Abstract
BACKGROUND & AIMS
Intestinal homeostasis and regeneration after injury is controlled by 2 different types of cells–slow cycling, injury-resistant reserve intestinal stem cells (ISC) and actively proliferative ISC. Putative reserve ISCs have been identified using a variety of methods, including CreER insertions at Hopx or Bmi1 loci in mice and DNA label retention. Label-retaining cells (LRCs) include dormant stem cells in several tissues; in the intestine, LRCs appear to share some properties with reserve ISCs, which can be marked by reporter alleles. We investigated the relationships between these populations.
METHODS
Studies were performed in Lgr5–EGFP-IRES-creERT2, Bmi1-CreERT2, Hopx-CreERT2, and TRE-H2BGFP::Hopx-CreERT2::lox-stop-lox-tdTomato mice. Intestinal epithelial cell populations were purified; we compared reporter allele-marked reserve ISCs and several LRC populations (marked by H2B-GFP retention) using histologic, flow cytometry and functional and single-cell gene expression assays.
RESULTS
LRCs were dynamic and their cellular composition changed with time. Short-term LRCs had properties of secretory progenitor cells undergoing commitment to the Paneth or enteroendocrine lineages while retaining some stem cell activity. Long-term LRCs lost stem cell activity and were a homogenous population of terminally differentiated Paneth cells. Reserve ISCs marked with HopxCreER were primarily quiescent (in G0), with inactive Wnt signaling and robust stem cell activity. In contrast, most LRCs were in G1 arrest and expressed genes that are regulated by the Wnt pathway or are in the secretory lineage.
Conclusions
LRCs are molecularly and functionally distinct from reporter-marked reserve intestinal stem cells. This information provides an important basis for future studies of relationships among intestinal stem cell populations.
Keywords: quiescence, epithelium, differentiation, crypt
Introduction
Emerging data provides compelling evidence of the existence of reserve stem cells in the intestinal epithelium. Functionally, we define the reserve ISC as an indispensible stem cell capable of giving rise to all intestinal cell types that is injury-resistant and indispensible for tissue maintenance, although this definition is arbitrary and a potential source of contention in the literature. Recently, a burgeoning body of literature using a plethora of proxy markers has identified cells with all of these functional qualities, or subsets thereof. These studies employ primarily two technical approaches: the use of proxy-reporter alleles (either targeted or randomly integrated transgenes) for cell isolation, ablation, and lineage tracing (via CreER), and the use of label retention assays employed under the assumption that reserve ISCs should be slow cycling and thus retain DNA label (although methods for how this label is initially introduced vary widely).
The most extensively studied proxy knockin reporter alleles used to mark reserve ISCs include CreER insertions into the endogenous Hopx and Bmi1 loci1-6. Lineage tracing from these alleles verified the presence of bona fide stem cells within marked populations capable of giving rise to all cell types of the intestinal epithelium, including the active crypt base columnar ISC (CBC)2, 4-6. Further, single cell gene expression analysis of Hopx- and Bmi1-CreER-marked cells revealed these populations to be largely overlapping with one another, but distinct from CBCs marked by Lgr5-eGFP-IRES-CreER 2, 7. These reserve ISCs are also resistant to high doses of gamma irradiation that ablate actively cycling CBCs (12Gy and higher), although recent evidence indicates that a rare population of Lgr5lowWntlow cells above the crypt base but still marked by the Lgr5-eGFP-IRES-CreER reporter are also radioresistant6, 8. Further functional evidence illustrating the critical importance of Bmi1-/Hopx-CreER-marked ISCs within the intestinal stem cell niche come from diphtheria toxin cell ablation experiments driven by Bmi1-CreER that demonstrate that these reserve ISCs are required for tissue fidelity and maintenance of normal crypt-villus architecture, while, in contrast, Lgr5-expressing cells are completely dispensable3, 5. The de novo generation of Lgr5-expressing cells is, however, required for regeneration after radiation injury9. These studies collectively demonstrate the existence of an indispensable, Wnt-negative, radioresistant reserve ISC that gives rise to active, WntHigh CBCs. It is important to point out here that these functional assays were all performed using CreER knockin reporter alleles, and that the populations marked by these alleles are not equivalent to those containing endogenous Bmi1 or Hopx mRNAs, both of which can be found non-specifically throughout the crypt base and thus cannot serve as proxies for specific stem cell identity2, 10, 11. Numerous other proxy alleles have been described that almost certainly mark populations overlapping to various degrees with the Hopx-/Bmi1-CreER-marked reserve ISC population, however the rigorous molecular and functional assays applied to the Hopx-/Bmi1-CreER-marked cells are incomplete for many of these other populations (specifically lineage tracing to verify long-term self-renewal and differentiation capacity, cell ablation to determine requirement for tissue fidelity, and single cell profiling to address heterogeneity) and thus we do not describe this literature here in detail12-16. To date, no non-genetic means exist for the prospective identification and isolation of these reserve ISCs.
Beside the use of genetic proxy reporter alleles for reserve ISC identification, it has been proposed that such a stem cell should cycle slowly, and thus retain DNA label if the label is introduced during a period prior to the induction of dormancy (i.e., in the newborn or after injury). The presence label retaining cells (LRCs) was first proposed by Potten et al. to be evidence of the immortal strand hypothesis, although support for this idea lacks consensus in the face of conflicting data17-21. Nonetheless, DNA labeling in young mice or in mature animals after injury revealed the presence of long-term LRCs in the intestine (up to one month chase) that consisted of Paneth cells (long-lived cells intercalated between CBCs) as well as non-Paneth cells residing around the +4 to +15 position from the crypt base22. These non-Paneth LRCs were posited to be ISCs, but no functional evidence for this existed at the time. Over a decade later, these findings were reproduced using inducible H2B fusion proteins rather than the DNA radiolabeling used in the original studies13, 23, 24. Interestingly, these studies incorporated label not in newly born mice or after injury, but in mature mice under basal conditions, suggesting that the LRCs may not be dormant stem cells upstream of CBCs, but rather a downstream cell that incorporates label while undergoing cell cycle exit and differentiation (i.e., the label should not incorporate into dormant stem cells in the absence of DNA replication, either induced post-injury, or during development).
The use of H2B-eGFP fusion proteins to identify these LRCs enabled for the first time their prospective isolation and molecular profiling. These studies, performed on shorter-term LRCs (usually around 8-12 days), revealed that the LRC population expresses direct target genes of the canonical Wnt/β-catenin pathway such as Sox9 and Lgr5 and markers of the secretory lineages13, 23, 24. Single cell profiling of 10 day LRCs, however, found them to be a highly heterogeneous population24. Remarkably, the use of an H2B-split-Cre reporter allele that enables lineage tracing from LRCs revealed stem cell activity from at least some cells contained within this population24. Further, these 10-day LRCs could give rise to clonal lineage tracing events after exposure to mid-dose gamma irradiation (6Gy), although the frequency of these events was vanishingly small, with fewer than 10 tracing events observed along the entire length of the intestine24. Taken together, these studies suggested that non-Paneth LRCs are a secretory progenitor cell population that can serve as a reserve intestinal stem cell. These observations, coupled with their location above the crypt base and slow cycling nature, has lead researchers to posit that the short term LRCs and reserve ISCs marked by the Bmi1- and Hopx-CreER proxy alleles are one in the same1, although no cell ablation evidence exists demonstrating a functional importance for LRCs as it does for the proxy allele-marked reserve ISCs.
