Abstract
Neurotransmission requires precise control of neurotransmitter release from axon terminals. This process is regulated by glial cells; however, underlying mechanisms are not fully understood. Here we report that glutamate release in the brain is impaired in mice lacking low density lipoprotein receptor-related protein 4 (Lrp4), a protein critical for neuromuscular junction formation. Electrophysiological studies indicate compromised release probability in astrocyte-specific Lrp4 knockout mice. Lrp4 mutant astrocytes suppress glutamate transmission by enhancing the release of ATP, whose levels are elevated in the hippocampus of Lrp4 mutant mice. Consequently, the mutant mice are impaired in locomotor activity and spatial memory and are resistant to seizure induction. These impairments could be ameliorated by adenosine A1 receptor antagonist. The results reveal a critical role of Lrp4, in response to agrin, in modulating astrocytic ATP release and synaptic transmission. Our study provides insight into the interaction between neurons and astrocytes for synaptic homeostasis and/or plasticity.
Introduction
In the nervous system, neurons communicate with each other by synapses, a tripartite structure consisting of axon terminal of one neuron, postsynaptic membrane of another neuron, and surrounding glial cell processes1–3. Synaptic transmission is critical for perception, thinking, learning and memory and response to environmental changes. Synapse dysfunction is implicated in various neuropsychiatric disorders including epilepsy, addiction, schizophrenia and autism. Neurotransmission requires precise control of neurotransmitter release from presynaptic terminals and responsiveness of neurotransmitter receptors on postsynaptic membrane. Increasing evidence suggests that this process is tightly regulated by glial cells1–4. Astrocytes, which account for more than half of the cells in the human brain5, ensheath a majority of excitatory synapses in the hippocampus, for example6. They regulate neuronal transmission by releasing soluble factors such as ATP7, 8. Astrocytic ATP could rapidly degrade to adenosine, which suppresses glutamatergic transmission by activating presynaptic purinergic receptors9, 10. However, little is known about signaling pathways that control the release of gliotransmitters.
Lrp4 is a type I single transmembrane protein of the LDL receptor family11, 12. Recent studies indicate that Lrp4 serves as a receptor for agrin, a motor nerve-derived factor, and is critical for the formation and maintenance of the neuromuscular junction (NMJ), a peripheral cholinergic synapse between motor neurons and skeletal muscle fibers13. In the current working model, agrin binds to Lrp4 on the post-junctional membrane, and resulting tetrameric complex activates the receptor-like tyrosine kinase MuSK in muscle fibers and downstream signaling pathways for postjunctional membrane differentiation13–19. Lrp4 in muscle fibers may also direct a retrograde signal for presynaptic differentiation at NMJ20, 21.
Being expressed in the brain22, 23, Lrp4 has been implicated in hippocampal synaptic plasticity23, 24. However, little is known about underlying mechanisms. Previous studies indicate that Lrp4 is a protein present at the postsynaptic density of pyramidal neurons22, 23. We did not observe abnormal neurotransmission in mutant mice where Lrp4 was ablated in excitatory neurons in preliminary studies. Intriguingly, glutamate release was impaired in astrocyte-specific Lrp4 mutant mice, as well as in GFAP-Lrp4−/− mice where the Lrp4 gene was ablated in both neurons and astrocytes. These results suggest a role of Lrp4 in astrocytes for glutamatergic transmission. We have investigated underlying mechanisms using a combination of cell biology, electrophysiology, and pharmacology techniques. Our results suggest that astrocytic Lrp4, in response to agrin, controls the release of ATP from astrocytes and thus maintain glutamatergic transmission.
Results
Reduced sEPSC frequency in brain-specific Lrp4 mutant mice
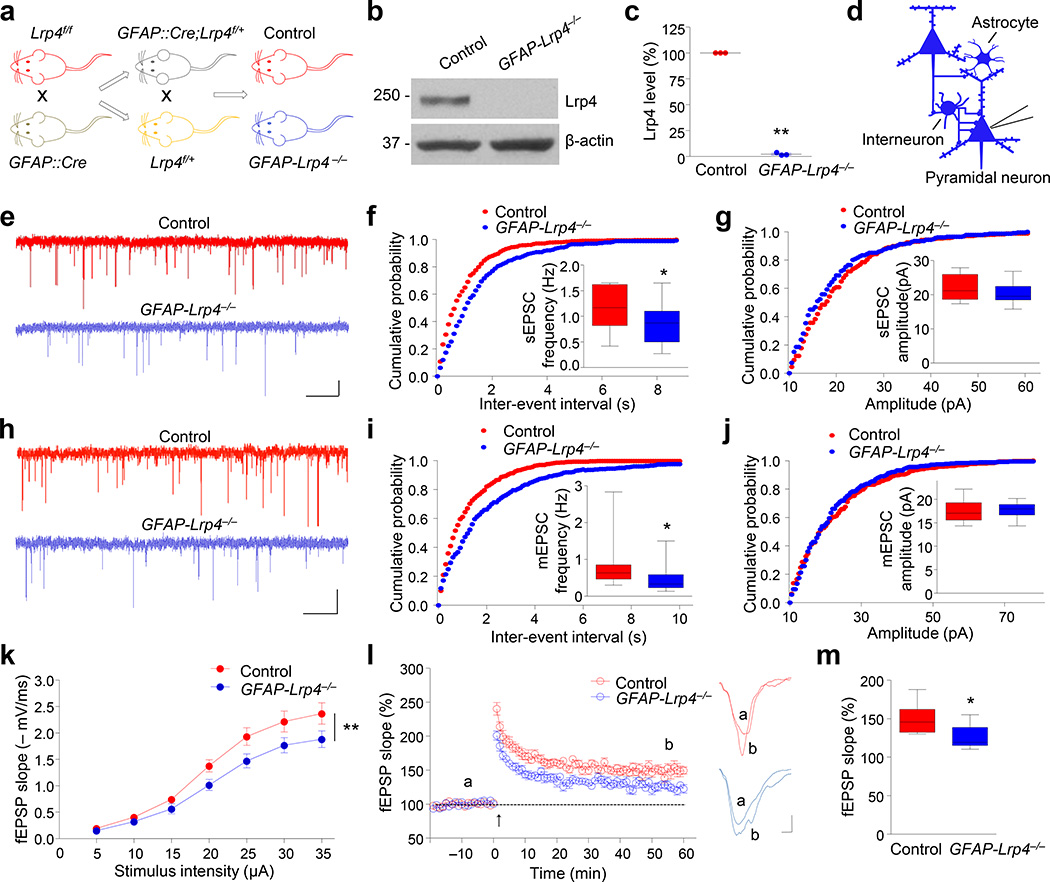
Lrp4 was expressed in the brain including the hippocampus22, 23 and its expression is regulated developmentally (Supplementary Fig. 1a, b). To study whether Lrp4 plays a role in the brain, we generated brain-specific Lrp4 mutant mice by crossing Lrp4f/f mic21 with GFAP::Cre mice where Cre is under control of the human glial fibrillary acidic protein (GFAP) promoter whose expression is restricted in neural progenitor cells that give rise to neurons and glial cells in the brain25 (Fig. 1a). GFAP::Cre;Lrp4f/f (GFAP-Lrp4−/−, for short) mice were viable and showed no difference in body weight, compared to control mice (i.e., Lrp4f/f, unless otherwise indicated) (data not shown). Western blot analysis indicated that Lrp4 was hardly detectable in the hippocampus of GFAP-Lrp4−/− mice (Fig. 1b, c). We recorded, in whole-cell configuration, spontaneous and miniature excitatory postsynaptic current (sEPSC and mEPSC, respectively) of pyramidal neurons in CA1 region of hippocampus. sEPSC and mEPSC frequency was decreased in GFAP-Lrp4−/− hippocampus, compared with control (Fig. 1d–f, h and i). No change was observed in sEPSC and mEPSC amplitude (Fig. 1g and j). In addition, there was a downward shift of input–output (I/O) curves of fEPSPs (field excitatory postsynaptic potentials) at Schaffer collateral (SC)–CA1 synapses in GFAP-Lrp4−/− hippocampus, compared with controls (Fig. 1k). These results suggest hypofunction of glutamatergic transmission in Lrp4 mutant mice. In agreement, long-term potentiation in the hippocampus was impaired (Fig. 1l and m)23, 24. These observations indicate that Lrp4 is involved in proper glutamatergic transmission and synaptic plasticity in the brain.
Figure 1. Reduced EPSC frequency and impaired synaptic plasticity in GFAP-Lrp4−/− mice.
(a) Lrp4f/f mice were crossed with GFAP::Cre mice; resulting GFAP::Cre;Lrp4f/+ mice were crossed with Lrp4f/+ mice to generate GFAP::Cre;Lrp4f/f (GFAP-Lrp4−/−) and Lrp4f/f (Control) mice.
(b) Lrp4 was not detectable in hippocampus of one-month-old GFAP-Lrp4−/− mouse. Shown were representative blots of three independent experiments with similar results. Full-length blots/gels are presented in Supplementary Figure 12.
(c) Quantitative analysis of data in (b). n = 3 pairs of mice. Lrp4 band density was normalized by the loading control β-actin; values of control mice were taken as 100%. Paired Student’s t test, t(2) = 128.3, p < 0.0001.
(d) Recording diagrams. Pyramidal neurons were recorded in whole-cell configuration. Blue color denotes Lrp4 mutant.
(e) Representative traces of sEPSCs in CA1 pyramidal neurons from control and GFAP-Lrp4−/− mice. Scale bar: 2 s, 10 pA.
(f, g) Cumulative probability plots of sEPSC inter-event intervals and histograms of sEPSC frequency (f) and amplitude (g). n = 15 neurons of 4 mice for both genotypes. For (f), student’s t test, t(28) = 2.538, p = 0.017; for (g), student’s t test, t(28) = 0.9343, p = 0.3581.
(h) Representative traces of mEPSCs in CA1 pyramidal neurons from control and GFAP-Lrp4−/− mice. Scale bar: 2 s, 10 pA.
(i, j) Cumulative probability plots of mEPSC inter-event intervals and histograms of mEPSC frequency (i) and amplitude (j). n = 29 neurons, 5 control mice; n = 22 neurons, 5 GFAP-Lrp4−/− mice. For (i), student’s t test, t(49) = 2.12, p = 0.0391; for (j), student’s t test, t(49) = 0.6891, p = 0.494.
(k) Depressed I/O curves in the hippocampus of GFAP-Lrp4−/− mice. fEPSPs were recorded by stimulating SC-CA1 pathway with gradual increasing intensities. n = 9 slices from 4 mice for both genotypes. Two-way ANOVA, F(1, 112) = 20.35, p < 0.0001.
(l) Impaired LTP at SC-CA1 synapses in the hippocampus of GFAP-Lrp4−/− mice. Normalized fEPSP slopes were plotted every 1 min. Arrow denotes LTP induction. Shown on the right were representative traces taken before (a) and 50 min after high frequency stimulation (b). Scale bar: 2 ms, 0.2 mV.
(m) Quantitative analysis of LTP level in (l). n = 9 slices from 4 mice for both genotypes. Student’s t test, t(16) = 2.879, p = 0.0109.
Data in (c and k) were presented as mean ± s.e.m; data in (f, g, i, j and m) were presented as median with interquartile range, whiskers are the minimum and maximum; *, p < 0.05; **, p <0.01.
To investigate underlying mechanisms, we first characterized the morphology of the brain. The mutant mice showed no detectable defects in the laminar structure of the cortex and hippocampus, or in the number of neurons (Supplementary Fig. 1c–f). Similar levels of synaptic proteins were found in homogenates of mutant and control hippocampus (Supplementary Fig. 2a, b). Dendritic arborization of CA1 neurons in control and Lrp4 mutant mice was comparable and quantitative Sholl analysis showed no difference in number of dendritic intersections between genotypes (Supplementary Fig. 2c, d). In addition, Lrp4 mutation had no effect on intrinsic excitability of pyramidal neurons, I/V curves of AMPA and NMDA receptor-mediated currents, or AMPA/NMDA ratio (Supplementary Fig. 3), in agreement with little change in sEPSC and mEPSC amplitude in GFAP-Lrp4−/− mice (Fig. 1g, j).
