Abstract
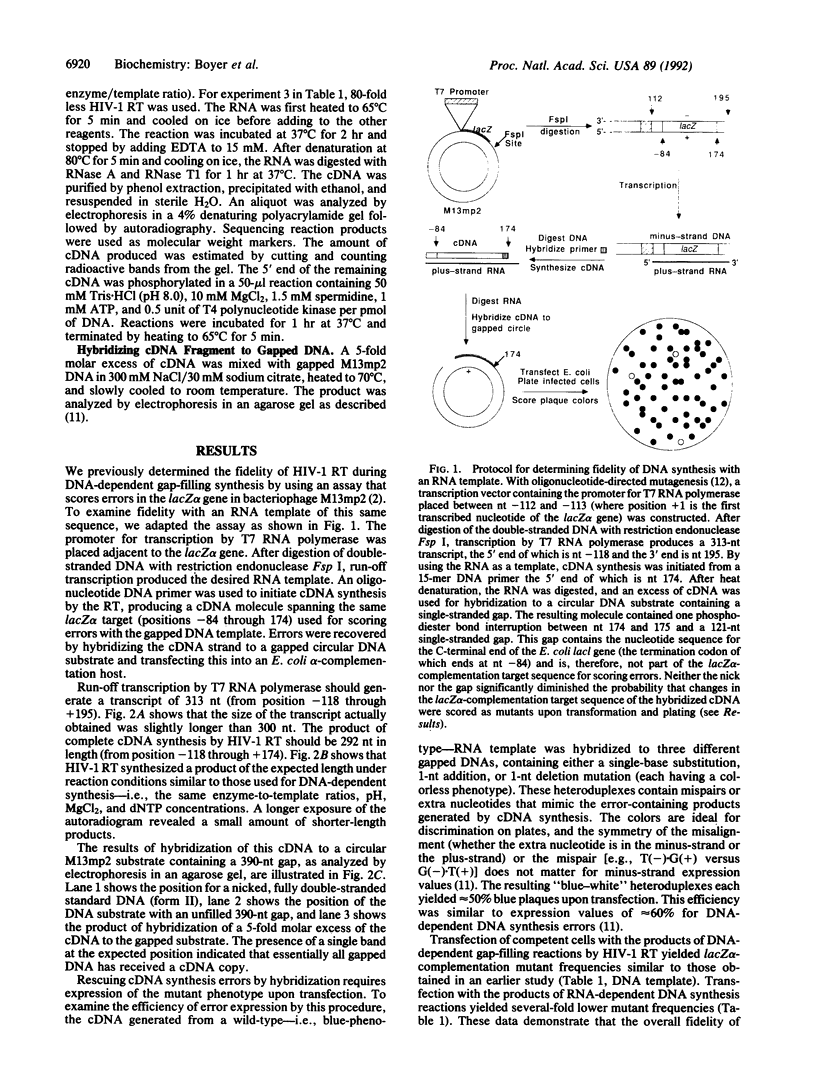
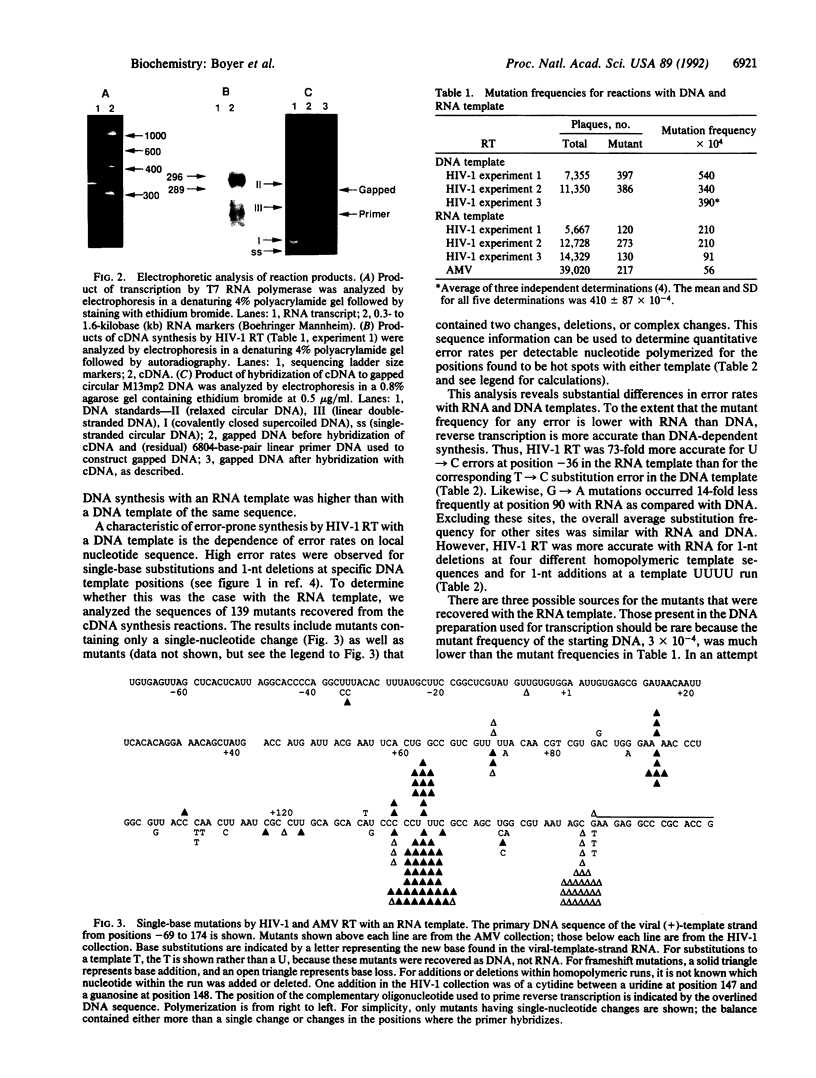
Sequence variation in the type 1 human immunodeficiency virus (HIV-1) results, in part, from inaccurate replication by reverse transcriptase. Although this enzyme is error-prone during synthesis in vitro with DNA templates, the fidelity of RNA-dependent DNA synthesis relevant to minus-strand replication in the virus life cycle has not been examined extensively. In the present study, we have developed a system to determine the fidelity of transcription and reverse transcription and have used it to compare the fidelity of DNA synthesis by the HIV-1 reverse transcriptase with RNA and DNA templates of the same sequence. Overall, fidelity was several-fold higher with RNA than with DNA. Sequence analysis of mutants generated with the two substrates revealed that differences in error rates were substantial for specific errors. Fidelity with RNA was greater than 10-fold higher for substitution and minus-one nucleotide errors at five different homopolymeric positions. Because such errors likely result from template-primer slippage, this result suggests that misaligned intermediates are formed and/or used less frequently with an RNA template-DNA primer than with a DNA template-DNA primer. The results also suggest that HIV-1 reverse transcriptase synthesis with an RNA template-DNA primer was error-prone during incorporation of the first two nucleotides, perhaps due to aberrant enzyme-substrate interactions as synthesis initiates. The unequal error rates with RNA and DNA templates suggest that mistakes during minus- and plus-strand DNA synthesis may not contribute equally to the mutation rate of HIV-1. The data also provide estimates of substitution and frameshift error rates during transcription by T7 RNA polymerase.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bebenek K., Abbotts J., Roberts J. D., Wilson S. H., Kunkel T. A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989 Oct 5;264(28):16948–16956. [PubMed] [Google Scholar]
- Drake J. W. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner A., Kruhoffer M., Grosse F., Krauss G. Fidelity of human immunodeficiency virus type I reverse transcriptase in copying natural RNA. J Mol Biol. 1992 Feb 5;223(3):595–600. doi: 10.1016/0022-2836(92)90975-p. [DOI] [PubMed] [Google Scholar]
- Jacques J. P., Kolakofsky D. Pseudo-templated transcription in prokaryotic and eukaryotic organisms. Genes Dev. 1991 May;5(5):707–713. doi: 10.1101/gad.5.5.707. [DOI] [PubMed] [Google Scholar]
- Ji J. P., Loeb L. A. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry. 1992 Feb 4;31(4):954–958. doi: 10.1021/bi00119a002. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Bebenek K., McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Misalignment-mediated DNA synthesis errors. Biochemistry. 1990 Sep 4;29(35):8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Soni A. Exonucleolytic proofreading enhances the fidelity of DNA synthesis by chick embryo DNA polymerase-gamma. J Biol Chem. 1988 Mar 25;263(9):4450–4459. [PubMed] [Google Scholar]