In order to understand the relationship between intestinal LRCs and proxy-reporter allele-marked ISCs, the current study undertakes a comprehensive and direct comparison of single cells within these two populations, including both short- and long-term LRCs (10 days, 1 month, 3 months), and reserve ISCs marked by Hopx-/Bmi1-CreER.
Methods
Mouse Strains
Lgr5–EGFP-IRES-creERT2 (JAX strain 008875) and Bmi1-CreERT2 (JAX strain, 010531) mice were obtained from the Jackson Laboratory. Hopx-CreERT2 (JAX strain 017606) mice were a kind gift from Dr. Jon Epstein. TRE-H2BGFP mice were obtained from Jackson Laboratory (JAX strain 016836). Mice were maintained on a C57/BL6N background. Mice (including the TRE-H2BGFP) received a single intraperitoneal (i.p.) injection of 100μL Tamoxifen (Tam) (Sigma T5648, 10mg/ml in corn oil). All mouse protocols were approved by the IACUC at the University of Pennsylvania under protocol 803415 to Dr. Lengner.
H2B-GFP labeling
TRE-H2BGFP::Hopx-CreERT2::lox-stop-lox-tdTomato mice were maintained on Dox (Sigma D9891, 1mg/ml in 1% sucrose) for six weeks starting at postnatal day 14 in order to fully label nuclei with GFP. Dox was withdrawn when mice reached 8 weeks of age and mice were sacrificed 10 days, 1month, or 3 months after Dox withdrawal and initiation of tracing. Hopx-CreERT2::lox-stop-loxtdTomato activity was initiated with one dose of Tam 18 hours before sacrifice.
EdU Labeling, RNA Content Staining, Flow Cytometry, Single Cell FACS
The intestine was cut open longitudinally and incubated with 5mM EDTA-HBSS solution at 4 °c for 30min to isolate epithelial cells. To generate a single cell suspension, cells were incubated with Accutase (BD Biosciences, San Jose, CA) at 37°c for 10min. Flow cytometry analysis was performed with BD LSRFortessa cell analyzer (BD Biosciences, San Jose, CA). DAPI negative cells were selected, then gated for single cell based on Forward-scatter height versus forward-scatter width (FSC-H vs FSC-W) and side-scatter height vs side-scatter width (SSC-H vs. SSC-W) profiles. Single-cell sorting experiments was performed with BD FACSAriaII cell sorter, each single cell was sorted into a different well of a 96-well PCR plate, using the FACSAriaII flow cytometer software package (FACSDiva) with single cell precision mode. Paneth cell isolation was done based on CD24 (eBioscience, 12-0242081)) and c-Kit (eBioscience, 25-1171-81) double staining. The size of the nozzle for all sorting is 100 μm (20 psi).
For analysis of S-phase, mice were injected i.p. with 300 μg/ 10 g body weight of EdU dissolved in PBS and the first 2-12cm of the jejunum were harvested 2 hours after injection. For flow cytometric analysis, single cell suspensions were prepared as described above. EdU staining was performed according to the manufacturer's instructions (Life technologies, C10634). In the context of radiation injury, mice were subjected to 12Gy of whole-body gamma-irradiation, then, after 54 hours, were given a single dose of Tam. EdU was administered 70hours post-injury, and mice were sacrificed and analyzed at 72 hours post-injury. It is important to point out here that the 72-hour post-injury timepoint was utilized to evaluate EdU incorporation during S-phase because analysis of EdU incorporation at earlier timepoints would be confounded by our inability to distinguish S-phase from the DNA repair response, as both of these processes result in EdU incorporation.
For quiescence (G0) analysis Tomato+ and GFP+ cells were FACS-sorted into 100% ethanol and fixed in 4 °C overnight. The cells were then staine d with Hoechst 33342 (Thermo Fisher Scientific, H3570, 10 μg/ ml at final) for 30 min at RT, followed by Pyronin Y (Sigma, P9172, 1 μg/ml at final) staining for 15min at RT before flow cytometric analysis. All samples were recorded in 1 hour.
Single Cell RT-PCR
The two-step single-cell gene expression protocol (advanced development protocol 33) from Fluidigm was adopted for this study. Briefly, 5 μL of RT Mix Solution which includes 1.2 μL 5X VILO™ Reaction Mix (Life Technologies, 11754-250), 0.3 μL SUPERase-In (Life Technologies, AM2696) and 0.25 μL 10% NP40 (Thermo Scientific, 28324) was dispensed into each well of 96-well-plate. Single cells were sorted into the well directly. The plate was vortexed and immediately frozen on dry ice. For RT cycling, the plate was thawed on ice and RNA denatured by incubating at 65 °C for 90 sec, then chilled on ice for 5 min. Each well was supplemented with 1 μl mixture of 10X SuperScript® Enzyme Mix (Life Technologies, 11754-250) and T4 Gene 32 Protein (New England BioLabs, PN M0300S). mRNA was reverse transcribed into cDNA following the thermal cycling conditions below: 25 °C, 5 min/ 50 °C, 30 min/ 55 °C, 25 min/60 °C, 5 min/ 70 °C, 10 min. Resulting cDNA was pre-amplified with 50 nM primer mix for 23 PCR cycles (96 °C for 5 sec, 60 °C for 4 min), then treated with ExoI for 30 min to remove unincorporated primers. The final product was diluted 1:3 with TE buffer. For each chip sample inlet, 2.25 μl diluted cDNA, 2.5 μl 2X Sso Fast EvaGreen supermix with low ROX and 0.25 μl of Fluidigm sample loading agent were added. Individual gene-specific DELTAgene assays were diluted at 1:10 ratios with TE buffer. 2.5 μl of each primer was then mixed with 2.5 μl assay loading agent inserted into chip “assay” inlets. Chip loading and PCR was performed according to the manufacturer's protocol. The data were analyzed by Fluidigm Gene Expression Analysis Package.
Genes analyzed include: Areg, Ascl2, Atoh1, Axin2, Bmi1, Bmpr1a, Ccnd1, Cdkn1a, Cdx1, Chga, Cubn, Dll4, Dvl2, Efnb1, Epas1, Ephb2, Ereg, Fut2, Gapdh, Gsk3b, Gusb, H6pd, Hes1, Hes5, Hif1a, Hopx, Jag1, Lgr5, Lrig1, Lyz2, Msi1, Msi2, Myb, Myc, Notch1, Numb, Olfm4, Pcna, Ppargc1b, Rhoa, Saa2, Sirt3, Sox9, Tat, Tcf4, Tert, Wnt3, and Wnt6.
Hopx-CreER lineage tracing after irradiation
To determine the contribution of Hopx-CreER ISCs to regeneration after irradiation, Hopx-CreER::Lox-Stop-Lox-tdTomato mice were given 3 daily doses of Tamoxifen, followed by 72 hours of chase to allow residual Tam to leave the system and avoid de novo labeling of any newly regenerated cells post-IR (as the half-life of Tam is 12-18 hours). Mice were then subjected to 12Gy of whole-body gamma-irradiation, followed by 4 days of post-injury recovery prior to analysis of Hopx-CreER progeny in the regenerated epithelium of the jejunum by staining for tdTomato+ cells.