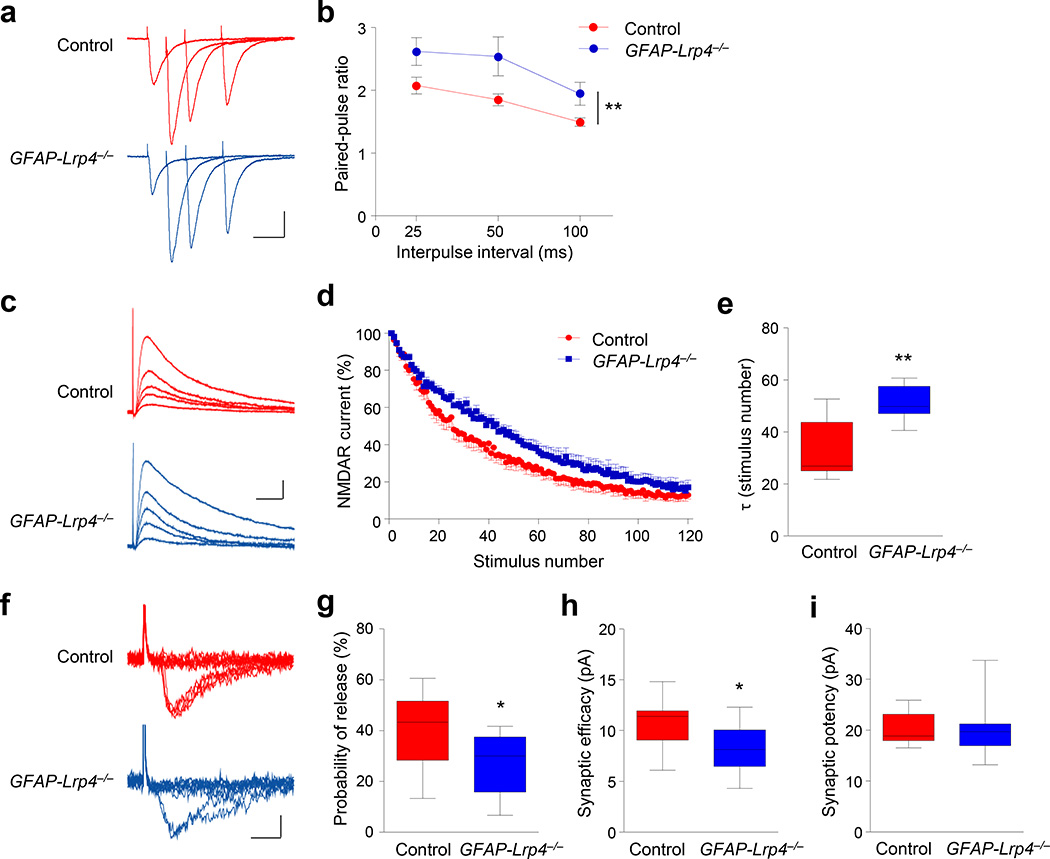
CA1 neurons of GFAP-Lrp4−/− mice displayed fewer spines, compared with control mice (Supplementary Fig. 4a, b), in agreement with a recent report23. To determine whether Lrp4 mutation alters glutamate release, we performed the following three sets of experiments. First, we characterized the paired-pulse ratios (PPR) of eEPSCs in hippocampal CA1 pyramidal neurons in response to two consecutive stimulations. At excitatory synapses, the second stimulation generates larger eEPSC because of residual calcium concentration of the first eEPSC26. Shown in Figure 2a were averaged traces of 7 consecutive eEPSCs induced by paired stimuli at different inter-pulse intervals. Statistically, PPRs in GFAP-Lrp4−/− slices were increased, compared with those in control slices (Fig. 2b), suggesting that Lrp4 mutation modulates glutamatergic transmission by a presynaptic mechanism. Second, NMDAR-mediated synaptic currents were measured in the presence of MK-801, an irreversible open-channel blocker. Due to the channel usage-dependent blockage, eEPSC amplitudes of CA1 pyramidal neurons were progressively reduced during repetitive stimulation of the SC pathway27, 28. The decay of the NMDAR currents in the presence of MK-801 was slower in GFAP-Lrp4−/− slices than that in control slices (Fig. 2c–e). This result suggests a reduction of presynaptic release probability. Finally, we characterized eEPSCs in CA1 pyramidal neurons in response to minimal stimulation, which is thought to activate only a single axon29. The rate of successful response was lower in GFAP-Lrp4−/− mice, compared with control (Fig. 2f, g). Accordingly, the synaptic efficacy (amplitudes of failed and successful eEPSCs) was decreased (Fig. 2h). No change was observed in synaptic potency (amplitudes of successful eEPSCs) between the two genotypes (Fig. 2i). Moreover, little difference was observed in PSD length, or the number and diameter of presynaptic vesicles between control and GFAP-Lrp4−/− mice (Supplementary Fig. 4c–f). Together, these results indicate that sEPSC frequency reduction in GFAP-Lrp4−/− mice may also be due to diminished probability of glutamate release.
Figure 2. Reduced excitatory vesicle release probability in the absence of Lrp4.
(a) Representative superimposed sweeps with three different inter-stimulus intervals of pair-pulse stimulations from control and GFAP-Lrp4−/− mice. Scale bar, 50 ms, 100 pA.
(b) Paired-pulse ratios plotted against inter-stimulus intervals. n = 11 neurons, 3 mice for both control and GFAP-Lrp4−/− genotypes. Two-way ANOVA, F(1,60) = 13.55, p = 0.0005
(c) Representative traces of NMDAR currents in the presence of MK-801 (40 µm). Scale bar, 20 ms, 40 pA.
(d) Normalized NMDAR currents plotted against stimulus number. The first pulse given in the presence of MK-801 was considered as 100%.
(e) Increased τ values in GFAP-Lrp4−/− mice. n = 7 neurons, 3 mice for both genotypes. Student’s t test, t(12) = 3.594, p = 0.0037.
(f) Representative 10 successive individual sweeps of EPSCs evoked by minimal stimulation. Scale bar, 5 ms, 10 pA.
(g, h and i) Reduced release probability, reduced synaptic efficacy, and no change in synaptic potency. n = 13 neurons, 4 control mice; n = 14 neurons, 4 GFAP-Lrp4−/− mice. For (g), student’s t test, t(25) = 2.682, p = 0.0128; for (h), student’s t test, t(25) = 2.615, p = 0.0149; for (i), student’s t test, t(25) = 0.1254, p = 0.9012.
Data in (b) were presented as mean ± s.e.m; data in (e and g-i) were presented as median with interquartile range, whiskers are the minimum and maximum; *, p < 0.05; **, p < 0.01.
Modulation of glutamate release by Lrp4 in astrocytes
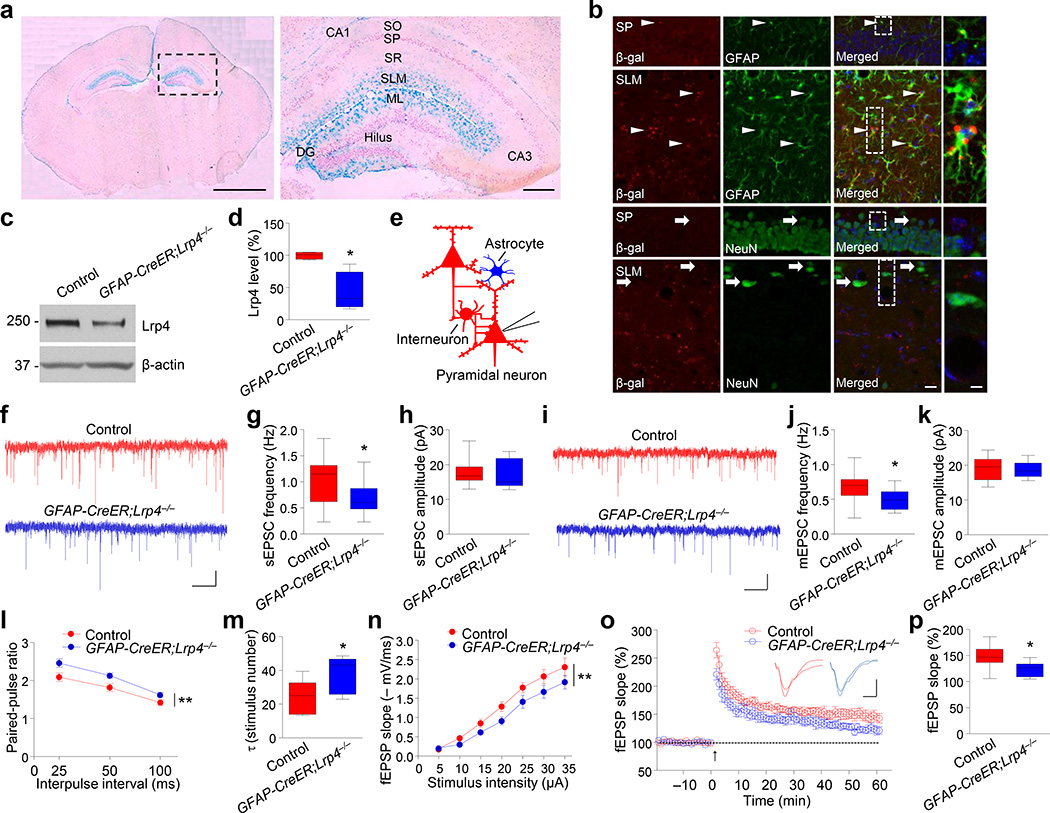
Because of the lack of a reliable Lrp4 antibody for immunostaining, to determine where Lrp4 is expressed, we generated Lrp4-lacZ mice where the Lrp4 gene was replaced with a cassette encoding a β-galactosidase (β-gal) fusion protein. Under the control of the endogenous promoter, β-gal expression is thought to faithfully indicate where Lrp4 is expressed. As shown in Figure 3a, X-gal staining showed that β-gal activity was enriched in stratum lacunosum-moleculare layer (SLM) and molecules layer (ML) of the hippocampus, areas where astrocytes were abundant. The staining was weak in stratum pyramidale layer (SP) where somas of pyramidal neurons were located. Moreover, β-gal-labeled cells were positive for astrocyte marker GFAP whereas its co-location with the neuronal marker NeuN was hardly detectable (Fig. 3b), suggesting that Lrp4 is expressed abundantly in astrocytes. This notion was confirmed by western blotting of astrocyte cultures from hippocampus (Supplementary Fig. 5a).
Figure 3. Astrocytic Lrp4 is critical for glutamatergic transmission.
(a) Enriched β-gal activity in SLM and ML layers. X-gal staining was carried out with coronal brain sections of muscle rescued Lrp4-LacZ homozygous mice. Left, whole brain section; right, enlarged dotted area of the left image. SO: Stratum oriens; SP: Stratum pyramidale; SR: Stratum radiatum; SLM: stratum lacunosum- moleculare; ML: molecular layer. Scale bars, 2 mm (left), 0.2 mm (right). Shown were representative images of more than three independent experiments with similar results.
(b) Co-localization of β-gal with astrocytic marker GFAP. Sections were stained with antibodies against β-gal and GFAP or neuronal marker NeuN. Arrow heads, cells positive for β-gal and GFAP; arrows, cells positive for NeuN, but not β-gal. Scale bar, 20 µm (left), 5 µm (right). Shown were representative images of three independent experiments with similar results.
(c) Reduced Lrp4 level in the hippocampus in GFAP-CreER;Lrp4−/− mice, two weeks after tamoxifen injection. Shown were representative blots of three independent experiments with similar results. Full-length blots/gels are presented in Supplementary Figure 12.
(d) Quantitative analysis of data in (c). n = 4 pairs of mice. Lrp4 band density was normalized by the loading control β-actin; values of control mice were taken as 100%. Paired Student’s t test, t(3) = 3.727, p = 0.0337.
(e) Recording diagrams. Pyramidal neurons were recorded in whole-cell configuration. Blue color denotes Lrp4 mutant; red color indicate control.
(f) Representative sEPCS traces of CA1 pyramidal neurons of control and GFAP-CreER;Lrp4−/− mice. Scale bar: 1 s, 10 pA.
(g) Reduced sEPSC frequency in CA1 pyramidal neurons of GFAP:: CreER;Lrp4−/− mice. n = 13 neurons, 4 control mice; n = 14 neurons, 4 GFAP-CreER;Lrp4−/− mice. Student’s t test, t(25) = 2.145, p = 0.0419.
(h) sEPSC amplitude was comparable between control and GFAP-CreER;Lrp4−/− hippocampus. n = 13 neurons, 4 control mice; n = 14 neurons, 4 GFAP-CreER;Lrp4−/− mice. Student’s t test, t(25) = 0.3502, p = 0.7291.
(i) Representative traces of mEPSCs in CA1 pyramidal neurons from Control and GFAP-CreER;Lrp4−/− mice. Scale bar: 2 s, 10 pA.
(j, k) Reduction in mEPSC frequency (j), but not amplitude (k). n = 13 neurons, 3 control mice; n = 12 neurons, 3 GFAP-CreER;Lrp4−/− mice. For (j), student’s t test, t(23) = 2.368, p = 0.0266; for (k), t(23) = 0.2528, p = 0.8027.
(l) Paired-pulse ratios plotted against inter-stimulus intervals. n = 10 neurons, 4 control mice; n = 11 neurons, 4 GFAP-CreER;Lrp4−/− mice. Two-way ANOVA, F(1,57) = 16.15, p = 0.0002.
(m) Increased τ values in GFAP-CreER;Lrp4−/− mice. n = 7 neurons, 4 control mice; n = 8 neurons, 4 GFAP-CreER;Lrp4−/− mice. Student’s t test, t(13) = 2.32, p = 0.0373.
(n) Depressed I/O curves in the hippocampus of GFAP-CreER;Lrp4−/− mice. fEPSPs were recorded by stimulating SC-CA1 pathway with gradual increasing intensities. n = 8 slices from 4 control mice; n = 7 from 4 GFAP-CreER;Lrp4−/− mice. Two-way ANOVA, F(1, 91) = 14.52, p = 0.0003.
