.001 t = 9.538 df = 6; Two-tailed unpaired Student’s t-test).
(i) The mean peak amplitudes of CMAPs in sciatic nerves of control and Zeb2 cKO mice at P42. Data are presented as mean ± S.E.M (n = 4 control and mutant tissues; *P < 0.05; P = 0.0417 t = 2.582 df = 6; Two-tailed unpaired Student’s t-test).
(j) The average durations of CMAPs in sciatic nerves of control and Zeb2 cKO mice at P42. Data are presented as mean ± S.E.M (n = 4 control and mutant tissues s; *P < 0.05; P = 0.0155 t = 3.348 df = 6; Two-tailed unpaired Student’s t-test).
To further define Zeb2 levels during SC lineage progression, we examined Zeb2 mRNA expression using SC-enriched sciatic nerves at different stages by RT-quantitative PCR (qPCR). Consistently, Zeb2 transcripts were detected at the neonatal stage P0 and peaked at the perinatal stage P10, but then was reduced in adulthood (Fig. 1b).
Zeb2 mutant mice display severe motor deficits
To determine the cell-autonomous role of Zeb2 in SC development, we generated conditional mutant mice lacking functional Zeb2 in SCs by breeding mice carrying a Zeb2 floxed allele (Zeb2flox/flox) mice 21,22 with a SC-expressing Cre line driven by the Desert-hedgehog (Dhh)-promoter 17 (Fig. 1c). We used Zeb2fl/+;Dhh-cre or Zeb2fl/fl littermates as controls since they were phenotypically normal in SC development (Supplementary Fig. 1a–g).
Cre-mediated inactivation of Zeb2 efficiently abrogated Zeb2 presence in Sox10+ SCs in peripheral nerves (Fig. 1d). Zeb2fl/fl;Dhh-cre (hereafter named Zeb2 cKO) mice were born in predicted numbers according to Mendelian ratios. During the first postnatal week, mutant mice were essentially indistinguishable from littermate controls. Starting in the second week, however, all Zeb2 cKO mice began to develop severe tremors, unsteady gait and hindlimb paralysis, and the majority died by 8 weeks (Supplementary Fig. 2a).
Upon tail suspension, mutant mice at P21 showed abnormal hindlimb clasping (Fig. 1e). Zeb2 cKO sciatic nerves at P7 exhibited a thin and translucent appearance, which stood in stark contrast to the thick and opaque control nerves, suggesting a severe deficit at the onset of myelinogenesis (Fig. 1f). Consistent with the overt clinical signs of hypomyelination in the Zeb2 cKO mutants, the motor unit function of mutant mice was severely impaired, as reflected by a marked reduction in conduction velocity (Fig. 1g) measured by electrophysiological recordings of compound muscle action potentials (CMAPs) during electrical stimulation of the sciatic nerve in vivo. The nerve conduction velocity was reduced from 27.7 ± 2.7 ms−1 in littermate controls to 1.8 ± 0.1 ms−1 in the mutants at the young adult stage (P42), which is close to the level of the slower thinly myelinated Aδ or unmyelinated C fiber conduction velocity in mice 23 (Fig. 1h). Furthermore, the mean peak amplitudes and duration of CMAPs were also severely affected in Zeb2 cKO mice (Fig. 1i,j), accounting for the defect in motor function in Zeb2 mutants.
Zeb2-deficient SCs fail to myelinate in peripheral nerves
We evaluated myelinogenesis in Zeb2 cKO peripheral nerves by myelin basic protein (MBP) immunolabeling. In contrast to the robust expression in control nerves, MBP was scarcely expressed in the mutant sciatic nerves at P14 (Fig. 2a). Consistently, expression of major myelin genes in the sciatic nerves, Mbp and Mpz, was virtually undetectable in mutants by in situ hybridization (Fig. 2b) and qPCR (Fig. 2c). Ultrastructural examination by electron microscopy (EM) revealed that, whereas by P4 control SCs have established a 1:1 relationship with large-caliber axons and began wrapping myelin membrane around axons, the majority of mutant SCs were associated with multiple axons and failed to initiate myelination, indicative of incomplete axonal sorting and a dysmyelination phenotype (Fig. 2d). At P21, axon bundles ensheathed by Zeb2 cKO SCs remained essentially unsorted and unmyelinated (Fig. 2e,f), despite apparent normal SC process extension around associated axons (Fig. 2f). Moreover, small diameter axons remained unsegregated in axon bundles with large diameter axons, further confirming sorting defects in Zeb2 cKO (Supplementary Fig. 1c). However, while testing for cutaneous thermal hypersensitivity, Zeb2 cKO showed a similar decrease in withdrawal latency to 50° C water following tissue inflammation compared to their baseline responsiveness as was seen with littermate controls (Supplementary Fig. 1h).
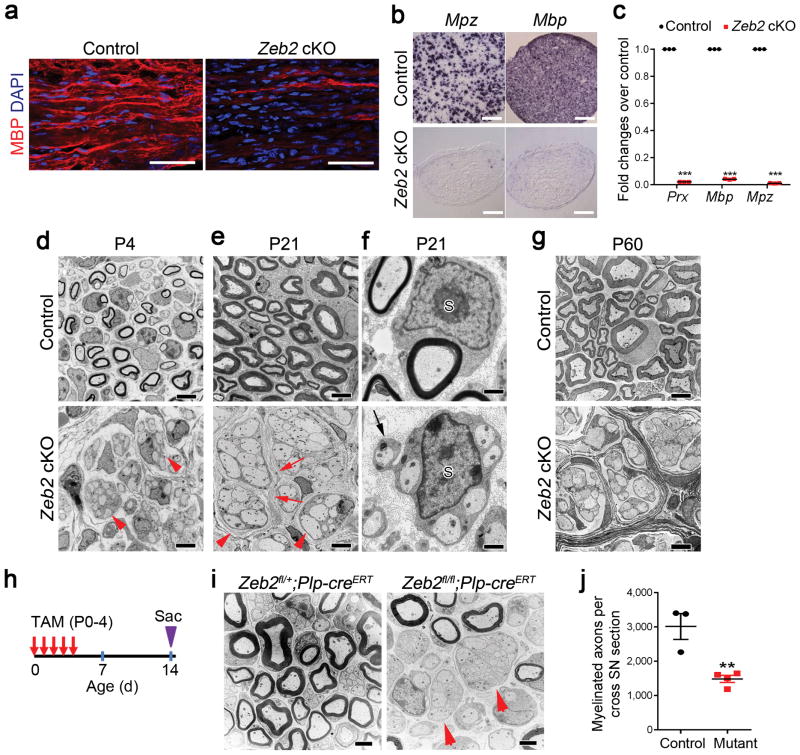
Figure 2. Zeb2 is required for SC myelination.
(a) Immunofluorescence labeling showing MBP staining in P14 control and Zeb2 cKO longitudinal sciatic nerves (n = 3 control and mutant tissues). Scale bars: 50 μm.
(b) Expression of Mbp and Mpz in transverse sections of sciatic nerves from control and Zeb2 cKO mice (n = 3 control and mutant tissues) at P21 by in situ hybridization. Scale bars: 200 μm.
(c) qRT-PCR showing the levels of myelin-related genes in sciatic nerves of P7 control and Zeb2 cKO mice. Data are presented as mean ± S.E.M (***P < 0.0001, n = 3 control and 3 mutant tissues, P(Prx) < 0.0001 t = 2750 df = 4; P(Mbp) < 0.0001 t = 512.5 df = 4 ; P(Mpz) < 0.0001 t = 1088 df = 4; Two-tailed unpaired Student’s t-test).
(d–e) Representative EM images showing ultrastructure of control and Zeb2 cKO sciatic nerves at P4 (d) and P21 (e). n = 3 control and mutant tissues. Arrowheads indicate axons and SC-axon-units respectively. Scale bars: 2 μm.
(f) EM images showing a 1:1 SC-axon unit in a P21 control sciatic nerve (upper) and a SC (S) associating with multiple axons in a mutant nerve (bottom). n = 3 control and mutant tissues. Arrow indicates mutant SC process surrounding an axon. Scale bars: 500 nm.
(g) Representative EM images of P60 control and Zeb2 cKO sciatic nerves. n = 3 control and mutant tissues. Scale bar: 2 μm.
(h) A diagram showing the tamoxifen (TAM) administration scheme. Pups were injected with TAM via i.p. into lactating dams once per day from P0 to P4, and sciatic nerves were harvested on P14.
(i) EM images of cross sections of P14 sciatic nerves from control (Zeb2fl/+;Plp-creERT; n = 3) and Zeb2 iKO mice (Zeb2fl/fl;Plp-creERT; n = 4) induced by TAM from P0–4. Arrows denote incompletely sorted axon bundles. Scale bar: 2 μm.
(j) Quantification of myelinated axon number per cross section of P14 sciatic nerves from control and TAM-treated Zeb2 iKO. Data are presented as mean ± S.E.M, n = 3 controls and 4 mutants. Two-tailed unpaired Student’s t-test. **P < 0.01; P = 0.0062 t = 4.529; df = 5.
Very few mutant mice were able to survive to adulthood (Supplementary Fig. 2a). The gross structure of peripheral nerves in rare Zeb2 cKO survivors at 2 months showed minifascicles of SCs and axons with no apparent sign of myelination, suggesting that Zeb2 loss in SCs leads to a sustained myelination block in peripheral nerves (Fig. 2g).
To bypass the possible influence of the Zeb2 loss during early SC development, we inactivated Zeb2 in the peripheral nerves of neonates by using a tamoxifen-inducible Plp-creERT driver 24 with tamoxifen administration from P0 to P4 (Fig. 2h). By P14, more than half of the large axons were not properly sorted and remained unmyelinated in Zeb2 iKO (Zeb2fl/fl;Plp-creERT) mutant nerves (Fig. 2i,j). The ultrastructural observations seen upon Zeb2 deletion in immature SCs suggest that Zeb2 is required for proper initiation of SC differentiation and myelination.
The diminished expression of Zeb2 in mature nerves (Fig. 1a,b) suggested its nonobligatory function in myelin maintenance. Consistently, tamoxifen-induced deletion of Zeb2 in adult mice did neither alter myelin sheath thickness nor the integrity in peripheral nerves (Supplementary Fig. 2b), suggesting a crucial role for Zeb2 in initiating or priming SC differentiation rather than maintaining myelin sheaths.
Zeb2 controls myelinogenic programs for SC differentiation
In Zeb2 cKO sciatic nerves, we detected reduced levels of mRNAs encoding SC pro-differentiation regulators including Sox10, Krox20 and Oct6 at P7 (Fig. 3a). In contrast, among known negative regulators, we observed an upregulation of Jagged1, encoding a ligand for Notch, and Hes1 (Fig. 3b). In-step with their transcript levels, Oct6 and Krox20 proteins were essentially absent in mutant SCs in these Zeb2 cKO sciatic nerves (Fig. 3c–e). We further investigated Zeb2 functions in SC differentiation by Zeb2 siRNA knockdown in rat SC cultures, either in proliferation or differentiation media. The expression of Oct6, Krox20 and major myelin genes, as well as their corresponding proteins was significantly reduced in SCs that underwent Zeb2 knockdown (Supplementary Fig. 3a–d), suggesting a cell-autonomous requirement of Zeb2 in SC differentiation. To determine whether Zeb2 could regulate Oct6 directly, we identified two Zeb2 consensus-binding sites [CACCT(G)] 25 in the Oct6 promoter, and detected genomic occupancy of Zeb2 to the Oct6 promoter in differentiating SCs, but not in proliferating cells (Supplementary Fig. 3e).
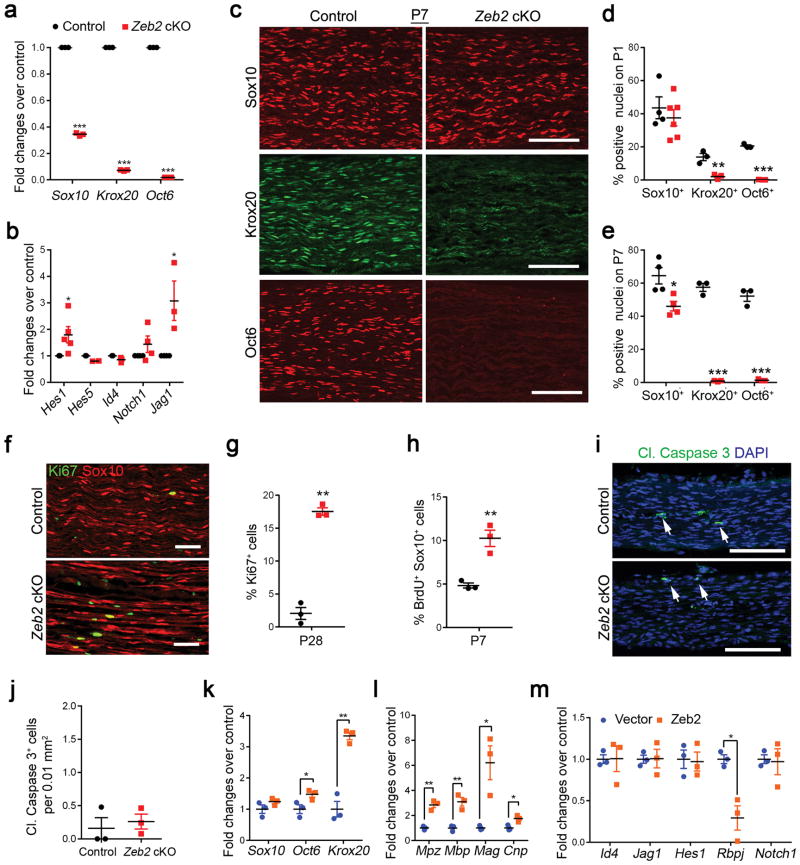
Figure 3. Zeb2 deletion in SCs inhibits SC differentiation and myelination.
