Abstract
Here we report the development of two rapid real-time quantitative PCR assays with TaqMan® probes to detect the MCR-1 plasmid-mediated colistin resistance gene from bacterial isolates and faecal samples from chickens. Specificity and sensitivity of the assay were 100% on bacterial isolates including 18 colistin-resistant isolates carrying the mcr-1 gene (six Klebsiella pneumoniae and 12 Escherichia coli) with a calibration curve that was linear from 101 to 108 DNA copies. Five out of 833 faecal samples from chickens from Algeria were positive, from which three E. coli strains were isolated and confirmed to harbour the mcr-1 gene by standard PCR and sequencing.
Keywords: Antibiotic resistance surveillance; colistin resistance; mcr-1 gene; PCR detection, Taqman probe
Introduction
The increasing prevalence of infections caused by multidrug-resistant Gram-negative bacteria combined with few antimicrobial agents being in development has led to a resurgence in interest in colistin as a last-line therapy with the inevitable risk of emerging resistance [1], [2], [3]. MCR-1 plasmid-mediated colistin resistance is a member of the phosphoethanolamine transferase enzyme family, with expression in Escherichia coli resulting in the addition of phosphoethanolamine to lipid A and resistance to colistin [4]. This plasmid-mediated colistin resistance is an emerging concern that has already spread worldwide [5] in E. coli and Klebsiella pneumoniae from pigs, chicken, retail meat (pork, chicken), humans [4]. In animal health, colistin is used to prevent infections from E. coli isolates that are known to cause serious adverse effects such as diarrhoea, sepsis and colibacillosis, which result in huge economic losses [6]. The extensive use of antibiotics in food-animal production has been shown to increase the risk of transferring resistant bacteria to humans [7].
There is a need to screen for colistin resistance even in patients without a history of colistin usage for the timely detection and isolation of patients harbouring such resistant strains to prevent clonal transmission [8]. For this reason the aim of this study was to develop rapid real-time quantitative PCR (qPCR) to detect the MCR-1 plasmid-mediated colistin resistance and to evaluate its sensitivity and specificity both from strains and stool samples.
Materials and methods
Specific primers and probes design
Primers and probes design. We designed specific primers and probes to develop two real-time qPCR assays (PE1 and PE2) for the detection of MCR-1-encoding gene (Table 1). Specificity of the primers and probes were verified in silico by blastN analysis on the National Center for Biotechnology Information (NCBI) database.
Table 1.
Primers and probe designed to target the plasmid-mediated colistin resistance (MCR-1)
| Primer/probe name | Sequence | PCR product size (bp) | References |
|---|---|---|---|
| Real-time PCR | |||
| PE_F1 | GCAGCATACTTCTGTGTGGTAC | 145 | This study |
| PE_R1 | ACAAAGCCGAGATTGTCCGCG | ||
| PE_Probe 1 | 6 FAM –GACCGCGACCGCCAATCTTACC-TAMRA | ||
| PE_F2 | GGGTGTGCTACCAAGTTTGCTT | ||
| PE_R3 | TATGCACGCGAAAGAAACTGGC | ||
| PE_Probe | 6 FAM –GCGCTGATTTTACTGCCTGTGGTG-TAMRA | ||
| Standard PCR | |||
| PE_F1 | GCAGCATACTTCTGTGTGGTAC | 554 | This study |
| PE_R3 | TATGCACGCGAAAGAAACTGGC | ||
| CLR5-F | 5′-CGGTCAGTCCGTTTGTTC-3′ | [4] | |
| CLR5-R | 5′-CTTGGTCGGTCTGTA GGG-3′ | ||
Sample collection
Bacterial strains. A total of 100 strains from humans and animals were used in this study including 18 colistin-resistant isolates carrying the mcr-1 gene (six K. pneumoniae and 12 E. coli). Phenotypic and genotypic features of these strains are summarized in Table 2.
Table 2.