(o) Impaired LTP at SC-CA1 synapses in the hippocampus of GFAP-CreER;Lrp4−/− mice. Normalized fEPSP slopes were plotted every 1 min, Arrow denotes LTP induction. Shown on the right were representative traces taken before (a) and 50 min after high frequency stimulation (b). Scale bar: 4 ms, 0.4 mV.
(p) Quantitative analysis of LTP level in (o). n = 8 slices from 4 control mice; n = 7 from 4 GFAP-CreER;Lrp4−/− mice. Student’s t test, t(13) = 2.354, p = 0.035.
Data in (d, g, h, j, k, m and p) were presented as median with interquartile range, whiskers are the minimum and maximum; data in (l and n) were presented as mean ± s.e.m; *, p < 0.05; **, p < 0.01.
To determine whether astrocytic Lrp4 is critical for glutamatergic transmission, we used GFAP::CreER mice where the expression of CreER is driven by a 2-kb minimal promoter of the human GFAP gene30, 31. The Cre recombinase is inactive but could be induced by tamoxifen (Supplementary Fig. 5b). To verify the specificity of this line, GFAP::CreER mice were crossed with Rosa::LSL-tdTomato reporter mice to generate GFAP::CreER;tdTomato mice. As shown in Supplementary Figure 5c, tdTomato co-stained with the astrocytic marker GFAP, but not the neuronal marker NeuN in hippocampal slices of tamoxifen-treated GFAP::CreER;tdTomato mice. These results demonstrate that Cre activity was specifically activated in GFAP+ cells in GFAP::CreER mice, in agreement with previous reports30, 31. Next, we crossed GFAP::CreER mice with Lrp4f/f mice to generate GFAP::CreER;Lrp4−/− mice. Lrp4 level was decreased in these mice two weeks after tamoxifen treatment, compared with control mice (Fig. 3c, d). Spine numbers of CA1 neurons were similar between control and GFAP-CreER;Lrp4−/− mice (Supplementary Fig. 5d, e), indicating that spines were not altered by astrocyte-specific knockout of Lrp4. However, CA1 pyramidal neurons displayed a reduction in sEPSC and mEPSC frequency, but not amplitude (Fig. 3e–k); increased PPR (Fig. 3l); decreased τ value in GFAP-CreER;Lrp4−/− mice in MK-801 block assay (Fig. 3m), down shifted I/O curve of fEPSPs (Fig. 3n) and deficient LTP (Fig. 3o, p), compared with control mice. These results demonstrate that astrocytic ablation of Lrp4 impairs presynaptic glutamate release and synaptic plasticity without altering spines, suggesting Lrp4 in astrocytes is critical for glutamatergic transmission. In agreement, Lrp4 mutation specifically in CA1 pyramidal neurons (by Camk2α::Cre) had no apparent effect on Lrp4 level, sEPSC frequency, amplitude or PPR (Supplementary Fig. 6).
Increased astrocytic ATP release by Lrp4 mutation
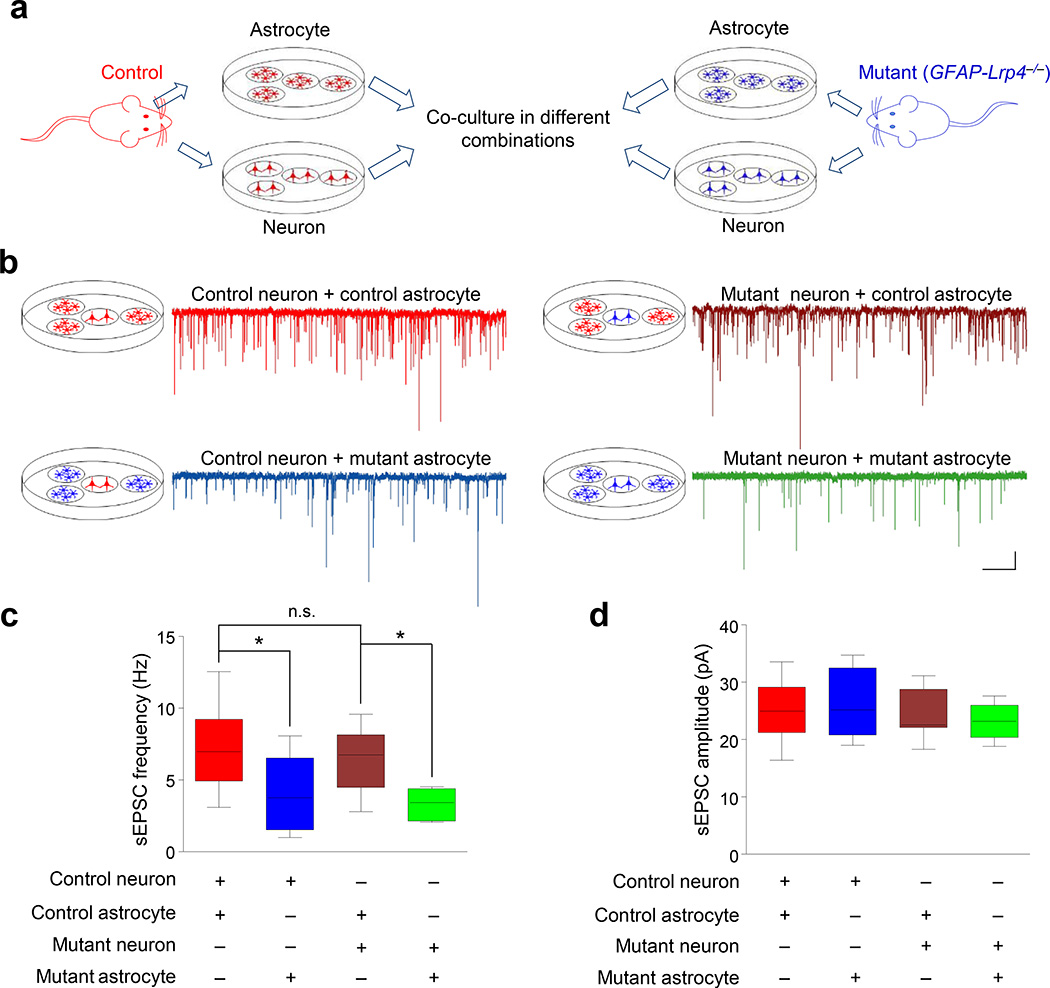
Lrp4 mutation did not change the number of astrocytes or their morphology including the distance between astrocytic and synaptic membranes (Supplementary Figs 4g, 7a–f). To investigate how Lrp4 in astrocytes modulates glutamatergic transmission, we cultured hippocampal neurons of E18 embryos and astrocytes from P3 hippocampus and grew them in separate dishes. Lrp4 mutation seemed to have little effect on the number of cultured neurons and astrocytes (Supplementary Fig. 7g–j). At days-in-vitro (DIV10), one coverslip of neurons and three coverslips of confluent astrocytes were co-cultured, without contact, in a 35-mm dish for 24 h before recording9. The sEPSC frequency was decreased in control neurons that were co-cultured with Lrp4 mutant astrocytes, compared with that of control neuron-control astrocyte co-culture (Fig. 4a–c), indicating that Lrp4 in astrocytes is critical for glutamatergic transmission. sEPSC frequency of Lrp4 mutant neurons that were co-cultured with control astrocytes was similar to that of control neuron-control astrocyte co-culture (Fig. 4c). This suggests a dispensable role of the Lrp4 gene in neurons, in agreement with study of neuron-specific mutant mice. Moreover, mutant neurons co-cultured with mutant astrocytes showed similar reduction in sEPSC frequency (Fig. 4c). Notice that sEPSC amplitudes were similar in all combinations of co-culture, suggesting Lrp4 mutation has no effect on sEPSC amplitudes (Fig. 4d). These experiments provide evidence that Lrp4 in astrocytes modulated glutamatergic transmission through a secretable factor. Factors released from astrocytes that regulate neuronal transmission include D-serine and ATP4, 7, 8. D-serine activates NMDA receptor on presynaptic terminal to promote glutamate release and that on postsynaptic membrane to potentiate NMDA response32, 33. We treated hippocampal slices with the NMDA receptor antagonist DL-AP5 (100 µM), which caused a dose-dependent reduction in sEPSC frequency (Supplementary Fig. 8a). However, similar reduction was observed in GFAP-Lrp4−/− slices (Fig. 5a). These results suggested a limited role of D-serine as an Lrp4-modulated factor to regulate glutamate release.
Figure 4. Astrocytic Lrp4 affects glutamatergic transmission in a non-contact way.
(a) Experimental design for co-culture. Neurons and astrocytes were isolated from control (red) or mutant (blue; GFAP-Lrp4−/−) mice and cultured in separate dishes. After maturation, one cover slip of neurons was co-cultured with three cover slips of astrocytes in different combinations.
(b) Representative sEPSC traces of neurons co-cultured with astrocytes. Genotypes of neurons and astrocytes are color-coded: red, control; blue, mutant. Scale bar: 2 s, 10 pA.
(c, d) Quantitative analysis of sEPSC frequency (c) and amplitude (d). 3 times of cell culture; n = 11 control neurons co-cultured with control astrocytes; n = 9 control neurons co-cultured with mutant astrocytes; n = 8 mutant neurons co-cultured with control astrocytes; n = 8 mutant neurons co-cultured with mutant astrocytes. For (c), one-way ANOVA, F(3,32) = 5.652, p = 0.0032. Control neuron + control astrocyte vs control neuron + mutant astrocyte, student’s t test, t(18) = 2.532, p = 0.0209; control neuron + control astrocyte vs mutant neuron + control astrocyte, student’s t test, t(17) = 0.6137, p = 0.5475; mutant neuron + control astrocyte vs mutant neuron + mutant astrocyte, student’s t test, t(14) = 3.573, p = 0.0031. For (d), one-way ANOVA, F(3,32) = 0.8197, p = 0.4926.
Data in (c and d) were presented as median with interquartile range, whiskers are the minimum and maximum; n.s., not significant; *, p < 0.05; **, p < 0.01.
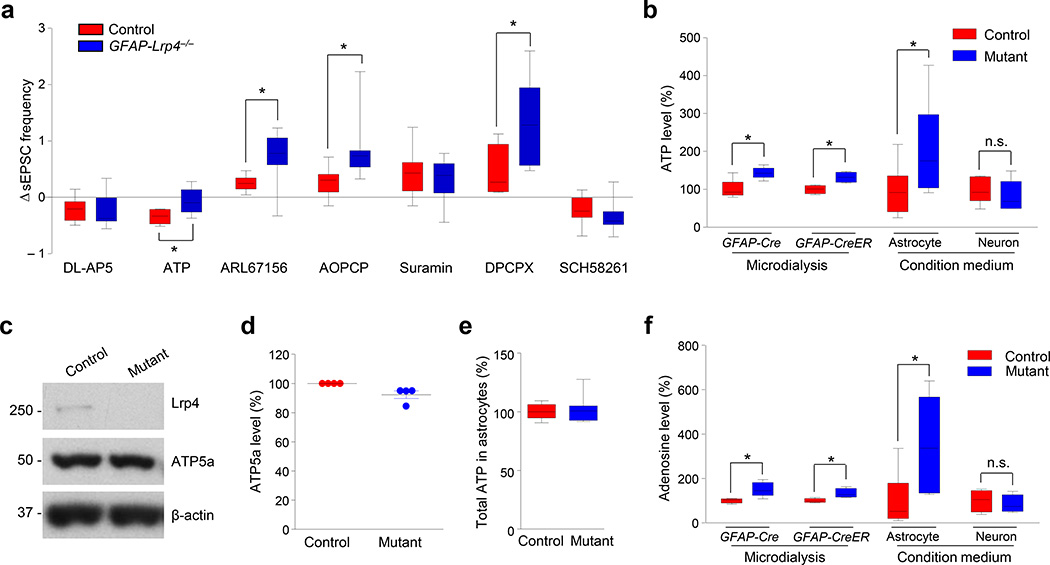
Figure 5. Modulation of astrocytic ATP release by Lrp4.
(a) Effects of chemicals on sEPSCs of CA1 pyramidal neurons in Control and GFAP-Lrp4−/− mice. ΔsEPSC frequency was calculated by (f2 - f1)/f1, where f1 and f2 were the frequencies of sEPSCs recorded before and after drug treatment, respectively. The number of recorded neurons and mice were as follows: 8 neurons of 3 control mice and 9 neurons of 3 GFAP-Lrp4−/− mice for DL-AP5 (100 µM). Student’s t test, t(15) = 0.2202, p = 0.8287; 7 neurons of 3 mice of each genotype for ATP (10 µM). Student’s t test, t(12) = 2.944, p = 0.0123; 8 neurons of 4 mice of each genotype for ARL67156 (100 µM). Student’s t test, t(14) = 2.675, p = 0.0181; 8 neurons of 3 mice of each genotype for AOPCP (300 µM). Student’s t test, t(14) = 2.483, p = 0.0263; 9 neurons of 3 mice of each genotype for Suramin (10 µM). Student’s t test, t(16) = 0.4528, p = 0.6567; 7 neurons of 3 control mice and 8 neurons of 3 GFAP-Lrp4−/− mice for DPCPX (800 nM). Student’s t test, t(13) = 2.451, p = 0.0291; 8 neurons of 3 control mice and 7 neurons of 3 GFAP-Lrp4−/− mice for SCH58261 (5 µM). Student’s t test, t(13) = 0.8303, p = 0.4213.