(a) qRT-PCR showing the levels of promyelinating transcriptional regulators in sciatic nerves of P7 control and Zeb2 cKO mice. Data are presented as mean ± S.E.M (***P < 0.001, n = 3 control and mutant tissues, P(Sox10) < 0.0001 t = 101.4 df = 4; P(Krox20) < 0.0001 t = 291.8 df = 4; P(Oct6) < 0.0001 t = 14865 df = 4; Two-tailed unpaired Student’s t-test).
(b) qRT-PCR showing the levels of negative myelination regulators in control and mutant sciatic nerves at P7 (*P < 0.05, n = 3 control and mutant tissues; P(Hes1) = 0.0308 t = 2.617 df = 8; P(Hes5) = 0.0081 t = 4.885 df = 4; P(Id4) = 0.0466 t = 2.846 df = 8; P(Notch1) = 0.2181 t = 1.376 df = 6; P(Jag1) = 0.0496 t = 2.784 df = 4; Two-tailed unpaired Student’s t-test).
(c) Immunolabeling of pro-myelinating transcription factors Sox10, Krox20 and Oct6 in longitudinal sections of control and Zeb2 cKO sciatic nerves (n = 3 animals) at P7. Scale bars: 100 μm.
(d) Quantification of Sox10+, Krox20+ and Oct6+ cells relative to DAPI+ nuclei in control and Zeb2 cKO sciatic nerves (longitudinal sections) at P1. Data are presented as mean ± S.E.M (**P < 0.01; ***P < 0.001, n = 4 control and 6 mutant tissues for Sox10; n = 3 control and mutant tissues for Krox20 and Oct6, P(Sox10) = 0.4801 t = 0.7406 df = 8; P(Krox20) = 0.0072 t = 5.052 df = 4; P(Oct6) < 0.0001 t = 27.75 df = 4; Two-tailed unpaired Student’s t-test).
(e) Quantification of Sox10+, Krox20+ and Oct6+ cells relative to DAPI+ nuclei in control and Zeb2 cKO sciatic nerves at P7. Data are presented as mean ± S.E.M ((*P < 0.05; ***P < 0.001, n = 4 control and mutant tissues for Sox10; n = 3 control and mutant tissues for Krox20 and Oct6;, P(Sox10) = 0.0184 t = 3.120 df = 6; P(Krox20) < 0.0001 t = 23.3 df = 4; P(Oct60) < 0.0001 t = 16.4 df = 4; Two-tailed unpaired Student’s t-test).
(f) Immunofluorescence labeling for Ki67 in control and Zeb2 cKO sciatic nerves (n = 3 animals/genotype) at P28. Ki67 (green); Sox10 (red). Scale bar: 50 μm.
(g) Quantification of cell proliferation by Ki67 positive nuclei in control and mutant sciatic nerves at P28. (*P < 0.05, n = 3 control and mutant tissues; P = 0.0013 t = 11.82 df = 4, Two-tailed unpaired Student’s t-test).
(h) Quantification of the proliferation rate by BrdU incorporated cells per Sox10+ SC in control and mutant sciatic nerves P7. n = 3 control and mutant tissues; P = 0.0052 t = 5.527 df = 4; Two-tailed unpaired Student’s t-test.
(i) Immunofluorescence labeling for cleaved caspase 3 in P1 control and Zeb2 cKO sciatic nerves (n = 3 control and mutant tissues). Cleaved caspase 3 (cl. Caspase 3; white arrows); DAPI (blue). Scale bar: 100 μm.
(j) Quantification of apoptosis by cl. caspase 3-labeled cells per 0.01mm2 in P1 control and mutant sciatic nerves. Data are presented as mean ± S.E.M., n = 3 control and mutant tissues, P = 0.6334 t = 0.5155 df = 4; Two-tailed unpaired Student’s t-test.
(k) qRT-PCR showing Sox10, Oct6 and Krox20 expression in rat SCs rat SCs induced to differentiate following transfection with control and Zeb2 vectors. Data are presented as mean ± S.E.M (*P < 0.05, **P < 0.01; n = 3 independent experiments, P(Sox10) = 0.1826 t = 1.610 df = 4; P(Krox20) = 0.0011 t = 8.396 df = 4; P(Oct6) = 0.04811 t = 2.814 df = 4; Two-tailed unpaired Student’s t-test).
(l) qRT-PCR showing myelin gene expression in rat SCs transfected with control and Zeb2 vectors. Data are presented as mean ± S.E.M (*P < 0.05, **P < 0.01; n = 3 independent experiments, P(Mpz) = 0.0015 t = 7.775 df = 4; P(Mbp) = 0.0049 t = 5.626 df = 4; P(Mag) = 0.0184 t = 3.843 df = 4; P(Cnp) = 0.0169 t = 3.942 df = 4; Two-tailed unpaired Student’s t-test).
(m) qRT-PCR showing expression of negative regulatory genes in rat SCs transfected with control and Zeb2 vectors under differentiation conditions. Data are presented as mean ± S.E.M. (*P < 0.05, **P < 0.01; n = 3 independent experiments; P(Id4) = 0.9738 t = 0.06492 df = 4; P(Jag1) = 0.9568 t = 0.05771 df = 4; P(Hes1) = 0.8649 t = 0.1814 df = 4; P(Rbpj) = 0.0105 t = 4.538 df = 4; P(Notch1) = 0.8643 t = 0.1822 df = 4; Two-tailed unpaired Student’s t-test).
The proportion of Sox10+ SCs among DAPI+ cells in mutant nerves at P1 was comparable with that in control nerves (Fig. 3d), despite a slight reduction in SC number at P7 (Fig. 3e), which is likely due to the overall reduction in mature SCs. Accordingly, the SC proliferation rate revealed by Ki67 staining and BrdU incorporation appeared to increase substantially in Zeb2 mutant sciatic nerves (Fig. 3f–h). Apoptosis of mutant SCs, as determined by cleaved caspase-3 immunolabeling in P1 sciatic nerves, was not significantly altered (Fig. 3i,j). This indicates that Zeb2 deletion arrests SCs at their immature stage by blocking the transition from immature SCs to Oct6+/Krox20+ differentiating SCs.
Conversely, Zeb2 overexpression in SCs in vitro enhanced the induction of Oct6 and Krox20 (Fig. 3k), and myelin gene expression (Fig. 3l). Notably, we detected a substantial downregulation of the Notch effector Rbpj upon enforced Zeb2 expression in SCs (Fig. 3m). Thus, our data suggests that Zeb2 controls SC myelination by priming the transcriptional program that promotes SC differentiation, while attenuating the Rbpj-mediated Notch inhibitory pathway.
Zeb2 is required for SC differentiation during nerve repair
Although Zeb2 was maintained at minimal levels in mature SCs, it was highly upregulated in SCs in regenerating sciatic nerves after injury (Fig. 4a,b). We hypothesized that Zeb2 is required for SC differentiation and ultimately remyelination during nerve repair. To test this, we employed the peripheral nerve transection paradigm (Fig. 4a). After transection, SCs dedifferentiate, proliferate, establish a tissue bridge between the proximal and distal nerve stumps, and then guide regenerating axons across the bridge to reach the distal stump 26 with subsequent SC differentiation and remyelination. To determine the role of Zeb2 in SC differentiation following nerve injury, control (Zeb2fl/fl or Zeb2fl/+;Plp-creERT) or Zeb2fl/fl;Plp-creERT (Zeb2 iKO) mice at 4–6 weeks were treated with tamoxifen before and after injury, which efficiently removed Zeb2 from transected nerves in the Zeb2 iKO mice (Fig. 4c). Sciatic nerves were removed for analysis 14 days post-axotomy to assess SC differentiation, by which time a tissue bridge would have formed and axons guided by associated SCs as regeneration units would proceed to grow into the distal stump 26,27. Here we focused our analysis on the regenerating site defined as the SC-axon growth tip just proximal to the injury in the tissue bridge. In the regenerating region of Zeb2 iKO nerves, proliferation of mutant SCs (Ki67+Sox10+ cells) was indistinguishable from that of control SCs, however the number of Sox10+ SCs was significantly reduced (Fig. 4d–f). Proliferating SCs were not observed in either control or mutant contralateral sciatic nerves (data not shown). In the regenerating nerve region, mutant SCs failed to differentiate properly, as revealed by a substantial decrease in Oct6+ or Krox20+ pro-myelinating SCs in mutant nerves (Fig. 4g–i).
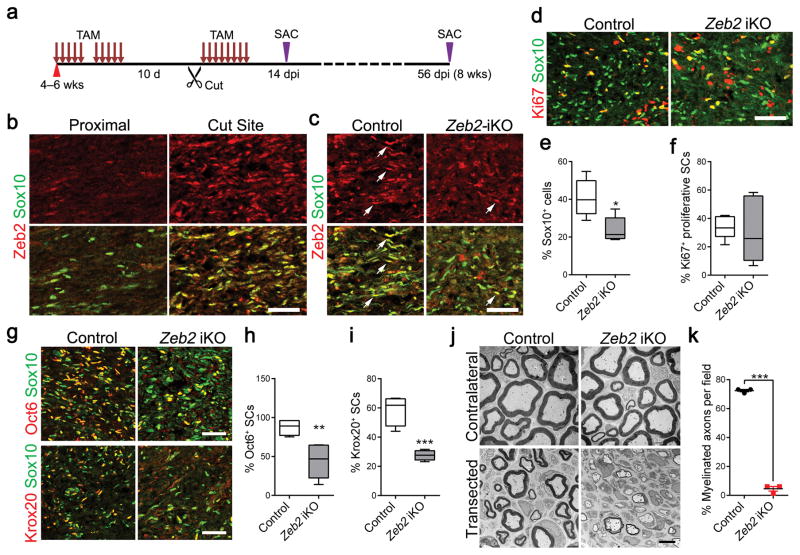
Figure 4. Zeb2 is required for SC differentiation during nerve repair.
(a) A diagram showing the nerve transection scheme in control and Zeb2 iKO. 4–6-week-old mice were treated with TAM via i.p. for 10 days following by nerve cut injury. Mice were then given TAM for 8 days, and nerves were analyzed at indicated days post injury (dpi).
(b) Immunofluorescence labeling for Zeb2 (red) and Sox10 (green) in lesions of control sciatic nerves at 14 dpi (n = 3 control and mutant tissues). SCs in the intact proximal nerves show negligible Zeb2 immunoreactivity. Scale bar: 50 μm.
(c) Immunofluorescence labeling for Zeb2 (red) and Sox10 (green) in the regenerating site of control and Zeb2 iKO sciatic nerves 14 dpi. Arrows: Sox10+/Zeb2+ SCs in control and Zeb2 iKO mutant nerves. Scale bar: 50 μm.
(d) Immunofluorescence labeling for Ki67 (red) and Sox10 (green) in the regenerating region of 14 dpi control and Zeb2 iKO sciatic nerves (n = 5 control and mutant tissues). Scale bar: 50 μm.
(e) Quantification of Sox10+ SCs in the regenerating area in 14 dpi control and Zeb2 iKO sciatic nerves. Data are presented as mean ± S.E.M; *P < 0.05; n = 5 control and mutant tissues; P = 0.0128 t = 3.192 df = 8; Two-tailed unpaired Student’s t-test. Whiskers show the minimum and maximum, boxes extend from the first to the third quartiles with cross lines at the medians.
(f) Quantification of Ki67+ proliferative SCs in the regenerating area in 14 dpi control and Zeb2-iKO sciatic nerves. Data are presented as mean ± S.E.M; not significant; n = 5 control and mutant tissues; P = 0.8308 t = 0.2207 df = 8; Two-tailed unpaired Student’s t-test. Whiskers show the minimum and maximum, boxes extend from the first to the third quartiles with cross lines at the medians.
(g) Immunofluorescence labeling of Sox10 (green) and Oct6 (red, upper) or Krox20 (red, bottom) in the regenerating region of 14 dpi control and Zeb2 iKO sciatic nerves (n = 5 control and mutant tissues). Scale bars: 50 μm.
(h) Quantification of the proportion of Oct6+ SCs in 14 dpi control and Zeb2 iKO sciatic nerves in the regenerating region (n = 4 control and 5 mutant tissues). Data are presented as mean ± S.E.M; ** P <0.01; P = 0.0087 t = 3.601 df = 7; Two-tailed unpaired Student’s t-test. Whiskers show the minimum and maximum, boxes extend from the first to the third quartiles with cross lines at the medians.
(i) Quantification of the proportion of Krox20+ SCs (right) in the regenerating region of 14 dpi control and Zeb2 iKO sciatic nerves (n = 4 control and 5 mutant tissues). Data are presented as mean ± S.E.M; ***P < 0.001; P = 0.0004 t = 6.402 df = 7; Two-tailed unpaired Student’s t-test. Whiskers show the minimum and maximum, boxes extend from the first to the third quartiles with cross lines at the medians.
(j) EM images of transverse sections of control and Zeb2 iKO 8 weeks after transection (n = 3 control and mutant tissues). Scale bar, 4 μm.
(k) Quantification of the proportion of myelinated axons per field from EM images of control vs. Zeb2 iKO 8 weeks after cut injury (n = 3 control and mutant tissues). Data are as mean ± S.E.M; ***P <0.0001; P < 0.0001 t = 37.64 df = 4; Two-tailed unpaired Student’s t-test. Whiskers show the minimum and maximum, boxes extend from the first to the third quartiles with cross lines at the medians.
We further assessed the extent of remyelination in control and Zeb2 iKO sciatic nerves by electron microscopy 8 weeks after surgery. During nerve regeneration, little, if any, radial sorting is required. In line with this, SCs lacking Zeb2 displayed normal association with axons in 1:1 ratios. Strikingly, while a majority of axons were remyelinated 8 weeks post injury in control nerves, Zeb2 iKO SCs were blocked at the promyelinating stage and failed to remyelinate (Fig. 4j,k). Myelin profiles in Zeb2 iKO nerves were scarce, among which the axons were primarily thinly myelinated. Together, our data suggest that Zeb2 is essential for SC myelination both in development and nerve regeneration.
