Abstract
Rationale
Vascular smooth muscle (SMC) phenotypic modulation is characterized by the down-regulation of SMC contractile genes. Platelet-derived growth factor-BB (PDGF-BB), a well-known stimulator of SMC phenotypic modulation, down-regulates SMC genes via posttranscriptional regulation. The underlying mechanisms, however, remain largely unknown.
Objective
To establish RNA editing as a novel mechanism controlling SMC phenotypic modulation.
Methods and Results
Precursor mRNAs (pre-mRNA) of SMC myosin heavy chain (SMMHC) and α-actin (α-SMA) were accumulated while their mature mRNAs were down-regulated during SMC phenotypic modulation, suggesting an abnormal splicing of the pre-mRNAs. The abnormal splicing resulted from SMC marker pre-mRNA editing that was facilitated by adenosine deaminase acting on RNA 1 (ADAR1), an enzyme converting adenosines to inosines (A→I editing) in RNA sequences. ADAR1 expression inversely correlated with SMMHC and α-SMA levels; knockdown of ADAR1 restored SMMHC and α-SMA expression in phenotypically-modulated SMC; and editase domain mutation diminished the ADAR1-mediated abnormal splicing of SMC marker pre-mRNAs. Moreover, the abnormal splicing/editing of SMMHC and α-SMA pre-mRNAs occurred during injury-induced vascular remodeling. Importantly, heterozygous knockout of ADAR1 dramatically inhibited injury-induced neointima formation and restored SMC marker expression, demonstrating a critical role of ADAR1 in SMC phenotypic modulation and vascular remodeling in vivo.
Conclusions
Our results unraveled a novel molecular mechanism, i.e., pre-mRNA editing, governing SMC phenotypic modulation.
Keywords: RNA editing, ADAR1, vascular smooth muscle, phenotypic modulation, vascular remodeling, smooth muscle differentiation
INTRODUCTION
The transition of smooth muscle (SMC) from a contractile phenotype to a synthetic state (phenotypic modulation) plays a critical role in the development of a number of cardiovascular diseases. 1 A hallmark feature of SMC phenotypic modulation is the down-regulation of SMC-specific genes. The underlying mechanisms, however, are poorly understood. Platelet-derived growth factor (PDGF)-BB plays a critical role in SMC phenotypic modulation. 2, 3 PDGF-BB inhibits SMC marker gene expression, but PDGF-BB-mediated repression of smooth muscle α-actin (α-SMA) promoter is cell density-dependent, in which PDGF-BB has no effect on α-SMA promoter activity in confluent culture condition. Moreover, nuclear run-on assays show no differences in α-SMA mRNA transcription between PDGF-BB and vehicle-treated SMC, 3 demonstrating that the decrease in α-SMA transcription is irrelevant to PDGF-BB-induced repression of α-SMA mRNA 3. These studies strongly support that PDGF-BB regulates SMC marker expression via a post-transcriptional mechanism. PDGF-BB may affect SMC marker mRNA stability 3, but virtually nothing is known about which post-transcriptional mechanism is involved.
RNA editing (A→I editing) is one of the posttranscriptional mechanisms for gene expression. It is catalyzed by adenosine deaminase acting on RNA (ADAR) causing nucleotide substitution in RNA substrates.4 ADAR1 catalyzes RNA editing by substituting A with I in RNA sequences. 4 Because I is recognized as G by both ribosomes and RNA polymerases, when A is converted to I in pre-mRNA, I pairs with C instead of U. The I•C pair adopts a stability and geometry that are similar to a G•C pair. 5 RNA editing largely occurs in 5’ or 3’ untranslated regions, introns, and non-coding microRNA precursors. The editing of protein-coding sequences results in recoding and subsequent alterations of their functions. A variety of disease phenotypes in humans are associated with either an increase or decrease in RNA editing levels including systemic lupus erythematosus, various cancers, neurological disorders, etc. 6 However, it is unknown if RNA editing is involved in SMC phenotypic modulation and vascular remodeling. In the present study, we found that ADAR1-mediated RNA editing is an essential mechanism for SMC phenotypic modulation. Blockade of ADAR1 markedly attenuates PDGF-BB-mediated down-regulation of SMC contractile proteins and injury-induced vascular remodeling.
METHODS
Animals
Male Sprague-Dawley rats weighing 450–500 g were purchased from Harlan. Male ADAR1+/− mice (B6.129(Cg)-ADARtm1.1phs, Stock # 034620-JAX) were purchased from Mutant Mouse Regional Resource Centers (MMRRC, USA). All animals received humane care in compliance with the Principles of Laboratory Animal Care formulated by the National Society for Medical Research and the Guide for the Care and Use of Laboratory Animals. All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Georgia.
Cell culture and transfection
Rat primary aortic SMCs (SMCs) were cultured by enzyme digestion method from rat thoracic aorta as described previously.7 SMC phenotype was confirmed by the expression of smooth muscle α-actin and SM22α. Plasmids were transfected into the SMC using Lipofectamine LTX reagents (Life Technology).