(b) Increased ATP levels in the dialysate from GFAP-Lrp4−/− and tamoxifen-treated GFAP-CreER;Lrp4−/− hippocampus and condition medium of mutant astrocytes, but not neurons. Cells were cultured from control and GFAP-Lrp4−/− (mutant) mice. n = 5 samples of dialysate from 5 control and 5 GFAP-Lrp4−/− mice, Student’s t test, t(8) = 3.33, p = 0.0104; n = 4 samples of dialysate from 4 control and 4 GFAP-CreER;Lrp4−/− mice; Student’s t test, t(6) = 3.565, p = 0.0119; n = 8 dishes (4 mice) for astrocytes, Student’s t test, t(14) = 2.276, p = 0.0391; n = 5 dishes (4 mice) for neurons, Student’s t test, t(8) = 0.7401, p = 0.4804
(c) Western blotting showing similar level of ATP5a in cultured astrocytes of control and mutant genotype. Shown were representative blots of two independent experiments with similar results. Full-length blots/gels are presented in Supplementary Figure 12.
(d) Quantitative analysis of data in (c). n = 4 dishes from 2 times of astrocyte culture for each genotype. ATP5a band density was normalized by the loading control β-actin; values of control mice were taken as 100%. Paired student’s t test, t(3) = 2.923, p = 0.0613.
(e) Similar levels of total ATP in mutant hippocampal astrocytes. Cultured astrocytes were homogenized. Supernatants after centrifugation were subjected to ATP measurement. n = 8 dishes from 2 times of culture for both genotypes. Paired student’s t test, t(3) = 0.4237, p = 0.6845.
(f) Increased adenosine levels in the dialysate from GFAP-Lrp4−/− and tamoxifen-treated GFAP-CreER;Lrp4−/− hippocampus and condition medium of mutant astrocytes, but not neurons. n = 5 samples of dialysate from 5 control and 5 GFAP-Lrp4−/− mice, Student’s t test, t(8) = 3.394, p = 0.0094; n = 4 samples of dialysate from 4 control and 4 GFAP-CreER;Lrp4−/− mice, Student’s t test, t(6) = 2.816, p = 0.0305; n = 6 dishes (4 mice) for astrocytes, Student’s t test, t(10) = 2.562, p = 0.0283; n = 4 dishes (4 mice) for neurons, Student’s t test, t(6) = 0.4766, p = 0.6505.
Data in (a, b, e and f) were presented as median with interquartile range, whiskers are the minimum and maximum; data in (d) were presented as mean ± s.e.m; n.s., not significant; *, p < 0.05.
On the other hand, ATP can directly or indirectly (by converting to adenosine) regulate glutamate release9, 10. To investigate whether ATP is regulated by Lrp4, we determined whether its response is altered by Lrp4 mutation. Treatment with ATP (10 µM) reduced sEPSC frequency in hippocampal slices of control mice (Supplementary Fig. 8b), in agreement with previous reports9. Interestingly, this effect was diminished in GFAP-Lrp4−/− slices (Fig. 5a). The inability of exogenous ATP to reduce sEPSC frequency suggests that endogenous ATP might have been elevated by Lrp4 mutation. To test this hypothesis, we measured ATP levels in the hippocampus by microdialysis. Indeed, ATP levels in GFAP-Lrp4−/− dialysates were higher than those in control mice (Fig. 5b), in support of the hypothesis. To identify the source of elevated ATP, we cultured hippocampal neurons and astrocytes of different genotypes and measured ATP in the condition medium. Higher ATP levels were observed in the condition medium of hippocampal mutant astrocytes, compared with controls. However, ATP level was similar between the condition media of mutant and control neurons (Fig. 5b). To further test this in vivo, we collected microdialysates from the hippocampus of tamoxifen-treated GFAP-CreER;Lrp4−/− mice. As shown in Figure 5b, compared with controls, ATP level was higher in the hippocampus where Lrp4 was specifically knocked out in astrocytes. These results suggest that increased ATP in the brain came from Lrp4 mutant astrocytes, not neurons. Notice that Lrp4 mutation had no effect on protein level of APT5a (a major ATP synthetase) or total ATP concentration in astrocytes (Fig. 5c–e). These results support a working model that Lrp4 may inhibit ATP release from astrocytes, and in the absence of Lrp4, ATP release from astrocytes was increased. In addition to ATP, the levels of its metabolite adenosine were also elevated in hippocampal dialysates of mutant mice and condition medium of mutant astrocytes (Fig. 5f).
To determine whether ATP or adenosine is involved, hippocampal slices were treated with ARL67156, an inhibitor of ecto-ATPase which catalyzes the hydrolysis of ATP to ADP9, 10, or adenosine 5’-(α, β-methylene) diphosphate (AOPCP), a blocker of AMP dephosphorylation to form adenosine34. Both chemicals increased sEPSC frequency in control slices, suggesting a role of adenosine in regulating glutamate release (Fig. 5a; Supplementary Fig. 8c). This effect was increased by Lrp4 mutation in GFAP-Lrp4−/− slices (Fig. 5a). These results indicated that inhibition of the ATP-to-adenosine reaction ameliorates the suppression effect of Lrp4 mutation on glutamate release and suggest the involvement of adenosine. To further test this hypothesis, hippocampal slices were treated with Suramin (an antagonist of P2 receptor), DPCPX (a blocker of A1 receptor) and SCH58261 (an A2A antagonist). As observed previously, sEPSC frequency was increased by Suramin and DPCPX, but reduced by SCH58261 (Supplementary Fig. 8d–f), and Lrp4 mutation had no effect on the effects of Suramin or SCH58261 (Fig. 5a). In contrast, the effect of DPCPX was significantly increased in GFAP-Lrp4−/− slices (Fig. 5a). These data are consistent with the idea that adenosine A1 receptor is the major target for elevated adenosine level to suppress glutamate release.
Modulation of astrocytic ATP release by agrin signaling
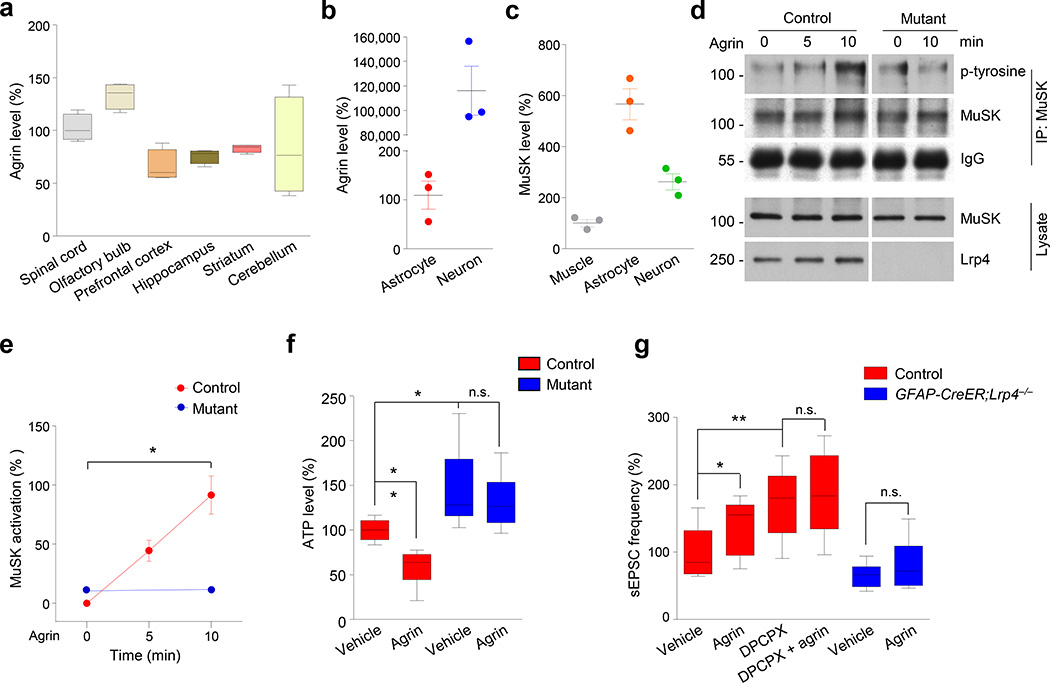
Lrp4 is a receptor of neuronal agrin for NMJ formation and maintenance14, 15, 18, 19. Being a surface protein in astrocytes (Supplementary Fig. 9a), Lrp4 may serve as an agrin receptor to regulate ATP release. Indeed, agrin is expressed in various regions in the brain35, 36 (Fig. 6a). As shown in Figure 6b, neuronal agrin (i.e., the isoform with B/z+ insert15, 35) was > 1000 folds higher in cultured hippocampal neurons than that in cultured astrocytes, indicating neurons as a major source of neuronal agrin in the brain (which is referred as agrin in this manuscript). MuSK was also detectable in the brain36, 37 and was present at higher levels in cultured astrocytes than in neurons or muscles (Fig. 6c). To determine whether agrin regulates ATP release, astrocytes were treated with agrin at a concentration that stimulates AChR clustering in muscle cells15, 21 (Supplementary Fig. 9b–e). Agrin had no effect on astrocyte proliferation (Supplementary Fig. 9f, g). However, as shown in Figure 6d, agrin stimulation caused an increase in MuSK phosphorylation in astrocytes. Concomitantly, ATP level in astrocytic condition medium was reduced (Fig. 6f), suggesting that agrin may inhibit ATP release by activating MuSK. These effects of agrin (to reduce ATP release and to activate MuSK) required Lrp4 (Fig. 6d–f). These observations demonstrate a role of agrin signaling in modulating ATP release from astrocytes.
Figure 6. Agrin regulates astrocytic ATP release.
(a, b) Agrin expression in brain regions and neurons. Total RNA of indicated regions (a), and from cultured astrocytes and neurons (b) was subjected to real-time quantitative PCR. GAPDH was used as the internal control. Expression in spinal cord (a) and astrocytes (b) was set as 100%. n = 4 times for (a); n = 3 times for (b).
(c) MuSK expression in astrocytes. Total RNA was isolated from cultured astrocytes, neurons and muscle cells. Expression level in muscle cells was set as 100%. n = 3 times.
(d) Agrin activated MuSK in astrocytes. Astrocytes were cultured from control and GFAP-Lrp4−/− (mutant) mice and treated with agrin (100 ng/ml) for different times. MuSK was isolated by immunoprecipitation and probed with p-tyrosine antibody to detect phosphorylated MuSK. Shown were representative blots of three independent experiments with similar results. Full-length blots/gels are presented in Supplementary Figure 12.
(e) Quantitative analysis of data in (d). p-tyrosine intensity was normalized by that of MuSK. n = 3 times. For control 0 vs 10 min groups, paired student’s t test, t(2) = 5.667, p = 0.0298; for mutant 0 vs 10 min groups, paired student’s t test, t(2) = 0.4547, p = 0.6939.
(f) Lrp4-depedent reduction of ATP release by agrin. Astrocytes of different genotypes were treated with vehicle or agrin (100 ng/ml) for 24 h. ATP in the condition medium was measured. n = 8 dishes of cultured astrocytes, 3 times. For control + vehicle vs control + agrin groups, student’s t test, t(14) = 5.217, p = 0.0001;for control + vehicle vs mutant + vehicle groups, student’s t test, t(14) = 2.902, p = 0.0116; for mutant + vehicle vs mutant + agrin groups, student’s t test, t(14) = 0.7957, p = 0.4395.
(g) Agrin increased sEPSC frequency in CA1 pyramidal neurons of control but not GFAP-CreER;Lrp4−/− mice and this effect was blocked by DPCPX. n = 8 neurons, 3 control mice for vehicle; n = 10 neurons, 3 control mice for agrin; n = 9 neurons, 3 control mice for DPCPX; n = 10 neurons, 3 control mice for DPCPX + agrin; n = 9 neurons, 3 GFAP-CreER;Lrp4−/− mice for vehicle; n = 9 neurons, 3 GFAP-Lrp4−/− mice for agrin. For control + vehicle vs control + agrin groups, student’s t test, t(16) = 2.168, p = 0.0456; for control + vehicle vs control + DPCPX groups, student’s t test, t(15) = 3.412, p = 0.0039; for control + vehicle vs control + agrin groups, student’s t test, t(16) = 2.168, p = 0.0456; for control + DPCPX vs control + DPCPX + agrin groups, student’s t test, t(17) = 0.5006, p = 0.6231; for GFAP-CreER;Lrp4−/− + vehicle vs GFAP-CreER;Lrp4−/− + agrin groups, student’s t test, t(16) = 1.059, p = 0.3052.
Data in (a, f and g) were presented as median with interquartile range, whiskers are the minimum and maximum; data in (b, c and e) were presented as mean ± s.e.m; n.s., not significant; *, p < 0.05;**, p < 0.01.