Zeb2 represses a SC differentiation inhibitor Sox2
Given severe SC myelination defects in both developing and regenerating nerves in Zeb2 mutants, we then sought to examine the expression of classic negative myelination regulators, such as Sox2 and c-Jun 28. At perinatal stages, in keeping with the severe reduction of myelinating SCs marked by Krox20, expression of an immature SC marker Sox2, which maintains SCs in an undifferentiated state 28, persisted even at P14, when Sox2 was rarely detectable in control nerves (Fig. 5a,b). Western blotting analysis corroborated Sox2 upregulation in postnatal mutant nerves (Fig. 5c). By contrast, c-Jun, another myelination inhibitor 29, appeared normally expressed in mutant sciatic nerves compared with controls (Fig. 5d,e).
Figure 5. Zeb2 represses Sox2 activity to promote SC differentiation.
(a) Immunofluorescence labeling of Sox2 (red) and Sox10 (green) on P7 (n = 3 control and mutant tissues) and P14 (n = 4 control and mutant tissues) control and Zeb2 cKO sciatic nerve longitudinal sections. Scale bar: 50 μm.
(b) Quantification of Sox2+ cells relative to Sox10+ SCs in sciatic nerves of Zeb2 cKO versus controls at P1, P7 and P14. Data are presented as mean ± S.E.M, ***P < 0.001, n = 3 control and mutant tissues for P1; n = 5 control and 6 mutant tissues for P7; n = 3 control and mutant tissues for P14. P(P1) < 0.05 t = 3.391 df = 17; P(P7) < 0.0001 t = 7.079 df = 17; P(P14) < 0.0001 t = 7.823 df = 17; Two-way ANOVA with Sidak’s multiple comparisons test.
(c) A western blot showing persistent expression of Sox2 in sciatic nerves of Zeb2 cKO compared with controls at P14. GAPDH was the loading control. The experiment was repeated 3 times. Full-length blots are presented in Supplementary Figure 9a.
(d) Immunofluorescence labeling for c-Jun (green) and Sox10 (red) in P7 control and mutant sciatic nerves (n = 3 animals/genotype). Scale bar: 50 μm.
(e) Quantification of c-Jun+ cells relative to DAPI+ cells in sciatic nerves of Zeb2 cKO versus controls at P1 and P7. Data are presented as mean ± S.E.M, n = 3 control and mutant tissues, P(P1) = 0.5188 t = 0.7066 df = 4; P(P7) = 0.2363 t = 1.392 df = 4; Two-tailed unpaired Student’s t-test.
(f) Immunostaining showing Krox20 expression in SCs transfected with control (pGFP) and Sox2 (pSox2) vectors in cAMP containing-differentiation medium for 3 days (n = 5 independent experiments). Scale bar: 20 μm.
(g) Quantification of Krox20+ among SCs transfected with pGFP and pSox2, differentiated for 3 days by cAMP induction. Data are presented as mean ± S.E.M, ***P < 0.001, n = 3 independent experiments, P = 0.0005 t = 7.922 df = 5; Two-tailed unpaired Student’s t-test.
(h) Quantification of proliferating Ki67+ cells among SCs transfected with pGFP and pSox2, cultured in neuregulin-containing proliferation medium. Data are presented as mean ± S.E.M, n = 4 independent experiments, P = 0.6864 t = 0.4202 df = 6; Two-tailed unpaired Student’s t-test.
(i) Diagram (left) showing the promoter of rat Sox2 carrying the consensus Zeb2 binding sites [CACCT(g)] (red bars). TSS denotes transcription start site. ChIP-qPCR assays (right) for Zeb2 enrichment to the Zeb2 binding sites (P1–P3) in the Sox2 promoter on chromatin prepared from primary SCs exposed in proliferation or differentiation (9 h) media. IgG IP was used as control. ***P < 0.0001; (Proliferation: P(P1) = 0.0612 t = 2.299 df = 6 n = 4; P(P2) = 0.1428 t = 1.625 df = 8 n = 5; P(P3) = 0.1353 t = 1.725 df = 6 n = 4; Differentiation: P(P1) = 0.0046 t = 3.34 df = 14 n = 8; P(P2) = 0.0002 t = 5.02 df = 14 n = 6; P(P3) = 0.0001 t = 18.4 df = 10 n = 6; Two-tailed unpaired Student’s t-tests; n: independent experiments. Whiskers show the minimum and maximum, boxes extend from the first to the third quartiles with cross lines at the medians.
(j) qRT-PCR showing Sox2 expression in Zeb2-overexpressing SCs induced to differentiate for 18 hours, compared with vector-expressing control. Data are presented as mean ± S.E.M (*P < 0.05; n = 3 independent experiments, P = 0.0211 t = 3.686 df = 6; Two-tailed unpaired Student’s t-test).
(k) Rat SCs transfected with luciferase reporter driven by wildtype Sox2 promoter (−2 kb from TSS) or mutant Sox2 promoter (Zeb2 binding site P1 deleted) together with Zeb2 expressing vectors. Values represent the average percentages of luciferase activity normalized to the corresponding vector expression controls from 5 independent experiments. Data are presented as mean ± S.E.M, **P < 0.01, P = 0.0013 t = 4.582 df = 9; Two-tailed unpaired Student’s t-test.
Sox2 overexpression in SCs in vitro markedly suppressed expression of Krox20 upon induction of differentiation by dibutyl cyclic AMP (Fig. 5f,g), though no apparent alterations of SC proliferation were detected (Fig. 5h). These observations suggest that upregulation of the differentiation inhibitor Sox2 caused by Zeb2 deletion, inhibits SC differentiation.
Since Zeb2 can function as a transcriptional repressor 25, we then hypothesized that it might directly target the promoter-proximal regulatory elements of Sox2 and inhibit its expression. We identified three conserved Zeb2 consensus-binding sites in the promoter region of Sox2 within 5 kb upstream of the transcription start site (TSS) (Fig. 5i). ChIP assays with an anti-Zeb2 specific antibody showed a strong enrichment of endogenous Zeb2-binding to these sites in the Sox2 promoter region in differentiating SCs, but not in their proliferating immature state (Fig. 5i). In contrast, no occupancy of Zeb2 to its candidate binding site in c-Jun promoter was detected (Supplementary Fig. 4a). There was also no enrichment of Zeb2 binding to Sox2 distal control regions (ChIP-PCR; Supplementary Fig. 4b). Furthermore, Zeb2 overexpression in SCs significantly reduced Sox2 (Fig. 5j). Consistently, enforced Zeb2 expression in SCs effectively repressed the Sox2 promoter activity by luciferase reporter assays (Fig. 5k). To address the specificity of Zeb2 action, we co-transfected the Zeb2 vector together with the Sox2 promoter in which the Zeb2 binding site was mutated. Zeb2 repressive activity was significantly diminished and luciferase reporter activity was restored to control levels (Fig. 5k). Together, our data indicate that Zeb2 targets directly to the promoter of Sox2 and inhibits Sox2 expression to promote terminal SC differentiation.
Zeb2 regulates multiple SC differentiation pathways
To identify potential additional Zeb2 targets that regulate SC differentiation, we performed RNA sequencing analysis (RNA-Seq) of control and Zeb2 cKO sciatic nerves at P7. Gene ontology analysis of differentially regulated genes indicated that the functions of down-regulated genes were particularly pertinent to lipid biosynthesis, myelination and SC differentiation (Fig. 6a), whereas those of up-regulated genes could be classified into categories involving cell proliferation control, adhesion and extracellular matrix organization (Fig. 6b). Consistent with our in situ hybridization and qPCR results, we detected a strong down-regulation of major myelin genes and differentiation regulatory genes such as Mbp, Mag, Mpz, Krox20 and Oct6, with a concomitant increase in myelination inhibitors, including Sox2, Jagged1 and Egr1 in Zeb2 cKO sciatic nerves (Fig. 6c,d). In keeping with the dramatic sorting defect in Zeb2 cKO nerves, we detected differential expression of selected genes involved in the regulation of radial sorting, including laminin α2, cdc42 and integrin α6 30 (Supplementary Fig. 5a,b). The RNA-Seq data demonstrated an upregulation of TGFβ signaling genes including Tgfb2, Bmper, Smad3 and Bmp7 in Zeb2 cKO sciatic nerves (Fig. 6c), however, in contrast to an activation of BMP receptor-activated Smads (Smad1/5/8) and downregulation of Smad7 observed in OLs lacking Zeb2 8, we did not detect Smad activation or BMP signaling inhibition, assayed by phosphorylation of Smad1/5/8 and gross Smad7 levels, respectively (Supplementary Fig. 6). These observations indicate that overall BMP-Smad signaling is not substantially altered in Zeb2-mutant SCs, suggesting that divergent mechanisms from those in CNS myelinogenesis 8 are operated by Zeb2 in SCs for the control of PNS myelination.
Figure 6. Zeb2 regulates the SC differentiation program and inhibits Notch effector Hey2 expression.
(a,b) Gene ontology analysis of downregulated (a) or upregulated (b) genes in P7 Zeb2 cKO sciatic nerves compared with controls. Each dot in the connecting lines represents the gene count of the corresponding biological function categories. Bars represent −log (P-value)..
(c) A heatmap showing representative differential expressed genes and their categories in control vs. Zeb2 cKO sciatic nerves at P7. n = 2 independent experiments.
(d) qRT-PCR analysis of the genes related to SC differentiation, neural crest, Notch and Tgfβ/BMP pathways in Zeb2 cKO sciatic nerves relative to controls at P7. Data are presented as mean ± S.E.M, n = 3 independent experiments. (*P < 0.05; **P < 0.01; ***P < 0.001; P(Prx) < 0.0001 t = 2750 df = 4; P(Mbp) < 0.0001 t = 512.5 df = 4; P(Sox10) < 0.0001 t = 101.4 df = 4; P(Krox20) < 0.0001 t = 291.8 df = 4; P(Oct6) < 0.0001 t = 14865 df = 4; P(Twist1) = 0.0003 t = 11.99 df = 4; P(Snail) = 0.0016 t = 4.45 df = 9; P(Slug) = 0.0007 t = 9.39 df = 4; P(Hes1) = 0.0974 t = 1.962 df = 6; P(Notch1) = 0.2186 t = 1.376 df = 6; P(Jag1) = 0.0496 t = 2.784 df = 4; P(Jag2) = 0.0179 t = 3.879 df = 4; P(Hey2) = 0.0284 t = 3.355 df = 4; P(Tgfb2) = 0.001 t = 6.000 df = 6; P(Bmper) = 0.0313 t = 3.254 df = 4; P(Bmp7) = 0.0013 t = 5.703 df = 6; Two-tailed unpaired Student’s t-test).
(e) qRT-PCR analysis of Hey2 levels in mouse sciatic nerves during development (n = 3 control and mutant tissues for each time point). Data are presented as mean ± S.E.M.
(f) Transcript levels of Hey2 in Zeb2 cKO sciatic nerves relative to those in control nerves at P7, 15 and 30. Data are presented as mean ± S.E.M (*P < 0.05; **P < 0.01; ***P < 0.001, n = 3 control and mutant tissues for each time point, P(P7) = 0.0136 t = 5.232 df = 4; P(P15) = 0.0101 t = 4.590 df = 4; P(P30) = 0.0097 t = 4.641 df = 4; Two-tailed unpaired Student’s t-test).
(g) Western blot showing Hey2 expression in control and Zeb2 cKO sciatic nerves at P14. GAPDH was the loading control. Data are from 3 independent experiments. Full-length blots are presented in Supplementary Fig. 9b.
(h) ChIP analysis of Zeb2 recruitment to the consensus Zeb2 binding site on the Hey2 promoter (red bar in upper panel) in primary SCs under proliferation or differentiation (9 hours) media. IgG IP was used as control. (Data are mean ± S.E.M; ***P < 0.001; Proliferation: P = 0.9367 t = 0.08283 df = 6 n = 4; Differentiation: P = 0.0005 t = 10.27 df = 4 n = 3; Two-tailed unpaired Student’s t-test). n: independent experiments.
(i) qRT-PCR analysis showing Hey2 gene expression in Zeb2-overexpressing SCs under differentiation media for 18 hours, compared with vector-expressing control (*P < 0.05; n = 3 independent experiments, P = 0.0272 t = 3.404 df = 4; Two-tailed unpaired Student’s t-test).
(j) Percentages of normalized luciferase activity in rat SCs transfected with luciferase reporter driven by wildtype or mutant Hey2 promoter together with Zeb2 expressing vector. Data are presented as mean ± S.E.M. n = 3 independent experiments. **P < 0.01, P = 0.0025 t = 6.747 df = 4; Two-tailed unpaired Student’s t-test.
(k) Immunostaining showing Krox20 expression in rat SCs transfected with control (pGFP) and Hey2-expressing (pHey2) vectors under differentiation condition for 3 days. The experiment was repeated 3 times. Scale bar: 20 μm.
(l) Quantification of the percentage of Krox20+ among transfected cells (n = 3 independent experiments). Data are presented as mean ± S.E.M; *P < 0.05; P = 0.0411 t = 2.733 df = 4; Two-tailed unpaired Student’s t-test.
(m) Immunostaining showing Ki67 expression in rat SCs transfected with control (pGFP) and Hey2-expressing (pHey2) vectors for 48h in proliferation medium. Scale bar: 20 μm. Data are from 5 independent experiments. (n) Quantification of the percentage of Ki67+ among transfected cells (n = 5 independent experiments). Data are presented as mean ± S.E.M; **P < 0.01; P = 0.0052 t = 3.800 df = 8; Two-tailed unpaired Student’s t-test.
