Abstract
The shallow-sea hydrothermal vents at White Point (WP) in Palos Verdes on the southern California coast support microbial mats and provide easily accessed settings in which to study chemolithoautotrophic sulfur cycling. Previous studies have cultured sulfur-oxidizing bacteria from the WP mats; however, almost nothing is known about the in situ diversity and activity of the microorganisms in these habitats. We studied the diversity, micron-scale spatial associations and metabolic activity of the mat community via sequence analysis of 16S rRNA and aprA genes, fluorescence in situ hybridization (FISH) microscopy and sulfate reduction rate (SRR) measurements. Sequence analysis revealed a diverse group of bacteria, dominated by sulfur cycling gamma-, epsilon-, and deltaproteobacterial lineages such as Marithrix, Sulfurovum, and Desulfuromusa. FISH microscopy suggests a close physical association between sulfur-oxidizing and sulfur-reducing genotypes, while radiotracer studies showed low, but detectable, SRR. Comparative 16S rRNA gene sequence analyses indicate the WP sulfur vent microbial mat community is similar, but distinct from other hydrothermal vent communities representing a range of biotopes and lithologic settings. These findings suggest a complete biological sulfur cycle is operating in the WP mat ecosystem mediated by diverse bacterial lineages, with some similarity with deep-sea hydrothermal vent communities.
Keywords: hydrothermal vents, microbial mat, Desulfuromusa, sulfur-cycling, sulfate reduction rates, pyrosequencing, fluorescence in situ hybridization (FISH)
Introduction
Hydrothermal vent ecosystems are considered biogeochemical hotspots due to their unique physico-chemical conditions. The variable geochemistry of vents produces distinct biotopes (Olenin and Ducrotoy, 2006), which select for unique microbial communities (Kelley et al., 2002; Kormas et al., 2006; Perner et al., 2007; Nakamura et al., 2009; Campbell et al., 2013). Many vents support chemosynthetic microbial mat populations (Reysenbach and Shock, 2002; Nakamura et al., 2009; Emerson and Moyer, 2010) dominated by sulfide-oxidizing bacteria (SOxB) (Brazelton et al., 2006; Hügler et al., 2010; Flores et al., 2011; Jaeschke et al., 2012; Fleming et al., 2013).
Common phylotypes identified from these studies are mat forming, sulfur-oxidizing Epsilonproteobacteria (e.g., Sulfurovum, Sulfurimonas) and Gammaproteobacteria, especially filamentous forms responsible for the visually conspicuous nature of the white-colored microbial mats (e.g., Beggiatoa, Thiothrix) (Muyzer et al., 1995; Crépeau et al., 2011; Yamamoto and Takai, 2011; Kato et al., 2012). In addition to SOxB, sulfate-reducing bacteria (SRB), especially members of the Deltaproteobacteria, are common in hydrothermal sites (Jannasch et al., 1988; Houghton et al., 2007; Frank et al., 2013). Endogenous sulfate reduction rates (SRR) at hydrothermal environments are typically higher (Weber and Jørgensen, 2002; Frank et al., 2013) than cold marine sediments (Thamdrup and Canfield, 1996; Ferdelman et al., 1997; Weber and Jørgensen, 2002; Frank et al., 2013). This raises the possibility of a complete biological sulfur cycle mediated by partnerships between oxidative and reductive sulfur bacteria. Tightly coupled (cryptic) sulfur cycling, where physically associated oxidative and reductive metabolic types co-exist, has been identified in phototrophic microbial mats and consortia (Fike et al., 2008; Wilbanks et al., 2014), and has recently been demonstrated in a companion investigation to this one (Dawson et al., 2016) in the White Point (WP) chemosynthetic vent mat communities found in the Palos Verdes (PV) hydrothermal vent field in San Pedro, CA, USA. However, it is not clear how significant a role biological sulfur/sulfate reduction plays in the WP mats.
Diverse, sulfur-cycling microbial mats have been observed in a range of marine, hydrothermal settings including mid-ocean ridges (Gerasimchuk et al., 2010; Lanzén et al., 2011; Urich et al., 2014), back arc spreading centers (Kato et al., 2009) and arc volcanoes (Emerson and Moyer, 2002; Murdock et al., 2010). The composition and structure of the chemoautotrophic population has been shown to vary across hydrothermal systems with varying geochemical energy sources (Desbruyères et al., 2001; Kelley and Shank, 2010; Amend et al., 2011; Flores et al., 2011; Nakamura and Takai, 2014) and lithologic composition (Baker and German, 2004). Most identified hydrothermal ecosystems in the ocean can be characterized within five major lithologic classifications (ultramafic, mafic/basaltic, andesitic, felsic or sediment) (Buatier et al., 1995; Embley et al., 2007; McCaig et al., 2007; Pašava et al., 2007; Tivey, 2007; Kakegawa et al., 2008; Zielinski et al., 2011) or as hybrid systems (e.g., basalt-sediment) (Nakamura et al., 2009; Amend et al., 2011).
Although deep-sea hydrothermal vents have been studied for decades (Corliss et al., 1979; Jannasch and Mottl, 1985; Tunnicliffe et al., 1986), their remote nature makes investigations challenging and expensive. Shallow-sea hydrothermal vent systems such as those identified from Italy, Greece and Mexico (Sievert et al., 1999; Amend et al., 2003; Forrest et al., 2005; Rusch et al., 2005; Price et al., 2013) as well as PV in the USA (Jacq et al., 1989; Kalanetra et al., 2004), represent more easily accessed analogs to study chemosynthetic microbial communities, although it is largely unknown how they compare to their deep-sea counterparts (Tarasov et al., 2005) where environmental conditions such as light, pressure, temperature and geochemistry are known to differ. This study focuses on the microbial mats that inhabit the intertidal region of WP, a hybrid basalt-sediment-hosted system in the PV hydrothermal vent field. Past studies of the WP mats have been limited, using microscopy, fatty acid characterization and cultivation approaches to investigate the large, filamentous sulfur-oxidizing Gammaproteobacteria found there (Jacq et al., 1989; Kalanetra et al., 2004). Another study investigated grazing of the mats by abalone (Stein, 1984); however, almost nothing is known about the overall diversity of the microbial mat community, the microorganisms involved or their biogeochemical interactions. Here, we combined molecular sequencing, fluorescence in situ hybridization (FISH) and SRR activity measurements to characterize this shallow-sea hydrothermal vent ecosystem.
Materials and Methods
Sample Collection
Microbial mat samples were collected repeatedly over 2 years (2012–2013) from the WP rocky intertidal hydrothermal vent field of the PV Peninsula (33.7159° N, 118.319° W) (Figure 1A) using a range of methods for different analyses. Replicate samples (e.g., duplicate sequencing) were always collected from individual rocks from within the same intertidal pool at WP (Figure 1A). Intertidal WP vents emit warm (∼28°C), sulfide-rich water (up to 650 μM/L) (Dawson et al., unpublished). White-colored microbial mats and streamers indicate diffuse venting in rocky substrates (Figures 1A,B); while over sediments, mats and blackened (sulfidic) sediment patches (Figure 1C) indicate venting. In the field, mat samples for DNA extraction were collected from colonized rock by scraping with a sterile razor and transferred into sterile 1.5 mL tubes. Duplicate rock scrapings were collected in June, 2012, for Sanger sequencing and two more rock scrapings were collected in February, 2013, for pyrosequencing (see below). All samples for molecular analyses were immediately frozen on dry ice for transport, then stored at -80°C in the laboratory until further analysis.
FIGURE 1.

Photographs of WP rocky intertidal hydrothermal field site and collection apparati. (A) Field site, (B), white bacterial mats and streamers covering rocks and (C) associated blackened (sulfidic) sediment patches. (D) PVC tubes containing natural fiber strings and glass microscope slides prior to deployment at field site. (E) Field collected samples of colonized strings in stoppered bottles and (F) colonized glass microscope slide in conical tube.
Natural fiber strings and glass microscope slides were mounted inside PVC pipes using water-resistant epoxy putty (J-B Weld, Sulfur Springs, TX, USA; Figure 1D) and deployed near the vents for 3 weeks at a time in August, October, and December 2013. String samples were collected for SRR and sealed with the hydrothermal effluent in 15 mL serum bottles (Figure 1E; Bellco, Vineland, NJ, USA), then transported to the lab for incubation (see below). Mat samples that colonized deployed glass slides in Aug. were preserved for FISH by placing the slide into 50 ml conical tubes (BD Biosciences, Franklin Lakes, NJ, USA) containing 1X phosphate-buffered saline (PBS) (130 mM sodium chloride, 10 mM sodium phosphate buffer [pH 7.2]) and then kept on ice prior to fixation with 4% paraformaldehyde (PFA) (Figure 1F) in the laboratory within 1.5 h (Daims et al., 2005).
Sulfate Reduction Rate Assays
A preliminary, time course experiment was conducted to determine a suitable incubation time for microbial sulfate reduction. This experiment indicated that SRR increased linearly over a 96 h period (Slope: 116.04, Intercept: 950.47, R2= 0.86229); therefore, 72 h incubations were used for subsequent experiments. On two different dates (September, December 2013) replicate experiments were performed. For each, the collected colonized strings (n = 5–6 sample replicates) were placed in 15 ml serum bottles containing 5 ml of hydrothermal effluent, were injected with 0.37 MBq (10 μCi) of carrier-free Na2[35SO4] (American Radiolabeled Chemicals, St. Louis, MO, USA) and incubated at room temperature for 72 h. In addition, two control bottles were prepared as above with the addition of sodium molybdate (Mo; 20 mM) to inhibit microbially mediated sulfate reduction (Oremland and Capone, 1988). Two additional negative controls were killed with 20% zinc acetate and 37% formaldehyde immediately following the Na2[35SO4] addition. Following the 72 h incubation, reactions in the live and Mo control samples were terminated in the same manner. The samples were centrifuged at 3,220 g for 10 min. Pelleted mat samples were processed following a slightly modified version of the passive extraction procedure of Ulrich et al. (1997), adapted from the single-step chromium reduction method (Fossing and Jørgensen, 1989). The supernatant (unreduced sulfate fraction) was decanted into 15 ml conical tubes and stored at room temperature until analysis with a scintillation counter (LSC 6500, Beckman Instruments, Irvine, CA, USA). All oxygen-sensitive procedures were conducted in an anaerobic chamber (Coy Laboratory Products Inc., Grass Lake, MI, USA). SRR were calculated following the equation of Fossing and Jørgensen (1989).
DNA Extraction and PCR Amplification, Cloning and Sequencing of 16S rRNA and aprA Genes
Total genomic DNA was isolated from 0.5 g of each of the rock scrapings described above in Sample Collection (2 for Sanger, 2 for Pyrosequencing) using the FASTDNA SPIN Kit for Soils (MP Biomedicals, Solon, OH, USA) according to the manufacturer’s instructions. Two of these independent extracts were used for bacterial 16S rRNA gene PCR amplifications performed using the GM3F and GM4R (Muyzer et al., 1995) primer pair in 50 μL reactions. Reaction mixtures contained 10 μL 5X PCR Buffer, 5 μL 25 mM MgCl2, 0.5 U of GoTaq DNA Polymerase (Promega, Madison, WI, USA), 2.5 μL of 0.4% (w/v) Bovine Serum Albumin, 2.5 μL of each 10 μM primer (Eurofins MWG Operon, Huntsville, AL, USA), 1.5 μL of 10 mM dNTP mixture (Promega), and ∼20 ng of extracted nucleic acids as template. PCR was performed with a Mastercycler Pro Gradient PCR machine (Eppendorf, Hauppauge, NY, USA) under the following conditions: 5 min initial denaturation at 94°C, followed by 34 cycles of denaturation (94°C for 30 s), annealing (53°C for 30 s), and elongation (72°C for 90 s). Amplification was completed by a final elongation step at 72°C for 10 min. Additional PCR reactions using the Desulfobacteraceae-specific 16S rRNA gene DSS-658 probe as a reverse primer (to assess taxonomic specificity of this probe) and primers targeting the aprA gene were performed as above, except with use of the following forward and reverse primer sets: GM3F/DSS-658 (Manz et al., 1998) and the AprA-1-FW/AprA-10-RV (Meyer and Kuever, 2007) respectively. PCR products were visually analyzed by electrophoresis on 1% agarose gels run in 1X TAE buffer to verify correct amplicon size.