Results
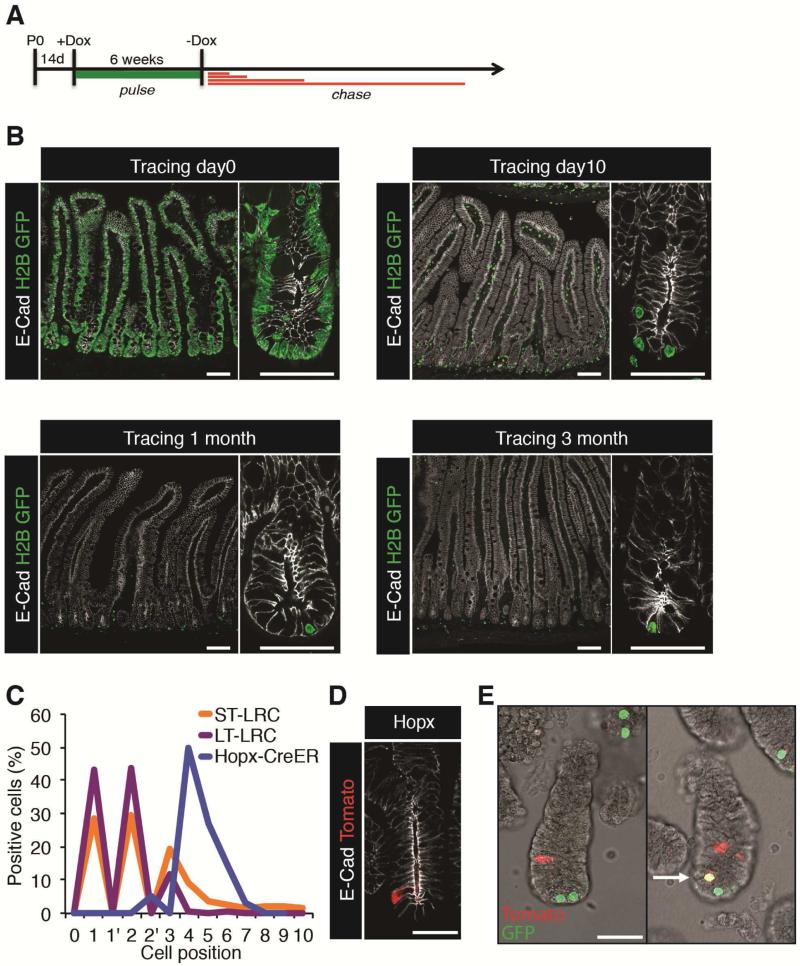
We adopted a genetic system to mark LRCs: a doxycycline (Dox)-inducible histone H2B-GFP mouse model in which a single copy of the Dox-inducible TRE-H2B-GFP cassette is targeted to safe-haven chromatin downstream of the Col1a1 locus and a reverse modified tetracycline transactivator (M2rtTA) is targeted to the ROSA26 locus25. H2B-GFP was induced with Dox in juvenile mice during a 6-week pulse period after which fluorescence loss was monitored in response to Dox withdrawal for up to 3 months (Figure 1A). Epithelial cells were uniformly labeled after the pulse period, and most cells had lost H2B-GFP within 10 days (Figure 1B). After one or three months chase, LRCs appeared less frequently (Figure 1B and Supplementary Figure 1A). One- and three-month LRCs (referred to collectively as long-term, or LT-LRCs) were primarily observed at the crypt base, while 10 day LRCs (referred to as short-term, or ST-LRCs) could routinely be found above the crypt base (Figure 1B). Quantification of positional information confirmed that the majority of LRCs (long- and short-term) reside at the crypt base intercalated between CBCs, however a portion of the ST-LRC population was located at the +4 through +6 position from the crypt base, and this population was lost in the LT-LRCs (Figure 1C). These findings are largely consistent with prior studies13, 23, 24. We were unable to identify epithelial LRCs in the colon (Supplemental Figure 1B).
Figure 1. Location of label-retaining cells and reserve ISC in small intestinal crypts.
A. Schematic of H2B-GFP labeling in TRE-H2BGFP mice. B. Micrographs of H2B-GFP+ LRCs 0 days to 3 months after Dox withdrawal. Scale=50μM and 10 μM. C. Quantification of position for indicated populations (1’ and 2’ represent CBC location) D. ISCs marked by Hopx-CreER::R26-Lox-Stop-LoxtdTomato 18 hours after a single Tamoxifen dose. Scale=10μM. E. Whole mount direct fluorescence showing crypts containing both ST-LRCs and Hopx-CreER ISCs, and rare double positive cells (arrow).
We compared LRCs with reserve ISCs marked by Hopx-CreER::R26Lox-Stop-Lox-tdTomato (referred to as Hopx-CreER) 18 hours after a single tamoxifen (Tam) injection. This strategy marks a reserve ISC population largely overlapping, but more homogenous than that marked by Bmi1-CreER2, 4. As expected, Hopx-CreER marked rare single cells around the +4 to +7 position from the crypt base (Figure 1C, D and Supplementary Figure 1A). The apparent overlap at the +4 to +6 positions from the crypt base between Hopx-CreER ISCs and LRCs suggests that these may be overlapping cell types1. To address this directly, we intercrossed TRE-H2B-GFP and Hopx-CreER::R26Lox-Stop-Lox-tdTomato mice and visualized Tomato and GFP by direct fluorescence in whole mount crypt preparations. While most GFP- and Tomato-positive cells appeared mutually exclusive, we were able to detect very rare double-positive cells (Figure 1E).
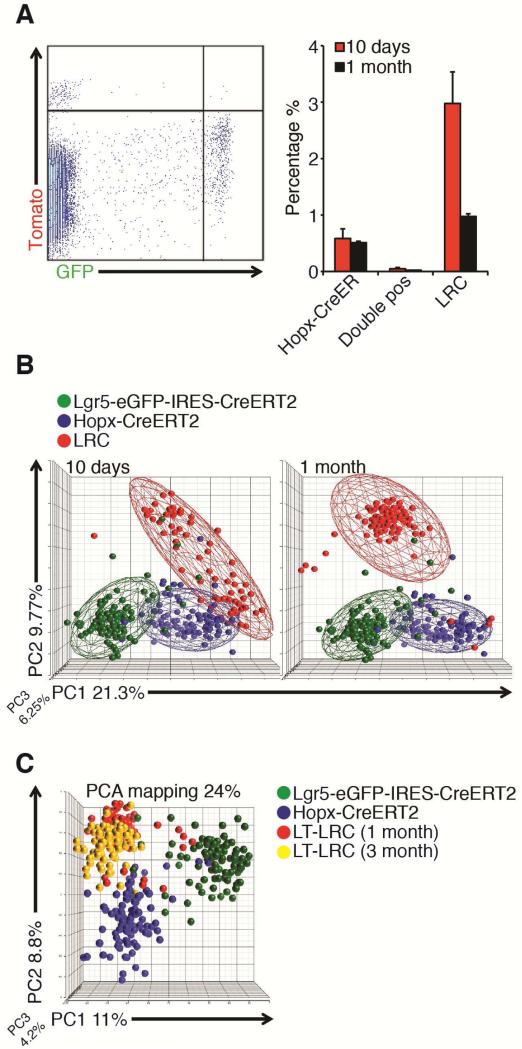
To quantitatively compare ISCs with LRCs, we prepared crypt epithelial preparations from Lgr5-eGFP-IRES-CreER, Hopx-CreER, and TRE-H2BGFP mice for flow cytometric analysis (Supplementary Figure 2A-D). We initially quantified the degree of overlap between LRCs and Hopx-CreER ISCs that we observed histologically in Figure 1E. Despite the localization of these cells in similar positions above the crypt base, flow cytometry revealed these to be nearly totally mutual exclusive populations (1.9% of ST-LRCs and 1.2% of LT-LRCs were also Tomato+, Figure 2A), demonstrating that these are primarily distinct epithelial populations. This analysis also revealed an approximately 60% decrease in the frequency of LRCs between 10 days and one month of chase, indicating that most of this population loses label due either to cell division or epithelial turnover during this period.