In accord, sEPSC frequency was enhanced in agrin-treated hippocampal slices (Fig. 6g), indicating a role of agrin in modulating glutamatergic transmission. This effect was not observed in GFAP-CreER;Lrp4−/− slices (Fig. 6g), suggesting its dependence on astrocytic Lrp4. Notice that in the presence of the A1 antagonist DPCPX, agrin was unable to further increase sEPSC frequency (Fig. 6g), suggest its dependence on A1 receptor. Together, these observations support a working model that agrin-Lrp4 signaling controls astrocytic ATP release that maintains homeostasis of glutamatergic activity.
Behavioral deficits in GFAP-Lrp4−/− mice
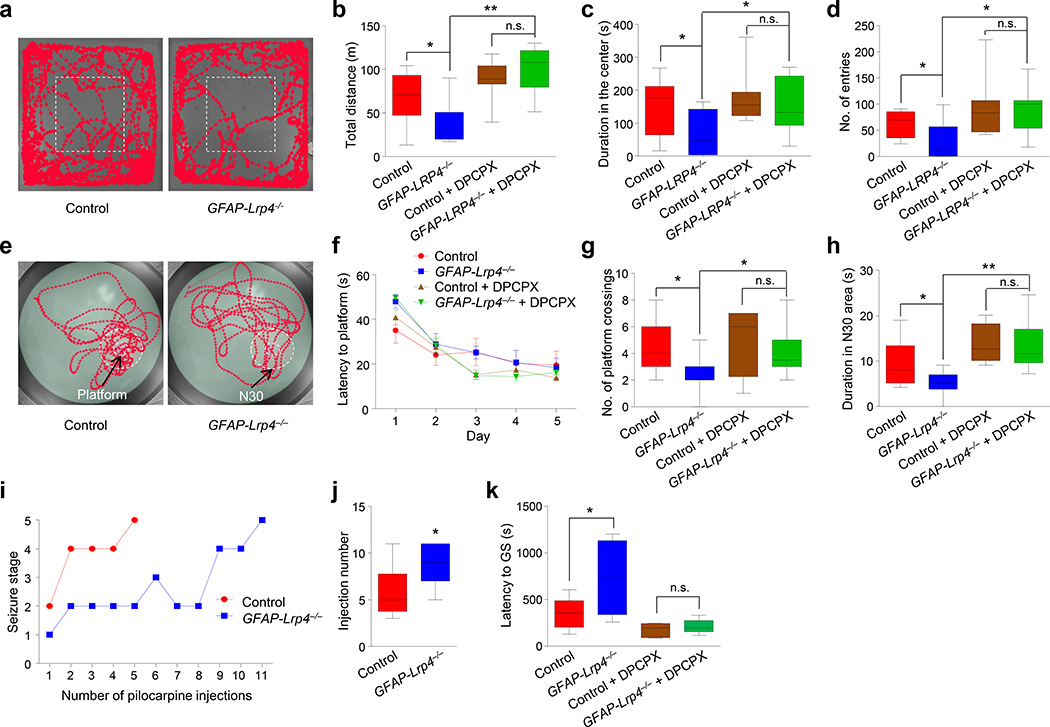
Synapse dysfunction is implicated in various neurological and psychiatric disorders. To further investigate the function of Lrp4 in the CNS, we tested locomotor activity of GFAP-Lrp4−/− mice in an open field and found that mutant mice displayed a pronounced decrease in total travel distance, compared with control mice (Fig. 7a and b), implying a hypoactive state in GFAP-Lrp4−/− mice. In addition, the mutant mice had fewer entries into the central area and spent less time there (Fig. 7c and d). However, no difference was observed between control and mutant mice in elevated plus maze and light/dark box tests, suggesting little anxiogenic effect of the mutation (Supplementary Fig. 10a–h). To investigate whether Lrp4 mutation causes deficits in learning and memory, we performed Morris water maze test38. The mutant mice exhibited similar swimming velocity to control mice (Supplementary Fig. 10i). During the training phase, the escape latency for GFAP-Lrp4−/− mice to locate the hidden platform was also similar to that of control mice (Fig. 7e, f), suggesting their learning ability may be normal. However, during the probe test, when the platform had been removed, GFAP-Lrp4−/− mice exhibited fewer crosses over the absent platform and spent less time in the 30-cm area (N30) surrounding the absent platform, compared with control mice (Fig. 7g, h). To determine whether the Lrp4 mutant mice are "smarter" at reversal learning when the platform was absent, we analyzed the number of crosses over the absent platform and duration in the N30 area every 10 s for 60 s. No difference was found in the interaction between genotype and time (Supplementary Fig. 10j, k), which suggests that Lrp4 mutant mice may not adapt faster than control mice after the platform removal. Together, these results suggest that Lrp4 mutant mice may be impaired in spatial memory consolidation. To determine whether the behavioral impairment was mediated by A1 receptor activation, we injected mice with DPCPX 30 min prior to tests. DPCPX-treated GFAP-Lrp4−/− and control mice showed no difference in locomotor activity (Fig. 7b–d) and spatial memory (Fig. 7g and h). These observations demonstrate a role of Lrp4 for proper behavior and cognitive function.
Figure 7. Ablation of Lrp4 caused abnormal behavior.
(a) Representative traces of first 5 min in the open field test.
(b) Reduced total distance traveled by mutant mice in 30 min. Mice were pretreated without or with DPCPX 30 min before test. n = 9 mice for each genotype without DPCPX; n = 11 mice for each genotype with DPCPX. For control vs GFAP-Lrp4−/− groups, student’s t test, t(16) = 2.417, p = 0.0279; for GFAP-Lrp4−/− vs GFAP-Lrp4−/− + DPCPX groups, student’s t test, t(18) = 5.543, p < 0.0001; for control + DPCPX vs GFAP-Lrp4−/− + DPCPX groups, student’s t test, t(20) = 1.089, p = 0.2891.
(c) Reduced time mutant mice spent in the center. n = 9 mice for each genotype without DPCPX; n = 11 mice for each genotype with DPCPX. For control vs GFAP-Lrp4−/− groups, student’s t test, t(16) = 2.178, p = 0.0447; for GFAP-Lrp4−/− vs GFAP-Lrp4−/− + DPCPX groups, student’s t test, t(18) = 2.411, p = 0.0268; for control + DPCPX vs GFAP-Lrp4−/− + DPCPX groups, student’s t test, t(20) = 0.8403, p = 0.4107.
(d) Reduced center entry by GFAP-Lrp4−/− mice. n = 9 mice for each genotype without DPCPX; n = 11 mice for each genotype with DPCPX. For control vs GFAP-Lrp4−/− groups, student’s t test, t(16) = 2.246, p = 0.0391; for GFAP-Lrp4−/− vs GFAP-Lrp4−/− + DPCPX groups, student’s t test, t(18) = 3.258, p = 0.0044; for control + DPCPX vs GFAP-Lrp4−/− + DPCPX groups, student’s t test, t(20) = 0.1557, p = 0.8779.
(e) Representative traces in the probe test of Morris water maze.
(f) No difference in latency to the platform during the hidden platform task between the two genotypes. Mice were pretreated without or with DPCPX 30 min before behavioral test. n = 9 mice for each genotype without DPCPX; n = 12 mice for each genotype with DPCPX. Two-way ANOVA, F(3,190) = 1.757, p = 0.1568.
(g) Reduced number of plateform crossings. n = 9 mice for each genotype without DPCPX; n = 12 mice for each genotype with DPCPX. For control vs GFAP-Lrp4−/− groups, student’s t test, t(16) = 2.49, p = 0.0241; for GFAP-Lrp4−/− vs GFAP-Lrp4−/− + DPCPX groups, student’s t test, t(19) = 2.513, p = 0.0211; for control + DPCPX vs GFAP-Lrp4−/− + DPCPX groups, student’s t test, t(22) = 0.9893, p = 0.3333.
(h) Reduced time spent in the N30 area. n = 9 mice for each genotype without DPCPX; n = 12 mice for each genotype with DPCPX. For control vs GFAP-Lrp4−/− groups, student’s t test, t(16) = 2.318, p = 0.034; for GFAP-Lrp4−/− vs GFAP-Lrp4−/− + DPCPX groups, student’s t test, t(19) = 4.625, p = 0.0002; for control + DPCPX vs GFAP-Lrp4−/− + DPCPX groups, student’s t test, t(22) = 0.1478, p = 0.8839.
(i) Representative time courses of seizure development by repeated pilocarpine injection. Mice of two genotypes were subjected to pilocarpine injection every 30 min and scored for seizure stage.
(j) Increased number of pilocarpine injection needed to reach stage-5 seizure for mutant mice. n = 10 mice for each genotype. Student’s t test, t(18) = 2.834, p = 0.011.
(k) Increased latency of mutant mice to generalized convulsive seizure in response to PTZ. Mice were pretreated without or with DPCPX 30 min before PTZ injection. Latency to generalized convulsive seizure was recorded. n = 8 mice for each genotype without DPCPX; n = 6 mice for each genotype with DPCPX. For control vs GFAP-Lrp4−/− groups, student’s t test, t(14) = 2.561, p = 0.0226; for control + DPCPX vs GFAP-Lrp4−/− + DPCPX groups, student’s t test, t(10) = 0.798, p = 0.4434.
Data in (b–d, g, h, j and k) were presented as median with interquartile range, whiskers are the minimum and maximum; data in (f) were presented as mean ± s.e.m; n.s., not significant; *, p < 0.05;**, p < 0.01.
Reduced glutamate release by Lrp4 mutation may alter the vulnerability to epilepsy. To test this hypothesis, mice were injected with pilocarpine to generate status epilepsy39. Most control mice developed status epilepticus after the 5th injection (Fig. 7i). In contrast, a majority of GFAP-Lrp4−/− mice would not develop status epilepticus even after 8th injection (Fig. 7j). These data indicate a raised threshold to seizure in GFAP-Lrp4−/− mice. To further test this notion, we injected mice with pentylenetetrazol (PTZ), a GABAA receptor antagonist that induces seizure via mechanisms different from pilocarpine40. The latency to the onset of generalized convulsive seizures (GS) was higher in GFAP-Lrp4−/− mice than that in control mice (Fig. 7k). The increased vulnerability in Lrp4 mutant mice was diminished by DPCPX treatment (Fig. 7k), suggesting again the involvement of the A1 receptor.
Discussion
Astrocytes are intimately involved in many aspects of brain development, function and disease1–3. Our study identifies a novel function of Lrp4 to modulate glutamatergic transmission. When Lrp4 is mutated in all cells in developing brain (i.e., by GFAP::Cre), mutant mice display a reduction in both spine density and mEPSC frequency. However, ablation of Lrp4 specifically in astrocytes (i.e., in tomaxifen-treated GFAP-CreER;Lrp4−/− mice) has no effect on spine numbers of hippocampal CA1 neurons, but reduces mEPSC frequency and glutamate release probability (Supplementary Fig. 5d, e; Fig. 3j and m). These observations suggest that Lrp4 mutation may alter glutamate release. In accord, electrophysiological studies reveal increased PPR, decreased decay of NMDAR currents in the presence of MK-801 and reduced successful rate of EPSCs by minimal stimulation in these mutant mice, suggesting compromised probability of glutamate release. Together, these results reveal that astrocytic Lrp4 modulates glutamate release.
The experiments with non-contact neuron-astrocyte co-culture provide evidence that Lrp4 mutant astrocytes impair glutamatergic transmission by a secretable factor. Previously implicated factors include glutamate and co-agonist D-serine4, 7, 8, 41. They have been shown to promote glutamate release by activating presynaptic NMDA receptors32, 42. Postsynaptically, activation of NMDA receptors is expected to change the amplitude of NMDAR-mediated EPSCs and AMPA/NMDA ratio32, 43. However, neither was changed in Lrp4 mutant mice (Supplementary Fig. 3h) (data not shown), suggesting similar NMDAR content at synapses. In addition, GFAP-Lrp4−/− CA1 neurons displayed similar I/V curves (Supplementary Fig. 3g). Therefore, the factor regulated by Lrp4 did not appear to be glutamate or D-serine.
The following evidence suggests that this factor may be ATP. First, levels of ATP and its derivative adenosine were higher in the dialysates from Lrp4 mutant hippocampus and in the condition medium of culture Lrp4 mutant astrocytes, compared with controls. Second, Lrp4 mutation-mediated reduction of sEPSC frequency could not be inhibited by suramine, an antagonist of P2 receptor for ATP; but it can be inhibited by ARL67156 or AOPCP (to inhibit ATP hydrolysis) or by DPCPX (to block adenosine A1 receptor). These observations suggest the involvement of A1 receptor in suppressing glutamatergic transmission. Third, DPCPX also diminished behavioral deficits in GFAP-Lrp4−/− mice including decreased locomotor activity, impaired spatial memory and increased threshold to seizure induction. These observations suggest that glutamatergic transmission in Lrp4 mutant mice is likely suppressed by A1 receptor activation by adenosine, a product of ATP hydrolysis.