(o) qRT-PCR showing the efficiency of siRNA-mediated knockdown of Zeb2, Sox2 and Hey2. Data are presented as mean ± S.E.M; *** P < 0.001; n = 5 independent experiments; P(Zeb2) < 0.0001 t = 23.37 df = 9; P(Hey2) < 0.0001 t = 11.43 df = 8; P(Sox2) < 0.0001 t = 11.47 df = 9; Two-tailed unpaired Student’s t-test. Whiskers show the minimum and maximum, boxes extend from the first to the third quartiles with cross lines at the medians.
(p) qRT-PCR showing myelin-associated gene expression in rat SCs with siRNA knockdown of Zeb2 and Sox2 or Hey2 compared with single Zeb2 siRNA knockdown. Data are presented as mean ± S.E.M; ** P < 0.01; *** P < 0.001; n = 5 independent experiments; One-way ANOVA with Tukey’s multiple comparisons test; P(Krox20) < 0.001: scrambled vs si-Zeb2, si-Zeb2 vs si-Zeb2,si-Hey2; P(Krox20) < 0.01: si-Zeb2 vs si-Zeb2,si-Sox2; F (3,16) = 345.8; P(Mpz) < 0.001: scrambled vs si-Zeb2, si-Zeb2 vs si-Zeb2,si-Sox2; P(Mpz) < 0.01: si-Zeb2 vs si-Zeb2,si-Hey2; F (3,10) = 29.67; P(Mbp) < 0.001: scrambled vs si-Zeb2, si-Zeb2 vs si-Zeb2,si-Hey2; P(Mbp) < 0.05: si-Zeb2 vs si-Zeb2,si-Sox2; F (3,10) = 20.59.
Zeb2 inhibits a novel Notch-Hey2 axis for SC differentiation
We next set out to investigate other potential Zeb2-regulated signaling pathways for SC differentiation based on transcriptome profiling analysis. We detected that a cohort of signaling components in the Notch pathway were significantly upregulated in Zeb2 cKO sciatic nerves (Fig. 6c). Among the up-regulated genes, Hey2, a Hes-related family basic helix-loop-helix (bHLH) effector of Notch 31, displayed the most substantial increase in mRNA level compared to control nerves (Fig. 6d). Because Hey2 functions as a transcriptional repressor in other systems, such as in cardiac and vascular development 31,32, we postulated that Hey2 might act as a blocker of SC differentiation. Hey2 transcript levels peaked at P5 and then rapidly declined to reach near-zero levels at P15, which closely paralleled the expression profile of myelination inhibitors, such as Sox2 28,33 (Fig. 6e). We validated the increase in Hey2 mRNA and protein levels in Zeb2 cKO nerves by qPCR (Fig. 6f) and western blotting (Fig. 6g), respectively.
To determine whether Zeb2 could regulate Hey2 directly, we identified an evolutionarily conserved Zeb2 binding site upstream of the TSS, and found that Zeb2 binding was enriched on the Hey2 promoter in differentiating SCs, but not in proliferating cells (Fig. 6h). ChIP of the distal control region of Hey2 showed no Zeb2 binding (Supplementary Fig. 4b). Moreover, enforced Zeb2 expression in SCs significantly down-regulated Hey2 (Fig. 6i). Zeb2 overexpression in SCs repressed Hey2 promoter activity, and its repression was partially abolished when the Zeb2-binding site in this promoter was mutated (Fig. 6j). Overexpression of Hey2 inhibited Krox20 induction in SCs cultured under differentiation conditions (Fig. 6k–l). In contrast, the proliferative capacity of SCs assessed by Ki67 expression with such constitutive Hey2 expression was significantly higher than that of control GFP-transfected SCs (Fig. 6m–n). These observations suggest that Hey2 inhibits SC differentiation by arresting SC in an undifferentiated state and driving their proliferation, and that Zeb2 targets directly to repress Hey2 to promote SC myelination.
To further confirm that Zeb2 down-regulates Hey2 or Sox2 transcription in order to promote SC differentiation, we performed sequential siRNA-mediated knockdown experiments, first by Zeb2 knockdown in SCs and then followed 24h later by either knockdown of Sox2 or Hey2 using siRNA. Subsequent knockdown of Sox2 or Hey2 in Zeb2-deficient SCs partially restored myelin-associated gene expression (Fig. 6o,p), indicating that Zeb2 drives SC differentiation via repression of inhibitory factor genes such as Sox2 and Hey2.
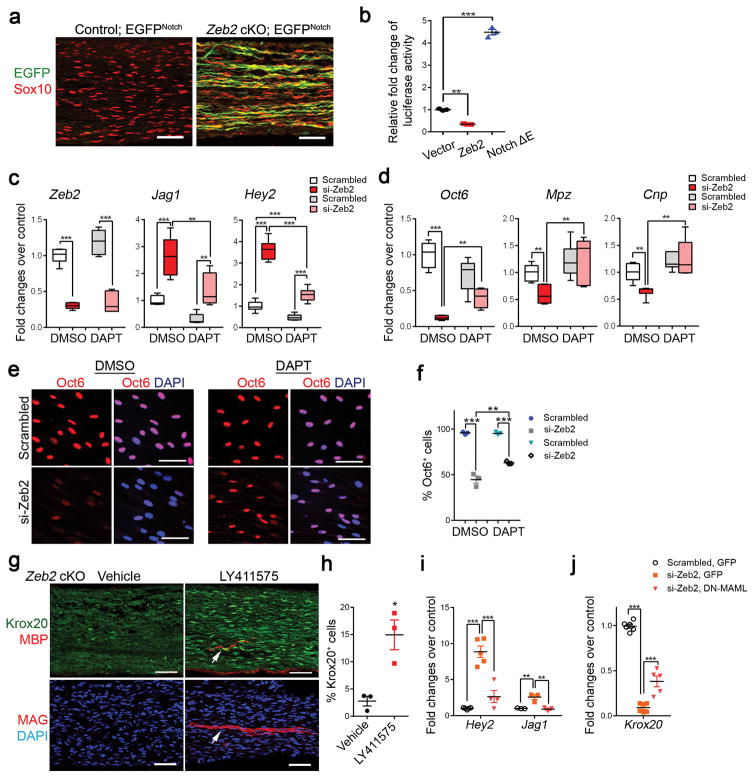
Considering that Notch signaling negatively controls SC myelination 16, we explored the possibility that Zeb2 promotes SC differentiation and myelination by opposing a Notch pathway-mediated differentiation block. Hence, we generated Zeb2 cKO mice carrying a Notch-responsive GFP reporter transgene (Supplementary Fig. 8a) 34. We detected a striking induction of Notch-dependent GFP expression in Zeb2 mutant nerves (Fig. 7a). Furthermore, overexpression of Zeb2 significantly repressed the reporter activity of the TP1 promoter carrying RBP-Jκ binding sites (Fig. 7b), which are highly specific to Notch signaling activation 35, suggesting that Zeb2 inhibits Notch signaling in SCs.
Figure 7. Zeb2 represses a Notch-Hey2 signaling axis.
(a) Representative immunolabeling of GFP and Sox10 in P7 sciatic nerves of control and Zeb2 cKO mice carrying a Notch-EGFP reporter (n = 3 for each group). Scale bar: 50 μm.
(b) Luciferase activity in HEK 293T cells transfected with a luciferase reporter driven by Notch-responsive TP-1 promoter and Zeb2 or Notch ΔE expressing vector. Data are presented as mean ± S.E.M, **P < 0.01; ***P < 0.001; P(Zeb2) < 0.001; P(Tp1) < 0.0001; F (2,6) = 890.1; n = 3 independent experiments; One-way ANOVA with Tukey’s multiple comparison tests.
(c) qRT-PCR showing Jagged1 and Hey2 expression in Zeb2 knockdown in SCs treated with 5μM DAPT over 4 days. Data are presented as mean ± S.E.M; *P < 0.05; **P < 0.01; ***P < 0.001, one-way ANOVA with Tukey’s multiple comparisons test; Zeb2: P(DMSO) < 0.0001, P(DAPT) < 0.0001; F (3,20) = 79.78; Jag1: P(DMSO) < 0.001, P(DAPT) < 0.01; P(si-Zeb2, DMSO vs DAPT) < 0.01; F (3,19) = 22.75; Hey2: P(DMSO) < 0.0001, P(DAPT) < 0.0001; P(scrambled, DMSO vs DAPT) < 0.05; P(si-Zeb2, DMSO vs DAPT) < 0.0001; F (3,20) = 119.5; Data are from 6 independent experiments.
(d) qRT-PCR showing Oct6, Mpz and Cnp expression in Zeb2 knockdown SCs treated with DAPT. Data are presented as mean ± S.E.M; **P < 0.01; ***P < 0.001; one-way ANOVA with Tukey’s multiple comparison’s test; Oct6: P(DMSO) < 0.0001; P(si-Zeb2, DMSO vs DAPT) < 0.01; F (3,18) = 33.29; Mpz: P(DMSO) < 0.01; P(si-Zeb2, DMSO vs DAPT) < 0.01; F (3,19) = 6.745; Cnp: P(DMSO) < 0.05; P(si-Zeb2, DMSO vs DAPT) < 0.001; F (3,19) = 10.16; Data are from 5 independent experiments.
(e) Immunostaining of Oct6 (red) in scrambled or si-Zeb2 knockdown SCs treated with DMSO or DAPT for 4 days in differentiation medium. DAPI, blue. The experiment was repeated 3 times. Scale bar: 50 μm.
(f) Quantification of Oct6-expressing cells among scrambled or si-Zeb2 knockdown SCs treated with DMSO or DAPT for 4 days in differentiation medium. Data are presented as mean ± S.E.M; **P < 0.01; ***P < 0.001; one-way ANOVA with Tukey’s multiple comparison’s test; P(DMSO) < 0.001, P(DAPT) < 0.001; P(si-Zeb2, DMSO vs DAPT) < 0.01; F (3,8) = 134.6. Data are from 3 independent experiments.
(g) Immunostaining of Krox20, MBP or MAG in sciatic nerves of Zeb2 cKO mice treated with vehicle or Notch-inhibitor LY411575 from P2–5 and harvested on P8. Arrows indicate MBP+ or MAG+ myelin segments. Data are from 3 individual mice from each group. Scale bars: 50 μm.
(h) Quantification of Krox20+ nuclei in Zeb2 cKO sciatic nerves treated with vehicle or LY411575. Data are presented as mean ± S.E.M; n = 3 individual mice from each group; *P < 0.05; P = 0.0135 t = 4.221 df = 4; Two-tailed unpaired Student’s t-test.
(i) qRT-PCR showing Jagged1 and Hey2 expression in Zeb2 knockdown SCs transduced with GFP or DNMAML-GFP-expressing lentivirus for 48 h in proliferation medium. Data are presented as mean ± S.E.M; **P < 0.01; ***P < 0.001; One-way ANOVA with Tukey’s multiple comparison test, Jag1: P(Scrambled GFP vs si-Zeb2 GFP) < 0.01, P(si-Zeb2 GFP vs si-Zeb2 DN-MAML GFP) < 0.01; F (2,6) = 18.91; Hey2: P(Scrambled GFP vs si-Zeb2 GFP) < 0.001, P(si-Zeb2 GFP vs si-Zeb2 DN-MAML GFP) < 0.001; F (2,9) = 30.74. Data are from 3 independent experiments.
(j) qRT-PCR showing Krox20 levels in Zeb2-knockdown SCs transduced with GFP or DNMAML-GFP-expressing lentivirus. Data are presented as mean ± S.E.M; ***P < 0.001; One-way ANOVA with Tukey’s multiple comparison test P(Scrambled GFP vs si-Zeb2 GFP) < 0.0001, P(si-Zeb2 GFP vs si-Zeb2 DN-MAML GFP) < 0.0001; F (2,16) = 244.8. Data are from 5 independent experiments.
Notch inhibition restores differentiation of Zeb2-mutant SCs
To test whether activation of Notch signaling is responsible for the differentiation failure in Zeb2-deficient SCs, we treated scramble and Zeb2-knockdown SCs over 4 days with a Notch inhibitor DAPT (5 μM), which inhibits γ-secretase, a protease required for Notch1 activation 36. The addition of DAPT efficiently inhibited gene expression of Jagged1 and Hey2 in scramble-transfected SCs (Fig. 7c), whereas this treatment did not alter Zeb2 levels. Consistently, SCs with Zeb2 knockdown showed increased levels of the Notch signaling genes Jagged1 and Hey2. DAPT treatment effectively reduced expression of these genes (Fig. 7c), and restored major myelin gene (e.g. Mpz, Cnp) expression to normal levels in Zeb2-knockdown SCs (Fig. 7d). Notch inhibition by DAPT also partially rescued the otherwise perturbed induction of Oct6 in differentiating SCs with Zeb2 knockdown (Fig. 7d–f). Strikingly, treatment of Notch inhibitor LY411575 in vivo led to a robust increase in Krox20+ SCs and appearance of MBP+ or MAG+ myelin segments in neonatal Zeb2 cKO mice (Fig. 7g–h). Furthermore, blocking Notch signaling transduction by delivering lentiviral virus expressing a dominant-negative form of a Notch coactivator, Mastermind (DN-MAML) 37, reduced Hey2 or Jag1 expression and partially restored expression of Krox20 in Zeb2-knockdown SCs (Fig. 7i–j). Collectively, our data suggest that Zeb2 inhibits Notch activation, at least in part, by suppressing its downstream effector Hey2, thereby promoting SC differentiation from their undifferentiated state.