Positive 16S rRNA (both bacterial GM3/GM4 and Desulfo bacteriaceae-specific GM3/DSS-658) and aprA gene amplicons were cloned and sequenced as previously described (Dillon et al., 2013). Sanger sequences were submitted to Genbank and have been assigned accession numbers KX422076 – KX422101 for aprA and KX422102 – KX422190 for 16S rRNA genes.
Sanger Sequence Analyses
The nucleotide sequence data from 16S rRNA and aprA gene Sanger sequences were trimmed and manually edited using 4Peaks Software1. Chimera detection for 16S rRNA gene sequences was performed using Mallard v.1.0 software (Ashelford et al., 2006) and putative chimeras confirmed by analyses with Pintail (Ashelford et al., 2005) were removed. Non-chimeric, full-length 16S rRNA gene sequences were aligned using the SINA aligner (Pruesse et al., 2012), imported into ARB software v5.2 (Ludwig et al., 2004) and manually refined with reference to close phylogenetic relatives. Full-length aprA sequences were initially aligned to all available gene sequences in the NCBI website using Clustal X (Larkin et al., 2007), and then imported into a custom-created ARB database. Custom lane masks of aligned sequences (both 16S rRNA and aprA genes) were created excluding hypervariable regions (16S rRNA gene sequences) and ambiguous nucleotide positions common to all sequences. A total of 1,185 (16S rRNA) and 1,273 (aprA) gene nucleotide positions, respectively were used to create maximum likelihood trees via the Blackbox RaxML tool on CIPRES Science Gateway v.7.2 (Miller et al., 2010). 1,000 bootstrap pseudo-replications, or fewer if stopped using the automated MRE bootstrapping criterion (e.g., 450, bacterial 16S rRNA gene tree), were performed.
Non-redundant DSS-658 16S rRNA gene sequences were first added to the full-length 16S rRNA gene tree using the parsimony tool in ARB to identify nearest relatives. Then a maximum likelihood tree including unique DSS-658 and relatives was constructed as above using 584 nucleotide positions.
16S rRNA Gene Pyrotag Analyses
Additional 16S rRNA gene pyrotag sequencing using the 530F/1100R primer pair (Lane, 1991; Dowd et al., 2008) was performed on two additional independent mat sample extracts (see above) by Research and Testing Laboratory (RTL), (Lubbock, TX, USA). These bacterial 16S rRNA gene 454 pyrosequences were denoised using RTL protocols and chimera-checked by the de novo method using UCHIIME (Edgar et al., 2011) with all low quality and possibly chimeric sequences removed. The remaining sequences were sorted and clustered into OTU clusters with 99% identity (1% divergence) using USEARCH (Edgar, 2010). These results were checked against the NCBI database (Altschul et al., 1997) using BLASTN+. Based upon the BLASTN+-derived sequence identities, the sequences were classified at the appropriate taxonomic level. More information on the RTL data analysis methodology can be found at http://www.researchandtesting.com. Alpha diversity was analyzed through rarefaction curves and both the Shannon–Weaver and the Simpson diversity indices were calculated. Beta diversity comparisons were performed in QIIME (Caporaso et al., 2010) using a Monte Carlo procedure. These sequences have been submitted to the SRA of NCBI and have been assigned project number SRP076744.
Comparative Hydrothermal Vent Dataset Analyses
16S rRNA gene pyrosequencing data from a total of 13 publically available datasets (NCBI) from a range of hydrothermal environments/biotopes were compared to our two 454 libraries (Table 1). Sequences used were generated using commonly used V4-V6 region primer sets and had at least 100 bp overlap with our 530F-1100R dataset. The combined sequence datasets were uploaded and analyzed using the QIIME software pipeline (Caporaso et al., 2010). All hydrothermal microbial community datasets were rarefied to 2,500 sequences and randomized to eliminate sampling size bias among datasets in the analysis. UniFrac (Lozupone and Knight, 2005) distance matrices were calculated using both weighted and unweighted parameters in QIIME and exported for analysis in PRIMER v6.2 (Primer-E Ltd., Plymouth, UK). Primer was used to construct non-metric multidimensional scaling (MDS) plots and to perform analyses of similarity (ANOSIM) community comparisons among the vent pyrotag datasets. Comparisons were made after coding sequence datasets based on their associated vent parameters including depth (shallow, deep), system lithology (basalt, andesite, basalt-andesite hybrid) and hydrothermal biotope (vent fluids, mats, sediments + mats, sulfide chimneys and non-sulfide chimneys). Hydrothermal microbial communities were categorized as either above (shallow) or below (deep) 200 m depth, a cut-off previously used in describing hydrothermal systems (Prol-Ledesma et al., 2005; Tarasov et al., 2005).
Table 1.
Hydrothermal systems used for comparative analysis in this study.
| Region | Location | Study site | Depth | Geologic setting | System lithology | Biotope | References |
|---|---|---|---|---|---|---|---|
| North Pacific | Juan De Fuca- | Needles | Deep | MOR | Sediment | Sulfide Chimney/Deposits | Frank et al., 2013 |
| Middle Valley | Dead Dog | Deep | MOR | Sediment | Sulfide Chimney/Deposits | Frank et al., 2013 | |
| Chowder Hill | Deep | MOR | Sediment | Sulfide Chimney/Deposits | Frank et al., 2013 | ||
| Mariana Arc | Nikko | Deep | Arc Volcano | Andesite | Hydrothermal fluids | Embley et al., 2007; | |
| Huber et al., 2010 | |||||||
| NW Eifuku | Deep | Arc Volcano | Basalt | Hydrothermal fluids | Embley et al., 2007; | ||
| Huber et al., 2010 | |||||||
| Daikoku | Deep | Arc Volcano | Andesite | Hydrothermal fluids | Embley et al., 2007; | ||
| Huber et al., 2010 | |||||||
| NW Rota 1 | Deep | Arc Volcano | Basalt-Andesite | Hydrothermal fluids | Embley et al., 2007; | ||
| Huber et al., 2010 | |||||||
| Forecast | Deep | Arc Volcano | Basalt | Hydrothermal fluids | Embley et al., 2007; | ||
| Huber et al., 2010 | |||||||
| Okinawa Trough | Kueishantao | Shallow | BASC | Andesite | Hydrothermal fluids | Tang et al., 2013 | |
| Mediterranean | Hellenic Arc | Kolumbo | Shallow | Arc Volcano | Felsic | Sulfide Chimney/Deposits | Kilias et al., 2013 |
| Arctic | Knipovich Ridge | Loki’s Castle | Deep | MOR | Basalt | Sulfide Chimney/Deposits | Jaeschke et al., 2012 |
| Jan Mayen | Troll Wall | Deep | MOR | Basalt | Sediments with mats | Lanzén et al., 2011 | |
| North Atlantic | Mid-Atlantic Ridge | Rainbow | Deep | MOR | Ultramafic | Sulfide Deposits | Flores et al., 2011 |
MOR, Mid-Ocean Ridge; BASC, Back-Arc Spreading Center.
Fluorescence In Situ Hybridization
Field-colonized slides were washed twice in 1X phosphate buffered saline (PBS) before being fixed by immersion in freshly prepared 4% PFA solution in the field and placed on ice for transport to the laboratory. After 3 h on ice, the fixed slides were washed again in 1X PBS to remove residual PFA and then placed in 1X PBS: 96% ethanol (v:v) and stored at -20°C. FISH was performed following established protocols (Daims et al., 2005) using fluorescently labeled, group-specific oligonucleotide probes (Table 2). Hybridizations were performed using 1.5 ng μL-1 of HPLC-purified probe (Eurofins MWG Operon) with a buffer containing 0.01% sodium dodecyl sulfate and 35% formamide (Fisher Scientific) for 3 h at 46°C followed by a washing step at 48°C for 10 min. Washed and dried slides were counter stained with DAPI (1 μg mL-1) and mounted with citifluor AF-1 antifadent (CitiFluor, Leicester, England). Microscopic observation and documentation was performed using an epifluorescence microscope under 630X magnification (BX51, Olympus America Inc., Melville, NY, USA), fluorescent images were analyzed using ImageJ (Schneider et al., 2012).
Table 2.
Probes used in this study.
| Probe | Target group | ||
|---|---|---|---|
| EUB338-I | Most (90%) Bacteria | GCT-GCC-TCC-CGT-AGG-AGT | Amann et al., 1990 |
| DELTA495A | Most deltaproteobacteria | AGT-TAG-CCG-GTG-CTT-CCT | Loy et al., 2002 |
| GAM42A | Gammaproteobacteria | GCC-TTC-CCA-CAT-CGT-TT | Manz et al., 1998 |
| BET42 | Betaproteobacteria | GCC-TTC-CCA-CTT-CGT-TT | Manz et al., 1998 |
| SRB385 | Most desulfovibrionales | CGG-CGT-CGC-TGC-GTC-AGG | Amann et al., 1990 |
| DSS-658a | Desulfobacteraceae | TCC-ACT-TCC-CTC-TCC-CAT | Manz et al., 1998 |
| DSM651 | Desulfuromusa spp. | CCT-CTC-CCA-TAC-TCA-AG | This study |
| DSM651 competitor | Desulfuromusa spp. | CCT-CTC-CCA-TAC-TCT-AG | This study |
aUsed as probe and primer.
To visualize Desulfuromusa-related 16S rRNA phylotypes via FISH, a sequence-specific oligonucleotide probe (DSM651; Table 2) was designed manually using the ARB software package (Ludwig et al., 2004). Since probe DSM651 was a modification of the DSS658 probe previously described (Manz et al., 1998), the same formamide concentration (35%) was used. Probe DSM651 targets all Desulfuromusa sequences obtained from the clone libraries, but has at least one mismatch to all other sequences in the ARB database (SSURef_NR99_115_SILVA_20_7_13_opt.arb). FISH assays for Desulfuromusa were conducted using a DSM651 competitor probe (one base altered) to minimize non-specific hybridization non-targeted genotypes (Table 2).
Results
White Point Microbial Mat Phylogenetic Diversity
Cloning and Sanger sequencing of the bacterial 16S rRNA genes yielded 79 non-chimeric nearly full-length sequences that were grouped into 62 unique OTUs (at 3% dissimilarity level). Taxonomic evaluation places these OTUs in five main clusters (Figure 2). The most diverse and abundant phylotype (71% of clones) were members of the sulfur-oxidizing order Thiotrichales within the Gammaproteobacteria. The next most abundant group (16%) of phylotypes branched within the Epsilonproteobacteria; 80 and 20% of which affiliated with members of the genera Arcobacter and Sulfurovum, respectively. The two deltaproteobacterial sequences were greater than 98% similar to cultured Desulfuromusa kysingii (Liesack and Finster, 1994). The remaining sequences were distributed between the Alphaproteobacteria and the Bacteroidetes (Figure 2).