Presentation of strains of the study with the genes specificity
| Species | Presence of gene MCR-1 | COL MIC (mg/L) | Genes specificity | CT value with PE1 system | CT value with PE2 system | Origins | References |
|---|---|---|---|---|---|---|---|
| Escherichia coli (n = 25) | + (n = 12) − (n = 13) |
4–16 <1–16 |
None | 18–25 0 |
19–25 0 |
Thailand, Laos, Algeria, France, Nigeria. |
[5], [7] unpublished data |
| Klebsiella pneumoniae (n = 33) | + (n = 6) − (n = 27) |
4–32 <1–32 |
mgrB* (n = 2) pmrB*(n = 1) mgrB* (12) |
18–24 0 |
19–25 0 |
Thailand, Laos, France, Nigeria, Algeria |
[9] unpublished data |
| Klebsiella oxytoca (n = 2) | – | 6–12 | mgrB* (n = 1) | 0 | 0 | [9] | |
| Salmonella enterica subsp. enterica (n = 5) | – | 0.125–16 | pmrB* (n = 2) blaCTX-M-2 (n = 5) |
0 | 0 | France | [10] |
| Pseudomonas aeruginosa (n = 10) | – | <1 | blaVIM-2 (n = 10) | 0 | 0 | Lebanon | [11] |
| Acinetobacter baumannii (n = 10) | – | <1 | blaOXA23 (n = 2), blaOXA24 (n = 2), blaOXA58 (n = 1), blaVEB (n = 1) | 0 | 0 | Algeria | [12], [13], [14] |
| Providencia rettgeri (n = 2) | – | >256 | blaNDM-1 (n = 1) | 0 | 0 | Israel | [15] |
| Morganella morganii (n = 2) | – | >256 | blaNDM-1 (n = 1) | 0 | 0 | Israel | [16] |
| Enterobacter cloacae (n = 5) | – | <1 | None | 0 | 0 | Laos, Nigeria. | unpublished data |
| Proteus mirabilis (n = 2) | – | >256 | None | 0 | 0 | Algeria | unpublished data |
| Proteus vulgaris (n = 2) | – | >256 | None | 0 | 0 | Algeria | unpublished data |
| Serratia marcescens (n = 2) | – | >256 | None | 0 | 0 | Algeria | unpublished data |
* Mutation; +, positive; −, negative.
COL MIC, minimum inhibitory concentration of colistin.
Chicken stool collection. A total of 833 faeces samples from broilers were collected between August and February 2015 from eight regions in Algeria (El Tarf, Souk Ahras, Skikda, Setif, Jijel, Algiers, Biskra and Ourgla; n = 503) and in three slaughterhouses in Marseille (n = 330). All the extracted DNA from the 833 faeces of broilers was tested using our qPCR assay and positive samples were inoculated on agar for isolation of positive mcr-1 isolates.
Molecular analysis
Strategy for PCR amplification and sequencing. Standard PCR amplification and sequencing of the MCR-1-encoding gene was used as the gold standard and performed as previously described [4]. Quantification of the MCR-1-encoding gene using our two sets of primers and probes was performed using a quantitative CFX96™ Real Time system C1000™ Touch thermal cycler (Bio-Rad, Singapore). The qPCR conditions were as follows: the reaction mixtures were kept at 95°C for 15 min and subsequently put through 35 cycles of 95°C for 30 s and 60°C for 1 min.
Specificity and reproducibility of the new system of real-time PCR. The specificity of the primers and probes were verified in vitro using our local collection of 100 strains (Table 2). The sensitivity of our assays was determined using ten-fold serial dilutions (between 108 and 101 DNA copies) of E. coli strain P10 by triplicate amplification, the number of mcr-1 in each sample was calculated based on the DNA copy numbers. The obtained Ct values were used to generate the calibration curves compared with the number of bacteria quantified by standard bacterial count on agar plates. The standard curve was constructed on the basis of the concordance between Ct values and number of log CFU/mL. The limit of detection was based on the final dilution detected by PCR. Efficacy of qPCR was calculated from a standard curve according to Rutledge and Cote [17].