Construction of adenoviral vector
Adenovirus expressing ADAR1 shRNA (Ad-shADAR1) was generated and purified as described previously.8 Green fluorescent protein (GFP)-expressing adenovirus (Ad-GFP) was used as a control.
Reverse transcription (RT)-PCR and Western Blot
Total RNA was isolated from primary SMCs or artery tissues homogenized in Trizol reagents (Life Technologies). Reverse transcription was performed using iScript Select cDNA synthesis Kit (Bio-Rad). Precursor (Pre-) and mature- mRNA of genes interested was amplified using specific primers and Advantage Long polymerase mix (Clontech) and quantified by normalizing to the GAPDH level. The primer sequences were listed in Online Table I. RT-PCR and western blot analyzing ADAR1, SMMHC, and α-SMA expression were performed as described. 7
Cloning and sequencing of pre-mRNA Cdna
SMMHC and α-SMA pre-mRNAs were reverse transcribed, amplified, and purified using Gel Extraction kit (Qiagen). The purified fragments were cloned into pGEM-T easy vector (Promega) followed by sequencing to detect A-I editing. The I was recognized as G in reverse transcribed cDNA. The A-I edits were identified by comparing the reverse-transcribed cDNA sequence with the genomic DNA sequence for the same gene in the control cells, or the splicing inhibitor-treated contractile SMC.
RNA secondary structure prediction
Secondary structure of normal or edited pre-mRNA fragment was predicted based on the sequencing data using mfold web servers (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form). Free energy of thermodynamic ensemble of the normal or edited pre-mRNA was analyzed using Vienna RNA web servers (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
Contractility assay
SMCs were transduced with adenovirus expressing control or ADAR1 shRNA followed by PDGF-BB treatment for 2 days. Contractility of the cells stimulated by 75 mM KCl was measured as described previously. 9 The cell perimeters of 12 cells each treatment were measured for quantification.
Rat carotid artery injury model
Rat carotid artery balloon injury was performed as described previously. 8 The morphometric analyses were performed in a double-blinded manner.
Mouse wire injury model
ADAR1+/− mice and its littermates were anesthetized, and carotid artery injury was performed using 0.15 inches straight tipped wire catheter (Cook Medical Inc). 28 days later, the injured arteries were excised and fixed with 4% paraformaldehyde followed by morphometric analyses in a double-blinded manner.
Histomorphometric analysis and immunohistochemistry (IHC) staining
Artery segments were cut by serial sectioning (5 µm). Modified hematoxylin and eosin, Elastica van Gieson, and IHC staining were performed as described previously. 8 The areas of the lumen, internal elastic lamina, and external elastic lamina were measured using Image-pro Plus Software.
Site-directed mutagenesis of ADAR1 editase domain
ADAR1 editase motif mutant was generated by converting Lysine to Arginine at the amino acid 258 by replacing the A with G at nucleotide 776 in ADAR1 cDNA (pDONR-221-ADAR1, Biodesign Institute/Arizona State University) as described previously using Quick Change Mutagenesis Kit (Aglient). 10
Statistical analysis
