Abstract
Objective
The Cancer Genome Atlas (TCGA) identified four integrated clusters for endometrial cancer (EC): POLE, MSI, CNL and CNH. We evaluated differences in gene expression profiles of obese and non-obese women with EC and examined the association of body mass index (BMI) within the clusters identified in TCGA.
Methods
TCGA RNAseq data was used to identify genes related to increasing BMI among ECs. The POLE, MSI and CNL clusters were composed mostly of endometrioid EC. Patient BMI was compared between these three clusters with one-way ANOVA. Association between gene expression and BMI was also assessed while adjusting for confounding effects of potential confounding factors. p-values testing the association between gene expression and BMI were adjusted for multiple hypothesis testing over the 20,531 genes considered.
Results
Mean BMI was statistically different between the ECs in the CNL (35.8) versus POLE (29.8) cluster (p=0.006) and approached significance for the MSI (33.0) versus CNL (35.8) cluster (p=0.05). 181 genes were significantly up- or down-regulated with increasing BMI in endometrioid EC (q-value<0.01), including LPL, IRS-1, IGFBP4, IGFBP7 and the progesterone receptor. DAVID functional annotation analysis revealed significant enrichment in "cell cycle" (adjusted p-value=1.5E-5) and "DNA metabolic processes" (adjusted p-value=1E-3) for the identified genes.
Conclusions
Obesity related genes were found to be upregulated with increasing BMI among endometrioid ECs. Patients with POLE tumors have the lowest median BMI when compared to MSI and CNL. Given the heterogeneity amongst endometrioid EC, consideration should be giving to abandoning the Type I and II classification of EC tumors.
INTRODUCTION
Obesity has been linked to increased risk of many cancers, including breast, colon and endometrial, among others [1]. Currently, new cancer cases are in the order of 1.6 million annually with over half a million cancer deaths per year, and nearly one in five are estimated to be due to obesity [1, 2]. It is postulated that elevations in insulin, glucose, insulin growth factor-1 (IGF-1), adipokines and cytokines resulting from over-nutrition in obese patients may provide abundant nutrients and growth factors to cancer cells, creating the ideal environment for tumor initiation and promotion [3].
Endometrial cancer (EC) is the most common gynecologic malignancy in the United States, accounting for 52,360 cases and over 8,500 deaths a year [2]. Traditionally, EC has been subdivided into Type I and Type II categories based on clinical and histologic differences. Type I cases comprise the majority of the cases and typically arise in a setting of unopposed estrogenic states such as obesity. These tumors tend to be well-differentiated, of endometrioid histology, diagnosed at early stages, and associated with good prognosis. Meanwhile, Type II ECs have classically been presumed to be estrogen independent, poorly differentiated, frequently of advanced stage, and associated with worse prognosis. The majority of Type II ECs consist of serous histology tumors [4]. Recent genomic characterization of EC through the TCGA project classified EC into four integrated clusters: POLE ultramutated (POLE), microsatellite instability hypermutated (MSI), copy-number low (CNL) and copy-number high (CNH). The POLE, MSI and CNL clusters were composed mostly of endometrioid histology tumors, while most serous histology tumors were found in the CNH cluster [5]. Women with POLE tumors had the best progression free survival (PFS), while women with CNH tumors had the worst PFS [5].
Given the rise in prevalence of obesity and its association with EC, it is imperative to investigate obesity as a modifiable risk factor that may reduce risk and lead to prevention and/or improved patient outcomes for this disease. We hypothesize that the metabolic consequences of obesity may be crucial in the development of EC, resulting in biologically distinct cancers than those arising in normal weight women, which could alter treatment approaches. Thus, we sought to assess differences in the gene expression profiles of obese and non-obese women with EC and to examine the association of BMI with the integrated clusters identified in the TCGA project.
METHODS
From the TCGA database, we collected expression measurements for 20,531 genes (IlluminaGA RNA-seq level 3 data) for differential gene expression analysis among the EC samples. Detailed information of the data processing, quality control and normalization can be found on the TCGA open access download directories (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp). The RNA-seq gene expression level 3 data utilized in this study were TCGA normalized gene read counts, pre-corrected such that the 75th percentile of each patient’s set of gene expression measurements were normalized to a value of 1000.
We identified differentially expressed genes with respect to BMI while adjusting for potential confounders via multiple linear regression. Specifically, for a single gene we modeled the log of the observed normalized read count from each subject with respect to the following predictors: BMI, clinical stage, grade, age, race, residual tumor, five principal component score variables (PCs) and subject-specific RNA-seq normalization factor. The PCs were computed from the gene expression data to control for potential batch effects in the expression data. A value of 1 was added to each sample’s normalized read count value to avoid taking the log of a zero count. From our fitted multiple linear regression model for a given gene, we then tested the association between BMI and log transformed normalized gene read count via the Wald Test. Because we performed this hypothesis test for each of 20,531 genes in our dataset, the Benjamini–Hochberg procedure was applied to control for False Discovery Rates (FDR) [6] and to address multiple testing issues. The p-values that were adjusted for FDR using the Benjamini–Hochberg procedure are referred to as adjusted p-values, and a threshold of adjusted p-values < 0.01 was used to report the significant results to reduce the number of false positives. The genes with the adjusted p-values < 0.01 were considered as significantly associated with BMI status; functional clustering analysis was conducted on those genes using the website of The Database for Annotation, Visualization and Integrated Discovery (DAVID) [7, 8]. In addition, hierarchical clustering analysis was applied to generate a representative heatmap using R statistical software. One-way ANOVA was used to detect significant differences in BMI among the different clusters. Student’s t-test was used to compare BMI among the different clusters of samples in a pairwise manner. Cox proportional hazards regression was utilized to compare survival between obese and non-obese patients.