Despite an established role of ATP in regulating glutamatergic transmission, mechanisms of astrocytic ATP release have begun to be elucidated7, 8, 41. Astrocytic ATP release could be mediated by activation of metabotropic and ionotropic receptors44. It could also be modulated by anion channels, ATP-binding cassette proteins, gap junctions, P2×7 receptors and lysosome45–48. This paper provides evidence that ATP release from astrocytes is regulated by agrin signaling, a pathway critical for NMJ formation and maintenance14, 15, 19–21. Intriguingly, MuSK expression in astrocytes is more than 5 folds higher than in skeletal muscles (Fig. 6c). This paper thus reveals a novel function of the agrin signaling in the brain. How agrin regulates ATP release is unclear. Previous studies suggest that astrocytic ATP release involves SNARE-dependent exocytotic pathways10. However, Lrp4 did not appear to associate with SNARE proteins in co-precipitation experiments (data not shown). Agrin signaling is likely to regulate ATP release via a unique mechanism. The possible involvement of MuSK provides new leads for future investigation.
The spine deficits in GFAP-Lrp4−/− mice (Supplementary Fig. 4a, b) and Lrp4mitt (muscle-rescued) mice23 suggest a role of Lrp4 in spine development. Astrocytes have been implicated in synaptogenesis3. In light of Lrp4 astrocytic enrichment in brain regions in particular in the hippocampus (Fig. 3), the spine deficits may result from loss of secretable factors from astrocytes such as thrombospondins (TSPs)49, during development. Lrp4 regulation of spinogenesis may be age-dependent because of no apparent spine deficits by astrocytic ablation of Lrp4 in young adult (i.e., tamoxifen-treated GFAP-CreER;Lrp4−/−) mice (Supplementary Fig.5d, e). We noticed that PPR was found not changed in the hippocampus of muscle-rescued Lrp4mitt mice23. Lrp4mitt mice, however, could express a truncated, N-terminal fragment of Lrp4 containing eight LDLa repeats (and thus the mutant mice were called Lrp4mitt)18. Lrp4 mutant mice of this study should not express Lrp4 or a fragment of it in neurons or astrocytes. The difference between Gomez et al.’s results and ours may suggest a role of the eight LDLa repeats in regulating neurotransmission or synapse formation.
In conclusion, our results support a working model where Lrp4 in astrocytes, presumably in response to agrin, facilitates glutamatergic transmission by controlling ATP release (Supplementary Fig. 11). Disruption of this pathway impairs neural activity and synaptic plasticity (such as LTP) and behaviors. Our results revealed a critical role of astrocytic Lrp4 in regulating synaptic transmission and uncover a previously unappreciated pathway to regulate ATP release from astrocytes. Because agrin expression in neurons is hundreds time more in neurons than in astrocytes and Lrp4 is enriched in astrocytes, future studies are warranted to determine whether other functions of astrocytes are regulated by the agrin-Lrp4 signaling. Such studies may provide insight into the interaction between neurons and asytrocytes for synaptic homeostasis and/or plasticity and pathological mechanisms of neurological and psychiatric disorders.
Methods
Reagents and antibodies
Chemicals were purchased from Sigma-Aldrich unless otherwise indicated. SCH58261 (2270), DL-AP5 (0105), CNQX (0190) and BMI (0130) were purchased from Tocris Bioscience. Recombinant rat agrin protein (#550-AG-100) were purchased from R&D system. Information of antibodies was as follows: mouse anti-Lrp4 (ECD) (UC Davis/NIH NeuroMab Facility) (75–221; 1:1,000 for blotting); mouse anti-NeuN (EMD Millipore) (MAB377; 1:500 for staining); mouse anti-GFAP (EMD Millipore) (AB5804; 1;1000 for blotting; 1:500 for staining); rabbit anti-β-actin (EMD Millipore) (04–1116; 1:4000 for blotting); mouse anti-PSD-95 (Synaptic Systems) (124011; 1:1000 for blotting); mouse anti-gephyrin (Synatpic System) (147011; 1:1000 for blotting); rabbit anti-GluR1 (EMD Millipore) (04–885; 1:500 for blotting); mouse anti-GluR2 (EMD Millipore) (MABN71; 1:500 for blotting); rabbit anti- Synaptophysin (Abcam) (ab32127; 1:5000 for blotting); rabbit anti-S100β(Dako) (Z031129-2; 1:500 for staining); rabbit anti-BLBP (Abcam) (ab32423; 1:500 for staining); chicken anti-β-gal (Abcam) (ab9361; 1:1000 for staining); mouse anti-Phospho-Tyrosine (Cell Signaling Technology) (9411; 1:2000 for blotting); mouse anti-GluN1 (Synaptic Systems) (114011; 1:1000 for blotting); rabbit anti-GluN2A (R&D Systems) (PPS012; 1:1000 for blotting); mouse anti-GluN2B (My BioSource) (MBS801315; 1:2000 for blotting); mouse anti-Transferrin Receptor (TFR) (Invitrogen) (13–6890; 1:2000 for blotting). Anti-MuSK antibody was described previously1 (1:1000 for western blotting).
Animals
Used for experiments were male mice and embryos or new born pups of either sex. Mouse strains were described previously: Lrp4f/f1; GFAP::Cre2, Camk2a::Cre3, GFAP::CreER4, tdTomato (Jackson Labs, #007909). Genotyping procedures were as follows: Lrp4f/f, 5’-CTCTC CCAGC TAAGT CCAGG A-3’ and 5’-CCTCC ATACT GTCTG TGAAT G-3’; GFAP::Cre, 5’-ACT CCT TCA TAA AGC CCT-3’ and 5’-GCC AGC TAC GTT GCT CAC TA-3’; Camk2a::Cre, 5’-AGA TGT TCG CGA TTC TC-3’ and 5’-AGC TAC ACC AGA GAC GG-3’; GFAP::CreER, 5’-GGT CGA TGC AAC GAG TGA TGA GG-3’ and 5’-GCT AAG TGC CTT CTC TAC ACC TGC G-3’. To induce Cre activity in GFAP::CreER;tdTomato and GFAP::CreER;Lrp4−/− mice, tamoxifen (in 1:10 EtOH:sunflower oil, 100 mg/kg) was administered by gavage to mice at postnatal day 30. Unless otherwise indicated, GFAP::CreER;Lrp4−/− mice refer to those that are treated with tamoxifen. Lrp4-LacZ reporter mice were from KOMP (VG15248), whose primer sequences are: 5’-GGT AAA CTG GCT CGG ATT AGG G-3’ and 5’-TTG ACT GTA GCG GCT GAT GTT G-3’. In all studies, at least three pairs of mice from same litters were used. Significant efforts are also made to minimize the total number of animals used while maintaining statistically valid group numbers. Mice for experiments were group-housed no more than 5 per cage in a room with a 12 hr light/dark cycle with ad libitum access to water and rodent chow diet (Diet 1/4’’ 7097, Harlan Teklad). Experiments with animals were approved by Institutional Animal Care and Use Committee of the Georgia Regents University.
Cell culture
C2C12 cells were cultured as described previously5, 6. Hippocampal and cortical neurons were cultured as described previously7. Briefly, rodent hippocampi or cortices were dissected from E18 animals separately and digested in 0.25% trypsin at 37°C for 30 min. Dissociated cells were resuspended in plating media (DMEM/F-12 50:50 supplemented with N2 and 10% FBS) and plated onto poly-L-lysine–coated 8-mm cover slips in 12 well-plates for 4 h before replacing the medium with maintenance medium (neural basal medium supplemented with B27) and cytosine arabinoside (10 µM) to inhibit astrocyte proliferation. 50% of the medium was changed every 4–5 days. In some experiments, cultured neurons were transfected with control or Lrp4 miRNA on DIV 8 and subjected to recording from DIV11.
Astrocytes cultures were prepared as described previously8. Briefly, hippocampi were dissected from P2–3 mice and digested in 0.25% trypsin at 37°C for 30 min. Dissociated cells were resuspended in plating media (DMEM and 10% FBS) and plated into a culture flask for 7–9 d. The flask was shaken at 250 rpm for 2 h to remove microglial cells and shaken for overnight to remove oligodendrocytes. Astrocytes were then used for ATP test and co-culture.
For co-culture experiment, one cover slip of DIV 10 pure neurons and three cover slips of astrocytes were placed in one 35-mm dish and incubated in the maintenance medium without cytosine arabinoside. One day later, neurons were subjected to electrophysiological recording.
Western blot
Homogenates of brain tissues and cultured neurons and astrocytes were prepared in RIPA Buffer containing (in mM): 50 Tris-HCl, pH 7.4, 150 NaCl, 2 EDTA, 1 PMSF, 50 sodium fluoride, 1 sodium vanadate, 1 DTT with 1% sodium deoxycholate, 1% SDS and 1% protease inhibitors cocktails. Samples were resolved on SDS/PAGE and transferred to nitrocellulose membranes, which were incubated in the TBS buffer containing 0.1% Tween-20 and 5% milk for 1 h at room temperature before the addition of primary antibody for incubation overnight at 4 °C. After wash, the membranes were incubated with HRP-conjugated secondary antibody (Life Technology) in the same TBS buffer for 1 h at room temperature. Immunoreactive bands were visualized using enhanced chemiluminescence (Pierce). Films were scanned with an Epson 1680 scanner and analyzed with Image J (NIH). Band density of interested proteins was normalized in relation to loading control.
Cell surface biotinylation
Cultured astrocytes were incubated with Sulfo-NHS-biotin (ThermoFisher Scientific) in PBS for 45 min at 4°C, and treated with 10 mM glycine to quench all unreacted biotin. After rinse with PBS, cells were lysed in RIPA buffer. Biotinylated proteins were precipitated overnight with Avidin Agarose Beads (Pierce).
Electrophysiological recording
Hippocampal slices were prepared as described previously9. Mice (5–7 weeks-old, male) were anesthetized with ketamine/xylazine (Sigma, 100/20 mg/kg, respectively, ip), brains were quickly removed and chilled in ice-cold modified artificial cerebrospinal fluid (ACSF) containing (in mM): 250 glycerol, 2 KCl, 10 MgSO4, 0.2 CaCl2, 1.3 NaH2PO4, 26 NaHCO3, and 10 glucose. Coronal hippocampal slices (300 µm) were cut in ice-cold modified ACSF using a VT-1000S vibratome (Leica, Germany) and transferred to a storage chamber containing regular ACSF (in mM) (126 NaCl, 3 KCl, 1 MgSO4, 2 CaCl2, 1.25 NaH2PO4, 26 NaHCO3, and 10 glucose) at 34°C for 30 min and at room temperature (25±1 °C) for additional 1 h before recording. All solutions were saturated with 95% O2 / 5% CO2 (vol/vol).
Slices were placed in the recording chamber, which was superfused (2 ml/min) with ACSF at 32–34 °C. Whole-cell patch-clamp recording from CA1 pyramidal neurons were visualized with infrared optics using an upright microscope equipped with a 40× water-immersion lens (Axioskop 2 Plus, Zeiss) and infrared-sensitive CCD camera (C2400-75, Hamamatsu). The Pipettes were pulled by a micropipette puller (P-97, Sutter instrument) with a resistance of 3–5 MΩ. Recordings were made with MultiClamp 700B amplifier and 1440A digitizer (Molecular Device).
For sEPSC recording, pyramidal neurons were held at −70 mV in the presence of 20 µM BMI, with the pipette solution containing (in mM): 125 Cs-methanesulfonate, 5 CsCl, 10 Hepes, 0.2 EGTA, 1 MgCl2, 4 Mg-ATP, 0.3 Na-GTP, 10 phosphocreatine and 5 QX314 (pH 7.40, 285 mOsm). In co-culture experiments, neurons on the Cover slips were recorded for sEPSCs with bath solution contained (in mM): 140 NaCl, 5 KCl, 1 MgSO4, 2 CaCl2, 1.25 NaH2PO4, 26 NaHCO3, and 10 glucose (pH 7.4, 290 mOsm), in the presence of co-culture medium (20% of total solution). mEPSCs were recorded in the presence of 1 µm TTX.
To detect electric property of CA1 excitatory neurons, they were current-clamped and measured by injecting a series of depolarizing pulses (from 50 to 140 pA at a step of 10 pA) in the presence of 20 µM CNQX, 100 µM DL-AP5 and 20 µM BMI, with the pipette solution containing (in mM): 125 K-gluconate, 5 KCl, 10 Hepes, 0.2 EGTA, 1 MgCl2, 4 Mg-ATP, 0.3 Na-GTP and 10 phosphocreatine (pH 7.40, 285 mOsm). The spike threshold was calculated in response to 50 pA-current injection. Membrane input resistance was calculated in response to a series of hyperpolarizing pulses.
To investigate property of AMPAR and NMDAR currents, EPSCs were evoked by stimulating Schaffer Collaterals (SC)-CA1 pathway at holding potentials ranging from −60 to +60 mV at a step of 20 mV, in the presence of 20 µM BMI. AMPAR and NMDAR current were measured as the peak amplitudes and 50 ms after the peak amplitude, respectively.