Figure 2. Reserve ISCs are distinct from label-retaining cells.
A. Flow cytometry of ST-LRCs or LT-LRCs versus Hopx-CreER ISCs in TRE-H2BGFP::Hopx-CreER::LSL-tdTomato compound mice, quantified at right (5×104 cells recorded). B. PCA (37.3% of variation) of ST-LRCs or LT-LRCs, Hopx-CreER ISCs, and CBCs. Cages represent 1.5 standard deviations from the mean for that group. C. PCA mapping of 1 month versus 3 month LT-LRCs versus CBCs and Hopx-CreER ISCs.
To further examine the molecular identities of these populations, we employed single-cell gene expression profiling for genes representing targets and components of the Wnt and Notch pathways, ISC signature genes, proliferation/metabolism-related genes, and genes encoding markers of terminally differentiated intestinal cells, using two unique primer pairs for each gene2 (Supplementary Table 1). Because the only means available to mark reserve ISCs is through Tam-mediated CreER reporter activation, we validated that Tam did not induce any detectable apoptosis in FACS-purified populations (Supplementary Figure 2E). We also validated that the specific target preamplification protocol did not introduce non-linear amplification of target transcripts in single cells (Supplemental Figure 3A, B). Further, to verify that the 8-hour post-Tam period required for activation and detection of the R26Lox-Stop-Lox-tdTomato reporter allele did not allow for changes in cellular identity, we examined cells sorted using Lgr5-eGFP (time=0) versus Lgr5-CreER::R26Lox-Stop-Lox-tdTomato (time=18 hours post Tam) and found no changes to their molecular identity (Supplementary Figure 3C). An analogous experiment with the Hopx-CreER-marked population cannot be performed due to the requirement for Tam induction of CreER to activate the reporter. However, cells with reserve stem cell identity have been observed using reporters that do not rely on Tam, including a subpopulation of Hopx-eGFP cells and cells marked by an mTert-GFP transgene2, 14.
Single cell expression profiling was applied to short- and long-term LRCs and the resulting molecular identities were compare to those established for Lgr5-eGFP+ CBCs and Hopx-CreER reserve ISCs using principal component analysis (PCA)2, 4. This revealed that ST-LRCs are a heterogeneous population, largely distinct from both CBCs or reserve ISCs, and that over time, the identity of LRCs becomes increasingly homogenous (Figure 2B). Between one and three months, the molecular identity of LT-LRCs remained unchanged, indicating that this population stabilizes sometime between the 10-day chase period used to define ST-LRCs and one month (Figure 2C). Taken together, these findings support the histological and flow cytometric data, demonstrating that LRCs, CBCs, and reserve ISCs represent molecularly distinct populations.
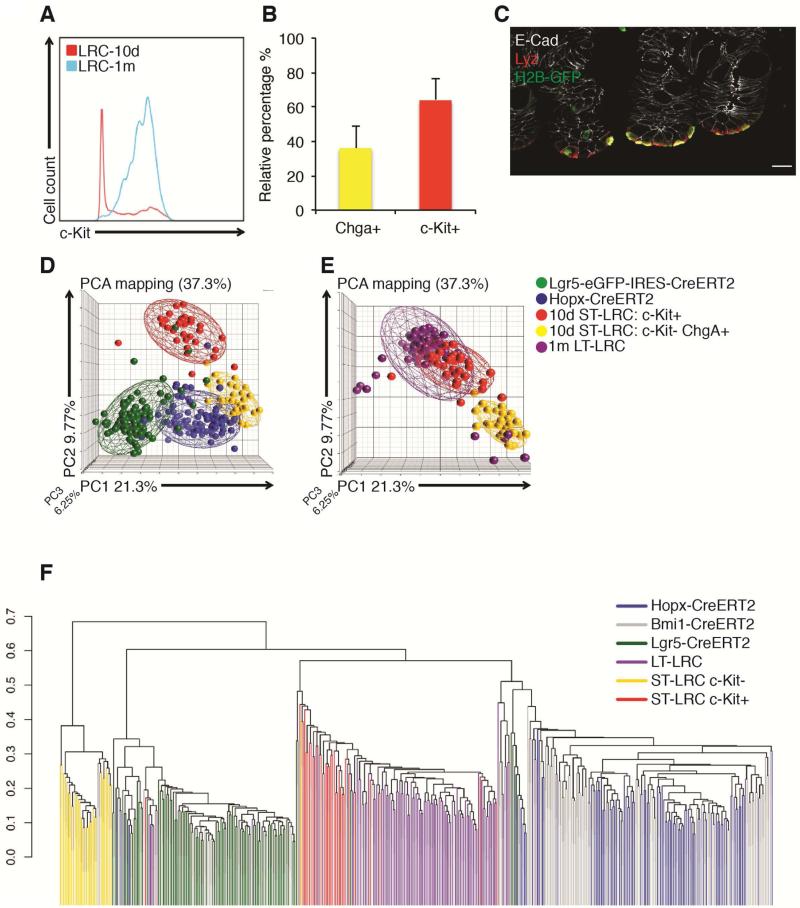
Paneth cells are long-lived secretory cells at the crypt base that have long been observed within LRC populations13, 23, 24. Given that LT-LRCs become increasingly localized to the crypt base relative to ST-LRCs, we examined how Paneth cells, marked by a c-Kit antibody previously shown to be Paneth cell-specific26 overlapped with LRCs. Consistent with their position within the crypt, ST-LRCs consisted of both a c-Kit+ Paneth and c-Kit− non-Paneth population, whereas LT-LRCs became almost uniformly c-Kit+, indicating that LT-LRCs are a homogenous population of Paneth cells (Figure 3A and Supplementary Figure 4A). Because ST-LRCs are known to express other secretory lineage-specific genes, we examined the expression of the enteroendocrine marker Chromogranin A (Chga) within this population. Interestingly, Chga+ and c-Kit+ ST-LRCs were mutually exclusive subpopulations, with the c-Kit+ cells making up the majority of the ST-LRC population and localizing to the crypt base (Figure 3B, C). To determine how the c-Kit+ vs. Chga+ cells within the heterogeneous ST-LRC population are distributed in the PCA analysis shown in Figure 2B, we color-coded ST-LRCs based on c-Kit versus Chga expression. This revealed a distinct separation of these two populations within the ST-LRC group (Figure 3D). As expected, the c-Kit+ population overlapped with the LT-LRC population, further confirming their Paneth cell identity (Figure 3E).
Figure 3. LRCs are a dynamic population molecularly distinct from reserve ISCs and CBCs.
A. Flow cytometric data assessing distribution of c-Kit immunoreactivity in ST- and LT-LRCs (2×105 cells recorded). B. Quantification of the relative abundance of Chga+ versus c-Kit+ ST-LRCs from flow cytometry. C. Costaining for ST-LRCs and the Paneth cell marker Lysozyme. D. PCA mapping of STLRCs, color-coded by ChgA expression versus c-Kit immunoreactivity, relative to Hopx-CreER and Lgr5High ISCs. E. PCA mapping of ST-LRCs and LT-LRCs. F. Hierarchical clustering of individual cells within indicated proxy reporter-marked populations.