Given the strong association between obesity and endometrioid ECs, in addition to the observation that three of the four integrated clusters (i.e. POLE, MSI, and CNL) were composed mostly of endometrioid histology [5], we limited our analysis to patients with endometrioid histology and excluded patients with serous and mixed histologies. Mean BMIs were compared amongst the POLE, MSI and CNL sub-types using Spearman’s Correlation.
RESULTS
Table 1 summarizes the demographics between the EC tumors from non-obese (BMI < 30) and obese women (BMI ≥ 30). There were no significant differences in the demographics between the EC tumors from non-obese (BMI < 30) and obese women (BMI ≥ 30) (Table 1), except for age being younger in the obese group. The mean age of the obese group was 60.8 years versus 64.5 years in the non-obese group. Table 2 summarizes the number of deaths and median overall survival in months for obese and non-obese patients. We foud that obese women had lower median survival compared to non-obese women (332.5 days versus 582.0 days); however, there was no significant difference in overall survival between groups (p=0.18, Hazard Ratio = 1.9), most likely driven by the small number of deaths overall in our set of patients.
Table 1.
Comparison of the demographics between the endometrial cancer patients from obese and non-obese women.
| Obese (n = 185) |
Non-obese (n = 105) |
p-value | ||
|---|---|---|---|---|
| Age (mean) | 60.8 | 64.5 | 0.0089 | |
| Race | 0.8964 | |||
| White | 83% | 81% | ||
| Black | 9% | 9% | ||
| Other | 8% | 10% | ||
| Stage | 0.2962 | |||
| I | 69% | 78% | ||
| II | 8% | 5% | ||
| III | 17% | 15% | ||
| IV | 5% | 2% | ||
| Grade | 0.2817 | |||
| G1 | 30% | 28% | ||
| G2 | 35% | 28% | ||
| G3 | 35% | 44% | ||
Table 2.
Median overall survival of obese and non-obese women with endometrial cancer.
| Number of events | Median of Overall Survival (days) |
|
|---|---|---|
| Obese | 16 | 332.5 |
| Non-obese | 6 | 582.0 |
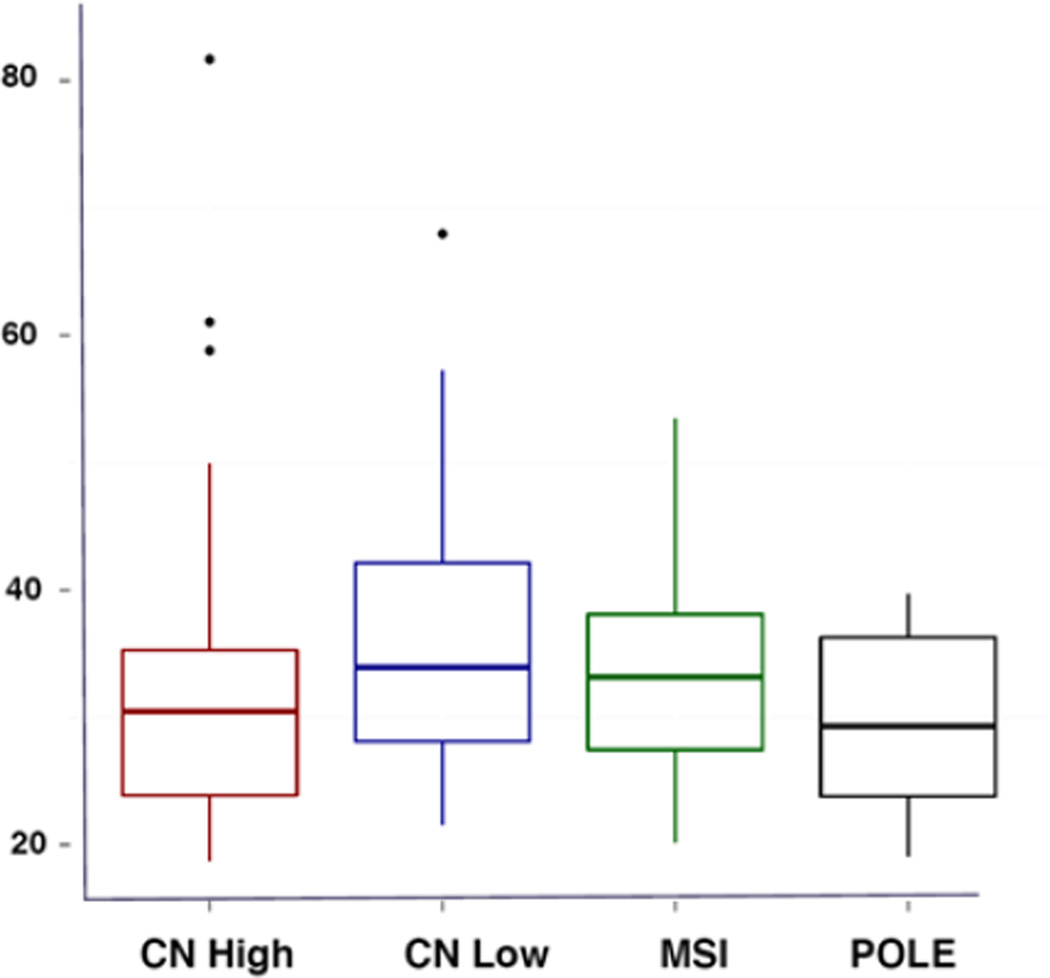
Of the EC clusters, POLE patients had a mean BMI of 29.8 (non-obese, but overweight), MSI had a mean BMI of 33.0 (obesity I classification), while CNL patients had a mean BMI of 35.8 (obesity II classification). CNH patients, comprised mostly of serous as opposed to endometriod histology tumors, had a BMI of 32.2 (obesity 1 classification). BMI was found to be statistically different between the ECs in the CNL (mean 35.8) versus POLE (mean 29.8) cluster (p=0.006) and approached statistical significance for the MSI (mean 33.0) versus CNL (mean 35.8) cluster (p=0.05). As illustrated in Figure 1, women with POLE tumors had the lowest median BMI. POLE tumors have been previously found to have significantly better PFS than MSI, CNL and CNH tumors [5].
Figure 1. Boxplots illustrating BMI means among endometrial cancer subtypes.