- Libby R. T., Gallant J. A. The role of RNA polymerase in transcriptional fidelity. Mol Microbiol. 1991 May;5(5):999–1004. doi: 10.1111/j.1365-2958.1991.tb01872.x. [DOI] [PubMed] [Google Scholar]
- Majumdar C., Abbotts J., Broder S., Wilson S. H. Studies on the mechanism of human immunodeficiency virus reverse transcriptase. Steady-state kinetics, processivity, and polynucleotide inhibition. J Biol Chem. 1988 Oct 25;263(30):15657–15665. [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrino F. W., Preston B. D., Sandell L. L., Loeb L. A. Extension of mismatched 3' termini of DNA is a major determinant of the infidelity of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8343–8347. doi: 10.1073/pnas.86.21.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston B. D., Poiesz B. J., Loeb L. A. Fidelity of HIV-1 reverse transcriptase. Science. 1988 Nov 25;242(4882):1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- Reardon J. E., Furfine E. S., Cheng N. Human immunodeficiency virus reverse transcriptase. Effect of primer length on template-primer binding. J Biol Chem. 1991 Jul 25;266(21):14128–14134. [PubMed] [Google Scholar]
- Ricchetti M., Buc H. Reverse transcriptases and genomic variability: the accuracy of DNA replication is enzyme specific and sequence dependent. EMBO J. 1990 May;9(5):1583–1593. doi: 10.1002/j.1460-2075.1990.tb08278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. D., Bebenek K., Kunkel T. A. The accuracy of reverse transcriptase from HIV-1. Science. 1988 Nov 25;242(4882):1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- Saag M. S., Hahn B. H., Gibbons J., Li Y., Parks E. S., Parks W. P., Shaw G. M. Extensive variation of human immunodeficiency virus type-1 in vivo. Nature. 1988 Aug 4;334(6181):440–444. doi: 10.1038/334440a0. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y., Nagumo T., Hoshino H. Low fidelity of cell-free DNA synthesis by reverse transcriptase of human immunodeficiency virus. J Virol. 1988 Oct;62(10):3900–3902. doi: 10.1128/jvi.62.10.3900-3902.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian J. P., Meyerhans A., Asjö B., Wain-Hobson S. Selection, recombination, and G----A hypermutation of human immunodeficiency virus type 1 genomes. J Virol. 1991 Apr;65(4):1779–1788. doi: 10.1128/jvi.65.4.1779-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J., Grosse F. Fidelity of human immunodeficiency virus type I reverse transcriptase in copying natural DNA. Nucleic Acids Res. 1989 Feb 25;17(4):1379–1393. doi: 10.1093/nar/17.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]