For paired-pulse ratios recording, EPSCs were evoked by stimulating SC-CA1 pathway at holding potential of -70 mV in the presence of 20 µM BMI. Interval of paired stimulations was set 25, 50 and 100 ms respectively. Value of ratios was defined as [(p2 / p1] × 100, where p1 and p2 are the amplitude of the EPSCs evoked by the first and second pulse, respectively.
For MK-801 block assay, pyramidal neurons in the CA1 of hippocampus were clamped at +40 mV. Evoked NMDAR-mediated EPSCs were recorded in response to 0.1 Hz presynaptic stimulation in the presence of 20 µM CNQX and 20 µM BMI. Slices were treated with 40 µM MK801 for 5 min prior to evoking and recording EPSCs for 20 min. EPSC amplitudes were normalized to the first EPSC, and fitted with single-exponential functions to calculate decay constants (τ, in number of stimuli).
For minimal stimulation recording, CA1 pyramidal neurons were clamped at -70 mV. EPSCs were evoked by stimulating the SC-CA1 pathway (0.067 Hz) with ACSF-filled glass pipettes. Stimulus intensity was adjusted to fulfill the following four criteria: 1) all or none synaptic events were generated; 2) little or no variation in EPSC latency; 3) no change in mean size or shape of EPSCs by a small change in stimulus intensity, and 4) complete failure to evoke EPSCs by 10–20% reduction in stimulus intensity10. Responses that did not meet these criteria were rejected.
LTP was recorded as described previously9. Briefly, SC-CA1 pathway was stimulated with a concentric bipolar electrode (FHC), and field EPSPs were recorded in CA1 neurons in current-clamp with ACSF-filled glass pipettes (1–5 MΩ). Stimuli consisted of monophasic 100-µs pulses of constant currents (with intensity adjusted to produce ~30% of maximal amplitudes) at a frequency of 0.033 Hz. The strength of synaptic transmission was determined by measuring the initial (10–60% rising phase) slope of fEPSPs. LTP was induced by a train of 100 pulses at same intensity in 1 sec. The level of LTP was determined at an average of 50–60 min after tetanus stimulation.
In some experiments, after 10 min baseline recording, DL-AP5, ATP, ARL67156, AOPCP, suramin, SCH58261 or DPCPX was bath applied for 30–40 min. In all experiments, series resistance was controlled below 20 MΩ and not compensated. Cells would be rejected if membrane potentials were more positive than −60 mV; or if series resistance fluctuated more than 20% of initial values. All recordings were done at 32–34 °C. Data were filtered at 1 kHz and sampled at 10 kHz.
Brain morphological analysis
Anesthetized mice were perfused transcardially with 4% PFA in PBS and tissues were fixed in 4% PFA at 4 °C for 8 h. After dehydration by 30% sucrose, brain blocks were frozen and cut into 30-µm-thick sections on cryostat (HM550; Thermo Scientific). Sections were permeabilized with 0.3% Triton X-100 and 5% BSA in PBS and incubated with primary antibodies at 4 °C overnight. After washing with PBS for 3 times, samples were incubated with Alexa Fluor-conjugated secondary antibodies (1:1,000, Invitrogen) for 1 h at room temperature. Samples were mounted with Vectashield mounting medium (Vector lab) and images were taken by Zeiss LSM510 confocal microscope.
For in situ X-gal assay, brains were quickly isolated and embedded in OCT (Tissue-Tek). Coronal sections were cut at 20 µm interval and every fourth section was collected and mounted on slides. Sections were fixed for 2 min in PBS containing (in mM):, 2 MgCl2, 5 EGTA with 0.2% glutaraldehyde. Sections were washed in ice-cold PBS and stained in X-gal solution [1 mg/ml X-gal, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 0.02% NP-40, 0.01% deoxycholate, and 2 mM MgCl2 in PBS] at 37°C overnight. After washing with PBS, they were counterstained with nuclear Fast Red (Vector Labs, H-3403).
Golgi staining
Golgi staining was performed by using a kit (FD NeuroTechnologies). Images were randomly taken from dorsal hippocampus. Spines were counted on primary, secondary and tertiary branches of apical dendrites in the stratum radiatum of CA1 hippocampal region by using imageJ. To measure dendritic morphology, CA1 pyramidal neurons were traced with Reconstruct (SynapseWeb). Neurons with clear dendritic branches without breaks and staining artefacts/precipitates were subjected to Sholl Analysis. The investigator who performed spine analysis was blinded to genotypes.
Electron Microscopy Analysis
Electron microscopy studies were carried as described previously11. Briefly, anesthetized mice were perfused transcardially with 0.1 M phosphate buffer containing 2% glutaraldehyde and 2% paraformaldehyde. Brains were post-fixed for 1 hr at 25°C and 4°C overnight. Ultrathin sections of the CA1 region were examined with a JEM 1230 transmission electron microscope (JEOL USA) at 110 kV. Images were collected with an UltraScan 4000 CCD camera and First Light Digital Camera Controller (Gatan). Synapses were identified by ultrastructural specializations, including alignment of presynaptic and postsynaptic membranes, presynaptic and postsynaptic thickenings, and clusters of synaptic vesicles. The length of postsynaptic density (PSD), density of synaptic vesicles within 0.04 µm2 areas surrounding active zones or adjacent to terminals, synaptic vesicles diameter and distance between astrocytes and synaptic membrane were quantified by investigators unaware of genotypes using ImageJ.
qRT-PCR Analysis
qRT-PCR was performed as described previously1. Briefly, total RNA was isolated by using TRIzol reagent (15596-026, Invitrogen). After DNAseI (18068-015, Invitrogen) digestion, RNA (1 µg) was reverse-transcribed with oligo dT-primers using Maxima reverse transcriptase (EP 0742, Fermentas). 5% of resulting cDNA was used for Q-PCR by using SYBR Green detection (K 0222, Fermentas). Samples were assayed in triplicates, with each plate having loading standards in duplicate. mRNA levels of agrin and MuSK were normalized to those of GAPDH. Primer sequences were: agrin, 5’- CAC TGC GAG AAG GGG ATA GTT G-3’ and 5’-GGC TGG GAT CTC ATT GGT CAG -3’; Musk, 5’- ACC GTC ATC ATC TCC ATC GTG T-3’ and 5’- CTC AAT GTT ATT CCT CGG ATA CTC C-3’; GAPDH, 5’- GCA CAG TCA AGG CCG AGA AT-3’ and 5’- GCC TTC TCC ATG GTG GTG AA-3’.
In vivo microdialysis
Mice were anesthetized with ketamine/xylazine (Sigma, 100/20 mg/kg, i.p.), and implanted stereotaxically with guide cannula (CMA-7) into the right hippocampus (AP: −2.0 mm, ML: +1.5 mm, DV: −1.0 mm). After surgery, mice were individually housed for five days for recovery. On the sixth day, microdialysis was performed with a probe (CMAP000082, Harvard Apparatus) at the flowrate of 1.0 µl/min with ACSF containing (in Mm): 147 NaCl, 2.7 KCl, 1.2 CaCl2 and 0.85 MgCl2 (pH, 7.4). Samples were collected into a polypropylene tube in a refrigerated fraction collector (Harvard Apparatus) and stored at –80 °C before assay. After microdialysis, brain sections were examined to confirm probe placement in the hippocampus.
ATP and adenosine test
ATP is measured by a luciferase reaction where light (560 nm) was emitted when D-luciferin is converted to oxyluciferin using the ENLITEN ATP Assay System (FF2000, Promega)8, 12. To inhibit ATP hydrolysis, condition media or microdialysis samples were incubated with the ectonucleotidase inhibitor ARL 67156 (6-N, N-diethyl- β-γ-dibromo-methylene-d- adenosine- 5-triphosphate trisodium salt hydrate, FPL 67156). Luminescence was measured using a luminometer (PE Applied Biosystems, TR717). ATP in samples was calculated based on a calibration curve with standard ATP samples.
Adenosine is measured by using the Adenosine Assay Kit (K327-100, BioVision). To inhibit adenosine degradation, samples were incubated with adenosine deminase inhibitor EHNA hydrochloride (E114, Sigma). Fluorescence was measured using a luminometer (PE Applied Biosystems, TR717). Adenosine in samples was calculated based on a calibration curve from standard adenosine samples.
Behavioral analysis
Mice were handled by investigators for three days before any behavioral test. Locomotor activity was measured as described previously13. Mice were placed in a chamber (50 × 50 × 10 cm) and movement was monitored for 30 min using an overhead camera and tracking software (EthoVision, Noldus). The center 25 × 25 cm region was artificially defined as the center region and frequency and duration spent in the center region was recorded.
Elevated plus maze (EPM) test was performed as described previously14, in an EPM that has two opposing wall-closed arms and two open arms, each at 5 × 66 cm. The EPM was placed ~50 cm above the floor. Each mouse was placed in one of the closed arms and was recorded for 15 min using an overhead camera and tracking software (EthoVision, Noldus). The time mice spent in the open arms, the number of entries and the latency to first entry were quantified autonomously.
In the light/dark box test, mice were placed in the dark chamber (20 × 15 × 23 cm) that was connected with the light chamber (30 × 20 × 23 cm) through an open door (7 × 5 cm), and recorded for 10 min. The time mice spent in the light box, number of entries and latency to first entry to the light box were quantified by the computational software.
Morris water maze test was performed as described previously15. A 120 cm pool and 10 cm platform were utilized for water maze studies. The day before training, mice were placed in a pool and scored for ability to find the visible platform within 60 sec. Mice that failed to locate and climb onto the platform twice were eliminated from further testing. The platform was then moved to a new location and submerged 1 cm beneath surface of colored water. Mice were trained for 5 days with 4 trials per day and 60 sec per trial to locate the hidden platform. Seven start positions were used to ensure that visual spatial memory was used by mice to find the hidden platform. On the 6th day the platform was removed and mice were placed into the pool at a new start position and scored for time spent in platform area (N30: 30 cm as diameter) and number of platform crossings within 60 sec.
For the pilocarpine model16, mice were injected with scopolamine (2 mg kg−1, i.p.) 30 min before pilocarpine treatment to block peripheral side effects. Mice were then injected with pilocarpine in 0.9% saline (wt/vol, i.p., 200 mg kg−1), followed by injections every 30 min at a dosage of 100 mg kg−1. Behavioral seizures were scored based on the criteria by Racine17: stage 0, no seizure; stage 1, head nodding; stage 2, sporadic full-body shaking, spasms; stage 3, chronic full-body spasms; stage 4, jumping, shrieking, falling over; stage 5, violent convulsions, falling over, death. Some mice were injected with PTZ (i.p., 50 mg kg−1) as previously described18 and observed for the latency till the onset of generalized convulsive seizures (GS). If GS was not observed in 20 min, 20 min were scored.
In some experiments, mice were injected with DPCPX (1mg/kg, i.p) 30 min before behavioral tests. Genotypes of mice in all behavior tests were blinded to investigators.
Except Golgi staining, EM and behavioral analysis described above, data collection and analysis were not performed blind to the conditions of the experiments.
Statistical analysis
Statistical analysis was done by the GraphPad Prism version 5.0 (GraphPad Software). Sample size choice was based on previous studies1, 14, 19, not predetermined by a statistical method. No randomization method was used. Data distribution was assumed to be normal, but this was not formally tested. Two-way ANOVA was used in morphological and electrophysiological studies that analyze more than two parameters. One-way ANOVA was used for analysis of data from three or more groups. Student’s t test or paired Student’s t test was used to compare data from two groups. All tests were two-sided. P < 0.05 was considered to be statistically significant. A supplementary methods c0068ecklist is available.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We are grateful to Dr. Woo-Ping Ge (UT Southwestern Medical Center) and Dr. Ken D. McCarthy (University of North Carolina) for GFAP::CreER mice. We thank members of the Mei and Xiong laboratories for discussion. This work was supported in part by grants from NIH (L.M., W.-C.X.) and VA (L.M., W.-C.X.), “Thousand Talents” Innovation Project from Jiangxi Province (L.M.), National Natural Science foundation of China (NSFC, 81471116, B.-M.L), and NSFC (81329003; U1201225; 31430032, T.-M.G.), GSTP (201300000093, T.-M.G.) and SRFDPHEC (20134433130002, T.-M.G.).
Footnotes
Authors contributions
X.-D. Sun and L. Mei conceived of, designed and directed the project and wrote the manuscript; X.-D. Sun performed electrophysiological recordings and analysis; L. Li conducted immunoblots, quantitative reverse-transcription PCR (qRT-PCR) and co-immunoporecipitation; F. Liu, A. Barik. and A. Sathyamurthy conducted Golgi staining, X-gal staining, immunofluorescence staining and analysis; Z.-H. Huang conducted immunoblots and astrocyte culture experiments and analysis; J. Bean performed behavioral tests and microdialysis analysis, with assistance of Y.-J. Chen and D.-M. Yin; H.-F. Jiao, S.-M. Kim and Y. Tian conducted cell culture experiments and analysis; H. Wu and C. Shen, provided and assisted with characterization of Lrp4 mutant mice; T.W. Lin conducted spine and synapse analysis; L. Xiong, H.-P. Lin, J.-X.Hu assisted with breeding and genotyping various Lrp4 mutant lines; B.-M. Li, T.-M. Gao and W.-C. Xiong helped with data interpretation and provided instruction; L. Mei supervised the project.