To determine if gene expression of tumors was regulated by BMI, RNA-seq and pathway analysis was undertaken. The list of genes was similar regardless of whether BMI was treated as a continuous variable or dichotomized into the categories of non-obese (BMI < 30) versus obese (BMI ≥ 30); therefore, data is presented for BMI as a continuous variable. A total of 214 genes were significantly associated with BMI. 161 were up-regulated and 53 were down-regulated with BMI (BMI < 30 versus BMI ≥ 30) (adjusted p-values< 0.01) (Appendix 1). Many of the genes identified have metabolically relevant activities related to obesity, including lipoprotein lipase (4.0-fold elevated expression in tumors from obese women over non-obese women), insulin receptor substrate 1 (5.1-fold), insulin-like growth factor binding protein 7 (4.7-fold), insulin-like growth factor binding protein 4 (4.4-fold) and the progesterone receptor (5.0-fold). DAVID functional annotation analysis revealed significant enrichment in "cell cycle" (adjusted p-value = 1.5E-5) and "DNA metabolic processes" (adjusted p-value = 1E-3). Other pathways or annotations did not have significant p-values after multiple testing correction.
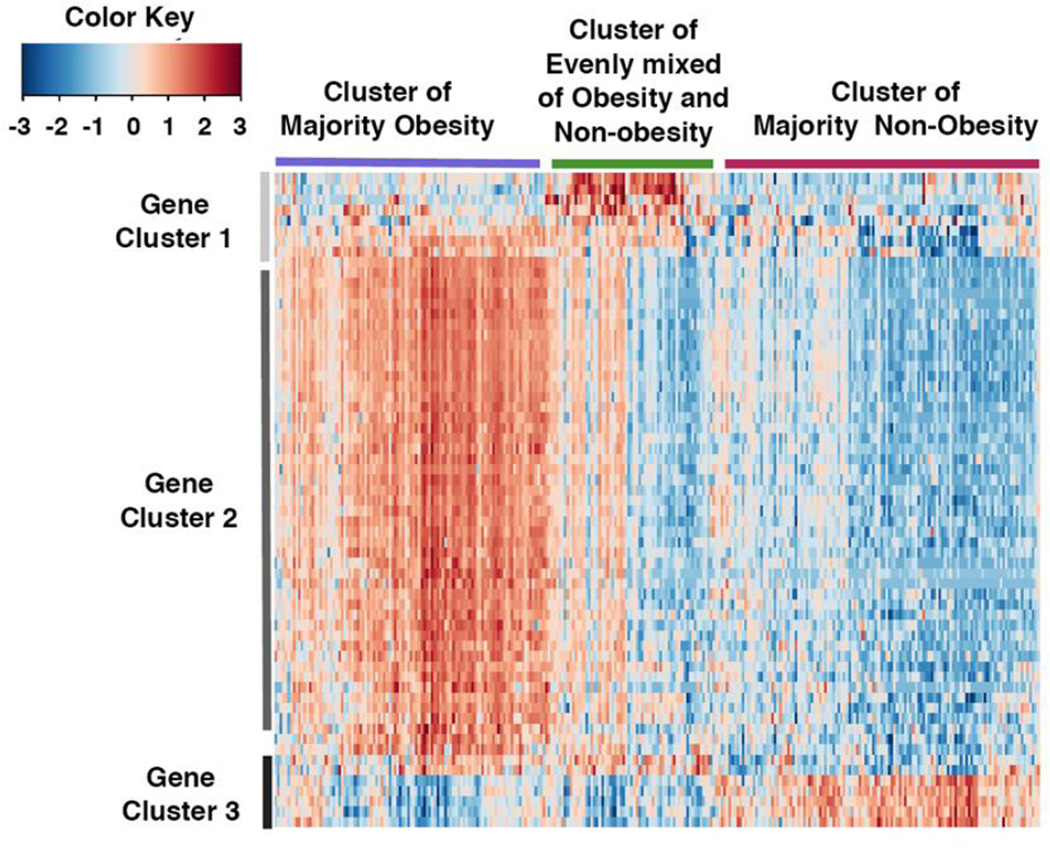
Hierarchical clustering was applied by using the 181 genes with a q value of <0.01, but the obtained heatmap did not show a sufficiently clear pattern. Therefore, a more stringent q value cut-off of 0.001 was applied to select genes for hierarchical clustering. The generated dendrogram identified three clusters for both individuals and genes (Figure 2). The three clusters were characterized by the proportions of obese and non-obese individuals. Specifically, the three clusters were named as “majority obese” because 69% of the patients were obese, “majority non-obese” contained 33% obese, and “mixed obese and non-obese” contained 59% obese patients. In addition, functional clustering analysis was conducted for the three clusters of genes. The first cluster of genes had significant enrichment in "cilium" (adjusted p-value = 3.4E-8) and "microtubule" (adjusted p-value = 5.1E-4) by DAVID functional annotation analysis. The second cluster of genes had significant enrichment in "cell cycle" (adjusted p-value = 4.7E-28) and "DNA metabolic processes" (adjusted p-value = 9E-14). The third cluster of genes did not show any functional enrichment by DAVID analysis. A summary of the genes in gene cluster 1 (cilium & microtubule cluster), 2 (cell cycle & DNA metabolic processes cluster) and 3 (no functional enrichment cluster) can be found in Table 3.
Figure 2. Genomic differences between endometrial cancer tumors from obese versus non-obese women reveal alterations in metabolically relevant genes.
Heat map representation of 181 genes found to be significantly up- or down-regulated with increasing BMI in endometroid histology tumors (q-value<0.01).
Table 3.
Differentially expressed genes between obese and non-obese endometrial cancer patients comprising Clusters 1 (Cilium/Microtubule), 2 (Cell cycle & DNA metabolic processes), and 3 (No functional enrichment)
| Cluster 1 (Cilium/Microtubule) |
Cluster 2 (Cell cycle & DNA metabolic processes) |
Cluster 2 Con't (Cell cycle & DNA metabolic processes) |
Cluster 3 (No functional enrichment) |
||||
|---|---|---|---|---|---|---|---|