Zeb2 recruits HDAC to antagonize differentiation inhibition
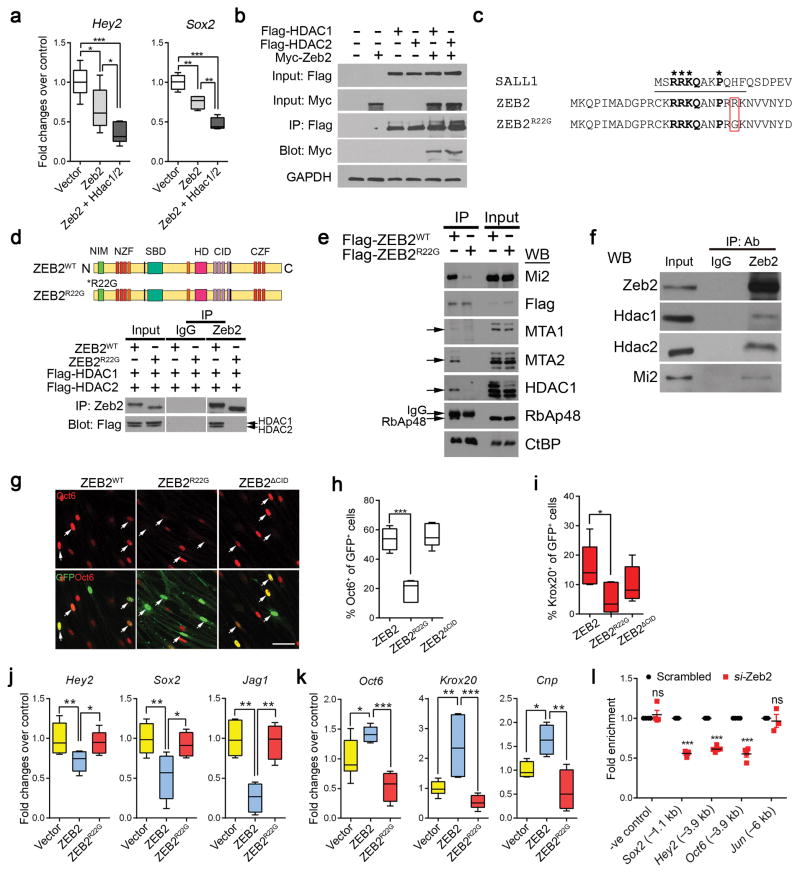
The observation that Zeb2 could effectively suppress multiple differentiation inhibitory pathways prompted us to postulate that Zeb2 mediates its function via forming a co-repressor complex. A previous study of the functional domain of Zeb2 has identified the NuRD-interacting motif (NIM) close to the N-terminus of Zeb2, which is similar to the minimal domain needed for NuRD recruitment identified in the transcription factor Sall1 10,38. Zeb2 associates with NuRD components including histone deacetylases HDAC1 and HDAC2 10. HDAC1/2 controls various stages of SC development; conditional inactivation of HDAC1 and HDAC2 in the SC lineage impairs radial sorting, SC development and myelination 39,40, which closely resembled the Zeb2 cKO phenotype. We therefore investigated the possibility of Zeb2 interaction with HDAC in a NuRD co-repressor complex context for achieving suppression of negative myelination regulators. Co-expression of Zeb2 together with HDAC1 or HDAC2 enhanced the repression of Hey2 and Sox2, to an extent much greater than Zeb2 overexpression alone, in SCs (Fig. 8a). By co-immunoprecipitation, we showed that HDAC1 and HDAC2 were present in a complex with Zeb2 (Fig. 8b).
Figure 8. Zeb2 associates with HDAC1 and HDAC2 to repress SC differentiation inhibitor expression.
(a) qRT-PCR showing the fold change in Hey2 and Sox2 expression in SCs overexpressing Zeb2 or coexpressing Zeb2, HDAC1 and HDAC2 relative to vector-expressing cells. Data are presented as mean ± S.E.M; *P < 0.05; **P < 0.01; ***P < 0.001. One-way ANOVA with Tukey’s multiple comparison tests; Hey2: P(vector vs Zeb2) < 0.05, P(Zeb2 vs Zeb2 + Hdac1/2) < 0.05; P(vector vs Zeb2 + Hdac1/2) < 0.0001; F (2,20) = 15.72; Sox2: P(vector vs Zeb2) < 0.001, P(Zeb2 vs Zeb2 + Hdac1/2) < 0.001; P(vector vs Zeb2 + Hdac1/2) < 0.0001; F (2,10) = 40.59. Data are from 3 independent experiments.
(b) Co-immunoprecipitation with Flag-tagged HDAC1 or HDAC2 in HEK 293T cells co-transfected with Myc-tagged Zeb2. IP, immunoprecipitation; GAPDH, loading control. Full-length blots are presented in Supplementary Figure 9c. The experiment was repeated 3 times.
(c) Alignment of Sall1, human wildtype and mutant ZEB2R22G in NuRD binding domain (underline). Amino acids in bold are critical for NuRD binding. The red box highlights the amino acid change in the ZEB2R22G mutant.
(d) (Upper) Schematic domains in Zeb2WTand Zeb2R22G proteins. NZF and CZF; N- and C-terminal zinc finger domains, NIM, NuRD interacting motif, SBD, Smad-binding domain, HD, homeodomain-like domain, and CID, the CtBP interacting domain. (Bottom). Coimmunoprecipitation of Zeb2 in HEK 293T cells transfected with Flag-tagged HDAC1 and HDAC2 together with Zeb2 or Zeb2R22G expressing vectors. IgG was used as the IP control. The experiment was repeated 3 times. Full-length blots are presented in Supplementary Figure 9d.
(e) Western-blot showing co-immunoprecipitation of endogenous NuRD subunits with ZEB2WT, but not ZEB2R22G in HEK293T cells transiently expressing Flag-ZEB2WT or Flag-ZEB2R22G. The experiment was repeated 3 times. Full-length blots are presented in Supplementary Figure 10b.
(f) Western blot showing co-immunoprecipitation with an antibody to Zeb2 and control IgG in rat SCs using antibodies to HDAC1. HDAC2 and Mi-2. The experiment was repeated 3 times. Full-length blots are presented in Supplementary Figure 10a.
(g) Immunolabeling of Oct6 or Krox20 in rat SCs transfected with Zeb2WT, Zeb2R22G or Zeb2ΔCID vectors carrying a GFP reporter for 48h under differentiation condition. Arrows, transfected cells. Scale bar: 50 μm. Data are from 5 independent experiments.
(h) Quantification of the proportion of Oct6+ cells among transfected GFP+ cells (n = 5 independent experiments). Data are presented as mean ± S.E.M; ***P < 0.001. One-way ANOVA with Tukey’s multiple comparison tests; P(Zeb2 vs Zeb2R22G) < 0.0001; P(Zeb2 vs Zeb2CID) = 0.5330; F (2,11) = 36.46.
(i) Quantification of the proportion of Krox20+ cells among transfected cells (n = 5 independent experiments). Data are presented as mean ± S.E.M; *P < 0.05. One-way ANOVA with Tukey’s multiple comparison tests; P(Zeb2 vs Zeb2R22G) < 0.05; P(Zeb2 vs Zeb2CID) = 0.2225; F (2,12) = 3.497.
(j) qRT-PCR analysis of Zeb2 targets, Jag1, Hey2 and Sox2 in rat SCs transfected with Zeb2 or Zeb2R22G mutant expression vectors. Data are presented as mean ± S.E.M; *P < 0.05; **P < 0.01; One-way ANOVA with Tukey’s multiple comparisons test; Hey2: P(vector vs Zeb2) < 0.01, P(Zeb2 vs Zeb2R22G) < 0.05; F (2,21) = 6.884; n = 7 independent experiments; Jag1: P(vector vs Zeb2) < 0.01, P(Zeb2 vs Zeb2R22G) < 0.01; F (2,10) = 16.24; n = 4 independent experiments; Sox2: P(vector vs Zeb2) < 0.01, P(Zeb2 vs Zeb2R22G) < 0.05; F (2,13) = 7.584; n = 5 independent experiments. Whiskers show the minimum and maximum, boxes extend from the first to the third quartiles with cross lines at the medians.
(k) qRT-PCR analysis of myelin-related genes Oct6, Krox20 and Cnp in rat SCs transfected with Zeb2 or Zeb2R22G mutant expression vectors. Data are presented as mean ± S.E.M; *P < 0.05; **P < 0.01; ***P < 0.01; One-way ANOVA with Tukey’s multiple comparisons tests; Oct6: P(vector vs Zeb2) < 0.05, P(Zeb2 vs Zeb2R22G) < 0.001; F (2,15) = 20.24; n = 5 independent experiments; Krox20: P(vector vs Zeb2) < 0.01, P(Zeb2 vs Zeb2R22G) < 0.001; F (2,14) = 12.31; n = 6 independent experiments; Cnp: P(vector vs Zeb2) < 0.05, P(Zeb2 vs Zeb2R22G) < 0.01; F (2,9) = 11.25; n = 4 independent experiments. Whiskers show the minimum and maximum, boxes extend from the first to the third quartiles with cross lines at the medians.
(l) HDAC1 occupancy by ChIP–PCR in rat SCs knocked down with control or Zeb2 siRNA on the promoters of Zeb2 target genes. Chromatin was from SCs induced to differentiate by cAMP-containing medium for 9 h. IgG IP was used as −ve control. Data are mean ± S.E.M; ***P < 0.001; n = at least 3 independent experiments; P(−ve control) = 0.4683 t = 0.7741 df = 6; P(Sox2, −1.1kb) < 0.0001 t = 32.85 df = 8; P(Hey2, −3.9kb) < 0.0001 t = 24.63 df = 8; P(Oct6, −3.9kb) < 0.0001 t = 11.26 df = 6; P(Jun, −6kb) = 0.6954 t = 0.4209 df = 4; Two-tailed unpaired Student’s t-test.
We identified a novel ZEB2 variant [c.64A>G; p.(Arg22Gly)] carrying a single residue ZEB2 mutation (R22G) in a patient with mild MOWS in the NuRD-interacting domain of ZEB2 (ZEB2R22G; Fig. 8c). Strikingly, we found that ZEB2R22G was sufficient to abolish HDAC1 and HDAC2 recruitment to the ZEB2 complex (Fig. 8d). Moreover, this missense mutation in ZEB2 markedly disrupted ZEB2 interaction with other NuRD subunits as determined by co-immunoprecipitation, whereas CtBP (C-terminal-binding protein 1) binding was unaffected (Fig. 8e; Supplementary Fig. 8b). We further confirmed the endogenous interaction of Zeb2 with NuRD complex components including HDAC1, HDAC2 and Mi2/Chd4 in rat SCs (Fig. 8f). Most notably, exogenous expression of ZEB2R22G markedly diminished the induction of Oct6 and Krox20 in SCs (Fig. 8g–i). However, overexpressing a Zeb2 mutant protein with deletion of CtBP-interacting domains (ZEB2ΔCID) 41 did neither alter Oct6 (Fig. 8g–h) nor Krox20 (Fig. 8i) induction in SCs. Consistently, enforced Zeb2R22G expression in SCs also lost the capacity to repress Zeb2 target genes, such as Hey2, Sox2, and Jag1, or to activate the transcription of myelin-related genes, Oct6, Krox20 and Cnp (Fig. 8j–k).
To determine whether HDAC1/2 recruitment by Zeb2 is required for target expression, we performed ChIP–qPCR in Zeb2-deficient SCs. The reduction of Zeb2 in SCs significantly compromised the enrichment of HDAC1 on the Zeb2-consensus binding regions in the promoters of Zeb2 target genes, such as Sox2, Oct6 and Hey2, in Zeb2-knockdown SCs (Fig. 8l). In contrast, HDAC1 occupancy on the promoter of a non-Zeb2 target, c-Jun, was comparable with IgG control (Fig. 8m). Similar results were also obtained for HDAC2 recruitment (data not shown). This suggests that the Zeb2-HDAC1/2 complex is required for the regulation of target gene transcription in SCs. Thus, our findings indicate that Zeb2 represses SC differentiation inhibitors by associating with the HDAC1/2-NuRD co-repressor complex to promote SC myelination.
Discussion
In this study, we provide the in vivo genetic evidence for the critical function of Zeb2 in SC precursor differentiation and peripheral nerve myelination, as well as remyelination during nerve regeneration, consistent with the observations by Quintes et al., 2016 42 [this issue of Nature Neuroscience]. We establish Zeb2 as a key regulatory switch that controls the timing for the onset of normal SC differentiation and remyelination after injury. Furthermore, our findings further reveal that Zeb2 cooperates with a HDAC1/2-NuRD chromatin-remodeling complex as repressor to target a network of inhibitory pathways, including Sox2 effects and Notch signaling to drive SC differentiation, in conjunction with acting as a transcriptional activator of other target genes such as Oct6 as well 43. Moreover, we identify a novel Zeb2-modulated Notch-Hey2 signaling axis that blocks SC differentiation. Inhibition of Notch signaling partially rescues the SC differentiation defect caused by Zeb2 deficiency. Thus, Zeb2 regulates SC differentiation and myelination at least in part through de-repression of Notch signaling (Supplementary Fig. 8c,d), pointing to the importance of HDAC1/2-NuRD-dependent Zeb2 activity in MOWS-associated neuropathies.
Zeb2 recruits HDAC1/2-NuRD to control SC differentiation
Delineation of differentially expressed genes by transcriptome analysis reveals that Zeb2 modulates inhibitory pathways, namely Sox2 effects and Notch-Hey2 signaling, during SC differentiation. Whether Zeb2 regulation of these individual inhibitory signals, exists in parallel, and whether Zeb2 mediates its actions through other unidentified co-factors of these or other pathways to control SC myelination, would require further investigation. Nonetheless, we find that Zeb2 engages the HDAC1/2-NuRD complex to exert repressive activity on myelination inhibitory factors. Several studies have reported radial sorting and myelination impairment in mouse mutants with SC-specific inactivation of NuRD subunits or associated factors including Mi-2/Chd4 and HDAC1/2 39,40,44 as well as interacting partners such as NAB proteins 33,45. Co-expression of Zeb2, HDAC1 and HDAC2 in SCs further augments repression in expression of inhibitory genes Sox2 and Hey2. Remarkably, a single missense mutation Zeb2R22G in the Zeb2 N-terminal NuRD-interacting domain completely abolishes its association with the HDAC1/2-NuRD and inhibits SC differentiation, which is likely due to inefficient repression of the aforementioned differentiation-inhibitory signals. Thus, by interacting with the HDAC1/2-NuRD complex 46, Zeb2 may mediate its repressive activity to fine-tune transcription of the genes that negatively impact on myelination. Taken together, our study indicates that Zeb2 acts as a key transcriptional repressor that effectively extinguishes multiple inhibitory pathways to direct the onset of SC differentiation and initiation of myelination.