Pre-mRNA, mature mRNA, and protein expression results were expressed as Mean±SEM. One way ANOVA was used for comparison among groups. Significance was confirmed by post-hoc analysis using Fisher’s Least Significant Difference (Fisher’s LSD) test. P<0.05 was considered statistical significant.
RESULTS
Abnormal pre-mRNA splicing of SMC contractile proteins occurred in SMC phenotypic modulation
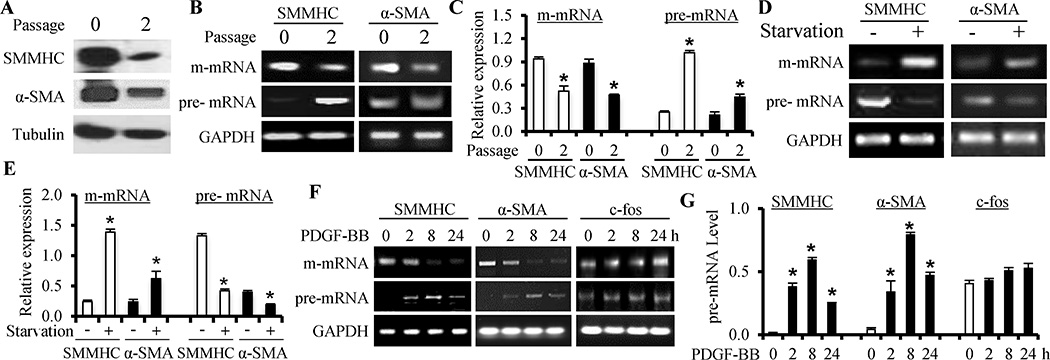
Artery media SMC rapidly modulates its phenotype from contractile to synthetic state once placed in culture. Thus, contractile proteins SMMHC and α-SMA were highly expressed in freshly isolated rat aortic artery SMC but significantly down-regulated after growing in culture for 2 passages (Figure 1A). The mature mRNA (m-mRNA) of both SMMHC and α-SMA was also significantly down-regulated (Figure 1B–1C). To explore the mechanism underlying the reduction of m-mRNA level, we amplified pre-mRNA transcripts from intron regions of both SMMHC and α-SMA, and found that the pre-mRNAs were barely detectable in artery SMC, indicating an effective splicing into m-mRNA. However, pre-mRNAs of SMMHC and α-SMA were accumulated in phenotypically modulated SMC (Figure 1B–1C and Online Figure I, A), indicative of an abnormal pre-mRNA splicing. Serum-starvation is known to induce re-differentiation of cultured SMCs. Indeed, serum-starvation significantly up-regulated the m-mRNA levels of SMMHC and α-SMA (Figure 1D–1E) while reducing their pre-mRNA levels (Figure 1D–1E and Online Figure I, B), further supporting the abnormal pre-mRNA splicing in SMC phenotypic modulation. Furthermore, PDGF-BB blocked serum-starved SMC to express SMMHC and α-SMA m-mRNA in a time-dependent manner (Figure 1F–1G and Online Figure I, D) while inducing the accumulation of their pre-mRNAs (Figure 1F and Online Figure I, C), indicating that PDGF-BB also caused abnormal pre-mRNA splicing. This effect is specific because c-fos pre-mRNA was not affected although its m-mRNA was altered by PDGF-BB (Figure 1F–1G and Online Figure I, D). The abnormal pre-mRNA splicing also occurred in phenotypically modulated human SMC (Online Figure I, E). Interestingly, several other factors important for SMC phenotype appeared not to undergo abnormal pre-mRNA splicing (Online Figure II), consistent with the current understanding that SMC phenotypes are regulated by multiple different mechanisms.
Figure 1. Abnormal RNA splicing occurred during SMC phenotypic modulation.
(A) SMMHC and α-SMA proteins were down-regulated after SMCs were cultured for two passages (passage 2 vs 0), indicative of SMC phenotypic modulation. (B) SMMHC and α-SMA mature mRNA (m-mRNA) were down-regulated while their precursor mRNA (pre-mRNA) levels (transcripts from SMMHC intron #12 and α-SMA intron #1 regions) increased when media SMCs were cultured for 2 passages. (C) Quantification of SMMHC and α-SMA m- and pre-mRNA expression shown in B by normalizing to GAPDH. *P<0.05 compared to the freshly isolated SMCs (passage 0) in each group, respectively (n=4). (D) Serum starvation of cultured SMC (+) for 48 h increased SMMHC and α-SMA m-mRNA levels while decreased their pre-mRNA levels, indicating a reversal of SMC phenotypic modulation. (E) Quantification of SMMHC and α-SMA m- and pre-mRNA expression shown in D by normalizing to GAPDH. *P<0.05 compared to the non-starved SMCs (−) in each group, respectively (n=4). (F) PDGF-BB induced a time-dependent abnormal splicing of SMMHC and α-SMA pre-mRNAs, but not the c-fos pre-mRNA. (G) Quantification of the pre-mRNA levels shown in F by normalizing to GAPDH. *P<0.05 compared to the vehicle-treated group (0 h) for each gene (n=4).