| ADAMTSL2 | Similar to ADAMTS-like 2; ADAMTS- like 2 |
AKAP14 | A kinase (PRKA) anchor protein 14 |
NAPSB | Napsin B aspartic peptidase pseudogene |
CDC6 | Cell division cycle 6 homolog (S. cerevisiae) |
| ANKRD34C | Ankyrin repeat domain 34C |
APOBEC4 | Apolipoprotein B mRNA editing enzyme |
NXNL2 | Nucleoredoxin- like 2 |
CENPN | Centromere protein N |
| APCDD1 | Adenomatosis polyposis coli down-regulated 1 |
CAPSL | Calcyphosine- like |
ODF3B | Outer dense fiber of sperm tails 3B |
CKAP2 | Cytoskeleton associated protein 2 |
| CST3 | Cystatin C | CCDC19 | Coiled-coil domain containing 19 |
PGR | Progesterone receptor |
EXO1 | Exonuclease 1 |
| IRS1 | Insulin receptor substrate 1 |
CCDC33 | Coiled-coil domain containing 33 |
RAET1E | Retinoic acid early transcript 1E |
FANCD2 | Fanconi anemia |
| CDHR3 | Hypothetical protein FLJ23834 |
ROPN1L | Ropporin 1-like | ||||
| CDHR4 | Cadherin-like 29 |
SERPINA6 | Serpin peptidase inhibitor |
||||
| CNGA4 | Cyclic nucleotide gated channel alpha 4 |
SERPINI2 | Serpin peptidase inhibitor |
||||
| DNAI1 | Dynein | SNTN | Sentan | ||||
| FAM92B | Family with sequence similarity 92 |
SPAG8 | Sperm associated antigen 8 |
||||
| GSTA3 | Glutathione S- transferase alpha 3 |
SRD5A2 | Steroid-5-alpha- reductase |
||||
| IGFBP7 | Insulin-like growth factor binding protein 7 |
STOML3 | Stomatin (EPB72)-like 3 |
||||
| KCNE1 | Potassium voltage-gated channel |
TPPP3 | Tubulin polymerization- promoting protein family member 3 |
||||
| KNCN | Kinocilin | TRIM55 | Tripartite motif- containing 55 |
||||
| LOC100287718 | Similar to ankyrin repeat domain 2 |
TTC23L | Tetratricopeptide repeat domain 23-like |
||||
| LRRC10B | Leucine rich repeat containing 10- like |
VWA3A | von Willebrand factor A domain containing 3A |
||||
| LRRC18 | Leucine rich repeat containing 18 |
VWA3B | von Willebrand factor A domain containing 3B |
||||
| MAOB | Monoamine oxidase B |
WDR38 | WD repeat domain 38 |
||||
| MORN5 | MORN repeat containing 5 |
WDR49 | WD repeat domain 49 |
||||
| MS4A8B | Membrane- spanning 4- domains |
ZMYND10 | Zinc finger | ||||
DISCUSSION
Obesity is a well-established risk factor for EC, with each increase in BMI of 5 kg/m2 accounting for a significant increased risk of developing EC (RR 1.59; 95% CI 1.50–1.68) [9]. Higher BMI has also been associated with the development of EC before age 45 [10]. In addition, in patients with endometrial adenocarcinoma, severe obesity (BMI > 40 kg/m2) carries a 6.25-fold increased risk of death compared to normal weight women (BMI 18.5 – 24.9) [1]. Obesity is a high-energy, pro-inflammatory condition that culminates in increased growth factor signaling via the insulin/IGF-1 axis, as well as a nutrient-saturated environment via increased glucose (and other nutrients), ultimately aiding in the development of endometrial cancer [3]. Therefore, obesity may create a unique environment that can be exploited by a therapeutic approach as a strategy to improve EC outcomes.