Ultimately, we performed hierarchical clustering of single cells from numerous ISC and LRC populations based on their expression profiles, including Lgr5-eGFP+ CBCs, Hopx-CreER-marked ISCs, Bmi1-CreER-marked ISCs, one month LT-LRCs, and 10 day ST-LRCs (color-coded based on c-Kit+ and c-Kit−, Chga+ identity) (Figure 3F). This analysis confirmed that Hopx- and Bmi-CreER-marked ISC populations are largely overlapping with one another2, and that they are molecularly distinct from all other populations. Next, it confirms the overlap in identity between c-Kit+ ST-LRCs and LT-LRCs (Paneth cells). Finally, it confirms the presence of a unique c-Kit− ST-LRC (Chga+) population with distinct molecular identity.
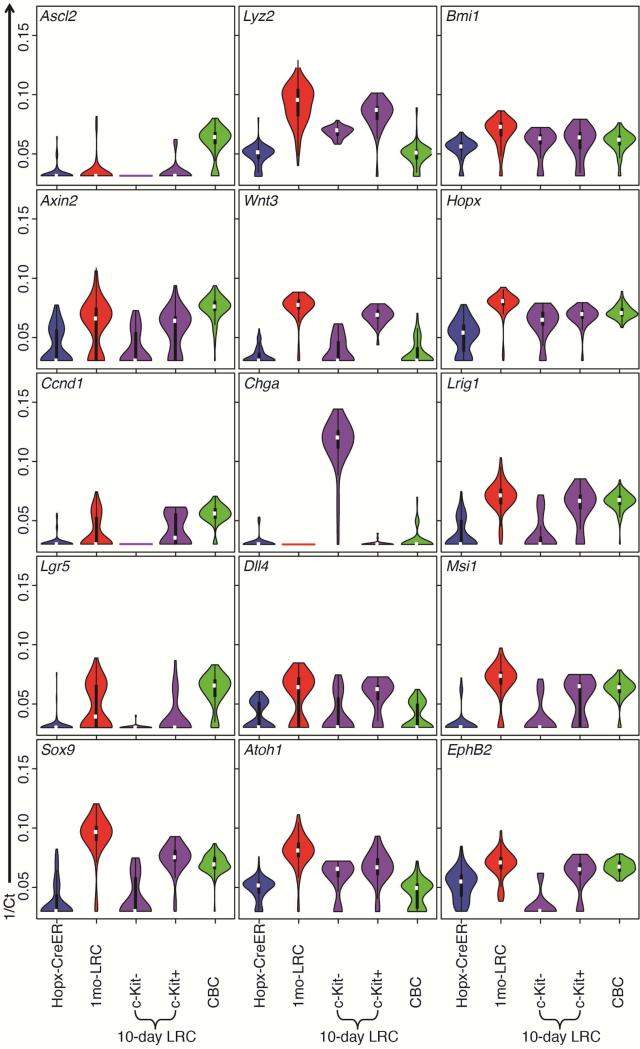
Examination of the expression of individual genes further revealed differences between these populations. As anticipated, LT-LRCs and c-Kit+ ST-LRCs exhibited an expression pattern characteristic of Paneth cells, including very high expression of Lysozyme (Lyz2), Wnt3, the Wnt/β-catenin target genes Axin2, EphB2, and Sox9, as well as the master secretory lineage transcriptional regulator Atoh1 (Math1) (Figure 4). Interestingly, these cells also expressed high levels of Lrig1, Msi1, Bmi1, and Hopx transcripts. These have all been referred to as intestinal stem cell marker genes3, 4, 12, 27, however the presence of these transcripts in Paneth cells (as well as CBCs and reserve ISCs in the case of Bmi1 and Hopx), demonstrates that their distribution is not restricted to stem cells and thus is not a viable proxy for any specific stem cell identity. C-Kit− ST-LRCs uniquely expressed Chga, as well as Atoh1. As expected, proliferation and CBC-associated Wnt/β-catenin target genes (Lgr5, Ascl2, Ccnd1) were selectively expressed in the Lgr5-eGFP+ population. Of note, the position-determining receptor gene EphB228 was expressed at levels well-correlated with the position of the marked populations in the crypt, with highest levels in cells at the crypt base (CBCs, LT-LRCs, and c-Kit+ ST-LRCs), decreasing in the Hopx-CreER ISCs above the crypt base, and decreasing even further in the c-Kit−, Chga+ ST-LRCs. The low EphB2 expression in Chga+ ST-LRCs may be a reflection of the upward movement of enteroendocrine cells along the crypt-villus axis and their ultimate loss prior to the 1-month label retention chase timepoint. Hopx-CreER ISCs are characterized mainly by their lack of expression of most of these Notch, Wnt, and proliferation-related genes. Indeed, the Cdkn1a transcript encoding the cell cycle inhibitor p21 is one of the few transcripts highly expressed in this population relative to CBCs2 (Supplementary Figure 4B).
Figure 4. Gene expression patterns in proxy reporter-marked populations.
Violin plots showing transcript levels of indicated genes in single reserve ISCs, LT-LRCs, ST-LRCs (separated into cKit+ and cKit−), and CBCs. The width of the violin is directly proportional to cell number. The white dot represents mean expression level for the population, the box represents the range of the middle 50% of the data, the whiskers represent the outer 50% of the data (each whisker represents the top and bottom quartile). Outliers fall outside of the whisker.
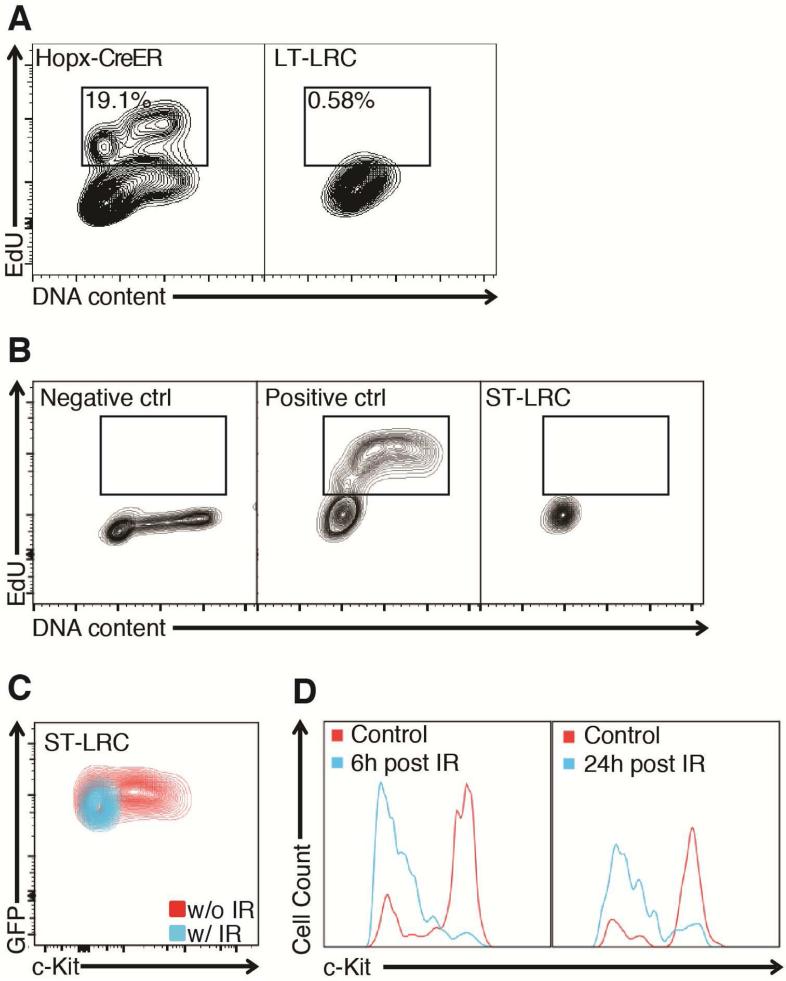
We next tested the ability of Hopx-CreER ISCs and LRCs to survive and enter the cell cycle in response to high-dose gamma irradiation (12 Gy IR). As expected, Hopx-CreER ISCs survived and incorporated EdU while LT-LRCs did not (Figure 5A). Some LT-LRCs survived radiation injury and persisted as single cells in the epithelium (Supplementary Figure 5A). We also performed EdU incorporation analysis on the ST-LRCs post-IR and found no evidence for entry of ST-LRCs into S-phase (Figure 5B). This is in contrast to the Hopx-CreER ISCs, which are not only capable of S-phase entry after high-dose radiation exposure, but broadly contribute progeny to the newly regenerated epithelium (Supplementary Figure 5B). Because this latter experiment is performed using multiple Tamoxifen doses and a washout period to minimize post-irradiation recombination induced by residual Tamoxifen in the bloodstream, we cannot discern what fraction of the radioresistant cells contributing to regeneration are the original Hopx-CreER ISCs versus their downstream progeny. Further, while we find that Hopx-CreER ISCs (or their progeny) contribute to all regenerated villi, we also observe significant contribution of unlabeled cells. Thus, either the Hopx-CreER allele marks only a subset of the quiescent ISC population, or another, distinct population is also contributing to regeneration. The latter notion is supported by recent data indicating that a radioresistant Lgr5Low population can revert to take on stem cell identity upon exposure to radiation 8, 29.