A novel Notch-Hey2 axis for inhibiting SC differentiation
Notch signaling regulates multiple stages of SC development, from promoting SC formation and proliferation to negatively regulating myelination 16. Strikingly, in the absence of Zeb2, Notch signaling is turned on constitutively in sciatic nerves, exerting a negative regulatory effect on SC differentiation and myelination. We observe a substantial increase of proliferating immature SCs in Zeb2 mutants, which is consistent with the notion that activation of Notch signaling leads to increased SC proliferation 16. The transcriptome profiling analysis further identifies Hey2, a transcriptional repressor downstream of Notch 47, as a Zeb2 target that suppresses Krox20 expression and blocks SC differentiation.
Nonetheless, unlike the sustained block of SC maturation in Zeb2 cKO nerves, genetically enforced Notch (i.e. NCID) expression in SCs does not completely prevent the initiation of myelinogenesis 16. Therefore, we cannot rule out the possibility that Hey2 regulation of SC development may also be independent of canonical Notch signaling, as reported in other contexts 48,49. If such is the case, Zeb2-regulated Hey2 would introduce an additional layer of repression to the already existing Notch inhibition on SC myelination.
Zeb2-deficient SCs exhibit persistent upregulation of Sox2 and a substantial increase in Sox2+ immature SCs in peripheral nerves. Sox2 has been shown to act downstream of Notch signaling in certain contexts. For instance, Notch activation induces Sox2 in sensory progenitors of the inner ear 50. Elevation of Sox2 levels maintains SCs in the undifferentiated state by suppressing expression of SC differentiation markers such as Krox20 33. Given that Hey2 overexpression inhibits Krox20 and increases the proliferation of immature SCs (Fig. 6k–n), Notch-Hey2 activation may induce or maintain Sox2 expression to inhibit SC differentiation.
Distinct mechanisms of Zeb2 in CNS and PNS myelination
Recent studies of Zeb2 revealed that Zeb2 activates Smad7 and inhibits the action of activated BMP-Smads on genes that would otherwise be inhibitory for OL myelinogenesis and hence promotes myelinogenesis in the CNS 8. In contrast, the present study shows that Zeb2 does not appear to control SC differentiation via regulation of BMP-Smad signaling or Smad7 expression in the PNS, suggesting that Zeb2 controls SC myelination via a unique mechanism. Here we find that Zeb2 directs SC differentiation by a disinhibition mechanism via antagonizing Notch-Hey2 signaling and Sox2 in the PNS, which was not observed in the CNS 8. Furthermore, a novel finding of this study in the PNS is that Zeb2, through recruitment of the HDAC1/2-NuRD, negatively regulates Sox2 and the Notch effector Hey2 to guard against the blockade of SC myelination mediated by these inhibitors.
In contrast to substantial down-regulation of Sox10 in OLs of the Zeb2 mutant spinal cord 8, Zeb2 does not seem to regulate Sox10 (Fig. 3d,k). However, we cannot exclude the possibility that Sox10 and Zeb2 coordinate together driving the SC differentiation program, similar to what has been described regarding their co-operative role in the enteric nervous system 51. Alternatively, we observed a preferential recruitment of Zeb2 to the promoter of Oct6 when SCs begin to differentiate (Supplemental Fig. 3e). Given the down-regulation of key pro-myelination regulators Oct6 and Krox20 in Zeb2-ablated sciatic nerves, our data suggests that Zeb2 may function as a dual-level switch control of SC differentiation through inactivating multiple differentiation inhibitors, including Notch signaling and Sox2, while promoting pro-myelinating factors Oct6 and Krox20 during SC development.
Zeb2 inactivation in SCs during nerve repair inhibits SC re-differentiation and thus remyelination, supporting the crucial role of Zeb2 in myelin regeneration in the PNS. However, we did not detect any difference in Sox2 or Hey2 mRNA expression in the mutant injured nerves 7 days post-transection, at which time regenerating mouse nerves will have reached the midpoint of axonal regrowth, as compared to the control injured sciatic nerves (Supplemental Fig. 7). Thus, we cannot rule out the possibility that Zeb2 may utilize other mechanisms after injury for SC regeneration and remyelination.
Deficient radial axonal sorting and myelination caused by Zeb2 loss could severely disrupt nerve conduction and, consequently, motor function in Zeb2 mutant mice, paralleling motor disabilities and delayed walking in MOWS patients. The phenotypic variation between the haplo-insufficient mutants in human could be attributed to a differential sensitivity to gene dosage across species or dominant-negative effects of human mutant proteins. Strikingly, we find that ZEB2R22G detected in a MOWS patient cannot associate with HDAC1/2-NuRD and blocks the SC differentiation program, suggesting that developmental defects caused by ZEB2 mutations affecting a functional domain may contribute to neuropathies and motor dysfunction seen in these patients. Given that the ZEB2-mediated functions are vital for SC development and myelination, Zeb2 may serve as a pivotal hub of the myelination regulatory network. Augmenting HDAC1/2-NuRD-dependent Zeb2 activity and inhibiting Notch signaling could potentially provide a novel therapeutic means of reversing adverse neuropathies seen in MOWS patients.
Methods
Animals
Zeb2lox/lox mice 22 were crossed with Dhh-cre 17 mice to obtain Zeb2lox/+; Dhh-cre+/− mice, which were then bred with Zeb2lox/lox mice to produce control (Zeb2lox/+;Dhh-cre+/−) and Zeb2 cKO offspring (Zeb2lox/lox;Dhh-cre+/−). Littermates Zeb2lox/lox or Zeb2lox/+;Dhh-cre+/− mice were used as controls. Transgenic Notch reporter (TNR) mice were obtained from the Jackson Laboratory (Stock 018322) and bred with Zeb2lox/+; Dhh-cre+/− mice to generate Zeb2lox/lox; Dhh-cre+/−;Notch-GFP mice. Recombination perinatally or after sciatic nerve transection was achieved by crossing Zeb2lox/lox mice with the inducible Cre recombinase Cre-ERT2 under the control of the PLP promoter (PlpCre-ERT2+/−) followed by tamoxifen injection 24. Zeb2lox/lox; PlpCre-ERT2+/− mice were then bred to mice containing a Cre-responsive Rosa26-floxed stop tdTomato reporter allele. Animals of either sex were used in the study and littermates were used as controls unless otherwise indicated. The mouse strains used in this study were generated and maintained on a mixed C57Bl/6;129Sv background and housed in a vivarium with a 12-hour light/dark cycle. No more than 4 adult mice are housed in the same cage per IACUC regulations at CCHMC. All animal experiments were conducted in mice of both genders. All animal use and studies were approved by the Institutional Animal Care and Use Committee at Cincinnati Children’s Hospital, USA.
Isolation, Growth and Differentiation of Primary Rat SCs
Rat SCs from sciatic nerves of newborn rats (1–2 days-old) were isolated as described previously52. SCs were grown routinely in DMEM/10% FBS (Life Technologies), supplemented with 10 ng/ml Neuregulin 1 (Nrg1; R&D Systems), and 5 μM forskolin (Sigma), plus L-glutamine and penicillin/streptomycin, hereafter termed SC proliferation medium. Cells between passages 2 and 6 were used in all experiments. >95% SC purity was achieved, assessed by Sox10 and S100β staining. To initiate differentiation, SCs were washed 4 times with DMEM and then cultured in differentiation medium containing DMEM/0.5% FBS and 1 mM dibutyl cyclic AMP (Sigma) with L-glutamine and penicillin/streptomycin, for the length of time indicated in the text, depending on the assays used. All tissue culture containers and coverslips were coated with 50 μg/ml poly-L-lysine (Sigma) in PBS for at least 30 min at room temperature and then rinsed in distilled water. Purified rat SCs seeded on coverslips were fixed in 4% paraformaldehyde (PFA) for 20 minutes and washed in 1x PBS 4 times prior to immunofluorescence staining.
Immunostaining, In Situ Hybridization and Electron Microscopy
The sciatic nerves of mice at defined ages were dissected and fixed for 45 min in 4% PFA, embedded in OCT, cryoprotected in 25% sucrose and sectioned at 9 μm as longitudinal sections. For BrdU pulse labeling, animals at P1 were injected subcutaneously with 100 mg BrdU/kg body weight 2 hours prior to sciatic nerve collection. For immunostaining, we used antibodies to Zeb2 (Santa Cruz Biotechnology, sc-48789), Sox10 (Goat, Santa Cruz Biotechnology, sc-17342; Rabbit, Millipore), Oct6 (Goat; Santa Cruz Biotechnology, sc-11661), Krox20 (Rabbit; Covance, PRB-236P), MBP (Goat; Santa Cruz Biotechnology), Sox2 (Goat; Santa Cruz Biotechnology, sc-17320), c-Jun (Rabbit; Epitomics, 1254-1), Hey2 (Rabbit; Proteintech, 10597-1-AP); Ki67 (Rabbit; Thermo Scientific, RM-9106), BrdU (Rat; Abcam, ab6326), Cleaved Caspase 3 (Rabbit; Cell signaling, #9661), S100β (Mouse; Sigma, SAB1402349); GFP (Rabbit; Life Technologies, A11122); c-MYC (Mouse; Santa Cruz Biotechnology, sc-40), Flag (Mouse; Cell Signaling, #8146) and HA-tag (Upstate, 50-171-890). Secondary antibodies conjugated to Cy2, Cy3 or Cy5 were from Jackson ImmunoResearch Laboratories. All images were acquired using a Zeiss LSM 510 or Nikon C2+ confocal microscope. RNA in situ hybridization was performed using digoxigenin-labeled riboprobes as described previously 53. The probes used were: murine Mbp and Mpz. Detailed protocols are available upon request. For electron microscopy, mice were perfused with 4% PFA, 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.2. Sciatic nerves were dissected and fixed in the same fixative solution overnight. Nerves were rinsed in PBS, postfixed in 1% OsO4 in PBS for 1 hour, dehydrated in graded ethanol, infiltrated with propylene oxide, and embedded in Epon. Semithin sections were stained with toluidine blue, and thin sections were stained with lead citrate.
Transient Transfections and Luciferase Assays
For plasmid transfections, rat SCs were transfected with expression vectors using Lipofectamine 3000 (Life Technologies) per the manufacturer’s protocol and assayed for immunocytochemistry or qRT-PCR analysis. pHA-Sox2 was purchased from Addgene.
For siRNA knockdown in SCs, we used Lipofectamine RNAiMAX (Life Technologies) per manufacturer’s instructions. SCs were induced to differentiate after 48 hours of transfection for 24 hours prior to harvesting for immunocytochemistry or qRT-PCR analysis. For sequential siRNA knockdown, SCs in proliferation medium was transfected with Zeb2-specific siRNA for 24 hours prior to Sox2 or Hey2 siRNA transfection. Triple knockdown was performed in the order of Zeb2-Hey2-Sox2 siRNA, each separated by a 24-hour interval. Zeb2 siRNA: SASI_Rn01_00044097 (gcaagaaauguauugguuu); SASI_Rn01_00044098 (guaugcaugugacuua ugu); SASI_Rn02_00203739 (cagaugaaccucugaauuu). Sox2 siRNA: ccaccuacagcauguccua[dt] [dt]; Hey2 siRNA: ggaucgaauaaauaacagu[dt][dt] and ccaugcagauucugcucuu[dt][dt].
For DAPT (Sigma) treatment, 48 hours subsequent to transfection with scramble or Zeb2 siRNA, SCs on coverslips or in 12-well culture dishes were induced to differentiate with 1 mM dbcAMP and treated with DMSO or 5 μM DAPT for 4 days, and then assayed for immunocytochemistry or qRT-PCT analysis. Proliferation was assessed by immunostaining for Ki67 in Sox10+ SCs seeded on PLL-coated coverslips for 2 days. Differentiation was assessed by immunostaining for Oct6 or Krox20. Multiple images were taken from each coverslip to obtain representative images from all areas of the coverslip, and at least 400 cells/coverslip were counted using ImageJ (NIH). For quantitation, GFP+ or fusion tag-expressing cells were counted, and the percentage of GFP+/fusion-tag+ cells that expressed the protein of interest was determined. At least 100 GFP+ or fusion-tag+ cells/coverslip were counted.
For reporter assays, purified rat SCs were transfected with pGL3-luciferase reporters containing inserts from 2 kb upstream of Sox2 TSS or 3.9 kb upstream of Hey2 TSS and expression vectors using using Lipofectamine 3000 (Life Technologies) per the manufacturer’s protocol and assayed 48 hours posttransfection for luciferase activity by using a Promega luciferase assay kit according to the manufacturer’s instructions. The pSV-β-Galactosidase control vector was included to control for variable transfection efficiencies between independent experiments (n ≥ 3). Mutant Sox2 promoter luciferase reporter was generated by mutating Zeb2 consensus binding site (P1; −1.1K from TSS; Fig. 5i) into an EcoRI site, and mutant Hey2 promoter luciferase reporter was generated by mutating a Zeb2 binding site into an EcoRI site.