Results
Specificity and technical sensitivity of the qPCR
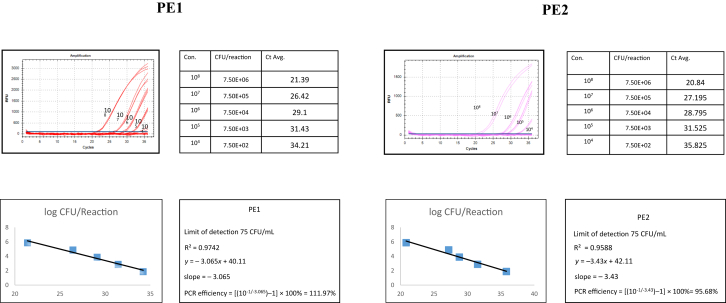
blastN analysis of the primers and probe designed for the development of the real-time PCR assay showed in silico a 100% homology with the MCR-1-encoding gene only. The sensitivity of the real-time PCR using the 18 mcr-1-positive strains using serial ten-fold dilutions of a calibrated inoculum was excellent with a calibration curve that was linear from 101–108 DNA copies corresponding to 35–21 Ct (Fig. 1). Regression formulae and PCR efficiency of the two real-time PCR assays are shown in Fig. 1. The reproducibility of the two qPCR assays was excellent, with a positive PCR at 21.4 ± 0.4 Ct for PE1 and 20.8 × 0.4 Ct for PE2 when testing one colony re-suspended in 200 μL of sterile water (Table 2). The specificity of the two qPCR assays in vitro against a panel of 82 clinically relevant bacteria negative for mcr-1 gene was 100% (all real-time PCR were negative, Table 2).
Fig. 1.
Real-time PCR sensitivity test to detect -encoding plasmid-mediated colistin resistance (MCR-1) encoding gene from Escherichia coli strain P10. PE1 and PE2 are two quantitative PCR assays developed.
Screening of faeces from broilers
Five of the 503 faecal samples from chickens from Algeria were positive, from which three E. coli strains were isolated and confirmed to harbour the mcr-1 gene by standard PCR and sequencing. None of the 330 samples from France were positive.
Discussion
The recent description and emergence of MCR-1 plasmid-mediated resistance to colistin in humans and animals is a major concern worldwide [5]. In this study, two new qPCR assays using Taqman probes were developed that demonstrate high sensitivity and specificity for confirmation of the presence of this gene in colistin-resistant bacterial isolates as well as for screening directly from stool samples. Indeed both systems have the same performance to screen for the presence of MCR-1-containing isolates and in stools. We recommend the use of PE1 as a first set of primers for the rapid screening of mcr-1 and PE2 system to confirm the positive results. Recently, Bontron et al. have reported a real-time PCR assay using SYBR green as fluorescent marker with similar sensitivity [18]. However it is well known that Taqman probes enhance specificity, which is a critical point when testing directly from biological samples. Our real-time PCR assays had advantages including sensitivity, specificity and the possibility of detecting MCR-1 plasmid-mediated colistin resistance very quickly (<2 h). We believe that these real-time PCR assays would be important and powerful tools that could be implemented easily in clinical microbiological laboratories that have molecular facilities, including at point of care, for identification of MCR-1 and implementation of healthcare policies.
Conflict of interest and financial disclosure
There is no potential conflict of interest or financial disclosure for all authors.
Acknowledgements
We are very grateful to IHU Mediterranean Infection.