Figure 5. LRCs and Hopx-CreER ISCs exhibit differential response to radiation injury.
A. Analysis of S-phase entry for LT-LRCs and Hopx-CreER ISCs 72hrs after exposure to 12Gy whole body γ-radiation. Hopx-CreER cells are marked with a single Tam injection 18hrs prior to harvest, and Edu is administered 2hrs prior to harvest (2.5×105 cells recorded, 2% contour plot). B. Analysis of S-phase entry for ST-LRCs 72hrs post-12Gy IR. Positive control is total epithelium under basal conditions. C. Analysis of c-Kit immunoreactivity in ST-LRCs 72hrs post-12Gy IR. A-C: 2.5×105 cells recorded, 2% contour plot. D. Flow cytometric data assessing distribution of c-Kit immunoreactivity in ST-LRCs 6h and 24h post 12 Gy IR (2.5×105 cells recorded).
Interestingly, exposure of ST-LRCs to 12 Gy IR resulted in a selective loss of the c-Kit+ population (Figure 5C). To determine the fate of the c-Kit+ ST-LRCs, we examined their presence at 6 and 24 hours post-IR. This revealed that the c-Kit+ ST-LRCs were lost as soon as 6 hrs post-IR, suggesting that these cells were not lost due to H2B-GFP dilution associated with proliferation, as the earliest proliferative events post-12Gy IR do not occur until after 24hrs6 (Figure 5D). This rapid loss of c-Kit+ ST-LRCs suggested radiation-induced apoptosis. We thus quantified apoptotic events in the ST-LRC population and observed nearly half of the c-Kit+ ST-LRC population is positive for cleaved caspase-3 within 6hrs of IR, consistent with c-Kit+ ST-LRC population being lost to apoptosis (Supplementary Figure 5C).
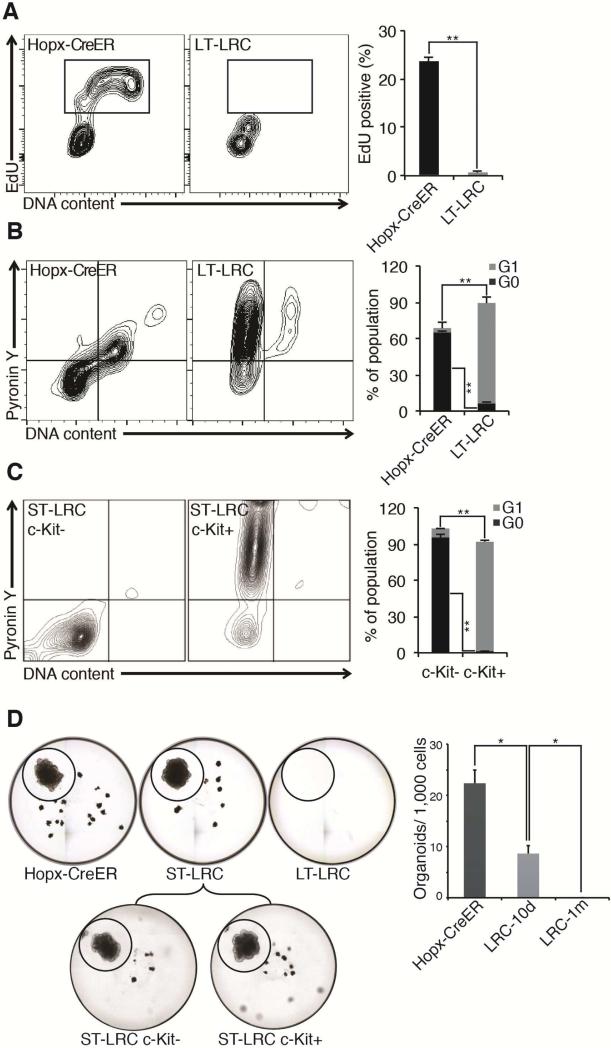
Finally, we compared the functional properties of Hopx-CreER ISCs and LRCs under basal conditions. LRCs are frequently referred to as being quiescent, although no published evidence for their residence in the G0 state of the cell cycle exists. We initially assessed EdU incorporation and found no LT-LRCs in S-phase, in contrast to the Hopx-CreER population in which more than 20% of cells incorporated EdU (consistent with their cycling at about half the rate of CBCs2) (Figure 6A and Supplementary 6A). We next assessed quiescence of these populations by measuring the fraction of cells in the G0 state outside of the cell cycle30-33. We find that the majority of Hopx-CreER ISCs reside in G0. This finding, coupled with the above observation that approximately one quarter of this population is in S-phase at any given time, is consistent with a model in which these cells periodically enter the cell cycle, divide, and return to the quiescent G0 state (Figure 6B). In contrast, the vast majority of LT-LRCs were arrested in G1, consistent with their identity as terminally differentiated Paneth cells with an active genome engaged in synthesis and secretion of anti-microbial factors and niche ligands for CBCs (Figure 6B). Analysis of quiescence in the ST-LRC population revealed that, similar to LT-LRCs, the cKit+ fraction was primarily in G1, however the cKit- fraction was entirely quiescent, consistent with its ability to survive radiation injury relative to the cKit+ fraction (Figure 6C). Analysis of the Lgr5-eGFP+ CBC population revealed, as expected, that the vast majority of these cell are not quiescent and reside in the cell cycle (Supplementary Figure 6B).
Figure 6. Hopx-CreER ISCs are functionally distinct from LRCs.
A. EdU incorporation in indicated populations under basal conditions, quantified at right. (**p<0.001). B. G0 analysis with Hoechst/PyroninY under basal conditions, quantified at right. (G1:**p<0.001, G0:**p<0.001). C. G0 analysis as in B in c-Kit− and c-Kit+ ST-LRCs under basal conditions. **p<0.001. A-C: 2.5×105 cells recorded, 2% contour plot. D. Ex vivo organoid-forming capacity of single flow-sorted Hopx-Tomato ISCs, ST-LRCs (split into c-Kit+ versus c-Kit−), and LT-LRCs, quantified at right. (*p<0.01).