ADAR1 mediated the pre-mRNA splicing of SMC marker genes
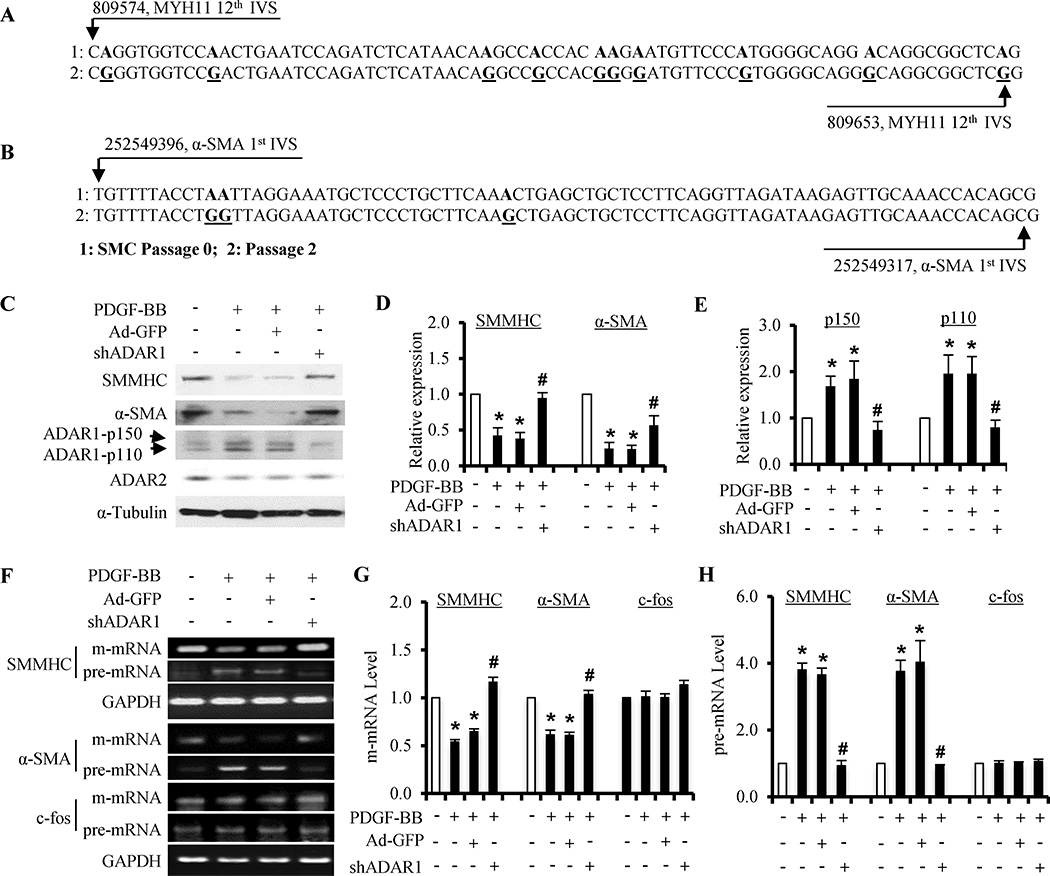
Sequencing of pre-mRNAs from SMMHC intron #12 (Figure 2A) and α-SMA intron #1 (Figure 2B) revealed multiple A to I editing sites. The edited SMMHC and α-SMA pre-mRNAs appeared to gain more stable secondary structures (Online Figure III). This type of pre-mRNA editing is mediated by ADAR1 or ADAR2.6 ADAR1, but not ADAR2, was induced in phenotypically modulated SMC (Fig 2C, 2E and Online Figure IV), suggesting that ADAR1, but not ADAR2, is involved in SMC phenotypic modulation. PDGF-BB induced the protein expression of two ADAR1 isoforms (p150 and p110) simultaneously while down-regulated SMC contractile proteins (Figure 2C–2E and Online Figure IV, D–F). Knockdown of ADAR1 by shRNA restored the contractile protein expression that was blocked by PDGF-BB (Figure 2C–2E). ADAR1 shRNA did not affect ADAR2 expression (Figure 2C and Online Figure IV, F). Importantly, ADAR1 shRNA blocked PDGF-BB-induced abnormal pre-mRNA splicing of SMMHC and α-SMA, but not c-fos (Figure 2F–2H). Moreover, ADAR1 knockdown converted flatten-shaped morphology of synthetic SMC to spindle-shaped contractile morphology (Online Figure V, A). ADAR1 shRNA-treated SMC also showed a contraction in response to the depolarization agent KCl (Online Figure V). These data demonstrate that ADAR1 played an essential role in SMC phenotypic modulation.
Figure 2. ADAR1 mediated PDGF-BB-induced SMC phenotypic modulation.
(A–B) Pre-mRNA transcripts from SMMHC intron #12 (A) and α-SMA intron #1 regions (B) were amplified from RNA isolated from cultured SMCs (passage 2), reverse transcribed, and cloned into pGEM-T vector followed by sequencing. The resulting cDNA sequences were compared to the genomic DNA sequences of artery media SMC (passage 0) to identify A–I editing. I was displayed as G when pre-mRNA was reverse-transcribed to cDNA. Representative A–I (G) editing sites in SMMHC intron #12 (A) and α-SMA intron #1 regions (B) are shown. (C) Knockdown of ADAR1 by its shRNA restored SMMHC and α-SMA protein expression that was down-regulated by PDGF-BB. (D–E) Quantification of the protein expression of SMMHC and α-SMA (D), and ADAR1 isoforms (E) shown in A by normalizing to α-tubulin. *P<0.01 compared to vehicle-treated group (−) for each corresponding protein; #P<0.05 compared to shRNA control group (Ad-GFP) of each corresponding protein (n=4). (F) Knockdown of ADAR1 blocked PDGF-BB-induced abnormal splicing of SMMHC and α-SMA pre-mRNA. (G–H) Quantification of SMMHC and α-SMA mature (m-) mRNA (G) and pre-mRNA (H) expression by normalizing to GAPDH. *P<0.01 compared to the vehicle-treated group (−), and #P<0.01 compared to the Ad-GFP/PDGF-BB treated group (+), for each corresponding gene or measurement, respectively (n=4).