To better understand the molecular basis for EC tumorigenesis in obese women, differential gene expression was compared between endometrial tumors of obese versus non-obese patients, using the TCGA database. Several genes were significantly up-regulated with BMI that are known to play important roles in EC pathogenesis. Some of these somatic aberrations included lipoprotein lipase (LPL); insulin receptor substrate 1 (IRS-1); insulin-like growth factor binding protein 7 (IGFBP7); insulin-like growth factor binding protein 4 (IGFBP4); and the progesterone receptor (PGR). DAVID analysis also identified proliferative pathways including cell cycle and DNA metabolic processes. How obesity regulated genes may be associated with EC is discussed as follows.
Lipoprotein lipase (LPL)
The expression of LPL was increased by 4-fold in the endometrial cancers from obese as compared to non-obese patients. LPL is involved in triglycerides hydrolysis and generation of fatty acids [11]. LPL appears to be upregulated in the presence of elevated glucose and certain fatty acids [11], as it is commonly the case in obese patients. Non-catalytic functions of LPL are carried on the protein C-terminus and include dimerization and bridging between lipoproteins and cell surface receptors such as heparin sulfate proteoglycans (HSPG) [12]. Bridging may involve different cell types, requiring LPL dimerization and HSPG on both surfaces [13]. Accordingly, expression of LPL by malignant cells may promote tumorigenesis by enabling the cell–stromal interactions that are critical for tumor maintenance [13, 14]. In fact, LPL is overexpressed in squamous cell carcinomas of the cervix and may be associated with increased tumor aggressiveness, likely secondary to the non-catalytic function of LPL [13]. Therefore, overexpression of LPL in the EC of obese women may be linked with the poorer outcomes that are associated with this patient population. If the increased tumor aggressiveness is in fact related to the non-catalytic C-terminal domain of LPL, that domain could potentially serve as a therapeutic target.
Insulin receptor substrate 1 (IRS-1)
Insulin receptor substrate 1 in endometrial tumors of obese women was increased 5.1-fold compared to tumors from non-obese patients. The insulin-like growth factor (IGF) signaling pathway is a complex system comprised of hormones, cell-surface receptors, as well as circulating and membrane bound IGF-binding proteins (IGFBP). This pathway plays a key role in the growth and development of many normal tissues but also plays a major role in the link between the metabolic syndrome/obesity and cancer [15]. Cell signaling occurs through the interaction of IGF-I, IGF-II and insulin with IGF-IR, IGF-IIR, the insulin receptor (IR) and hybrid receptors of IGF and insulin [16]. One of the substrates that interacts with the activated IGF-IR is the insulin receptor substrate (IRS)-1, which continues to transmit the IGF-IR signal downstream ultimately leading to activation of the type I phosphatidylinositol 3-kinase (PI3K)/Akt (PKB: protein kinase B), mammalian target of rapamycin (mTOR) pathway [17, 18]. Activation of the PI3k/Akt/mTOR pathway leads to cell proliferation and inhibition of apoptosis.