FIGURE 2.
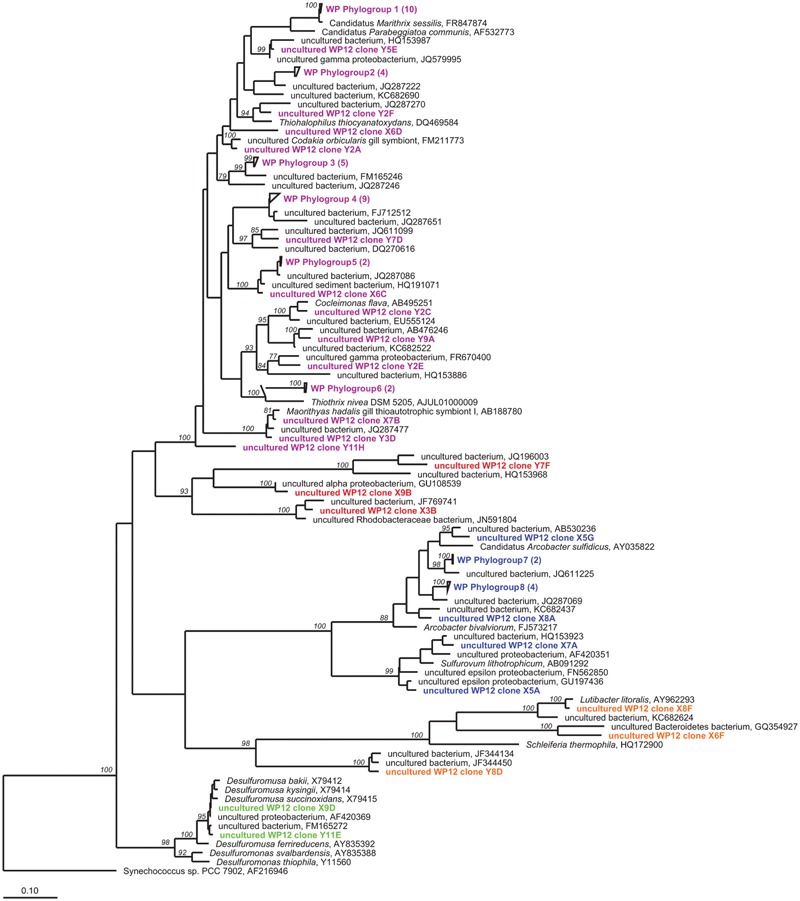
Unrooted maximum-likelihood dendrogram of 16S rRNA gene sequences amplified from WP microbial mat community, aligned with closest relatives (black) using ARB software. Colors indicate WP taxonomic groupings. Purple, gammaproteobacteria; red, alphaproteobacteria; blue, epsilonproteobacteria; orange, bacteroidetes, and green, deltaproteobacteria. Wedges indicate redundant sequences with number of sequences indicated in parentheses. Values at nodes indicate >75% bootstrap support. Scale bar shows a 10% estimated difference in nucleotide sequence positions.
In addition to the 16S rRNA gene sequencing, we also sequenced the functional gene, adenosine -5′-phosphosulfate reducates alpha subunit (aprA) gene to provide a targeted approach to investigate sulfur-cycling microorganisms in the WP mats. The aprA clone library resulted in a total of 39 clones with positive inserts, representing 26 novel, non-redundant genotypes all of which affiliated with oxidative gammaproteobacterial lineages (Figure 3). Despite the fact that this primer set is known to amplify reductive aprA gene copies, no deltaproteobacterial sequences were obtained.
FIGURE 3.
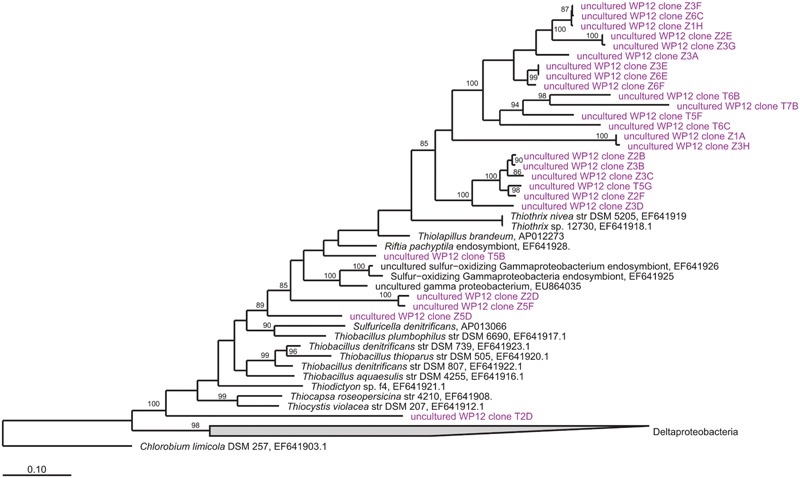
Unrooted maximum-likelihood dendrogram of apraA gene sequences amplified from WP microbial mat community (purple), aligned with closest relatives (black) from a custom database. Values at nodes indicate >85% bootstrap support. Scale bar shows a 10% estimated difference in nucleotide sequence positions.
Overall, the 454 pyrotag sequences displayed similar phylogenetic community composition patterns as the Sanger clones. The two pyrosequenced samples (WP-1, WP-2), generated 34,834 and 20,877 denoised, non-chimeric, high quality sequences, respectively and showed similar rarefaction curves indicating near-complete sampling (Supplementary Figure S1). Pyrotag sequences with abundances greater than 0.5% were clustered into 28,802 (WP-1) and 16,620 (WP-2) OTU’s at the 1% dissimilarity level (Figure 4). The Gammaproteobacteria and Epsilonproteobacteria classes accounted for approximately 69% (WP-1) and 66% (WP-2) of the sampled population. No significant differences in the overall community structure between the rarefied WP-1 and WP-2 datasets were detected (ANOSIM, p = 0.58). Monte Carlo analyses also found no significant differences between the samples when taken as a whole. However, differences in the dominant OTU (based on % composition) of the two libraries were observed, with uncultured Gammaproteobacteria of the Marithrix clade being most abundant in WP-1 and Epsilonproteobacteria most similar to Sulfurovum most abundant in the WP-2 library (compare Figures 5A,B). Overall, gammaproteobacterial pyrotags affiliated with the order Thiotrichales represented 31 and 11% of the communities, respectively. The most abundant members of the Thiotrichales in the two libraries had closest BLAST identities to Marithrix sessilis (Kalanetra et al., 2004) (WP-1) and to Thiothrix nivea (Stahl et al., 1987) (WP-2). Marithrix sessilis was the next most abundant gammaproteobacterium within WP-2 at 3%. Epsilon proteobacteria accounted for 32% (WP-1) and 43% (WP-2) of the library sequences. Interestingly, despite dominating the WP-2 library (19%), Sulfurovum lithotrophicum-like sequences only comprised 1.5% of the WP-1 library, which had greater prevalence of Sulfurimonas- (13%) and Arcobacter-like (10%) sequences (Figure 4).
FIGURE 4.
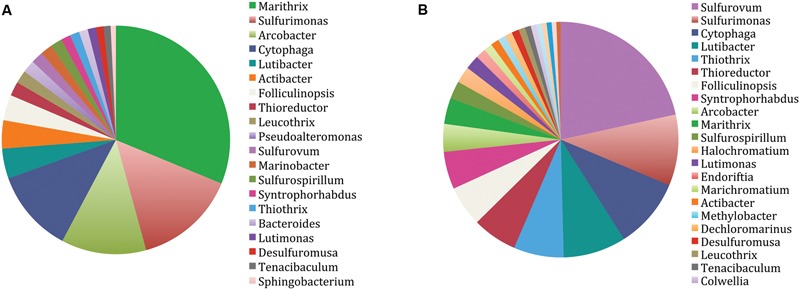
Taxonomic distribution of the two WP microbial mat community pyrosequence datasets, (A) WP-1, (B) WP-2, displaying the relative (%) incidence of identified genera. Total number of 1% dissimilarity OTUs assigned was (A) 28,802 and (B) 16,620.
FIGURE 5.
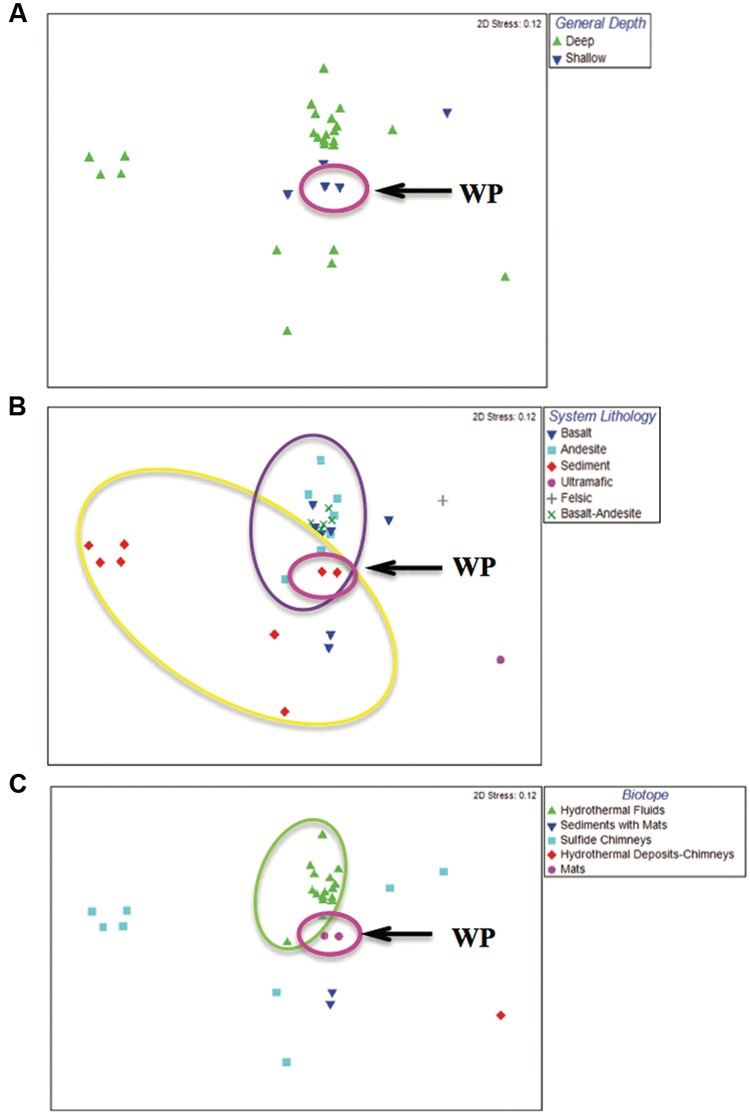
Multidimensional scaling (MDS) plots showing community variation, coded (colored symbols) according to depth, system lithology and biotope. Arrow and pink ellipse shows WP samples for all three panels, but does not denote statistical differences. (A) Depth. ‘Deep’ samples (>200 m) and ‘shallow’ samples (<200 m). (B) System lithology. Ellipses denote statistically significant communities as determined by ANOSIM (p < 0.005). Yellow denotes sediment-hosted sites; purple denotes Basalt, Andesite and Basalt-Andesite-hosted sites. (C) Biotope. Green ellipse denotes hydrothermal fluid sites that were significantly different from all others as determined by pair-wise ANOSIM comparisons (p < 0.01 for all).