ADAR1-mediated RNA editing was essential for the pre-mRNA splicing of SMC marker genes
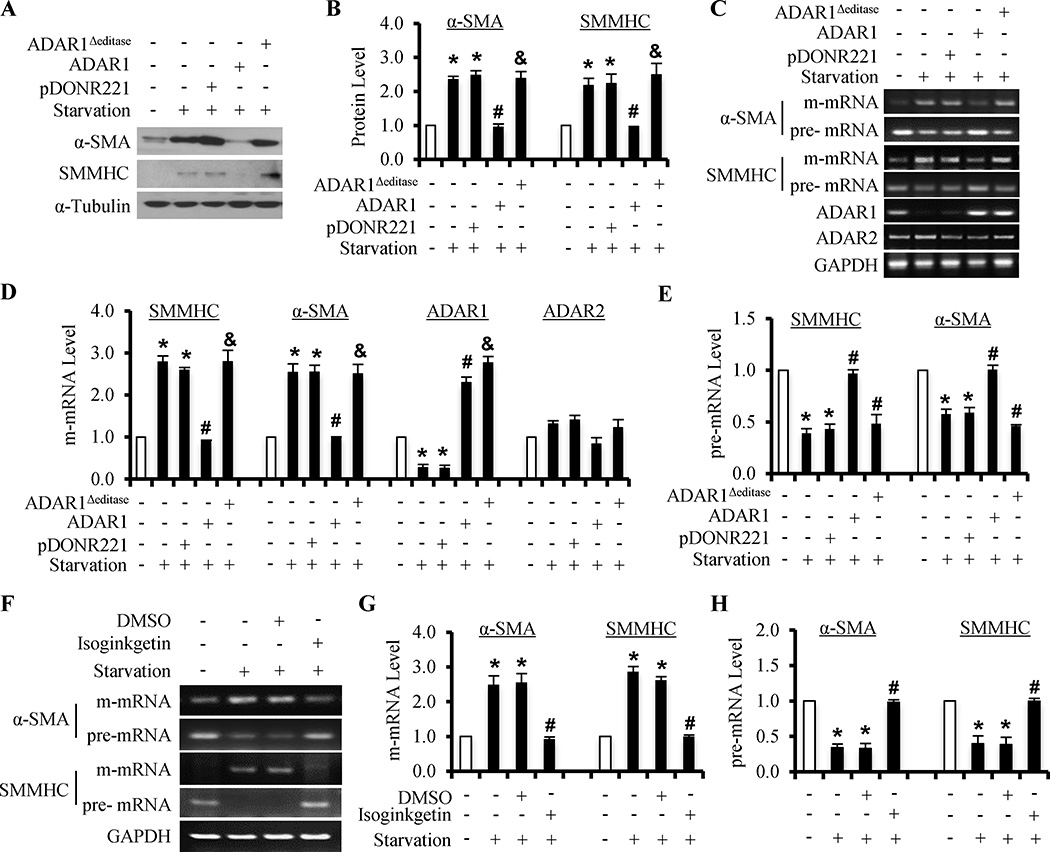
To determine if ADAR1-mediated RNA editing has caused the down-regulation of SMC marker proteins and the abnormal pre-mRNA splicing, we mutated the editase motif in ADAR1. An editase mutant form of ADAR1 was unable to block the serum starvation-induced increase in α-SMA and SMMHC protein expression that was blocked by the wild type ADAR1 (Figure 3A–3B). Of importance, editase motif mutation abolished the ADAR1-mediated reduction of m-mRNAs and accumulation of pre-mRNAs of α-SMA and SMMHC (Figure 3C–3E). These results indicated that ADAR1-mediated RNA editing is responsible for, at least in part, the down-regulation of a subset of SMC markers during SMC phenotypic modulation. To further establish if abnormal splicing contributed to the accumulation of pre-mRNA and reduction of m-mRNA, we used a general splicing inhibitor Isoginkgetin to treat serum-starved SMC and found that the splicing inhibitor blocked the starvation-induced increase in m-mRNA levels and the decrease in pre-mRNA levels of α-SMA and SMMHC genes (Figure 3F–3H), suggesting that the abnormal pre-mRNA splicing is an important mechanism regulating SMC phenotypic modulation. By using Isoginkgetin to accumulate pre-mRNA in contractile SMC, we confirmed the pre-mRNA editing occurred in phenotypically modulated SMC (induced by PDGF-BB), but not in the contractile SMC (Online Figure VI).
Figure 3. ADAR1-mediated editing caused the abnormal splicing of SMC marker pre-mRNAs in SMC phenotypic modulation.
(A) ADAR1 overexpression decreased α-SMA and SMMHC protein levels in serum-starved SMC. However, ADAR1Δeditase (ADAR1 editase motif mutation) was unable to alter the effect of serum starvation. (B) Quantification of α-SMA and SMMHC proteins shown in A by normalizing to α-Tubulin. (C) ADAR1, but not the ADAR1Δeditase, decreased the m-mRNA while increasing pre-mRNA levels of α-SMA and SMMHC in serum-starved SMC. (D–E) Quantification of m-mRNA (D) and pre-mRNA levels (E) shown in B by normalizing to the GAPDH level. *P<0.01 compared to non-starved group (−), #P<0.01 compared to pDONR221 (empty vector) transfected group, and &P<0.01compared to ADAR1-transfected group, for each individual genes, respectively (n=4). (F) General splicing inhibitor (Isoginkgetin, 33 µM) decreased the m-mRNA, but increased the pre-mRNA levels of α-SMA and SMMHC in starved SMC. (G–H) Quantification of m-mRNA (G) and pre-mRNA levels (H) shown in F by normalizing to GAPDH. *P<0.01 compared to non-starved group (−). #P<0.01 compared to vehicle (DMSO)-treated group for each corresponding gene, respectively (n=4).