Given the close relationship between obesity, hyperinsulinemia, the IGF signaling pathway and cancer, it is not surprising that IRS-1 was significantly over-expressed in the majority obese group. In general, IRS-1 exhibits increased expression in other malignancies including pancreatic, prostatic, and breast cancers [19–21]. However, overexpression of IRS-1 is not only a factor in tumorigenesis, but it also appears to increase cellular resistance to autophagy-dependent cell death caused by oxidative stress [22]. In addition, IRS-1 has an anti-apoptotic function that protects cells from apoptotic cell death [23]. Therefore, the significant overexpression of IRS-1 in the obese women may yet serve as another explanation for increased risk of developing EC. In addition, the anti-apoptotic and anti-autophagic functions associated with overexpression may also explain the difference in survival in this patient population.
Insulin-like growth factor binding protein 4 (IGFBP4)
Overexpression of IGFBP4 (4.4-fold) was detected in the obese EC group. Binding of IGF to IGFBPs prolongs IGF’s half-life. In addition, through high affinity binding, IGFBP can sequester IGFs away from their cell surface receptor, thus modulating IGFs biologic functions in target cells [24]. IGFBP4 appears to have IGF-independent anti-tumorigenic and anti-angiogenic activity in human glioma cells lines as well as in a glioblastoma xenograft model [25]. The anti-angiogenic and anti-tumorigenic inherent activity of IGFBP4 appears to be associated to the C-terminal domain, while the IGF-binding domain is localized to the N-terminus [25]. Whether or not IGFBP4 has similar anti-tumorigenic activity in EC is unknown. However, assuming that the IGF-independent activity translates to endometrial carcinoma, IGFBP4 could serve as yet another therapeutic target. The fact that it appears to be overexpressed in the mostly obese group may likely be a physiologic response to the increased circulating levels of insulin and IGF in this patient population. Thus, we would postulate that typically the IGF-dependent activity of IGFBP4 is dominating. However, if IGFBP4 does in fact block cancer progression in an IGF-independent fashion, we could potentially use the increased circulating levels to our advantage by blocking the IGF binding site and effectively activating the anti-tumorigenic and anti-angiogenic protein C-terminus.
Insulin-like growth factor binding protein 7 (IGFBP7)
IGFBP7 was upregulated 4.7-fold in obese compared to non-obese patient tumors. Unlike IGFBP 1–6, IGFBP7 has a relatively low affinity for IGF-1 while its affinity for insulin is up to 500-fold higher [26]. Elevated serum levels of IGFBP7 are linked to insulin resistance and Type II diabetes [26, 27]. Therefore, the increased expression of IGFBP7 in the mostly obese cohort may be secondary to their increased risk of insulin resistance rather than a direct or indirect effect from their tumor physiology. Currenlty, the role of IGFBP7 in EC pathogenesis remains poorly understood.
Progesterone receptor (PGR)
PGR was detected at 5-fold greater levels in obese compared to non-obese tumors. PGR expression by uterine cells is stimulated by estrogens via estrogen receptor-α and consequently progesterone responsiveness is dependent on the presence of an estrogenic drive [28, 29]. As the levels of progesterone increase during the secretory phase, activation of the PGR results in inhibition of endometrial proliferation and differentiation into a secretory phenotype. However, in hyper-estrogenic states, the effects of progesterone are disrupted mainly due to the decreased production of ovarian progesterone due to either anovulation or menopause [29]. This abnormal physiology represents one of the major risk factors for the development of endometrioid endometrial cancer. Obesity is associated with both higher estrogen levels and anovulation, thus explaining the higher expression of PGR in the tumors from women in the “mostly obese” group. The understanding that some endometrioid ECs have increased PGR expression, has led to the use of progestin agents as a therapeutic option for young women with low-grade endometrioid adenocarcinomas interested in preserving fertility. However, whether or not higher PGR expression affects outcomes remains a topic of debate. A few small studies have failed to show a positive relationship between survival and PGR expression [30, 31]. Whereas, a multicenter trial found decreased survival in patients with ER and PR receptor loss [32].
EC clusters from TCGA and relationship to BMI
Integrated genomic, transcriptomic, and proteomic characterization of 373 endometrial carcinomas from the TCGA led to classification of these tumors into four clusters based of their molecular characteristics [5]. The POLE cluster represented tumors with ultra-high mutation rates in the POLE gene, which is involved in nuclear DNA replication and repair. The MSI cluster was composed of hyper-mutated tumors with microsatellite instability, as well as reduced expression of the DNA mismatch repair gene MLH1, due to methylation of its promoter region. The CNL cluster consisted of tumors with lower mutations frequencies as well as the majority of microsatellite-stable cancers. Lastly, the CNH cluster primarily encompassed serous and many high-grade endometrioid tumors with overall low mutation rates but with frequent TP53 mutations. Surprisingly, the POLE cluster, despite having the highest mutation rate was associated with the best PFS, followed by MSI and CNL tumors. As expected, given the predominance of serous cancers in this cluster, CNH tumors were associated with the worst PFS out of the four clusters. Based on our analysis, there appeared to be an association between obesity and the EC integrated clusters. Women with CNL tumors had the highest BMI whereas women with POLE tumors had the lowest BMI. These findings further suggested that obesity affects tumor biology at the molecular level, and ultimately these changes may lead to more aggressive tumors and poorer outcomes.