Bacteroidetes, particularly members of the genus Lutibacter, represented the next most abundant group of pyrotag sequences recovered from the WP microbial mat samples (Figure 4). Members from three common classes of Bacteroidetes (Cytophaga, Flavobacteria, and Bacteroidia) accounted for roughly 24% (WP-1) and 22% (WP-2) of the sequences. Deltaproteobacteria accounted for 3% (WP-1) and 7% (WP-2) of the total number of pyrotags in the two WP microbial mat samples. The most abundant deltaproteobacterial genotype (1, 4% of the total 16S rRNA genes from the two pyrotag libraries, respectively) recovered was 99% similar to a deltaproteobacterial epibiont of the polychaete Alvinella pompejana, from deep-sea hydrothermal vents of the East Pacific Rise. As with the clone library, sequences affiliated with Desulfuromusa were detected, accounting for 30% (WP-1) and 12% (WP-2) of the Deltaproteobacterial pyrotags (∼1% of total sequences in ea. library). Among these, sequences with BLAST similarity to all four isolated species of the genus (D. kysingii, D. succinoxidans, D. ferrireducens, and D. bakii) (Liesack and Finster, 1994; Vandieken et al., 2006) were recovered. Additionally, sequences similar to a fifth, uncultured Desulfuromusa sp. were recovered from our samples. Minor additional phylotypes [cumulatively ∼3% (WP-1) and ∼4% (WP2) respectively] were identified as belonging to the Alpha- and Betaproteobacteria, Firmicutes, Tenericutes, Verrucomicrobia, and the Cyanobacteria (Figure 4).
Despite these differences noted above, the pyrotag sequence datasets had similar community richness. Rarefaction analysis revealed similar curves with high OTU richness from the two WP microbial mat samples (Supplementary Figure S1). Results of the Shannon–Weaver (H) and Simpson (D) diversity index calculations confirm the high levels of diversity within these two libraries (H = 8.67 and D = 0.95 for WP-1 and H = 9.51 and D = 0.98 for WP-2).
Comparative Analyses of Hydrothermal Microbial Communities
The 454 pyrotag sequences identified from the WP microbial mat community were compared collectively with 13 hydrothermal vent microbial community studies for which data were available in Genbank (Table 1). MDS analysis showed that the WP hydrothermal microbial community plotted centrally among samples, but appeared closest to samples from the Okinawa Trough (Tang et al., 2013) (Figure 5). Due to low sample replication in our study and many of these published datasets, we could not perform pair-wise comparisons between the WP samples and others. However, when datasets were grouped based on depth (shallow, deep), system lithology (basalt, andesite, basalt-andesite hybrid) and hydrothermal biotope (vent fluids, mats, sediments + mats, sulfide chimneys and non-sulfide chimneys), patterns of community associations across datasets were revealed.
Analyses of similarity comparisons between depth categories did not reveal significant differences between the shallow-sea hydrothermal microbial communities and the deep-sea hydrothermal microbial communities (Figure 5A). However, coding of samples based on hydrothermal system lithology revealed significant differences (Figure 5B). ANOSIM results indicated that the sediment-associated vent sites (i.e., Juan de Fuca, WP) were significantly different from the andesite-, basalt- and the basalt-andesite-hosted sites (p < 0.005 for all comparisons). Felsic and ultramafic site samples (Flores et al., 2011; Kilias et al., 2013) occurred as outliers and plotted furthest from the central clusters, although they were not significantly different from the other igneous lithologies. Finally, coding samples according to their hydrothermal biotope also revealed significant differences, as samples from hydrothermal fluids were significantly different from samples collected from the other three biotopes (p < 0.01 for all), although the non-fluid biotope communities were not significantly different from each other (p > 0.05 for all) (Figure 5C).
Fluorescence In Situ Hybridization
We performed FISH on six colonized slides to visualize the spatial orientation and relative abundance of the microbial populations inhabiting the WP microbial mat community. Very large (∼30–40 μm diameter) Marithrix-like filaments were consistently visualized with DAPI during our study. However, we failed to demonstrate a positive probe signal with the GAM42A (used with its competitor Bet42a) gammaproteobacterial-specific probe (Manz et al., 1998) or the more general bacterial probe EUB338 (Amann et al., 1990) on these large filaments during any of our FISH experiments (Figures 6A–C,F). Overall, we observed lower relative abundances of deltaproteobacteria-positive cells compared to Gammaproteobacteria-positive cells. Cells successfully probed with the DELTA495A and DSS658 probes targeting Deltaproteobacteria and the Desulfobacteraceae, respectively, (Manz et al., 1998; Loy et al., 2002) included single-cells (mean length 1.7 μm, n = 20 cells) (1–2 μm length) and aggregates of cocci to short chains of cells (range 10–20 μm length, n = 20 chains) (Figures 6B–D). The SRB385 probe, which specifically targets members of the Desulfovibrionales (Amann et al., 1990) rarely produced a signal; when positive probing occurred it was to small (mean length 5 μm, n = 5) aggregates of coccoid cells (Figure 6F). In some cases, these SRP385-positive aggregates were found associated with the Marithrix-size filaments. Unlike the Marithrix filaments a variety of gammaproteobacterial single-cells, tetrads, rods and short (20 μm length) filaments were detected using the GAM42A probe (Figures 6A,C–E). Of note were GAM42A-positive clusters of cells that were consistently observed throughout our study (Figure 6E).
FIGURE 6.
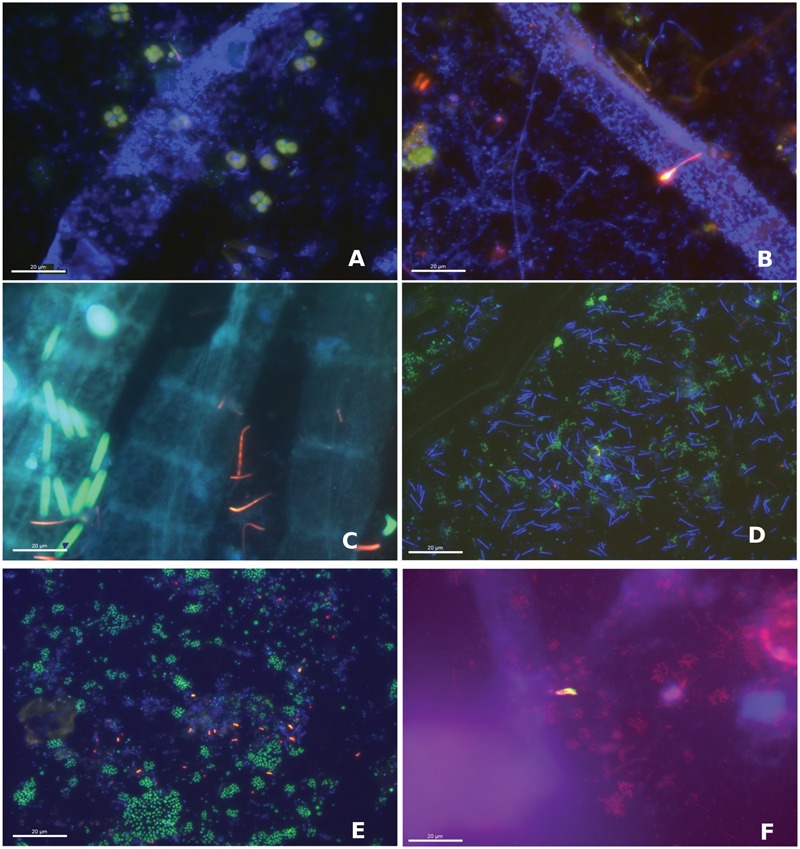
Fluorescence in situ hybridization (FISH) images of the WP microbial mat community showing different fields of view from two colonized glass slides collected in June 2014 at 630X magnification. (A) Large filament (blue) showing DAPI signal and tetrads of Gammaproteobacteria (green). (B) DAPI-stained filaments (blue) and Gammaproteobacteria (green) with chains, rods and cocci Deltaproteobacteria (red; Delta495). (C) Large filaments (blue) with rod-shaped Gammaproteobacteria (green) and chains of Deltaproteobacteria (red; DSS658). (D) Groups of gammaproteobacterial cocci (green) with DAPI-stained filaments (blue). (E) Gammaproteobacterial cocci (green), deltaproteobacterial curved rods (red, DSS658) and DAPI- stained cocci and filaments (blue). (F) Aggregates of Deltaproteobacteria (yellow; DSS658) with Gammaproteobacteria (red) and DAPI-stained filaments (blue).
The specificity of the DSS-658 probe was checked by PCR and sequencing using the probe as a reverse primer. Among the 26 non-redundant phylotypes, the majority (69 %) fell within the Deltaproteobacteria, the majority of which were most closely related (>96% similarity) to cultured Desulfuromusa species (Liesack and Finster, 1994) (Supplementary Figure S2). However, the remaining 31% of clones were affiliated with members of the Epsilonproteobacteria and the Gammaproteobacteria, including SOxB phylotypes (Moyer et al., 1995; Gros et al., 1996; Takai et al., 2006a). This suggests that the DSS658 probe was likely not specific to Deltaproteobacteria for the WP mat.
Since Desulfuromusa-like sequences were abundant in the DSS-658 primer clone library (although still rare in the overall 16S rRNA community) and curved rods diagnostic of this group were observed in the DSS-658 FISH (Figures 6C,E), we decided to target this group specifically via FISH using a newly designed, highly specific Desulfuromusa-targeted probe (DSM651) to verify their importance and to identify their association(s) within the mat community. Probe DSM651 produced a signal on all slides with the prevailing cell morphology being short rods forming chains up to 30 μm in length (Figure 7). In some cases, multiple filaments appeared to originate from a central axis (Figure 7B). Desulfuromusa were also often found in association with Gammaproteobacteria, including the large Marithrix-like filaments (Figure 7A), as well as thinner filamentous forms and clusters of unicells.
FIGURE 7.
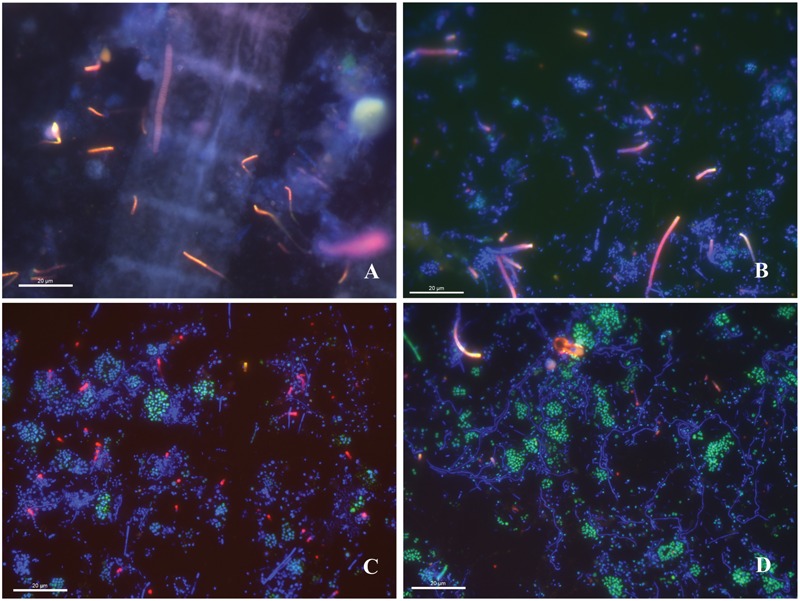
Fluorescence in situ hybridization images of the WP microbial mat community at 630X magnification hybridized using the DSM-651 Desulfuromusa-specific probe. (A) Large DAPI-stained filament (blue) and chains of Desulfuromusa rods (red). (B) DAPI-stained cells (blue) and Desulfuromusa (red) emerging from a central axis and chains. (C) DAPI-stained filaments and cocci (blue) with gammaproteobacterial cocci (green) and Desulfuromusa (red). (D) Gammaproteobacterial cocci clusters (green) with DAPI-stained filaments (blue) and Desulfuromusa cells (red).