ADAR1 was essential for vascular remodeling and SMC phenotypic modulation in vivo
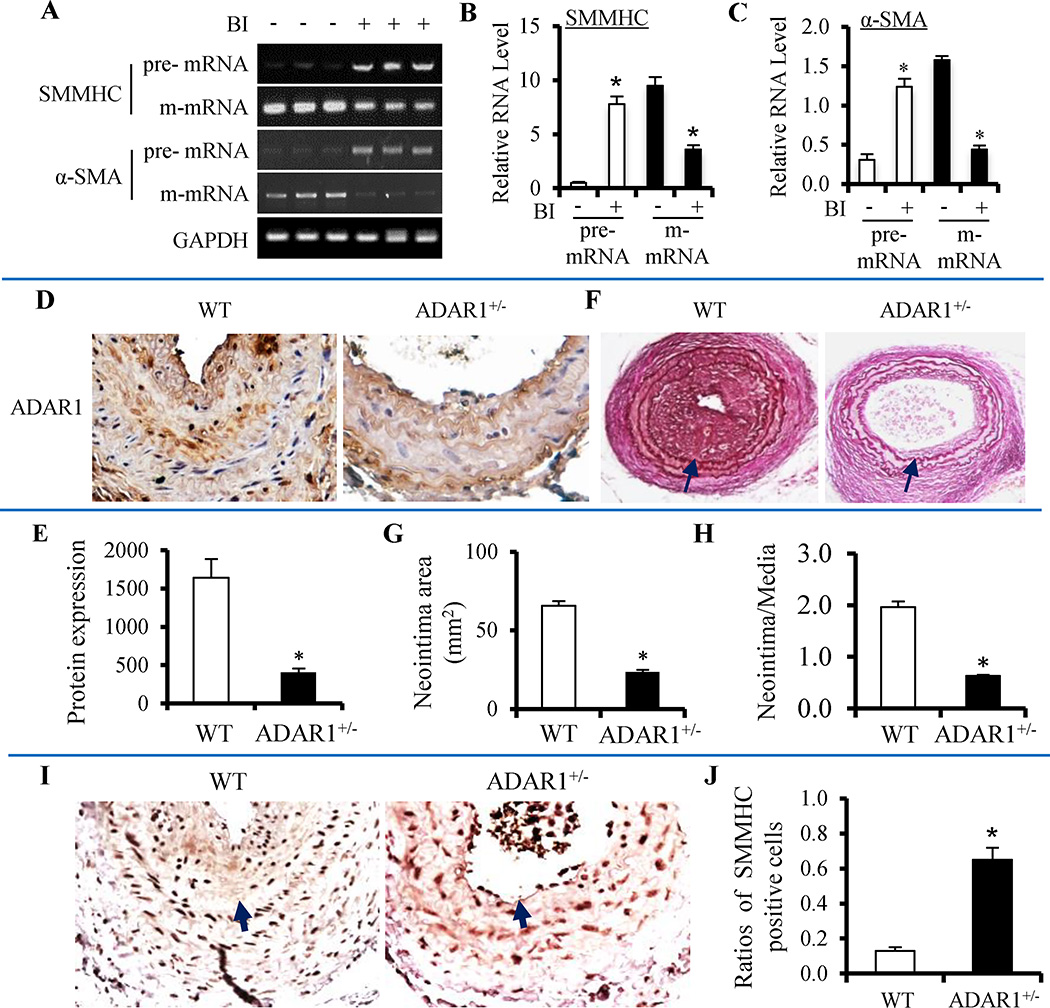
SMMHC and α-SMA pre-mRNAs were accumulated while their m-mRNAs down-regulated in balloon-injured rat carotid arteries (Figure 4A–4C). Consistently, RNA edits were observed in the SMMHC pre-mRNA of the injured, but not the control rat carotid arteries (Online Figure VII). Importantly, RNA editing caused a mutation in the binding site of a splicing factor Serine/arginine-rich (SR) proteins SRp55 (Online Figure VII).11 A shortening of the CA repeats in SMMHC pre-mRNA was also observed (Online Figure VII). CA repeats have been shown to be involved in pre-mRNA splicing.12 The SRp55 binding site mutation and the shortening of the CA repeats were also observed in the identical region of SMMHC pre-mRNA isolated from PDGF-BB-treated SMC (Data not shown), suggesting common editing/splicing mechanisms regulating the SMC phenotypic modulation in vitro and in vivo. Balloon injury also induced ADAR1 expression while down-regulating SMC contractile proteins in the injured carotid arteries (Online Figure VIII). To determine if ADAR1-mediated RNA editing plays a role in vascular remodeling, we performed a wire injury in carotid arteries of ADAR1 heterozygous knockout (ADAR+/−) mice (Figure 4D–4E). We used the ADAR+/− mice because ADAR1 homozygous knockout causes embryonic lethality. 13 As shown in Figure 4F–4H, the wire injury induced severe neointima formation in wild type mouse arteries. However, neointima formation was significantly blocked in ADAR+/− mice. These results demonstrated that ADAR1 is essential for injury-induced vascular remodeling. To test if ADAR1 is important for SMC phenotypic modulation in vivo, we detected the SMMHC expression in the injured artery, and found that ADAR+/− significantly increased the number of SMMHC-expressing SMC (Figure 4I–4J), indicating an accelerated reversal of SMC phenotypic modulation. These results indicated that ADAR1-mediated RNA editing plays a critical role in vascular remodeling and SMC phenotypic modulation in vivo.