Western Blotting and Co-Immunoprecipitation
For western blotting, the perineurium and epineurium were removed from sciatic nerves prior to snap-freezing and storage at −80°C. Sciatic nerves and rat SCs were lysed in RIPA buffer, containing protease and phosphatase inhibitors. Western blot analysis was performed as described previously 39. GAPDH was used as an input control. For immunoprecipitation, HEK293T cells cultured in 10% FBS/DMEM were transfected with expression vectors using PolyJet (Signagen) for 48 hours. Cells were lysed in NP-40 buffer (170 mM NaCl, 10 mM EDTA, 50 mM Tris pH 7.4, 50 mM NaF, and 0.5% NP-40) supplemented with protease inhibitor cocktail and phosphatase inhibitors (Roche Diagnostics Inc.). A total of 300 μg of cell lysate proteins were incubated with 2 μg IgG or appropriate antibodies and immunoprecipitated using Protein A/G beads (Santa Cruz Biotechnology). Western blotting was performed using chemiluminescence with the ECL kit (Pierce). The antibodies used were anti-Zeb2 (Millipore, ABT-332), anti-Flag (Rabbit #14793 or mouse #8146, Cell Signaling), anti-Myc (Santa Cruz, sc-40). For co-immunoprecipitation of endogenous NuRD subunits, extracts of HEK293T cells transiently expressing Flag-tagged ZEB2WT or Flag-tagged ZEB2R22G were precipitated in a Flag-dependent manner and eluted using 3XFlag peptides according to 10. Co-immunoprecipitation experiments showing endogenous interaction between Zeb2 and HDAC1/2 was performed by incubating protein lysates from purified rat SCs with anti-Zeb2 antibody followed by immunoprecipitation using Protein A/G beads and detection of HDAC1/2 using HDAC1/2-specific antibodies. Antibodies used were HDAC1 (Santa Cruz, sc-7872), HDAC2 (Abcam #7029), Mi2 (Santa Cruz, sc-11378), MTA-1 (Santa Cruz, sc-9446), MTA-2 (Santa Cruz, sc-28731), RbAp48 (Abcam, 38185) and CtBP (Sigma, C8741). Secondary antibodies conjugated to HRP were from Jackson ImmunoResearch Laboratories.
RNA-Sequencing and Data Analysis
RNA from P7 control and Zeb2 cKO sciatic nerves were extracted using TRIZOL (Life Technologies) followed by purification using an RNeasy Mini Kit (Qiagen). RNA-seq libraries were prepared using Illumina RNA-Seq Preparation Kit and sequenced by HiSeq 2000 sequencer. RNA-seq reads were mapped using TopHat with default settings (http://tophat.cbcb.umd.edu). TopHat output data were then analyzed by Cufflinks to (1) calculate FPKM values for known transcripts in mouse genome reference, and (2) test the changes of gene expression between Zeb2 cKO and control. Gene Ontology (GO) analysis was performed using The Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov). The heatmap was generated based on log2 [FPKM] by AltAnalyze 54,55 with normalization of rows relative to row mean.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were performed as described with minor modifications 39,56. Briefly, purified rat SCs grown in proliferation and differentiation (9 hours in 1 mM cAMP-containing medium conditions (<20 million cells) were fixed for 10 min at room temperature with 1% formaldehyde-containing medium. Nuclei were pelleted and sonicated in sonication buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, and protease inhibitor cocktail). Sonicated chromatin (<300 μg) was used for immunoprecipitation by incubation with IgG or Rabbit anti-Zeb2 antibody (H-260; Santa Cruz Biotechnology sc-48789; 4 μg) overnight at 4°C. 10% of chromatin used for each ChIP reaction was kept as input DNA. Pre-rinsed protein A/G plus agarose beads (50 μl) was added to each ChIP reaction and incubated for 1 hour at 4°C. The beads were then incubated in 200 μl elution buffer at 65°C for 20 minutes to elute immunoprecipitated materials. For ChIP, SCs at 70–90% confluency were transfected with control or Zeb2 siRNA for 48 hours using Lipofectamine RNAimax per manufacturer’s instructions and induced to differentiate for 9 hours with 1 mM cAMP. ChIP was performed using anti-HDAC1 (Active Motif #40967) and HDAC2 antibody (Abcam #7029). Real-time PCR was performed using quantitative SYBR green PCR mix (BioRad). The relative fold-enrichments were determined by the 2−ΔCT methods as described 39. Samples were normalized to input chromatin. Primers used for ChIP-qPCR analysis: Sox2 proximal promoter (P1): Forward attggtggaggagggagtta; Reverse tatggctgactgtagagctagg; Sox2 proximal promoter (P2): Forward ctgcacctgcacctctg; Reverse cagcgttcccaggatgg; Sox2 proximal promoter (P3): Forward ctccacccgtgtttcctaaat; Reverse cttctgccctctaagtcacaaa; Hey2 proximal promoter (−3.9 kb from TSS): Forward: gctcacaatagcctgtagtt; Reverse: aagtcagtctctgtcctggt. Oct6 proximal promoter: (−0.8 kb from TSS) Forward: tcctagagctcgcatctcc; Reverse: atcggcttgggcttcac. Oct6 proximal promoter: (−3.9 kb from TSS) Forward: gtgtccgtatgtcttcgagatg; Reverse: acacggtgctattcacacaa. cJun proximal promoter (−6 kb from TSS): Forward: cccaagacctgtgtgagaat; Reverse: ctcacagtttgattggctgaaa. Distal control region ChIP-PCR: Sox2 (+25 kb TSS): Forward gaacagatgagtgttgcctttag; Reverse actccctggaccaaacatac; Hey2 (+20 kb TSS): Forward ggaaactgtatacggcctctg; Reverse gatctgcatcgccctttct; Oct6 (+18 kb TSS): Forward gtagaggaaagcagaggaaagg; Reverse gaatcccatagccagacatgat.
RNA Isolation and Quantitative Real Time-Polymerase Chain Reaction
RNA from purified rat SCs or control and Zeb2 cKO mouse sciatic nerves was extracted using TRIZOL (Life Technologies). cDNA was synthesized from 1 μg RNA using iScript Reverse Transcription Supermix (BioRad) according to the manufacturer’s instructions. QRT-PCR was performed using the ABI Prism 7700 Sequence Detector System (Perkin-Elmer Applied Biosystems). qRT-PCR was performed using quantitative SYBR green PCR mix (BioRad) as previously described 57. PCR primer sequences are available upon request.
RNA-seq and data analysis
RNA-seq libraries were prepared using Illumina RNA-Seq Preparation Kit and sequenced by a HiSeq 2000 sequencer. All RNA-Seq data were aligned to mm9 using TopHat 58 with default settings. We used Cuff-diff 59 to (1) estimate FPKM values for known transcripts and (2) analyze differentially expressed transcripts. In all differential expression tests, a difference was considered significant if the q value was less than 0.05 (Cuff-diff default). Heatmap of gene expression was generated using R language (http://www.r-project.org). GO-analysis of gene expression changes was performed using Gene Set Enrichment (GSEA, http://www.broadinstitute.org/gsea/index.jsp). We used ToppCluster (https://toppcluster.cchmc.org/) to construct the network of genes belonging to over-represented GO-term categories.
Tamoxifen Injections
Tamoxifen (Sigma) dissolved to a stock concentration of 20 mg/ml in a vehicle of ethanol and sunflower seed oil (1:9 v/v). For perinatal tamoxifen injections, tamoxifen stock was injected i.p. at 2 mg/100 μl for 5 consecutive days to lactating mothers, thus administering tamoxifen to pups, beginning at P0. Sciatic nerves of pups were analyzed on P14 for electron microscopy. For adult tamoxifen treatment, 100 μl (2 mg/100 μl) was administered by i.p. injection once daily for 5 consecutive days to 4–6 week-old mice. Mice were treated again for 5 days after a 2-day rest period. Control mice were treated identically.
In vivo drug treatment
Control and Zeb2 cKO were injected on P2, P3, P5 and P7 with the Notch signaling (γ-secretase) inhibitor LY411575 (Selleckchem) at 0.30 mg kg−1 per day. Sciatic nerves were fixed in 4% PFA on P8 for immunostaining. LY411575 was diluted in a mixture of corn oil and EtOH (95:5 v/v). Vehicle was corn oil and EtOH (95:5 v/v).
Generation and transduction of lentivirus
Lentivirus was produced by transfection of DN-MAML lentivirus construct (MAML1 1–305 subcloned into BamHI and XhoI sites of PLU-ires-GFP mod vector, a kind gift from A.J. Capobianco) into HEK293T cells with the packaging vectors psPAX2 and pMD2.G using Polyjet in vitro DNA transfection reagent (Sigmagen) according to the instructions of the manufacturer. The supernatant was collected 72 hours after transfection and frozen in liquid nitrogen. A titer of 107/μl was achieved. SCs previously transfected with control or Zeb2 siRNA for 24 hours were infected with control GFP lentivirus or DNMAML-GFP lentivirus in the presence of 4 μg/ml polybrene for 48 hr before harvesting for RNA.
ZEB2 variant identification
The novel ZEB2 variant c. 64A>G [p.(Arg22Gly)] was identified by targeted diagnostic sequencing of the coding exons of ZEB2 in a patient with a facial gestalt reminiscent of MOWS (OMIM 235730). The patient had mild manifestation of MOWS phenotype. This mutation was not detected in any of other parents and is not contained in the ExAC database. This variant is atypical since missense mutations in ZEB2 are very rare in MOWS while usually truncating mutations are found. Due to his young age (4 years old), peripheral nerve related phenotype was clinically impossible to test for and therefore we could not assess the peripheral nerve related phenotype.
Sciatic Nerve Transection Surgeries
Sciatic nerve transection was performed 10 days after the last tamoxifen injection in 6–8 week-old mice. Under general anesthesia after injection of a mixture of ketamine (90 mg/kg) and xylazine (10 mg/kg), right sciatic nerves were exposed and transected at the midthigh level. Exposed left sciatic nerves were used as uncut controls. Mice were treated with tamoxifen for an additional 8 days following injury. Nerves were collected 14 days after surgery and processed for immunohistochemistry.
Compound Muscle Action Potential Recording
To analyze nerve conduction and motor unit function, single compound muscle action potentials (CMAPs) were recorded in vivo from the lateral gastrocnemius muscles of wildtype littermate controls or Zeb2 cKO mice during electrical stimulation of the sciatic nerve under sodium pentobarbital anesthesia (50 mg/kg, i.p.). The lateral gastrocnemius muscle was first exposed from the knee to approximately 6 mm above the ankle. A small opening was then made in the overlying fascia to facilitate implantation of the recording wires into the muscles. Next, the sciatic nerve was exposed near the biceps femoris muscles. Polymide-coated steel recording wires (California Fine Wire Inc. Stainless Steel 304: size = 0.0092 and insulated with polyimide; Material #: 100192) were then implanted into the lateral gastrocnemius muscles and reference wires were inserted under the nearby skin. To induce electrical activation of the nerve in vivo, a concentric bipolar stimulating electrode was placed on the sciatic nerve. CMAPs were amplified and obtained using a Micro 1401 data acquisition unit and analyzed using Spike2 software (Cambridge Electronic Design, Cambridge, UK). 3 successive electrical stimulations of the sciatic nerve at 2 mA (0.25–0.5 Hz, 0.1 ms duration) were initiated immediately proximal to the tibial, sural and common peroneal branches via a stimulus isolator (WPI Inc.) connected to the Micro 1401. After recording, the sciatic nerve was axotomized distal to the site of concentric electrode placement and stimulation of the proximal end of the sciatic nerve was performed to ensure CMAPs were generated from direct nerve stimulation. Conduction velocity was calculated after determining the latency of CMAP onset relative to the stimulus artifact induced by electrical stimulation of the sciatic nerve and the distance between recording and stimulating electrodes measured directly on the nerve. Peak CMAP amplitude and CMAP duration were calculated from each stimulation paradigm. The average of the stimulations of the sciatic nerve for each paradigm were obtained and then averaged across animals.
Heat hypersensitivity assessments
Mice at 3–4 weeks old were acclimatized individually in a box of at least 15 × 12 × 10 cm3 in dimensions for 15 min. Mice were then gently held and both hindpaws were immersed into a 50°C hot water bath. Latency (in s) to withdrawal from the heat stimulus was determined. 3 trials were performed with a 5-min interval in between each trial. The average of these three trials was then determined to obtain the baseline withdrawal latency. Next, mice were anesthetized under 3% isofluorane in order to induce cutaneous inflammation. Using a syringe with a 30 G needle, approximately 10 μL of 3% carrageenan (in 0.9% NaCl) was injected into the right hairy hindpaw skin. 24 hours later, thermal withdrawal latency using the methods described above were measured again to determine heat hypersensitivity (withdrawal latency) post inflammation60,61.
Morphometric analysis
The morphometric measurements in the early postnatal Zeb2 iKO experiment and the axon regeneration/remyelination assays were performed in Toluidine blue–stained semithin sections (0.5 μm). An entire sciatic nerve cross section per animal was reconstructed by merging several high magnification photographs taken by bright field microscopy at 100 × magnification. The quantification of the 1:1 promyelinating cells, myelinated fibers, and axon number was performed by manually counting all of the figures present in the entire sciatic nerves cross section using the cell counter plugin from ImageJ software (National Institutes of Health).
The number of myelinated axons per nerve, the g-ratio and the number of unmyelinated axons in Remak bundles were analyzed in ultrathin sections using a JEOL 1200 EXII or Hitachi H7650 electron microscope. Quantification was done by manually counting myelinated or unmyelinated axons present in the sciatic nerves using the cell counter plugin from ImageJ software. For g-ratio analysis, images were calibrated based on the scale bar corresponding to the magnification used for each micrograph. The axon (without myelin) and fiber areas (including myelin) of randomly chosen fibers were measured, except for those surrounded by a Schmidt Lanterman incisure. The axon and fiber diameters were estimated as 2·√(area/π) and g-ratio was calculated as axon diameter/fiber diameter. At least 100 nerve fibers per animal (n = 3 for each group) were analyzed.
For quantification of labeled cells in transfection studies, since the ZEB2, ZEB2R22G and ZEBΔCtBP expression plasmids carry a GFP-reporter, GFP+ cells represent ZEB2 or ZEB2-mutant transfected cells. The quantification was performed by manually counting antibody co-labeled cells based on the total number of transfected cells indicated by GFP expression.