Sulfate Reduction Rates
Very similar rates were observed during two subsequent SRR experiments with WP mat samples in September and December 2013. During both field samplings, the mean SRR for the intertidal mats were significantly above zero (1-sample T-test, p < 0.05 for both) at 46.7 and 36.5 nmol SO4 cm-3 d-1, respectively (Figure 8). SRR rates measured from our sodium molybdate inhibited and Zn-formaldehyde killed controls were less than 1% of the rates measured in the live controls (<1 nmol SO4 cm-3 d-1).
FIGURE 8.
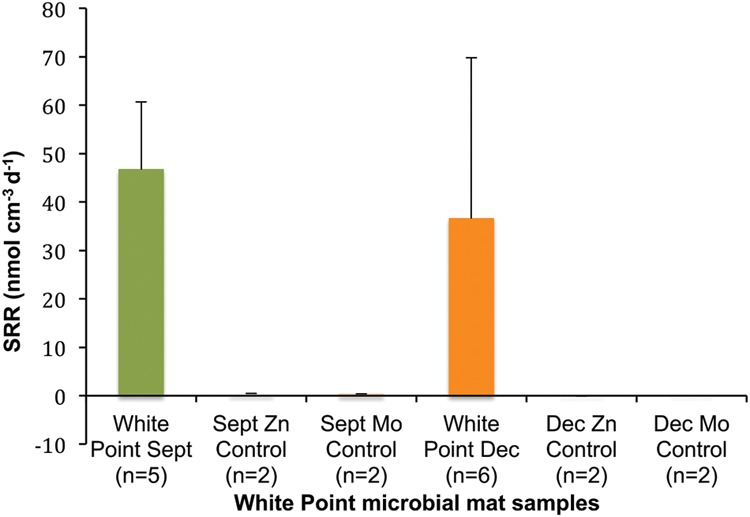
Mean SRR for WP microbial mat samples. Negative controls (Mo and kill) were always less than 1% of live rates (<1 nmol SO4 cm-3 day-1). Error bars = 1 SD.
Discussion
Deep-sea hydrothermal systems have long been known to support chemosynthetic sulfur-cycling microbial populations (Corliss et al., 1979; Karl et al., 1980; Jannasch and Mottl, 1985), although relatively fewer studies have been performed on shallow hydrothermal vent systems (Sievert et al., 2000; Giovannelli et al., 2013; Tang et al., 2013; Wang et al., 2015). Based on results from previous studies of the WP sulfur vent microbial mat community (microscopy, fatty acid characterization and cultivation-dependent approaches) (Jacq et al., 1989; Kalanetra et al., 2004), we hypothesized that a sulfur-cycling consortium comprised of SOxB converting geologically derived vent sulfide to elemental sulfur and sulfate, and SRB converting oxidized, biologically derived sulfur species back to sulfide, would be present in the mats.
Our hypothesis was supported using multiple approaches to study the WP microbial mats. Both microscopy and gene sequencing revealed the importance of filamentous gammaproteobacterial sulfur-oxidizing lineages such as Marithrix and Thiothrix. Marithrix-like filamentous bacteria have also been detected in 16S rRNA gene surveys of mats associated with cold seeps in the Eastern Mediterranean and the Barents Sea in the Eastern Atlantic suggesting they may be common in lower temperature vent mats such as those found at WP (Heijs et al., 2005; Grunke et al., 2012). These large filamentous microorganisms did not hybridize with any of the general bacterial probes (e.g., EUB338), possibly due to the thick sheath enclosing the cells. However, the DAPI staining pattern, large size and distinct morphology is consistent with Marithrix. These conspicuous Marithrix-like filaments comprise a significant fraction of the biomass in these samples, and also appear to be a dominant member of the mat community based on aprA and 16S rRNA gene libraries. Remarkably, there were also dozens of novel phylotypes affiliated with the Thiotrichales detected using 16S rRNA as well as aprA gene sequencing. Other studies of shallow vents have found members of the Thiotrichales to be key members of the chemoautotrophic vent communities (Brinkhoff et al., 1999; Dawson et al., 2016). However, a number of studies in both shallow and deep-sea vents systems have found mesophilic Epsilonproteobacteria such as Sulfurovum and Sulfurimonas to be the most common groups of SOxB recovered by molecular means (Flores et al., 2012; Sylvan et al., 2012; Wang et al., 2015) and cultivation (Inagaki et al., 2003, 2004). Our 16S rRNA gene sequencing revealed several epsilonproteobacterial lineages closely associated with Sulfurovum, Sulfurimonas, and Arcobacter, three known groups of SOxB to be among the most abundant OTUs. Microbial sulfur oxidation pathways differ between epsilon- and gammaproteobacterial sulfur oxidizers; these differences have been hypothesized to represent distinct ecophysiological strategies, for example Epsilonproteobacteria are also known to be able to use sulfur and thiosulfate as alternative electron donors (Yamamoto and Takai, 2011). The prevalence of the two groups of sulfur bacteria may be related to the geochemical setting. For example, the predominance of Gamma- vs. Epsilonproteobacteria in molecular libraries has been found to vary with distance from the vent source, which may be due to varying tolerance to anoxia (Tang et al., 2013; Wang et al., 2015; Zbinden et al., 2015). Additionally, the influence of differences in vent geochemistry (largely driven by compositional differences of underlying rocks) on sulfur cyclers as well as other common vent groups has been noted in numerous studies (Huber et al., 2003; Schrenk et al., 2004; Kelley et al., 2005; Nakagawa et al., 2005; Campbell et al., 2006, 2013; Amend et al., 2011; Flores et al., 2011, 2012) although biogeographic isolation has also been identified as a potential contributing factor to variation in vent community structure (Huber et al., 2010).
The next most abundant OTUs recovered were affiliated with the phylum Bacteroidetes, which were dominated by lineages from the genus Lutibacter. Cultured relatives include Lutibacter profundi, a microaerophilic heterotroph, first isolated from a microbial mat on a black smoker chimney at Loki’s castle hydrothermal vent in the Arctic mid-ocean ridge (Le Moine Bauer et al., 2016). Members of the Bacteroidetes are typically rare in molecular surveys of deep-sea vent systems including Loki’s castle (Flores et al., 2012; Jaeschke et al., 2012), however, they have been previously described from shallow vent systems (∼10–20% of OTUs) (Sievert et al., 2000; Giovannelli et al., 2013; Wang et al., 2015). Members of the genus Lutibacter were not noted as being important in other shallow systems, suggesting a unique aspect to the WP mat community.
In contrast to the diverse SOxB proteobacterial lineages described above, we detected much lower relative sequence abundances and diversity of likely sulfate-/sulfur-reducing deltaproteobacterial lineages, representing 1–4% of the overall community. The most abundant groups of deltaproteobacterial 16S rRNA phylotypes were affiliated with the genus Desulfuromusa and an uncultured epibiont on Alvinella pompejana tubeworms, using cloning and Sanger sequencing and pyrosequencing, respectively. The importance of these groups contrasts with other deep-sea vent systems where other genera of Deltaproteobacteria such as Desulfobulbus and Hippea have been more commonly found (Flores et al., 2011, 2012). No Deltaproteobacteria were recovered using the aprA gene, which may be due to under sampling and the low abundance of SRB relative to the diverse SOxB phylotypes in the sample. Cultures of Desulfuromusa are only known to be sulfur reducers, not sulfate reducers (Liesack and Finster, 1994; Vandieken et al., 2006), so these mat microbes would not be predicted to possess aprA genes. Our FISH analyses using group-specific oligonucleotide probes also revealed low abundances of Deltaproteobacteria cells (∼2–5% of hybridized cells) compared to Gammaproteobacteria-positive cells (always greater than 20%). Desulfuromusa cells were occasionally observed to be physically associated with the large filamentous Marithrix – like filamentous bacteria suggesting that they may form epibiotic associations. This is reminiscent of previous reports of sulfate-reducing Desulfonema found in association with members of the ensheathed cyanobacterium Coleofasciculus (formerly Microcoleus) in hypersaline microbial mats (Fike et al., 2008). The Desulfuromusa may be metabolizing elemental sulfur produced by the SOxB.
The 16S rRNA gene-based analyses did reveal a relatively minor presence (<1% of both pyrotag libraries) of phylotypes related to sulfate-reducing Deltaproteobacteria (e.g., Desulfomonile-like sequences), suggesting the metabolic potential for a complete sulfur cycle within the WP mat. This potential was confirmed by our 35SO42- radiotracer measurements of SRR that measurable biological sulfate reduction occurs in the WP mats. These SRR are among the first reported from shallow hydrothermal sulfur-vent systems, and are comparable to those typically reported in cold, coastal marine sediments (Howarth and Jørgensen, 1984; Jørgensen and Bak, 1991; Jørgensen and Nelson, 2004) as well as rates measured in sediments from a high temperature, deep-sea hydrothermal vent (Jørgensen et al., 1992). Our SRR are likely to be underestimates of the total reductive sulfur metabolism in the WP mat system. The SRR assay relies on the reduction of the labeled sulfate substrate and would not measure activity for groups only capable of reducing elemental sulfur. Thus, sulfur reduction may be of greater significance in these mats compared to typical marine sediments due to the relative importance of groups like Desulfuromusa.
One goal of this study was to compare the microbial diversity from this warm, shallow WP hydrothermal vent field to its hard-to-access deep-sea counterparts. Comparisons with database sequences revealed many similar phylotypes in WP to those identified from deep-sea hydrothermal vents, especially a number of sulfur-cycling lineages (Moyer et al., 1995; Takai et al., 2006b; Murdock et al., 2010; Sylvan et al., 2013). When the WP pyrosequences were compared with other large hydrothermal vent 16S rRNA datasets, a number of trends with regards to vent system depth, lithology and biotope emerged. The two other shallow samples tended to cluster near the WP samples in our MDS plots; however, no significant differences were observed between shallow and deep communities via ANOSIM. This finding may have been impacted by the relative paucity of datasets from shallow systems compared to their deep-sea counterparts and the low number of sample replicates (see discussion of this study limitation below). Greater variation was observed for the deeper vent communities, potentially resulting from additional factors such as system lithology and biotope, factors that have been noted to affect hydrothermal vent communities in past studies (Desbruyères et al., 2001; Kato et al., 2010; Amend et al., 2011; Flores et al., 2011, 2012; Sylvan et al., 2013). Microbial communities from hydrothermal fluids were found to be significantly different from all other biotope categories and were least similar to the microbial mat/chimney-biofilm biotope communities including the WP samples. This is consistent with past findings that attachment-associated and biofilm populations were more diverse than planktonic fluid communities in hydrothermal environments at a deep-sea volcano (Huber et al., 2003; Edwards et al., 2005) and that mixing with surrounding seawater alters vent plume communities (Sheik et al., 2015). We also observed variation within a given biotope, which may be explained by system lithology. For example, differences were observed between the two clusters of sulfide chimneys in our MDS plots, which may reflect the fact that the sediment-hosted MOR samples support different communities than the two igneous sites (basaltic MOR and a felsic arc volcano) (Jaeschke et al., 2012; Kilias et al., 2013). Overall, our ANOSIM comparisons found communities from sediment-hosted vents were significantly different from all communities found on andesite- and basalt-hosted lithologies.