Figure 4. ADAR1 mediated injury-induced vascular remodeling and SMC phenotypic modulation in vivo.
(A) Abnormal splicing of SMMHC and α-SMA pre-mRNAs occurred in injured rat carotid artery 14 days after the balloon injury (BI). SMMHC and α-SMA pre-mRNAs was accumulated while their m-mRNA levels decreased. (B–C) Quantification of pre- and m-mRNA levels of SMMHC (B) and α-SMA (C) by normalizing to GAPDH. *P < 0.05 compared to vessels without balloon injury (−) in each corresponding group (n=5). (D) Left carotid arteries of wild type (WT) littermates or ADAR1 heterozygous knockout mice (ADAR1+/−) were wire-injured for 28 days. ADAR1 expression was detected by immunohistochemistry staining. (E) Quantification of ADAR1 staining in D showed significantly lower expression of ADAR1 in ADAR1+/− mice. *P<0.01 compared to WT (n=7). (F) ADAR1 heterozygous knockout (ADAR1+/−) blocked neointima formation in mouse carotid artery with wire injury for 28 days as shown by elastin staining. Arrow indicates internal elastic lamina. (G–H) Quantification of the neointima area (G) and neointima/media ratio (H) shown in F. *P<0.01 compared to the WT arteries (n=7). (I) ADAR+/− enhanced SMC re-differentiation in injured artery as shown by the increased number of SMMHC-expressing SMC. Arrow indicates the internal elastic lamina. (J) Quantification of SMMHC-positive SMC by normalizing to the total SMC in neointima and media layers. *P<0.01 compared to WT (n=7).
DISCUSSION
SMC phenotypic modulation is a very complicated process. The underlying mechanisms have been largely focused on the transcription regulation of SMC contractile proteins. We have identified a novel posttranscriptional mechanism controlling this process, i.e., RNA editing/splicing, which causes the accumulation of the pre-mRNAs along with the reduction of their mature mRNAs of at least a subset of SMC contractile proteins. The RNA editing occurs when arterial SMCs are placed in culture, or serum-starved SMCs are treated with PDGF-BB. However, serum-starvation can reverse the editing process because starvation induces the contractile protein expression. Importantly, the abnormal SMC marker pre-mRNA editing/splicing also occurs in injury-induced vascular remodeling, indicating that RNA editing is involved in SMC phenotypic modulation in vivo. Although there are two enzymatically functional ADARs in mammalian, it appears that ADAR1 is responsible for the RNA editing in SMC phenotypic modulation because ADAR1, but not ADAR2, is induced by PDGF-BB. ADAR1 indeed mediates PDGF-BB-induced SMC phenotypic modulation because knockdown of ADAR1 blocks PDGF-BB-induced RNA editing/splicing and restores the SMC contractile protein expression. Moreover, the editase motif mutation also diminishes the ADAR1-mediated accumulation of pre-mRNA of SMC contractile proteins, further demonstrating that RNA editing plays an important role in SMC phenotypic modulation.
The abnormal splicing of SMC marker pre-mRNAs appears to involve multiple mechanisms. First, the A-I substitution could alter the second structures of SMC marker pre-mRNAs and thus decreases their thermodynamic free energy and hinders the normal RNA splicing. Secondly, the pre-mRNA editing may cause mutations in the binding elements of splicing factors (e.g., SRp55 site in SMMHC pre-mRNA), and thus block their activities. Thirdly, the deletion of the CA repeats in SMMHC pre-mRNA indicates that the alternative splicing in phenotypically-modulated SMC may also be affected by PDGF-BB. Intronic CA RNA elements has been found to function as either splicing enhancers or splicing silencers.12 In addition, an alteration in splicing complexes may also cause an accumulation of SMC marker pre-mRNAs in phenotypically-modulated SMC. Whether or not PDGF-BB affects the expression or assembly of the splicing complexes will be an interesting subject for the future investigation. Nevertheless, RNA editing is clearly a previously-unidentified mechanism regulating SMC phenotypic modulation.
In summary, we have identified an important novel mechanism, i.e., RNA editing, regulating SMC phenotypic modulation. Our results suggest that multiple mechanisms including transcription regulation and posttranscriptional modification among others may work together to regulate the complex process of SMC phenotypic modulation.
Supplementary Material
Novelty and Significance.
What Is Known?
Smooth muscle cell (SMC) phenotypic modulation is a complex process initiating vascular remodeling/neointima formation in proliferative vascular diseases.
Down-regulation of SMC contractile proteins is a hallmark of SMC phenotypic modulation.
SMC marker expression is regulated at both transcriptional and post-transcriptional levels.