We next tested stem cell potential in these populations using organoid formation assays. As expected2 single Hopx-CreER ISCs robustly formed organoids in the presence of BMP inhibitors and Wnt/Notch agonists (Figure 6D). Single LT-LRCs had no stem cell activity, consistent with these cells being terminally differentiated Paneth cells. In contrast, ST-LRCs gave rise to organoids with less than half the efficiency of Hopx-CreER ISCs. When we analyzed stem cell activity in the c-Kit−, Chga+, quiescent ST-LRCs in comparison to the c-Kit+ ST-LRCs, we found these populations to have roughly equivalent stem cell activity, possibly indicating that, despite their high expression of markers of terminal secretory differentiation, ST-LRCs retain stem cell potential that is lost upon further differentiation and establishment of the LT-LRC state.
Discussion
Since the first identification of genetic markers of ISC populations nearly a decade ago (Lgr5-eGFP-IRES-CreER and Bmi1-CreER 3, 7) there have been a torrent of publications describing an ever-expanding list of proxy reporter alleles reported to mark ISCs with various functional properties3, 4, 6, 12-16, 24, 26, 34, 35. This has lead to conflicting reports regarding the identity of specific ISC populations, with confusion likely attributable to the unknown heterogeity of cells within these populations and the erroneous assumption that functional ISCs validated by employing inducible CreER alleles for lineage tracing and cell ablation can be similarly identified by GFP reporters or the corresponding endogenous mRNAs. For example, studies have used the presence of Bmi1, Hopx, or Lrig1 mRNA as evidence for stem cell activity, which was initially functionally established using CreER knockin reporters at these loci, however single cell analyses from several independent groups, including this current study, have revealed that the presence of these mRNAs does not correlate with CreER activity, thus invalidating the use of endogenous mRNA as a proxy for stem cell activity2, 10, 11. Why these CreER knockin reporters mark rare populations with stem cell activity while the endogenous mRNA emanating from these loci can be found non-specifically throughout the crypt is a point of speculation, however there is precedent for stem cells transcribing genes whose mRNA is then stored for inheritance into daughter cells36, and thus it is tempting to speculate that the CreER knockin alleles report on cells where the gene of interest is actively transcribed, while the endogenous mRNAs are distributed throughout daughter cells (due possibly to the endogenous mRNAs containing large 3’ untranslated regions with posttranscriptional regulatory activity which are disrupted upon insertion of CreER cassettes). It is thus important to keep in mind when evaluating the literature that functional assays demonstrating stemness in these ISC populations almost always rely on CreER alleles- no functional data exists examining stem cell activity in populations prospectively identified or purified based on the presence of any particular mRNA.
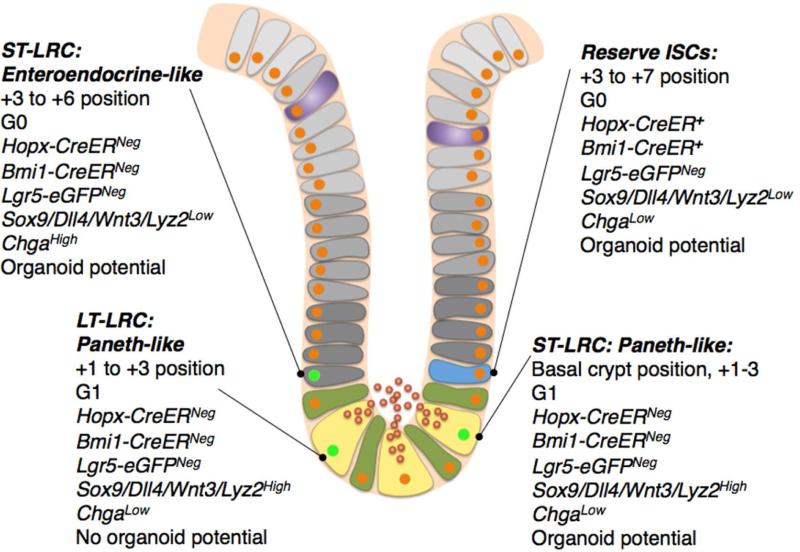
In the current we directly compare two epithelial populations that have often been referred to interchangeably in the literature, the so-called +4 label retaining cell and the +4 Hopx-/Bmi1-CreER marked reserve ISC (Figure 7). We focus on these populations due to published data indicating that both of these populations contain cells with stem cell activity, including in vitro organoid-forming capabilities, radioresistance, and lineage tracing, as well as the finding that both of these populations reside in a similar position within the crypt. We have previously demonstrated that the populations marked by Bmi1-CreER and Hopx-CreER are largely overlapping2, and to date this is the only population that has been demonstrated to be indispensable for intestinal homeostasis using cell ablation techniques3. Lgr5+ CBCs are dispensable for tissue maintenance5, and the necessity of LRCs has not been tested. Here we analyze LRCs at several chase timepoints after administration of a uniform pulse of H2B-GFP label. These findings reveal, perhaps not unexpectedly, that LRCs are a dynamic population. Our findings with long-term LRCs (defined here as one or three months chase) indicate that this is a stable, homogenous population of terminally differentiated Paneth cells lacking stem cell activity. This finding is largely in agreement with the data presented in the studies of Potten et al. and subsequent studies of LRCs using longer chase periods22, 23. We find that these long-term LRCs are molecularly and functionally distinct from the Hopx-/Bmi1-CreER marked reserve ISCs, and thus conclude that the terminology LRC and reserve ISC cannot be used interchangeably (Figure 7).
Figure 7.
Integrated model for understanding stem cell and label retaining cell identity within intestinal crypts.
Less unambiguous is the LRC population marked by shorter chase periods (usually around 10 days). Both our current study and prior studies found this population to contain a majority of cells at the crypt base with expression of the Paneth cell markers13, 22-24, as well as a fraction of cells exhibiting expression of secretory lineage genes13, 23, 24. Two recent studies posited that this ‘non-Paneth’ short-term LRC population had reserve stem cell activity in response to radiation injury in vivo. The first study found non-Paneth ST-LRCs exhibited high expression of the canonical Wnt target gene Sox913, then using a Sox9-CreER allele found that cells recombining a Lox-Stop-Lox reporter were broadly present after high dose radiation. Sox9 and the Sox9-CreER allele are, however, broadly expressed at the crypt base marking a variety of cell types (shown in the current study and 13, 37, 38). The heterogeneity within the Sox9-marked population, coupled with the proximity of Tamoxifen treatment to the time of irradiation (24 hours) make it difficult to interpret the relationship between Sox9-CreER-expressing cells, ST-LRCs, and radioresistant reserve ISC, as one cannot rule out that Tam-induced recombination in these experiments is occurring in de novo generated Wnt-active stem cells after radiation injury (the in vivo half life of Tamoxifen being about 12-18hrs39, leaving ample drug in the system to induce recombination post-injury).