The WP samples did not perfectly cluster with any one biotope or lithology, nor did they cluster with all of the other shallow vent samples. This may reflect the complex lithology of the WP system as vent fluids pass through three distinct lithologic sequences: the mafic-derived Catalina Schist basement rock, the organic-rich Monterey Formation, and also Quaternary sediments (Woodring et al., 1946; Bebout and Barton, 1989; Grove et al., 2008). We hypothesize that this hybrid lithologic nature of WP generates a geochemical environment that selects for a diverse assemblage of microorganisms with similarities to a broad range of globally distributed hydrothermal communities, although this requires more detailed investigation. One limitation of our comparative analyses is a lack of replication in the pyrosequencing datasets we analyzed, including our own, many of which had only a single sample replicate from a given vent location or even vent field. These studies and our own have likely undersampled the variation in microbial diversity in these locations, as was suggested by our finding of a difference in the most abundant SOxB lineage in our two libraries. The lack of sample replicates meant that we could not perform direct site-to-site comparisons, so instead we compared across pooled categories of samples from multiple datasets. The reduced statistical power may have also reduced our ability to discern differences between and among hydrothermal systems (e.g., shallow vs. deep vents where we expected to observe greater differences due to the differences in temperature, pressure, light and geochemistry). Some recent studies have utilized a greater sampling effort to examine variability in deep-sea vent communities and found both intra- and inter-field variability in communities potentially driven by large scale differences in geological and geochemical processes, while in other comparisons, communities were indistinguishable (Flores et al., 2011, 2012). Ongoing work in the WP vent system will address this issue of biogeographic variation (Roussos et al., unpublished).
Overall, our results confirm the importance of both oxidative and reductive sulfur cyclers in the WP shallow water hydrothermal vent mats and demonstrate that biological sulfate reduction contributes sulfide to the WP system in addition to the abundant geologically derived sulfide. The high level of taxonomic diversity of both gamma- and epsilonproteobacterial SOxB lineages in the WP mats was somewhat surprising, as these lineages should all be competing for similar resources in these habitats, suggesting a degree of niche differentiation that is not fully understood at present. This study complements a recent report that used FISH and stable isotope probing to show that multiple groups of Gammaproteobacteria and a single coherent group of Deltaproteobacteria in the WP mat could be distinguished ecophysiologically based on their utilization of labeled acetate, ammonia and sulfate (Dawson et al., 2016). More broadly, our findings suggest that there is an underlying bacterial community structure that is shared among hydrothermal vent communities, although it also highlights that no two vent communities are identical, suggesting that variation in factors such as vent depth, biotope, lithology as well as perhaps other untested ecological or geological parameters combine to determine microbial habitation.
Author Contributions
Conception or design of the work: PM, RH, VO, and JD. Data acquisition, analysis and interpretation: PM, NM, RH, and JD. Drafting the article: PM, and JD. Critical revision of the article: PM, NM, RH, VO, and JD. All authors have read and approved this submission.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Christine Whitcraft and Lora Stevens for project advising and Katherine Dawson for providing the WP sulfide measurements.
Funding. This study was performed with support from the National Science Foundation (EAR-1124398 to JD, EAR-112391 to VO) and CSU-COAST student research grant to PM. Partial support was additionally provided by the Gordon and Betty Moore Foundation through grant GBMF3306 to VO.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01163
References
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amend J. P., Mccollom T. M., Hentscher M., Bach W. (2011). Catabolic and anabolic energy for chemolithoautotrophs in deep-sea hydrothermal systems hosted in different rock types. Geochim. Cosmochim. Acta 75 5736–5748. 10.1016/j.gca.2011.07.041 [DOI] [Google Scholar]
- Amend J. P., Rogers K. L., Shock E. L., Gurrieri S., Inguaggiato S. (2003). Energetics of chemolithoautotrophy in the hydrothermal system of Vulcano Island, southern Italy. Geobiology 1 37–58. 10.1046/j.1472-4669.2003.00006.x [DOI] [Google Scholar]
- Ashelford K. E., Chuzhanova N. A., Fry J. C., Jones A. J., Weightman A. J. (2005). At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71 7724–7736. 10.1128/AEM.71.12.7724-7736.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford K. E., Chuzhanova N. A., Fry J. C., Jones A. J., Weightman A. J. (2006). New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72 5734–5741. 10.1128/AEM.00556-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E. T., German C. R. (2004). “On the global distribution of hydrothermal vent fields,” in Mid-Ocean Ridges eds German C. R., Lin J., Parson L. M. (Washington, DC: American Geophysical Union; ) 245–266. [Google Scholar]
- Bebout G. E., Barton M. D. (1989). Fluid flow and metasomatism in a subduction zone hydrothermal system: catalina schist terrane, California. Geology 17 976–980. [DOI] [Google Scholar]
- Brazelton W. J., Schrenk M. O., Kelley D. S., Baross J. A. (2006). Methane-and sulfur-metabolizing microbial communities dominate the lost city hydrothermal field ecosystem. Appl. Environ. Microbiol. 72 6257–6270. 10.1128/AEM.00574-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkhoff T., Sievert S. M., Kuever J., Muyzer G. (1999). Distribution and diversity of sulfur-oxidizing Thiomicrospira spp. at a shallow-water hydrothermal vent in the Aegean Sea (Milos, Greece). Appl. Environ. Microbiol. 65 3843–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buatier M. D., Früh-Green G. L., Karpoff A. M. (1995). Mechanisms of Mg-phyllosilicate formation in a hydrothermal system at a sedimented ridge (Middle Valley, Juan de Fuca). Contrib. Mineral. Petr. 122 134–151. 10.1007/s004100050117 [DOI] [Google Scholar]
- Campbell B. J., Engel A. S., Porter M. L., Takai K. (2006). The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4 458–468. 10.1038/nrmicro1414 [DOI] [PubMed] [Google Scholar]
- Campbell B. J., Polson S. W., Allen L. Z., Williamson S. J., Lee C. K., Wommack K. E., et al. (2013). Diffuse flow environments within basalt-and sediment-based hydrothermal vent ecosystems harbor specialized microbial communities. Front. Microbiol. 4:182 10.3389/fmicb.2013.00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corliss J. B., Dymond J., Gordon L. I., Edmond J. M., von Herzen R. P., Ballard R. D., et al. (1979). Submarine thermal sprirngs on the galapagos rift. Science 203 1073–1083. [DOI] [PubMed] [Google Scholar]
- Crépeau V., Cambon Bonavita M. A., Lesongeur F., Randrianalivelo H., Sarradin P. M., Sarrazin J., et al. (2011). Diversity and function in microbial mats from the Lucky Strike hydrothermal vent field. FEMS Microbiol. Ecol. 76 524–540. 10.1111/j.1574-6941.2011.01070.x [DOI] [PubMed] [Google Scholar]
- Daims H., Stoecker K., Wagner M. (2005). “Fluorescence in situ hybridization for the detection of prokaryotes,” in Advanced Methods in Molecular Microbial Ecology eds Osborn A. M., Smith C. J. (Abingdon: Bios-Garland; ) 213–239. [Google Scholar]
- Dawson K. S., Scheller S., Dillon J. G., Orphan V. J. (2016). Stable isotope phenotyping via cluster analysis of NanoSIMS data as a method for characterizing distinct microbial ecophysiologies and sulfur-cycling in the environment. Front. Microbiol. 7:774 10.3389/fmicb.2016.00774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbruyères D., Biscoito M., Caprais J.-C., Colaço A., Comtet T., Crassous P., et al. (2001). Variations in deep-sea hydrothermal vent communities on the Mid-Atlantic Ridge near the Azores plateau. Deep-Sea Res. Pt. 48 1325–1346. 10.1016/S0967-0637(00)00083-2 [DOI] [Google Scholar]
- Dillon J. G., Carlin M., Gutierrez A., Nguyen V., Mclain N. (2013). Patterns of microbial diversity along a salinity gradient in the Guerrero Negro solar saltern, Baja CA Sur, Mexico. Front. Microbiol. 4:399 10.3389/fmicb.2013.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd S. E., Callaway T. R., Wolcott R. D., Sun Y., Mckeehan T., Hagevoort R. G., et al. (2008). Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8:125 10.1186/1471-2180-8-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K. J., Bach W., Mccollom T. M. (2005). Geomicrobiology in oceanography: microbe–mineral interactions at and below the seafloor. Trends Microbiol. 13 449–456. 10.1016/j.tim.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Embley R. W., Baker E. T., Butterfield D. A., Chadwick W., Lupton J. E., Resing J. A., et al. (2007). Exploring the submarine ring of fire: mariana Arc-Western Pacific. Oceanography 20 68 10.5670/oceanog.2007.07 [DOI] [Google Scholar]
- Emerson D., Moyer C. L. (2002). Neutrophilic Fe-oxidizing bacteria are abundant at the Loihi Seamount hydrothermal vents and play a major role in Fe oxide deposition. Appl. Environ. Microbiol. 68 3085–3093. 10.1128/AEM.68.6.3085-3093.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D., Moyer C. L. (2010). Microbiology of seamounts: common patterns observed in community structure. Oceanography 23 148–163. [Google Scholar]
- Ferdelman T. G., Lee C., Pantoja S., Harder J., Bebout B. M., Fossing H. (1997). Sulfate reduction and methanogenesis in a Thioploca-dominated sediment off the coast of Chile. Geochim. Cosmochim. Acta 61 3065–3079. 10.1016/S0016-7037(97)00158-0 [DOI] [Google Scholar]
- Fike D. A., Gammon C. L., Ziebis W., Orphan V. J. (2008). Micron-scale mapping of sulfur cycling across the oxycline of a cyanobacterial mat: a paired nanoSIMS and CARD-FISH approach. ISME J. 2 749–759. 10.1038/ismej.2008.39 [DOI] [PubMed] [Google Scholar]
- Fleming E. J., Davis R. E., Mcallister S. M., Chan C. S., Moyer C. L., Tebo B. M., et al. (2013). Hidden in plain sight: discovery of sheath-forming, iron-oxidizing Zetaproteobacteria at Loihi Seamount, Hawaii, USA. FEMS Microbiol. Ecol. 85 116–127. 10.1111/1574-6941.12104 [DOI] [PubMed] [Google Scholar]
- Flores G., Shakya M., Meneghin J., Yang Z., Seewald J., Geoff Wheat C., et al. (2012). Inter-field variability in the microbial communities of hydrothermal vent deposits from a back-arc basin. Geobiology 10 333–346. 10.1111/j.1472-4669.2012.00325.x [DOI] [PubMed] [Google Scholar]