What New Information Does This Article Contribute?
Abnormal splicing of SMC marker pre-mRNAs contributes to the down-regulation of the marker expression during SMC phenotypic modulation.
The adenosine deaminase acting on RNA 1 (ADAR1)-mediated A to I editing is one of the mechanisms underlying the abnormal splicing of SMC marker pre-mRNAs.
ADAR1 plays a critical role in SMC phenotypic modulation and vascular remodeling in vivo.
Prior studies have shown that SMC phenotypic modulation plays an important role in injury-induced vascular remodeling, and platelet-derived growth factor-BB (PDGF-BB), a well-known stimulator of SMC phenotypic modulation, down-regulates SMC genes via posttranscriptional regulation. The underlying mechanisms, however, remain largely unknown. Here, we use an ADAR1 deficient mouse model, molecular, and cellular analyses to identify a novel mechanism underlying SMC phenotypic modulation. Our studies demonstrate for the first time that abnormal SMC marker pre-mRNA splicing due to RNA editing is a novel mechanism controlling SMC phenotypic modulation and injury-induced vascular remodeling. The pre-mRNA editing is facilitated by ADAR1. PDGF-BB induces ADAR1 expression while down-regulating the expression of smooth muscle myosin heavy chain (SMMHC) and smooth muscle alpha-actin (α-SMA). ADAR1 deficiency or mutation of the editase prevents the reduction of SMC markers, demonstrating an essential role of ADAR1 in SMC phenotypic modulation in vitro. Animal studies show that SMMHC and α-SMA pre-mRNA accumulate while their mature mRNAs decrease along with the expression of ADAR1 in balloon-injured rat carotid arteries. Heterozygous ADAR1 deficiency dramatically inhibits injury-induced neointima formation with a down-regulation of SMC markers, demonstrating a critical role of ADAR1 in SMC phenotypic modulation and vascular remodeling in vivo.
Acknowledgments
SOURCES OF FUNDING
This work was supported by grants from National Institutes of Health (HL107526, HL119053, and HL123302). X.B.C. is supported by an American Heart Association Postdoctoral Fellowship (14POST20480015).
Nonstandard Abbreviations and Acronyms
- SMC
vascular smooth muscle cell
- SMMHC
smooth muscle myosin heavy chain
- α-SMA
smooth muscle α-actin
- pre-mRNA
precursor mRNA
- ADAR
adenosine deaminase acting on RNA
- PDGF-BB
Platelet-derived growth factor
Footnotes
DISCLOSURES
None.
REFERENCES
- 1.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiological reviews. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 2.Hart CE, Kraiss LW, Vergel S, Gilbertson D, Kenagy R, Kirkman T, Crandall DL, Tickle S, Finney H, Yarranton G, Clowes AW. PDGFbeta receptor blockade inhibits intimal hyperplasia in the baboon. Circulation. 1999;99:564–569. doi: 10.1161/01.cir.99.4.564. [DOI] [PubMed] [Google Scholar]
- 3.Corjay MH, Blank RS, Owens GK. Platelet-derived growth factor-induced destabilization of smooth muscle alpha-actin mRNA. Journal of cellular physiology. 1990;145:391–397. doi: 10.1002/jcp.1041450302. [DOI] [PubMed] [Google Scholar]
- 4.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annual review of biochemistry. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vendeix FA, Munoz AM, Agris PF. Free energy calculation of modified base-pair formation in explicit solvent: A predictive model. RNA. 2009;15:2278–2287. doi: 10.1261/rna.1734309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maas S. Gene regulation through RNA editing. Discovery medicine. 2010;10:379–386. [PubMed] [Google Scholar]
- 7.Christen T, Bochaton-Piallat ML, Neuville P, Rensen S, Redard M, van Eys G, Gabbiani G. Cultured porcine coronary artery smooth muscle cells. A new model with advanced differentiation. Circ Res. 1999;85:99–107. doi: 10.1161/01.res.85.1.99. [DOI] [PubMed] [Google Scholar]
- 8.Wang JN, Shi N, Xie WB, Guo X, Chen SY. Response gene to complement 32 promotes vascular lesion formation through stimulation of smooth muscle cell proliferation and migration. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:e19–e26. doi: 10.1161/ATVBAHA.111.230706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo X, Stice SL, Boyd NL, Chen SY. A novel in vitro model system for smooth muscle differentiation from human embryonic stem cell-derived mesenchymal cells. Am J Physiol Cell Physiol. 2013;304:C289–C298. doi: 10.1152/ajpcell.00298.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci U S A. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran Q, Roesser JR. SRp55 is a regulator of calcitonin/CGRP alternative RNA splicing. Biochemistry. 2003;42:951–957. doi: 10.1021/bi026753a. [DOI] [PubMed] [Google Scholar]
- 12.Hui J, Hung LH, Heiner M, Schreiner S, Neumuller N, Reither G, Haas SA, Bindereif A. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinman RA, Wang Q. ADAR1 isoform involvement in embryonic lethality. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E199. doi: 10.1073/pnas.1105004108. author reply E200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.