Conclusions
Our study is the first to examine the association between BMI and the integrated EC clusters identified through analysis of the TCGA data. We demonstrate that increasing BMI was associated with metabolically relevant alterations in gene expression among endometrioid ECs, including genes related to the insulin/IGF-1 pathway among others. As previously reported by TCGA, integrated genomic characterization of endometrial carcinomas found POLE tumors to be associated with significantly better progression free survival than any of the other clusters [5]. Meanwhile, in our study, patients with POLE tumors had the lowest median BMI, suggesting a correlation between BMI and survival of EC integrated clusters. Our study had several limitations mainly stemming from the fact that it was a post-hoc analysis. Because of this, we were not able to control for other factors such as diabetes, insulin and metformin use, which may affect tumor behavior and outcomes.
In conclusion, our findings suggest that the metabolic consequences of obesity may play a role in tumor development and resultant tumor biology and aggressiveness. The genetic heterogeneity found among Type 1 cancers based on BMI in combination with that previously seen among the four integrated EC clusters in TCGA provides further support that the classification of endometrial cancers into the categories of Type I and II tumors is too simplistic. Lastly, further work is needed focusing on the identification of pathways and biomarkers unique to obese women, which could prove useful in the development of targeted therapies or diagnostic tools more specific to this high risk patient population and obesity-driven ECs.
Supplementary Material
Highlights.
Women with POLE endometrial tumors have the lowest average BMI when compared to women with MSI and CNL tumors.
Increasing BMI is associated with metabolically relevant alterations in gene expression among endometrioid endometrial carcinomas.
Differential gene expression was related to BMI, suggesting that categorizing endometrial cancers into Type I/II is too simplistic.
Acknowledgments
VBJ was supported by NIH/NCI 1K23CA143154-01A1.
LM was supported by the University Cancer Research Fund (LM)
VBJ and LM were both supported by a Department of Defense/Ovarian Cancer Research Program (DOD/OCRP) Translational Pilot Award
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presented as a featured poster presentation at the 2014 Annual Meeting of the Society of Gynecologic Oncology in Tampa, FL.
CONFLICT OF INTEREST STATEMENT
The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Khandekar MJ, Cohen P, Spiegelman BM. Molecular mechanisms of cancer development in obesity. Nature reviews Cancer. 2011;11:886–895. doi: 10.1038/nrc3174. [DOI] [PubMed] [Google Scholar]
- 4.Hendrickson M, Ross J, Eifel P, Martinez A, Kempson R. Uterine papillary serous carcinoma: a highly malignant form of endometrial adenocarcinoma. Am J Surg Pathol. 1982;6:93–108. doi: 10.1097/00000478-198203000-00002. [DOI] [PubMed] [Google Scholar]
- 5.The Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 7.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 8.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet. 2008;371:569–578. doi: 10.1016/S0140-6736(08)60269-X. [DOI] [PubMed] [Google Scholar]
- 10.Pellerin GP, Finan MA. Endometrial cancer in women 45 years of age or younger: a clinicopathological analysis. Am J Obstet Gynecol. 2005;193:1640–1644. doi: 10.1016/j.ajog.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Mead JR, Irvine SA, Ramji DP. Lipoprotein lipase: structure, function, regulation, and role in disease. Journal of molecular medicine. 2002;80:753–769. doi: 10.1007/s00109-002-0384-9. [DOI] [PubMed] [Google Scholar]
- 12.Merkel M, Kako Y, Radner H, Cho IS, Ramasamy R, Brunzell JD, et al. Catalytically inactive lipoprotein lipase expression in muscle of transgenic mice increases very low density lipoprotein uptake: direct evidence that lipoprotein lipase bridging occurs in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13841–13846. doi: 10.1073/pnas.95.23.13841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carter SA, Foster NA, Scarpini CG, Chattopadhyay A, Pett MR, Roberts I, et al. Lipoprotein lipase is frequently overexpressed or translocated in cervical squamous cell carcinoma and promotes invasiveness through the non-catalytic C terminus. Br J Cancer. 2012;107:739–747. doi: 10.1038/bjc.2012.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansouri M, Sevov M, Fahlgren E, Tobin G, Jondal M, Osorio L, et al. Lipoprotein lipase is differentially expressed in prognostic subsets of chronic lymphocytic leukemia but displays invariably low catalytical activity. Leukemia research. 2010;34:301–306. doi: 10.1016/j.leukres.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 15.Maki RG. Small is beautiful: insulin-like growth factors and their role in growth, development, and cancer. J Clin Oncol. 2010;28:4985–4995. doi: 10.1200/JCO.2009.27.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan PD, Goss PE. The emerging role of the insulin-like growth factor pathway as a therapeutic target in cancer. Oncologist. 2008;13:16–24. doi: 10.1634/theoncologist.2007-0199. [DOI] [PubMed] [Google Scholar]