- Flores G. E., Campbell J. H., Kirshtein J. D., Meneghin J., Podar M., Steinberg J. I., et al. (2011). Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge. Environ. Microbiol. 13 2158–2171. 10.1111/j.1462-2920.2011.02463.x [DOI] [PubMed] [Google Scholar]
- Forrest M. J., Ledesma-Vázquez J., Ussler W., Kulongoski J. T., Hilton D. R., Greene H. G. (2005). Gas geochemistry of a shallow submarine hydrothermal vent associated with the El Requesón fault zone, Bahía Concepción, Baja California Sur, México. Chem. Geol. 224 82–95. [Google Scholar]
- Fossing H., Jørgensen B. B. (1989). Measurement of bacterial sulfate reduction in sediments: evaluation of a single-step chromium reduction method. Biogeochemistry 8 205–222. 10.1007/BF00002889 [DOI] [Google Scholar]
- Frank K. L., Rogers D. R., Olins H. C., Vidoudez C., Girguis P. R. (2013). Characterizing the distribution and rates of microbial sulfate reduction at Middle Valley hydrothermal vents. ISME J. 7 1391–1401. 10.1038/ismej.2013.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimchuk A., Shatalov A., Novikov A., Butorova O., Pimenov N., Lein A. Y., et al. (2010). The search for sulfate-reducing bacteria in mat samples from the Lost City hydrothermal field by molecular cloning. Microbiology 79 96–105. 10.1134/S0026261710010133 [DOI] [PubMed] [Google Scholar]
- Giovannelli D., D’errico G., Manini E., Yakimov M., Vetriani C. (2013). Diversity and phylogenetic analyses of bacteria from a shallow-water hydrothermal vent in Milos island (Greece). Front. Microbiol. 4:184 10.3389/fmicb.2013.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros O., Darrasse A., Durand P., Frenkiel L., Moueza M. (1996). Environmental transmission of a Sulfur-Oxidizing bacterial gill endosymbiont in the tropical lucinid bivalve Codakia orbicularis. Appl. Environ. Microbiol. 62 2324–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove M., Bebout G., Jacobson C., Barth A., Kimbrough D., King R., et al. (2008). The Catalina Schist: evidence for middle cretaceous subduction erosion of southwestern North America. Geol. Soc. Am. Spec. Pap. 436 335–361. [Google Scholar]
- Grunke S., Lichtschlag A., De Beer D., Felden J., Salman V., Ramette A., et al. (2012). Mats of psychrophilic thiotrophic bacteria associated with cold seeps of the Barents Sea. Biogeosciences 9 2947–2960. 10.5194/bg-9-2947-2012 [DOI] [Google Scholar]
- Heijs S. K., Damste J. S., Forney L. J. (2005). Characterization of a deep-sea microbial mat from an active cold seep at the Milano mud volcano in the Eastern Mediterranean Sea. FEMS Microbiol. Ecol. 54 47–56. 10.1016/j.femsec.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Houghton J., Seyfried W., Jr., Banta A., Reysenbach A.-L. (2007). Continuous enrichment culturing of thermophiles under sulfate and nitrate-reducing conditions and at deep-sea hydrostatic pressures. Extremophiles 11 371–382. 10.1007/s00792-006-0049-7 [DOI] [PubMed] [Google Scholar]
- Howarth R. W., Jørgensen B. B. (1984). Formation of 35 S-labelled elemental sulfur and pyrite in coastal marine sediments (Limfjorden and Kysing Fjord, Denmark) during short-term 35 SO 4 2- reduction measurements. Geochim. Cosmochim. Acta 48 1807–1818. [Google Scholar]
- Huber J. A., Butterfield D. A., Baross J. A. (2003). Bacterial diversity in a subseafloor habitat following a deep-sea volcanic eruption. FEMS Microbiol. Ecol. 43 393–409. 10.1111/j.1574-6941.2003.tb01080.x [DOI] [PubMed] [Google Scholar]
- Huber J. A., Cantin H. V., Huse S. M., Mark Welch D. B., Sogin M. L., Butterfield D. A. (2010). Isolated communities of Epsilonproteobacteria in hydrothermal vent fluids of the Mariana Arc seamounts. FEMS Microbiol. Ecol. 73 538–549. 10.1111/j.1574-6941.2010.00910.x [DOI] [PubMed] [Google Scholar]
- Hügler M., Gärtner A., Imhoff J. F. (2010). Functional genes as markers for sulfur cycling and CO2 fixation in microbial communities of hydrothermal vents of the Logatchev field. FEMS Microbiol. Ecol. 73 526–537. 10.1111/j.1574-6941.2010.00919.x [DOI] [PubMed] [Google Scholar]
- Inagaki F., Takai K., Hideki K. I., Nealson K. H., Horikishi K. (2003). Sulfurimonas autotrophica gen. nov., sp nov., a novel sulfur-oxidizing epsilon-proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 53 1801–1805. 10.1099/ijs.0.02682-0 [DOI] [PubMed] [Google Scholar]
- Inagaki F., Takai K., Nealson K. H., Horikoshi K. (2004). Sulfurovum lithotrophicum gen. nov., sp nov., a novel sulfur-oxidizing chemolithoautotroph within the epsilon-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 54 1477–1482. 10.1099/ijs.0.03042-0 [DOI] [PubMed] [Google Scholar]
- Jacq E., Prieur D., Nichols P., White D., Porter T., Geesey G. (1989). Microscopic examination and fatty acid characterization of filamentous bacteria colonizing substrata around subtidal hydrothermal vents. Arch. Microbiol. 152 64–71. 10.1007/BF00447013 [DOI] [Google Scholar]
- Jaeschke A., Jørgensen S. L., Bernasconi S. M., Pedersen R. B., Thorseth I. H., Früh-Green G. L. (2012). Microbial diversity of loki’s castle black smokers at the arctic mid-ocean ridge. Geobiology 10 548–561. 10.1111/gbi.12009 [DOI] [PubMed] [Google Scholar]
- Jannasch H. W., Mottl M. J. (1985). Geomicrobiology of deep-sea hydrothermal vents. Science 229 717–725. 10.1126/science.229.4715.717 [DOI] [PubMed] [Google Scholar]
- Jannasch H. W., Wirsen C. O., Molyneaux S. J., Langworthy T. A. (1988). Extremely thermophilic fermentative archaebacteria of the genus Desulfurococcus from deep-sea hydrothermal vents. Appl. Environ. Microbiol. 54 1203–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen B. B., Bak F. (1991). Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (Kattegat, Denmark). Appl. Environ. Microbiol. 57 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen B. B., Isaksen M. F., Jannasch H. W. (1992). Bacterial sulfate reduction above 100°C in deep-sea hydrothermal vent sediments. Science 258 1756–1757. 10.1126/science.258.5089.1756 [DOI] [PubMed] [Google Scholar]
- Jørgensen B. B., Nelson D. C. (2004). “Sulfide oxidation in marine sediments: geochemistry meets microbiology,” in Sulfur Biogeochemistry—Past and Present eds Amend P., Edwards K. J., Lyons T. W. (Boulder, CO: Geological Society of America; ) 63–81. [Google Scholar]
- Kakegawa T., Utsumi M., Marumo K. (2008). Geochemistry of sulfide chimneys and basement pillow lavas at the Southern Mariana Trough (12.55 N–12.58 N). Resour. Geol. 58 249–266. 10.1111/j.1751-3928.2008.00060.x [DOI] [Google Scholar]
- Kalanetra K. M., Huston S. L., Nelson D. C. (2004). Novel, attached, sulfur-oxidizing bacteria at shallow hydrothermal vents possess vacuoles not involved in respiratory nitrate accumulation. Appl. Environ. Microbiol. 70 7487–7496. 10.1128/AEM.70.12.7487-7496.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl D., Wirsen C., Jannasch H. (1980). Deep-Sea primary production at the Galapagos hydrothermal vents. Science 207 1345–1347. 10.1186/1471-2148-11-96 [DOI] [Google Scholar]
- Kato S., Kobayashi C., Kakegawa T., Yamagishi A. (2009). Microbial communities in iron-silica-rich microbial mats at deep-sea hydrothermal fields of the Southern Mariana Trough. Environ. Microbiol. 11 2094–2111. 10.1111/j.1462-2920.2009.01930.x [DOI] [PubMed] [Google Scholar]
- Kato S., Nakamura K., Toki T., Ishibashi J.-I., Tsunogai U., Hirota A., et al. (2012). Iron-based microbial ecosystem on and below the seafloor: a case study of hydrothermal fields of the Southern Mariana Trough. Front. Microbiol. 3:89 10.3389/fmicb.2012.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S., Takano Y., Kakegawa T., Oba H., Inoue K., Kobayashi C., et al. (2010). Biogeography and biodiversity in sulfide structures of active and inactive vents at deep-sea hydrothermal fields of the Southern Mariana Trough. Appl. Environ. Microbiol. 76 2968–2979. 10.1128/AEM.00478-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D. S., Baross J. A., Delaney J. R. (2002). Volcanoes, fluids, and life at mid-ocean ridge spreading centers. Annu. Rev. Earth Planet. Sci. 30 385–491. 10.1146/annurev.earth.30.091201.141331 [DOI] [Google Scholar]
- Kelley D. S., Karson J. A., Früh-Green G. L., Yoerger D. R., Shank T. M., Butterfield D. A., et al. (2005). A serpentinite-hosted ecosystem: the Lost City hydrothermal field. Science 307 1428–1434. 10.1126/science.1102556 [DOI] [PubMed] [Google Scholar]
- Kelley D. S., Shank T. M. (2010). “Hydrothermal systems: a decade of discovery in slow spreading environments,” in Diversity of Hydrothermal Systems on Slow Spreading Ocean Ridges eds Rona P. A., Devey C. W., Dyment J., Murton B. J. (Wshington, DC: American Geophysical Union; ) 369–407. [Google Scholar]
- Kilias S. P., Nomikou P., Papanikolaou D., Polymenakou P. N., Godelitsas A., Argyraki A., et al. (2013). New insights into hydrothermal vent processes in the unique shallow-submarine arc-volcano, Kolumbo (Santorini), Greece. Sci. Rep. 3 2421 10.1038/srep02421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormas K. A., Tivey M. K., Von Damm K., Teske A. (2006). Bacterial and archaeal phylotypes associated with distinct mineralogical layers of a white smoker spire from a deep-sea hydrothermal vent site (9°N, East Pacific Rise). Environ. Microbiol. 8 909–920. [DOI] [PubMed] [Google Scholar]
- Lane D. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics eds Stackebrandt E., Goodfellow M. (New York, NY: Wiley; ) 115–175. [Google Scholar]
- Lanzén A., Jørgensen S. L., Bengtsson M. M., Jonassen I., Øvreås L., Urich T. (2011). Exploring the composition and diversity of microbial communities at the Jan Mayen hydrothermal vent field using RNA and DNA. FEMS Microbiol. Ecol. 77 577–589. 10.1111/j.1574-6941.2011.01138.x [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N., Chenna R., Mcgettigan P. A., Mcwilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Le Moine Bauer S., Roalkvam I., Steen I. H., Dahle H. (2016). Lutibacter profundi sp. nov., isolated from a deep-sea hydrothermal system on the Arctic Mid-Ocean Ridge and emended description of the genus Lutibacter. Int. J. Syst. Evol. Microbiol. 10.1099/ijsem.0.001105 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Liesack W., Finster K. (1994). Phylogenetic analysis of five strains of gram-negative, obligately anaerobic, sulfur-reducing bacteria and description of Desulfuromusa gen. nov., including Desulfuromusa kysingii sp. nov., Desulfuromusa bakii sp. nov., and Desulfuromusa succinoxidans sp. nov. Int. J. Syst. Bacteriol. 44 753–758. [Google Scholar]
- Loy A., Lehner A., Lee N., Adamczyk J., Meier H., Ernst J., et al. (2002). Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68 5064–5081. 10.1128/AEM.68.10.5064-5081.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71 8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W., Strunk O., Westram R., Richter L., Meier H., Buchner A., et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32 1363–1371. 10.1093/nar/gkh293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz W., Eisenbrecher M., Neu T. R., Szewzyk U. (1998). Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol. Ecol. 25 43–61. 10.1111/j.1574-6941.1998.tb00459.x [DOI] [Google Scholar]