Statistical analysis
All analyses were done using Microsoft Excel or GraphPad Prism 6.00 (San Diego California, www.graphpad.com). Data are shown in dot plots as mean ± s.e.m. or as Box-and-whisker plots, P < 0.05 is deemed statistically significant. Data distribution was assumed to be normal, but this was not formally tested. Count data was assumed to be non-parametric, and appropriate statistical tests were used. Statistical analysis was performed by two-tailed unpaired Student’s t tests, Mann-Whitney test, one-way ANOVA with post-hoc analysis by Tukey’s multiple comparison test, or Two-way repeated measures ANOVA with Sidak’s multiple comparisons test, or as indicated. Quantifications were performed from at least three experimental groups in a blinded fashion. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those generally employed in the field. No randomization was used to collect all the data, but they were quantified blindly. No animals or data points were excluded from analyses.
A methods checklist is available with the supplementary materials.
Data availability
All the RNA-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE74381. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
Authors would like to thank X. Chen and Z. Ma for technical support and initial observation of Zeb2 mutants. We thank A. Rauch for discussion ZEB2 missense mutations in MOWS patients, J. Svaren and E. Hurlock for suggestions, and G. Verstappen and L. van Grunsven for initial study of Zeb2R22G-NuRD interaction. We are grateful for K. Nave for communications of unpublished data. We also thank R. Kopan (Univ. Cincinnati) and A J. Capiobianco (Univ. of Miami) for TP-1 reporter and lentiviral DN-MAML constructs, and J. W. Schneider and E. N. Olson (UT Southwestern) for Notch-GFP reporter mice and Flag-HRT2/Hey2 vectors. This study was funded in part by grants from the US National Institutes of Health R01NS072427 and R01NS075243 to QRL, R01NS062796 to JRC and R01AR064551 to MPJ, and the National Multiple Sclerosis Society (NMSS-4727) to QRL and NMSS Postdoctoral Fellowship (FA 2045A1/T) to LMNW. The work was also supported by Belspo grant IAP7-07 DevRepair, the Research Council of KU Leuven (GOA-11/012) to DH, FWO-V (G.0782.14) to DH, the type 3 large-infrastructure support InfraMouse by the Hercules Foundation (ZW09-03) and Erasmus MC start-up funds to DH.
Footnotes
Accession codes: All the RNA-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE74381.
AUTHOR CONTRIBUTIONONS
L.M.N.W. and Q.R.L. designed the experiments. L.M.N.W. carried out the studies. J.W., A.C. and L.Z. assisted with co-immunoprecipitation biochemical experiments for HDAC-NuRD and BMP-Smads. C.Zh. assisted with ChIP experiments. H.W. assisted with Notch-TP1 reporter assays. C.Zw. provided ZEB2 variant identification data. Z.F. assisted with heat hypersensitivity experiments. M.P.J. assisted with CMAP recordings. B.G.A., P.M., A.Z., J.R.C. provided input and data interpretation. D.H. provided Zeb2 floxed mice and input on the study. L.M.N.W. and Q.R.L. wrote the manuscript.
COMPETETINGNG FINANCNCIAL INTENTENTERESTS
The authors declare no competing financial interests.
References
- 1.Pereira JA, Lebrun-Julien F, Suter U. Molecular mechanisms regulating myelination in the peripheral nervous system. Trends Neurosci. 2012;35:123–134. doi: 10.1016/j.tins.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Nave KA, Werner HB. Myelination of the nervous system: mechanisms and functions. Annu Rev Cell Dev Biol. 2014;30:503–533. doi: 10.1146/annurev-cellbio-100913-013101. [DOI] [PubMed] [Google Scholar]
- 3.Salzer JL. Schwann cell myelination. Cold Spring Harb Perspect Biol. 2015;7:a020529. doi: 10.1101/cshperspect.a020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dastot-Le Moal F, et al. ZFHX1B mutations in patients with Mowat-Wilson syndrome. Hum Mutat. 2007;28:313–321. doi: 10.1002/humu.20452. [DOI] [PubMed] [Google Scholar]
- 5.Mowat DR, Wilson MJ, Goossens M. Mowat-Wilson syndrome. J Med Genet. 2003;40:305–310. doi: 10.1136/jmg.40.5.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagaya M, Kato J, Niimi N, Tanaka S, Wakamatsu N. Clinical features of a form of Hirschsprung’s disease caused by a novel genetic abnormality. J Pediatr Surg. 2002;37:1117–1122. doi: 10.1053/jpsu.2002.34455. [DOI] [PubMed] [Google Scholar]
- 7.McKinsey GL, et al. Dlx1&2-dependent expression of Zfhx1b (Sip1, Zeb2) regulates the fate switch between cortical and striatal interneurons. Neuron. 2013;77:83–98. doi: 10.1016/j.neuron.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng Q, et al. Dual-mode modulation of Smad signaling by Smad-interacting protein Sip1 is required for myelination in the central nervous system. Neuron. 2012;73:713–728. doi: 10.1016/j.neuron.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Berghe V, et al. Directed migration of cortical interneurons depends on the cell-autonomous action of Sip1. Neuron. 2013;77:70–82. doi: 10.1016/j.neuron.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Verstappen G, et al. Atypical Mowat-Wilson patient confirms the importance of the novel association between ZFHX1B/SIP1 and NuRD corepressor complex. Hum Mol Genet. 2008;17:1175–1183. doi: 10.1093/hmg/ddn007. [DOI] [PubMed] [Google Scholar]
- 11.Michailov GV, et al. Axonal neuregulin-1 regulates myelin sheath thickness. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- 12.Taveggia C, et al. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron. 2005;47:681–694. doi: 10.1016/j.neuron.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feltri ML, Wrabetz L. Laminins and their receptors in Schwann cells and hereditary neuropathies. J Peripher Nerv Syst. 2005;10:128–143. doi: 10.1111/j.1085-9489.2005.0010204.x. [DOI] [PubMed] [Google Scholar]
- 14.Nodari A, et al. Alpha6beta4 integrin and dystroglycan cooperate to stabilize the myelin sheath. J Neurosci. 2008;28:6714–6719. doi: 10.1523/JNEUROSCI.0326-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monk KR, et al. A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science. 2009;325:1402–1405. doi: 10.1126/science.1173474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodhoo A, et al. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat Neurosci. 2009;12:839–847. doi: 10.1038/nn.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaegle M, et al. The POU proteins Brn-2 and Oct-6 share important functions in Schwann cell development. Genes & development. 2003;17:1380–1391. doi: 10.1101/gad.258203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Topilko P, et al. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- 19.He Y, et al. Yy1 as a molecular link between neuregulin and transcriptional modulation of peripheral myelination. Nat Neurosci. 2010;13:1472–1480. doi: 10.1038/nn.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svaren J, Meijer D. The molecular machinery of myelin gene transcription in Schwann cells. Glia. 2008;56:1541–1551. doi: 10.1002/glia.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng Q, et al. Dual-mode modulation of Smad signaling by Smad-interacting protein Sip1 is required for myelination in the central nervous system. Neuron. 2012;73:713–728. doi: 10.1016/j.neuron.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higashi Y, et al. Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis. 2002;32:82–84. doi: 10.1002/gene.10048. [DOI] [PubMed] [Google Scholar]
- 23.Douglas WW, Ritchie JM. Mammalian nonmyelinated nerve fibers. Physiological reviews. 1962;42:297–334. doi: 10.1152/physrev.1962.42.2.297. [DOI] [PubMed] [Google Scholar]
- 24.Doerflinger NH, Macklin WB, Popko B. Inducible site-specific recombination in myelinating cells. Genesis. 2003;35:63–72. doi: 10.1002/gene.10154. [DOI] [PubMed] [Google Scholar]
- 25.Verschueren K, et al. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J Biol Chem. 1999;274:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- 26.Jessen KR, Mirsky R, Lloyd AC. Schwann Cells: Development and Role in Nerve Repair. Cold Spring Harb Perspect Biol. 2015 doi: 10.1101/cshperspect.a020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parrinello S, et al. EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell sorting. Cell. 2010;143:145–155. doi: 10.1016/j.cell.2010.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. 2005;6:671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- 29.Parkinson DB, et al. c-Jun is a negative regulator of myelination. J Cell Biol. 2008;181:625–637. doi: 10.1083/jcb.200803013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feltri ML, Poitelon Y, Previtali SC. How Schwann Cells Sort Axons: New Concepts. Neuroscientist. 2015 doi: 10.1177/1073858415572361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes & development. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xin M, et al. Essential roles of the bHLH transcription factor Hrt2 in repression of atrial gene expression and maintenance of postnatal cardiac function. Proc Natl Acad Sci U S A. 2007;104:7975–7980. doi: 10.1073/pnas.0702447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le N, et al. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc Natl Acad Sci U S A. 2005;102:2596–2601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- 35.Souilhol C, et al. Nas transgenic mouse line allows visualization of Notch pathway activity in vivo. Genesis. 2006;44:277–286. doi: 10.1002/dvg.20208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 37.Pinnix CC, et al. Active Notch1 confers a transformed phenotype to primary human melanocytes. Cancer Res. 2009;69:5312–5320. doi: 10.1158/0008-5472.CAN-08-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lauberth SM, Rauchman M. A conserved 12-amino acid motif in Sall1 recruits the nucleosome remodeling and deacetylase corepressor complex. J Biol Chem. 2006;281:23922–23931. doi: 10.1074/jbc.M513461200. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, et al. HDAC-mediated deacetylation of NF-kappaB is critical for Schwann cell myelination. Nat Neurosci. 2011;14:437–441. doi: 10.1038/nn.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacob C, et al. HDAC1 and HDAC2 control the transcriptional program of myelination and the survival of Schwann cells. Nat Neurosci. 2011;14:429–436. doi: 10.1038/nn.2762. [DOI] [PubMed] [Google Scholar]
- 41.van Grunsven LA, et al. XSip1 neuralizing activity involves the co-repressor CtBP and occurs through BMP dependent and independent mechanisms. Dev Biol. 2007;306:34–49. doi: 10.1016/j.ydbio.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 42.Quintes S, et al. Zeb2 is essential for Schwann cell differentiation, myelination and nerve repair. Nature Neuroscience. 2016 doi: 10.1038/nn.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conidi A, et al. Few Smad proteins and many Smad-interacting proteins yield multiple functions and action modes in TGFbeta/BMP signaling in vivo. Cytokine Growth Factor Rev. 2011;22:287–300. doi: 10.1016/j.cytogfr.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Hung H, Kohnken R, Svaren J. The nucleosome remodeling and deacetylase chromatin remodeling (NuRD) complex is required for peripheral nerve myelination. J Neurosci. 2012;32:1517–1527. doi: 10.1523/JNEUROSCI.2895-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desmazieres A, Decker L, Vallat JM, Charnay P, Gilardi-Hebenstreit P. Disruption of Krox20-Nab interaction in the mouse leads to peripheral neuropathy with biphasic evolution. J Neurosci. 2008;28:5891–5900. doi: 10.1523/JNEUROSCI.5187-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynolds N, O’Shaughnessy A, Hendrich B. Transcriptional repressors: multifaceted regulators of gene expression. Development. 2013;140:505–512. doi: 10.1242/dev.083105. [DOI] [PubMed] [Google Scholar]
- 47.Fischer A, et al. Hey basic helix-loop-helix transcription factors are repressors of GATA4 and GATA6 and restrict expression of the GATA target gene ANF in fetal hearts. Mol Cell Biol. 2005;25:8960–8970. doi: 10.1128/MCB.25.20.8960-8970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doetzlhofer A, et al. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell. 2009;16:58–69. doi: 10.1016/j.devcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benito-Gonzalez A, Doetzlhofer A. Hey1 and Hey2 control the spatial and temporal pattern of mammalian auditory hair cell differentiation downstream of Hedgehog signaling. J Neurosci. 2014;34:12865–12876. doi: 10.1523/JNEUROSCI.1494-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Owen T, Fang J, Srinivasan RS, Zuo J. In vivo Notch reactivation in differentiating cochlear hair cells induces Sox2 and Prox1 expression but does not disrupt hair cell maturation. Dev Dyn. 2012;241:684–696. doi: 10.1002/dvdy.23754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanchina L, Van de Putte T, Goossens M, Huylebroeck D, Bondurand N. Genetic interaction between Sox10 and Zfhx1b during enteric nervous system development. Dev Biol. 2010;341:416–428. doi: 10.1016/j.ydbio.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 52.Brockes JP, Fields KL, Raff MC. Studies on cultured rat Schwann cells. I. Establishment of purified populations from cultures of peripheral nerve. Brain Res. 1979;165:105–118. doi: 10.1016/0006-8993(79)90048-9. [DOI] [PubMed] [Google Scholar]
- 53.Lu QR, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 54.Emig D, et al. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic acids research. 2010;38:W755–762. doi: 10.1093/nar/gkq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salomonis N, et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci U S A. 2010;107:10514–10519. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu Y, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Becker W. Recent insights into the function of DYRK1A. FEBS J. 2011;278:222. doi: 10.1111/j.1742-4658.2010.07953.x. [DOI] [PubMed] [Google Scholar]
- 58.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ririe DG, Bremner LR, Fitzgerald M. Comparison of the immediate effects of surgical incision on dorsal horn neuronal receptive field size and responses during postnatal development. Anesthesiology. 2008;109:698–706. doi: 10.1097/ALN.0b013e3181870a32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marsh D, Dickenson A, Hatch D, Fitzgerald M. Epidural opioid analgesia in infant rats I: mechanical and heat responses. Pain. 1999;82:23–32. doi: 10.1016/S0304-3959(99)00028-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the RNA-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE74381. The data that support the findings of this study are available from the corresponding author upon reasonable request.