- 17.Gunter MJ, Hoover DR, Yu H, Wassertheil-Smoller S, Manson JE, Li J, et al. A prospective evaluation of insulin and insulin-like growth factor-I as risk factors for endometrial cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:921–929. doi: 10.1158/1055-9965.EPI-07-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCampbell AS, Broaddus RR, Loose DS, Davies PJ. Overexpression of the insulin-like growth factor I receptor and activation of the AKT pathway in hyperplastic endometrium. Clin Cancer Res. 2006;12:6373–6378. doi: 10.1158/1078-0432.CCR-06-0912. [DOI] [PubMed] [Google Scholar]
- 19.Bergmann U, Funatomi H, Kornmann M, Beger HG, Korc M. Increased expression of insulin receptor substrate-1 in human pancreatic cancer. Biochemical and biophysical research communications. 1996;220:886–890. doi: 10.1006/bbrc.1996.0500. [DOI] [PubMed] [Google Scholar]
- 20.Hellawell GO, Turner GD, Davies DR, Poulsom R, Brewster SF, Macaulay VM. Expression of the type 1 insulin-like growth factor receptor is up-regulated in primary prostate cancer and commonly persists in metastatic disease. Cancer Res. 2002;62:2942–2950. [PubMed] [Google Scholar]
- 21.Rocha RL, Hilsenbeck SG, Jackson JG, VanDenBerg CL, Weng C, Lee AV, et al. Insulin-like growth factor binding protein-3 and insulin receptor substrate-1 in breast cancer: correlation with clinical parameters and disease-free survival. Clinical cancer research : an official journal of the American Association for Cancer Research. 1997;3:103–109. [PubMed] [Google Scholar]
- 22.Chan SH, Kikkawa U, Matsuzaki H, Chen JH, Chang WC. Insulin receptor substrate-1 prevents autophagy-dependent cell death caused by oxidative stress in mouse NIH/3T3 cells. Journal of biomedical science. 2012;19:64. doi: 10.1186/1423-0127-19-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Qi X, Williams M, Shi Y, Keegan AD. Overexpression of insulin receptor substrate-1, but not insulin receptor substrate-2, protects a T cell hybridoma from activation-induced cell death. J Immunol. 2002;168:6215–6223. doi: 10.4049/jimmunol.168.12.6215. [DOI] [PubMed] [Google Scholar]
- 24.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocrine reviews. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 25.Moreno MJ, Ball M, Rukhlova M, Slinn J, L'Abbe D, Iqbal U, et al. IGFBP-4 anti-angiogenic and anti-tumorigenic effects are associated with anti-cathepsin B activity. Neoplasia. 2013;15:554–567. doi: 10.1593/neo.13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutsukake M, Ishihara R, Momose K, Isaka K, Itokazu O, Higuma C, et al. Circulating IGF-binding protein 7 (IGFBP7) levels are elevated in patients with endometriosis or undergoing diabetic hemodialysis. Reproductive biology and endocrinology : RB&E. 2008;6:54. doi: 10.1186/1477-7827-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Bermejo A, Khosravi J, Fernandez-Real JM, Hwa V, Pratt KL, Casamitjana R, et al. Insulin resistance is associated with increased serum concentration of IGF-binding protein-related protein 1 (IGFBP-rP1/MAC25) Diabetes. 2006;55:2333–2339. doi: 10.2337/db05-1627. [DOI] [PubMed] [Google Scholar]
- 28.Tsai M-J, Clark JH, Schrader WT, BW OM. Mechanisms of action of hormones that act as transcription-regulatory factors. In: Wilson J, Foster D, Kronenberg H, P L, editors. Williams Textbook of Endocrinology. Philadelphia: WB: Saunders Company; 1998. pp. 55–94. [Google Scholar]
- 29.Patel B, Elguero S, Thakore S, Dahoud W, Bedaiwy M, Mesiano S. Role of nuclear progesterone receptor isoforms in uterine pathophysiology. Human reproduction update. 2014 doi: 10.1093/humupd/dmu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunderson CC, Dutta S, Fader AN, Maniar KP, Nasseri-Nik N, Bristow RE, et al. Pathologic features associated with resolution of complex atypical hyperplasia and grade 1 endometrial adenocarcinoma after progestin therapy. Gynecol Oncol. 2014;132:33–37. doi: 10.1016/j.ygyno.2013.11.033. [DOI] [PubMed] [Google Scholar]
- 31.Singh M, Zaino RJ, Filiaci VJ, Leslie KK. Relationship of estrogen and progesterone receptors to clinical outcome in metastatic endometrial carcinoma: a Gynecologic Oncology Group Study. Gynecol Oncol. 2007;106:325–333. doi: 10.1016/j.ygyno.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 32.Trovik J, Wik E, Werner HM, Krakstad C, Helland H, Vandenput I, et al. Hormone receptor loss in endometrial carcinoma curettage predicts lymph node metastasis and poor outcome in prospective multicentre trial. Eur J Cancer. 2013;49:3431–3441. doi: 10.1016/j.ejca.2013.06.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.