References
- 1.Biswas S., Brunel J.M., Dubus J.C., Reynaud-Gaubert M., Rolain J.M. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther. 2012;10:917–934. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 2.Rolain J.M., Olaitan A.O. Plasmid-mediated colistin resistance: the final blow to colistin? Int J Antimicrob Agents. 2016;47:4–5. doi: 10.1016/j.ijantimicag.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Olaitan A.O., Morand S., Rolain J.M. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 5.Olaitan A.O., Chabou S., Okdah L., Morand S., Rolain J.M. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16:147. doi: 10.1016/S1473-3099(15)00540-X. [DOI] [PubMed] [Google Scholar]
- 6.Kempf I., Fleury M.A., Drider D., Bruneau M., Sanders P., Chauvin C. What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? Int J Antimicrob Agents. 2013;42:379–383. doi: 10.1016/j.ijantimicag.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Olaitan A.O., Thongmalayvong B., Akkhavong K., Somphavong S., Paboriboune P., Khounsy S. Clonal transmission of a colistin-resistant Escherichia coli from a domesticated pig to a human in Laos. J Antimicrob Chemother. 2015;70:3402–3404. doi: 10.1093/jac/dkv252. [DOI] [PubMed] [Google Scholar]
- 8.Olaitan A.O., Morand S., Rolain J.M. Emergence of colistin-resistant bacteria in humans without colistin usage: a new worry and cause for vigilance. Int J Antimicrob Agents. 2016;47:1–3. doi: 10.1016/j.ijantimicag.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Olaitan A.O., Diene S.M., Kempf M., Berrazeg M., Bakour S., Gupta S.K. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents. 2014;44:500–507. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 10.Olaitan A.O., Dia N.M., Gautret P., Benkouiten S., Belhouchat K., Drali T. Acquisition of extended-spectrum cephalosporin- and colistin-resistant Salmonella enterica subsp. enterica serotype Newport by pilgrims during Hajj. Int J Antimicrob Agents. 2015;45:600–604. doi: 10.1016/j.ijantimicag.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Al Bayssari C., Dabboussi F., Hamze M., Rolain J.M. Emergence of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii in livestock animals in Lebanon. J Antimicrob Chemother. 2015;70:950–951. doi: 10.1093/jac/dku469. [DOI] [PubMed] [Google Scholar]
- 12.Bakour S., Kempf M., Touati A., Ait A.A., Haouchine D., Sahli F. Carbapenemase-producing Acinetobacter baumannii in two university hospitals in Algeria. J Med Microbiol. 2012;61:1341–1343. doi: 10.1099/jmm.0.045807-0. [DOI] [PubMed] [Google Scholar]
- 13.Bakour S., Touati A., Bachiri T., Sahli F., Tiouit D., Naim M. First report of 16S rRNA methylase ArmA-producing Acinetobacter baumannii and rapid spread of metallo-β-lactamase NDM-1 in Algerian hospitals. J Infect Chemother. 2014;20:696–701. doi: 10.1016/j.jiac.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Rolain J.M., Diene S.M., Kempf M., Gimenez G., Robert C., Raoult D. Real-time sequencing to decipher the molecular mechanism of resistance of a clinical pan-drug-resistant Acinetobacter baumannii isolate from Marseille, France. Antimicrob Agents Chemother. 2013;57:592–596. doi: 10.1128/AAC.01314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olaitan A.O., Diene S.M., Assous M.V., Rolain J.M. Genomic plasticity of multidrug-resistant NDM-1 positive clinical isolate of Providencia rettgeri. Genome Biol Evol. 2015;8:723–728. doi: 10.1093/gbe/evv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olaitan A.O., Diene S.M., Gupta S.K., Adler A., Assous M.V., Rolain J.M. Genome analysis of NDM-1 producing Morganella morganii clinical isolate. Expert Rev Anti Infect Ther. 2014;12:1297–1305. doi: 10.1586/14787210.2014.944504. [DOI] [PubMed] [Google Scholar]
- 17.Rutledge R.G., Cote C. Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res. 2003;31:e93. doi: 10.1093/nar/gng093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bontron S., Poirel L., Nordmann P. Real-time PCR for detection of plasmid-mediated polymyxin resistance (mcr-1) from cultured bacteria and stools. J Antimicrob Chemother. 2016 Apr 27 doi: 10.1093/jac/dkw139. [DOI] [PubMed] [Google Scholar]