Competing financial interests
The authors declare no competing financial interests.
References for main text
- 1.Haydon PG. GLIA: listening and talking to the synapse. Nature reviews. Neuroscience. 2001;2:185–193. doi: 10.1038/35058528. [DOI] [PubMed] [Google Scholar]
- 2.Perea G, Navarrete M, Araque A. Tripartite synapses: astrocytes process and control synaptic information. Trends in neurosciences. 2009;32:421–431. doi: 10.1016/j.tins.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Clarke LE, Barres BA. Emerging roles of astrocytes in neural circuit development. Nature reviews. Neuroscience. 2013;14:311–321. doi: 10.1038/nrn3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halassa MM, Haydon PG. Integrated brain circuits: astrocytic networks modulate neuronal activity and behavior. Annual review of physiology. 2010;72:335–355. doi: 10.1146/annurev-physiol-021909-135843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azevedo FA, et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. The Journal of comparative neurology. 2009;513:532–541. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- 6.Witcher MR, Kirov SA, Harris KM. Plasticity of perisynaptic astroglia during synaptogenesis in the mature rat hippocampus. Glia. 2007;55:13–23. doi: 10.1002/glia.20415. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton NB, Attwell D. Do astrocytes really exocytose neurotransmitters? Nature reviews. Neuroscience. 2010;11:227–238. doi: 10.1038/nrn2803. [DOI] [PubMed] [Google Scholar]
- 8.Araque A, et al. Gliotransmitters travel in time and space. Neuron. 2014;81:728–739. doi: 10.1016/j.neuron.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang JM, et al. ATP released by astrocytes mediates glutamatergic activity-dependent heterosynaptic suppression. Neuron. 2003;40:971–982. doi: 10.1016/s0896-6273(03)00717-7. [DOI] [PubMed] [Google Scholar]
- 10.Pascual O, et al. Astrocytic purinergic signaling coordinates synaptic networks. Science (New York, N.Y.) 2005;310:113–116. doi: 10.1126/science.1116916. [DOI] [PubMed] [Google Scholar]
- 11.Herz J, Strickland DK. LRP: a multifunctional scavenger and signaling receptor. The Journal of clinical investigation. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen C, Xiong WC, Mei L. LRP4 in neuromuscular junction and bone development and diseases. Bone. 2015 doi: 10.1016/j.bone.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Gautam M, et al. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell. 1996;85:525–535. doi: 10.1016/s0092-8674(00)81253-2. [DOI] [PubMed] [Google Scholar]
- 14.Kim N, et al. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell. 2008;135:334–342. doi: 10.1016/j.cell.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B, et al. LRP4 serves as a coreceptor of agrin. Neuron. 2008;60:285–297. doi: 10.1016/j.neuron.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zong Y, et al. Structural basis of agrin-LRP4-MuSK signaling. Genes & development. 2012;26:247–258. doi: 10.1101/gad.180885.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeChiara TM, et al. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell. 1996;85:501–512. doi: 10.1016/s0092-8674(00)81251-9. [DOI] [PubMed] [Google Scholar]
- 18.Weatherbee SD, Anderson KV, Niswander LA. LDL-receptor-related protein 4 is crucial for formation of the neuromuscular junction. Development (Cambridge, England) 2006;133:4993–5000. doi: 10.1242/dev.02696. [DOI] [PubMed] [Google Scholar]
- 19.Barik A, Lu Y, Sathyamurthy A. LRP4 Is Critical for Neuromuscular Junction Maintenance. 2014;34:13892–13905. doi: 10.1523/JNEUROSCI.1733-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yumoto N, Kim N, Burden SJ. Lrp4 is a retrograde signal for presynaptic differentiation at neuromuscular synapses. Nature. 2012;489:438–442. doi: 10.1038/nature11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, et al. Distinct roles of muscle and motoneuron LRP4 in neuromuscular junction formation. Neuron. 2012;75:94–107. doi: 10.1016/j.neuron.2012.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian QB, et al. Interaction of LDL receptor-related protein 4 (LRP4) with postsynaptic scaffold proteins via its C-terminal PDZ domain-binding motif, and its regulation by Ca/calmodulin-dependent protein kinase II. The European journal of neuroscience. 2006;23:2864–2876. doi: 10.1111/j.1460-9568.2006.04846.x. [DOI] [PubMed] [Google Scholar]
- 23.Gomez AM, Froemke RC, Burden SJ. Synaptic plasticity and cognitive function are disrupted in the absence of Lrp4. eLife. 2014;3:e04287. doi: 10.7554/eLife.04287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pohlkamp T, et al. Lrp4 domains differentially regulate limb/brain development and synaptic plasticity. PloS one. 2015;10:e0116701. doi: 10.1371/journal.pone.0116701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuo L, et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis (New York, N.Y. : 2000) 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- 26.Zucker RS, Regehr WG. Short-term synaptic plasticity. Annual review of physiology. 2002;64:355–405. doi: 10.1146/annurev.physiol.64.092501.114547. [DOI] [PubMed] [Google Scholar]
- 27.Rosenmund C, Clements JD, Westbrook GL. Nonuniform probability of glutamate release at a hippocampal synapse. Science (New York, N.Y.) 1993;262:754–757. doi: 10.1126/science.7901909. [DOI] [PubMed] [Google Scholar]
- 28.Hessler NA, Shirke AM, Malinow R. The probability of transmitter release at a mammalian central synapse. Nature. 1993;366:569–572. doi: 10.1038/366569a0. [DOI] [PubMed] [Google Scholar]
- 29.Stevens CF, Wang Y. Facilitation and depression at single central synapses. Neuron. 1995;14:795–802. doi: 10.1016/0896-6273(95)90223-6. [DOI] [PubMed] [Google Scholar]
- 30.Casper KB, Jones K, McCarthy KD. Characterization of astrocyte-specific conditional knockouts. Genesis (New York, N.Y. : 2000) 2007;45:292–299. doi: 10.1002/dvg.20287. [DOI] [PubMed] [Google Scholar]
- 31.Ge WP, Miyawaki A, Gage FH, Jan YN, Jan LY. Local generation of glia is a major astrocyte source in postnatal cortex. Nature. 2012;484:376–380. doi: 10.1038/nature10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panatier A, et al. Glia-derived D-serine controls NMDA receptor activity and synaptic memory. Cell. 2006;125:775–784. doi: 10.1016/j.cell.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, et al. Contribution of astrocytes to hippocampal long-term potentiation through release of D-serine. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15194–15199. doi: 10.1073/pnas.2431073100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu S, Xiong W, Parkinson FE. Effect of ecto-5'-nucleotidase (eN) in astrocytes on adenosine and inosine formation. Purinergic signalling. 2014;10:603–609. doi: 10.1007/s11302-014-9421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bezakova G, Ruegg MA. New insights into the roles of agrin. Nature reviews. Molecular cell biology. 2003;4:295–308. doi: 10.1038/nrm1074. [DOI] [PubMed] [Google Scholar]
- 36.Ksiazek I, et al. Synapse loss in cortex of agrin-deficient mice after genetic rescue of perinatal death. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:7183–7195. doi: 10.1523/JNEUROSCI.1609-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Osta A, et al. MuSK expressed in the brain mediates cholinergic responses, synaptic plasticity, and memory formation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:7919–7932. doi: 10.1523/JNEUROSCI.1674-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris RG, Garrud P, Rawlins JN, O'Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- 39.Curia G, Longo D, Biagini G, Jones RS, Avoli M. The pilocarpine model of temporal lobe epilepsy. Journal of neuroscience methods. 2008;172:143–157. doi: 10.1016/j.jneumeth.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki T, et al. Efhc1 deficiency causes spontaneous myoclonus and increased seizure susceptibility. Human molecular genetics. 2009;18:1099–1109. doi: 10.1093/hmg/ddp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hines DJ, Haydon PG. Astrocytic adenosine: from synapses to psychiatric disorders. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2014;369 doi: 10.1098/rstb.2013.0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jourdain P, et al. Glutamate exocytosis from astrocytes controls synaptic strength. Nature neuroscience. 2007;10:331–339. doi: 10.1038/nn1849. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Krupa B, Kang JS, Bolshakov VY, Liu G. Glycine site of NMDA receptor serves as a spatiotemporal detector of synaptic activity patterns. Journal of neurophysiology. 2009;102:578–589. doi: 10.1152/jn.91342.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coco S, et al. Storage and release of ATP from astrocytes in culture. J Biol Chem. 2003;278:1354–1362. doi: 10.1074/jbc.M209454200. [DOI] [PubMed] [Google Scholar]
- 45.Stout CE, Costantin JL, Naus CC, Charles AC. Intercellular calcium signaling in astrocytes via ATP release through connexin hemichannels. J Biol Chem. 2002;277:10482–10488. doi: 10.1074/jbc.M109902200. [DOI] [PubMed] [Google Scholar]
- 46.Suadicani SO, Brosnan CF, Scemes E. P2×7 receptors mediate ATP release and amplification of astrocytic intercellular Ca2+ signaling. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:1378–1385. doi: 10.1523/JNEUROSCI.3902-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roman RM, et al. Hepatocellular ATP-binding cassette protein expression enhances ATP release and autocrine regulation of cell volume. J Biol Chem. 1997;272:21970–21976. doi: 10.1074/jbc.272.35.21970. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z, et al. Regulated ATP release from astrocytes through lysosome exocytosis. Nature cell biology. 2007;9:945–953. doi: 10.1038/ncb1620. [DOI] [PubMed] [Google Scholar]
- 49.Christopherson KS, et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
Methods-only references
- 1.Wu H, et al. Distinct roles of muscle and motoneuron LRP4 in neuromuscular junction formation. Neuron. 2012;75:94–107. doi: 10.1016/j.neuron.2012.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhuo L, et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis (New York, N.Y. : 2000) 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- 3.Tsien JZ, et al. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- 4.Casper KB, Jones K, McCarthy KD. Characterization of astrocyte-specific conditional knockouts. Genesis (New York, N.Y. : 2000) 2007;45:292–299. doi: 10.1002/dvg.20287. [DOI] [PubMed] [Google Scholar]
- 5.Shen C, et al. Antibodies against low-density lipoprotein receptor-related protein 4 induce myasthenia gravis. The Journal of clinical investigation. 2013;123:5190–5202. doi: 10.1172/JCI66039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, et al. LRP4 serves as a coreceptor of agrin. Neuron. 2008;60:285–297. doi: 10.1016/j.neuron.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ting AK, et al. Neuregulin 1 promotes excitatory synapse development and function in GABAergic interneurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:15–25. doi: 10.1523/JNEUROSCI.2538-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao X, et al. Astrocyte-derived ATP modulates depressive-like behaviors. Nature medicine. 2013;19:773–777. doi: 10.1038/nm.3162. [DOI] [PubMed] [Google Scholar]
- 9.Chen YJ, et al. ErbB4 in parvalbumin-positive interneurons is critical for neuregulin 1 regulation of long-term potentiation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21818–21823. doi: 10.1073/pnas.1010669107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil Z, Connors BW, Amitai Y. Efficacy of thalamocortical and intracortical synaptic connections: quanta, innervation, and reliability. Neuron. 1999;23:385–397. doi: 10.1016/s0896-6273(00)80788-6. [DOI] [PubMed] [Google Scholar]
- 11.Yin DM, et al. Regulation of spine formation by ErbB4 in PV-positive interneurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:19295–19303. doi: 10.1523/JNEUROSCI.2090-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang JM, et al. ATP released by astrocytes mediates glutamatergic activity-dependent heterosynaptic suppression. Neuron. 2003;40:971–982. doi: 10.1016/s0896-6273(03)00717-7. [DOI] [PubMed] [Google Scholar]
- 13.Wen L, et al. Neuregulin 1 regulates pyramidal neuron activity via ErbB4 in parvalbumin-positive interneurons. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1211–1216. doi: 10.1073/pnas.0910302107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi LL, et al. Amygdala NRG1-ErbB4 is critical for the modulation of anxiety-like behaviors. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2015;40:974–986. doi: 10.1038/npp.2014.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin DM, et al. Reversal of behavioral deficits and synaptic dysfunction in mice overexpressing neuregulin 1. Neuron. 2013;78:644–657. doi: 10.1016/j.neuron.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curia G, Longo D, Biagini G, Jones RS, Avoli M. The pilocarpine model of temporal lobe epilepsy. Journal of neuroscience methods. 2008;172:143–157. doi: 10.1016/j.jneumeth.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Racine RJ. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalography and clinical neurophysiology. 1972;32:281–294. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki T, et al. Efhc1 deficiency causes spontaneous myoclonus and increased seizure susceptibility. Human molecular genetics. 2009;18:1099–1109. doi: 10.1093/hmg/ddp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y, et al. Maintenance of GABAergic activity by neuregulin 1-ErbB4 in amygdala for fear memory. Neuron. 2014;84:835–846. doi: 10.1016/j.neuron.2014.09.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.