- McCaig A. M., Cliff R. A., Escartin J., Fallick A. E., Macleod C. J. (2007). Oceanic detachment faults focus very large volumes of black smoker fluids. Geology 35 935–938. 10.1130/G23657A.1 [DOI] [Google Scholar]
- Meyer B., Kuever J. (2007). Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5’-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology 153 3478–3498. 10.1099/mic.0.2007/008250-0 [DOI] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W., Schwartz T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in proceedings of the Gateway Computing Environments Workshop (GCE) (New Orleans, LA: IEEE; ) 1–8. [Google Scholar]
- Moyer C. L., Dobbs F. C., Karl D. M. (1995). Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 61 1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdock S., Johnson H., Forget N., Juniper S. K. (2010). Composition and diversity of microbial mats at shallow hydrothermal vents on Volcano 1, South Tonga Arc. Cah. Biol. Mar. 51 407–413. [Google Scholar]
- Muyzer G., Teske A., Wirsen C. O., Jannasch H. W. (1995). Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164 165–172. 10.1007/BF02529967 [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Takai K., Inagaki F., Hirayama H., Nunoura T., Horikoshi K., et al. (2005). Distribution, phylogenetic diversity and physiological characteristics of Epsilonproteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 7 1619–1632. 10.1111/j.1462-2920.2005.00856.x [DOI] [PubMed] [Google Scholar]
- Nakamura K., Morishita T., Bach W., Klein F., Hara K., Okino K., et al. (2009). Serpentinized troctolites exposed near the kairei hydrothermal field, central indian ridge: insights into the origin of the kairei hydrothermal fluid supporting a unique microbial ecosystem. Earth Planet. Sci. Lett. 280 128–136. [Google Scholar]
- Nakamura K., Takai K. (2014). Theoretical constraints of physical and chemical properties of hydrothermal fluids on variations in chemolithotrophic microbial communities in seafloor hydrothermal systems. Prog. Earth Planet. Sci. 1 1–24. 10.1186/2197-4284-1-5 [DOI] [Google Scholar]
- Olenin S., Ducrotoy J.-P. (2006). The concept of biotope in marine ecology and coastal management. Mar. Poll. Bull. 53 20–29. 10.1016/j.marpolbul.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Oremland R. S., Capone D. G. (1988). “Use of “specific” inhibitors in biogeochemistry and microbial ecology,” in Advances in Microbial Ecology ed. Marshall K. C. (Boston, MA: Springer; ) 285–383. [Google Scholar]
- Pašava J., Vymazalová A., Petersen S. (2007). PGE fractionation in seafloor hydrothermal systems: examples from mafic-and ultramafic-hosted hydrothermal fields at the slow-spreading Mid-Atlantic Ridge. Miner. Deposita 42 423–431. 10.1007/s00126-006-0122-2 [DOI] [Google Scholar]
- Perner M., Kuever J., Seifert R., Pape T., Koschinsky A., Schmidt K., et al. (2007). The influence of ultramafic rocks on microbial communities at the Logatchev hydrothermal field, located 15°N on the Mid-Atlantic Ridge. FEMS Microbiol. Ecol. 61 97–109. 10.1111/j.1574-6941.2007.00325.x [DOI] [PubMed] [Google Scholar]
- Price R. E., Lesniewski R., Nitzsche K. S., Meyerdierks A., Saltikov C., Pichler T., et al. (2013). Archaeal and bacterial diversity in an arsenic-rich shallow-sea hydrothermal system undergoing phase separation. Front. Microbiol. 4:158 10.3389/fmicb.2013.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prol-Ledesma R., Dando P., De Ronde C. (2005). Special issue on “shallow-water hydrothermal venting”. Chem. Geol. 224 1–4. 10.1016/j.chemgeo.2005.07.012 [DOI] [Google Scholar]
- Pruesse E., Peplies J., Glöckner F. O. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28 1823–1829. 10.1093/bioinformatics/bts252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach A.-L., Shock E. (2002). Merging genomes with geochemistry in hydrothermal ecosystems. Science 296 1077–1082. 10.1126/science.1072483 [DOI] [PubMed] [Google Scholar]
- Rusch A., Walpersdorf E., Gurrieri S., Amend J. P. (2005). Microbial communities near the oxic/anoxic interface in the hydrothermal system of Vulcano Island, Italy. Chem. Geol. 224 169–182. 10.1016/j.chemgeo.2005.07.026 [DOI] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., Schindelin J., Arganda-Carreras I., Frise E., et al. (2012). NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrenk M. O., Kelley D. S., Bolton S. A., Baross J. A. (2004). Low archaeal diversity linked to subseafloor geochemical processes at the Lost City hydrothermal field, Mid-Atlantic Ridge. Environ. Microbiol. 6 1086–1095. 10.1111/j.1462-2920.2004.00650.x [DOI] [PubMed] [Google Scholar]
- Sheik C. S., Anantharaman K., Breier J. A., Sylvan J. B., Edwards K. J., Dick G. J. (2015). Spatially resolved sampling reveals dynamic microbial communities in rising hydrothermal plumes across a back-arc basin. ISME J. 9 1434–1445. 10.1038/ismej.2014.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert S. M., Brinkhoff T., Muyzer G., Ziebis W., Kuever J. (1999). Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 65 3834–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert S. M., Kuever J., Muyzer G. (2000). Identification of 16S ribosomal DNA-defined bacterial populations at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 66 3102–3109. 10.1128/AEM.66.7.3102-3109.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Lane D. J., Olsen G. J., Heller D. J., Schmidt T. M., Pace N. R. (1987). Phylogenetic analysis of certain sulfide-oxidizing and related morphologically conspicuous bacteria by 5S ribosomal ribonucleic acid sequences. Int. J. Syst. Bacteriol. 37 116–122. 10.1099/00207713-37-2-116 [DOI] [Google Scholar]
- Stein J. L. (1984). Subtidal gastropods consume sulfur-oxidizing bacteria: evidence from coastal hydrothermal vents. Science 223 696–698. 10.1126/science.223.4637.696 [DOI] [PubMed] [Google Scholar]
- Sylvan J. B., Sia T. Y., Haddad A. G., Briscoe L. J., Toner B. M., Girguis P. R., et al. (2013). Low temperature geomicrobiology follows host rock composition along a geochemical gradient in Lau Basin. Front. Microbiol. 4:61 10.3389/fmicb.2013.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvan J. B., Toner B. M., Edwards K. J. (2012). Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. mBio 3 e00279-11 10.1128/mBio.00279-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K., Miyazaki M., Nunoura T., Hirayama H., Oida H., Furushima Y., et al. (2006a). Sulfurivirga caldicuralii gen. nov., sp. nov., a novel microaerobic, thermophilic, thiosulfate-oxidizing chemolithoautotroph, isolated from a shallow marine hydrothermal system occurring in a coral reef. Japan. Int. J. Syst. Evol. Microbiol. 56 1921–1929. 10.1099/ijs.0.64297-0 [DOI] [PubMed] [Google Scholar]
- Takai K., Suzuki M., Nakagawa S., Miyazaki M., Suzuki Y., Inagaki F., et al. (2006b). Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen-and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 56 1725–1733. [DOI] [PubMed] [Google Scholar]
- Tang K., Liu K., Jiao N., Zhang Y., Chen C.-T. A. (2013). Functional metagenomic investigations of microbial communities in a shallow-sea hydrothermal system. PLoS ONE 8:e72958 10.1371/journal.pone.0072958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov V. G., Gebruk A. V., Mironov A. N., Moskalev L. I. (2005). Deep-sea and shallow-water hydrothermal vent communities: two different phenomena? Chem. Geol. 224 5–39. 10.1016/j.chemgeo.2005.07.021 [DOI] [Google Scholar]
- Thamdrup B., Canfield D. E. (1996). Pathways of carbon oxidation in continental margin sediments off central Chile. Limnol. Oceanogr. 41 1629–1650. 10.4319/lo.1996.41.8.1629 [DOI] [PubMed] [Google Scholar]
- Tivey M. K. (2007). Generation of seafloor hydrothermal vent fluids and associated mineral deposits. Oceanography 20 50–65. 10.5670/oceanog.2007.80 [DOI] [Google Scholar]
- Tunnicliffe V., Botros M., De Burgh M. E., Dinet A., Johnson H. P., Juniper S. K., et al. (1986). Hydrothermal vents of explorer ridge, northeast pacific. deep sea research part 1. Oceanogr. Res. Pap. 33 401–412. 10.1016/0198-0149(86)90100-7 [DOI] [Google Scholar]
- Ulrich G. A., Krumholz L. R., Suflita J. M. (1997). A rapid and simple method for estimating sulfate reduction activity and quantifying inorganic sulfides. Appl. Environ. Microbiol. 63 1627–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich T., Lanzén A., Stokke R., Pedersen R. B., Bayer C., Thorseth I. H., et al. (2014). Microbial community structure and functioning in marine sediments associated with diffuse hydrothermal venting assessed by integrated meta-omics. Environ. Microbiol. 16 2699–2710. 10.1111/1462-2920.12283 [DOI] [PubMed] [Google Scholar]
- Vandieken V., Mußmann M., Niemann H., Jørgensen B. B. (2006). Desulfuromonas svalbardensis sp. nov. and Desulfuromusa ferrireducens sp. nov., psychrophilic, Fe (III)-reducing bacteria isolated from Arctic sediments, Svalbard. Int. J. Syst. Evol. Microbiol. 56 1133–1139. [DOI] [PubMed] [Google Scholar]
- Wang L., Cheung M. K., Kwan H. S., Hwang J. S., Wong C. K. (2015). Microbial diversity in shallow-water hydrothermal sediments of Kueishan Island, Taiwan as revealed by pyrosequencing. J. Basic Microbiol. 55 1308–1318. 10.1002/jobm.201400811 [DOI] [PubMed] [Google Scholar]
- Weber A., Jørgensen B. B. (2002). Bacterial sulfate reduction in hydrothermal sediments of the Guaymas Basin, Gulf of California, Mexico. Deep-Sea Res. Pt. I 49 827–841. [Google Scholar]
- Wilbanks E. G., Jaekel U., Salman V., Humphrey P. T., Eisen J. A., Facciotti M. T., et al. (2014). Microscale sulfur cycling in the phototrophic pink berry consortia of the Sippewissett Salt Marsh. Environ. Microbiol. 16 3398–3415. 10.1111/1462-2920.12388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodring W. P., Bramlette M. N., Kew W. S. W. (1946). Geology and Paleontology of Palos Verdes Hills, California. Washington, DC: U.S. Govt. Print. Off. [Google Scholar]
- Yamamoto M., Takai K. (2011). Sulfur metabolisms in Epsilon-and Gamma-Proteobacteria in deep-sea hydrothermal fields. Front. Microbiol. 2:192 10.3389/fmicb.2011.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbinden M., Marqué L., Gaudron S. M., Ravaux J., Léger N., Duperron S. (2015). Epsilonproteobacteria as gill epibionts of the hydrothermal vent gastropod Cyathermia naticoides (North East-Pacific Rise). Mar. Biol. 162 435–448. 10.1007/s00227-014-2591-7 [DOI] [Google Scholar]
- Zielinski F. U., Gennerich H. H., Borowski C., Wenzhöfer F., Dubilier N. (2011). In situ measurements of hydrogen sulfide, oxygen, and temperature in diffuse fluids of an ultramafic-hosted hydrothermal vent field (Logatchev, 14° 45’ N, Mid-Atlantic Ridge): implications for chemosymbiotic Bathymodiolin mussels. Geochem. Geophys. Geosyst 12 Q0AE04. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