The second study providing evidence of reserve ISC activity in the ST-LRC population performed single cell profiling and found this population to be highly heterogeneous. At the bulk population level, these ST-LRCs express secretory lineage markers such as Chga at higher levels than Lgr5High CBCs24. Using an elegant H2B-split-Cre fusion protein, they went on to show that some cells within the ST-LRC population had reserve ISC activity in response to mid-dose (6Gy) gamma irradiation. This activity was extremely limited, however, with fewer than 10 clonal tracing events occurring along the entire length of the intestinal tract24. Thus, given the demonstrated heterogeneity of the ST-LRC population and the rare frequency of reserve ISC activity, coupled with the lack of cell ablation studies, it is impossible to determine which cells, if any, with the ST-LRC population are truly radioresistant reserve ISCs. Based on our current findings, around 1% of LRCs are also marked by Hopx-CreER, leading us to speculate that the very rare radioresistant ISC activity observed in the aforementioned study comes from this population. In the current study we find no evidence of cell cycle entry in short- or long-term LRC populations in response to high dose (12Gy) gamma irradiation, in contrast to Hopx-CreER ISCs that robustly contribute to post-IR regeneration.
Interestingly, the both the latter study and our study found in vitro organoid-forming stem cell activity within the ST-LRC population. Remarkably, we found that this activity was not limited to the non-Paneth ST-LRC population, but rather was equally distributed between ST-LRC with Paneth or enteroendocrine lineage gene expression. This finding, coupled with the observation that these LRCs take up label in adult animals in the absence of injury, lead us to a model in which the ST-LRC represents a cell undergoing cell cycle exit downstream of the CBC and is en route to a terminally differentiated Paneth or enteroendocrine fate, yet retains some plasticity in its ability to form organoids in vitro. Interestingly, we observed no molecular or histological distinction between the Paneth-like ST-LRCs capable of organoid formation and the LT-LRC Paneth cells lacking stem cell activity- both express high levels of Paneth cell marker genes, cluster together in PCA analysis, and reside in G1 (Figure 7).
Overall, we conclude that LRCs, either short- or long-term, are both molecularly and functionally distinct from the reserve ISCs found in the Bmi1-/Hopx-CreER-marked populations that have been shown to give rise to CBCs (Figure 7). These findings present a cautionary tale regarding the use of DNA label retention as a proxy marker for stem cell identity, as this population is highly dynamic in the intestinal epithelium. Our findings also highlight the importance of understanding the heterogeneity inherent in a cell population marked by proxy reporters and illustrate the impetus for applying single-cell resolution techniques as our understanding of stem cell biology moves forward.
Supplementary Material
Acknowledgments
This work was supported by R01 CA16865 from the NCI (C.J.L). This work was supported in part by the NIH/NIDDK Center for Molecular Studies in Digestive and Liver Diseases (P30DK050306) and its core facilities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no competing financial interests and no personal conflicts of interests to declare.
Author contributions:
N.L and C.J.L designed all experiments and wrote the manuscript. N.L. performed all of the experiments. A.D. performed mouse husbandry and genotyping analyses. S.T.J., J.T., and N. L. performed computational analyses of single-cell profiling data.
References
- 1.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–5. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li N, Yousefi M, Nakauka-Ddamba A, et al. Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Reports. 2014;3:876–91. doi: 10.1016/j.stemcr.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–20. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda N, Jain R, LeBoeuf MR, et al. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–4. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian H, Biehs B, Warming S, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–9. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan KS, Chia LA, Li X, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–71. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 8.Tao S, Tang D, Morita Y, et al. Wnt activity and basal niche position sensitize intestinal stem and progenitor cells to DNA damage. EMBO J. 2015;34:624–40. doi: 10.15252/embj.201490700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metcalfe C, Kljavin NM, Ybarra R, et al. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–59. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Munoz J, Stange DE, Schepers AG, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–91. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itzkovitz S, Lyubimova A, Blat IC, et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol. 2012;14:106–14. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powell AE, Wang Y, Li Y, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–58. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roche KC, Gracz AD, Liu XF, et al. SOX9 Maintains Reserve Stem Cells and Preserves Radioresistance in Mouse Small Intestine. Gastroenterology. 2015;149:1553–1563. e10. doi: 10.1053/j.gastro.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montgomery RK, Carlone DL, Richmond CA, et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 2011;108:179–84. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asfaha S, Hayakawa Y, Muley A, et al. Krt19(+)/Lgr5(−) Cells Are Radioresistant Cancer-Initiating Stem Cells in the Colon and Intestine. Cell Stem Cell. 2015;16:627–38. doi: 10.1016/j.stem.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westphalen CB, Asfaha S, Hayakawa Y, et al. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest. 2014;124:1283–95. doi: 10.1172/JCI73434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falconer E, Chavez EA, Henderson A, et al. Identification of sister chromatids by DNA template strand sequences. Nature. 2010;463:93–7. doi: 10.1038/nature08644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiel MJ, He S, Ashkenazi R, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–42. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potten CS, Hume WJ, Reid P, et al. The segregation of DNA in epithelial stem cells. Cell. 1978;15:899–906. doi: 10.1016/0092-8674(78)90274-x. [DOI] [PubMed] [Google Scholar]
- 20.Escobar M, Nicolas P, Sangar F, et al. Intestinal epithelial stem cells do not protect their genome by asymmetric chromosome segregation. Nat Commun. 2011;2:258. doi: 10.1038/ncomms1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schepers AG, Vries R, van den Born M, et al. Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J. 2011;30:1104–9. doi: 10.1038/emboj.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381–8. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 23.Hughes KR, Gandara RM, Javkar T, et al. Heterogeneity in histone 2B-green fluorescent protein-retaining putative small intestinal stem cells at cell position 4 and their absence in the colon. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1188–201. doi: 10.1152/ajpgi.00080.2012. [DOI] [PubMed] [Google Scholar]
- 24.Buczacki SJ, Zecchini HI, Nicholson AM, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–9. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 25.Foudi A, Hochedlinger K, Van Buren D, et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothenberg ME, Nusse Y, Kalisky T, et al. Identification of a cKit(+) colonic crypt base secretory cell that supports Lgr5(+) stem cells in mice. Gastroenterology. 2012;142:1195–1205. e6. doi: 10.1053/j.gastro.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potten CS, Booth C, Tudor GL, et al. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 28.Batlle E, Henderson JT, Beghtel H, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–63. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 29.Tetteh PW, Basak O, Farin HF, et al. Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell Stem Cell. 2016;18:203–13. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Shapiro HM. Flow cytometric estimation of DNA and RNA content in intact cells stained with Hoechst 33342 and pyronin Y. Cytometry. 1981;2:143–50. doi: 10.1002/cyto.990020302. [DOI] [PubMed] [Google Scholar]
- 31.Rodgers JT, King KY, Brett JO, et al. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature. 2014;510:393–6. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Elf SE, Miyata Y, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4:37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kharas MG, Lengner CJ, Al-Shahrour F, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16:903–8. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breault DT, Min IM, Carlone DL, et al. Generation of mTert-GFP mice as a model to identify and study tissue progenitor cells. Proc Natl Acad Sci U S A. 2008;105:10420–5. doi: 10.1073/pnas.0804800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maria Cambuli F, Rezza A, Nadjar J, et al. Brief report: musashi1-eGFP mice, a new tool for differential isolation of the intestinal stem cell populations. Stem Cells. 2013;31:2273–8. doi: 10.1002/stem.1428. [DOI] [PubMed] [Google Scholar]
- 36.Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11:118–26. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 37.Furuyama K, Kawaguchi Y, Akiyama H, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 38.Van Landeghem L, Santoro MA, Krebs AE, et al. Activation of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1111–32. doi: 10.1152/ajpgi.00519.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson SP, Langan-Fahey SM, Johnson DA, et al. Metabolites, pharmacodynamics, and pharmacokinetics of tamoxifen in rats and mice compared to the breast cancer patient. Drug Metab Dispos. 1991;19:36–43. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.