Abstract
Malarial parasite P. falciparum, an apicomplexan protozoan has a 23.3 MB nuclear genome and encodes ~ 5600 transcripts. The genetic diversity of the parasite within and across geographical zones is a challenge to gene expression studies which are essential for understanding of disease process, outcome and developing markers for diagnostics and prognostics. Here, we describe the strategy involved in designing a custom P. falciparum 15K array using the Agilent platform and Genotypic's Right Design methodology to study the transcriptome of Indian field isolates for which genome sequence information is limited. The array contains probes representing genome sequences of two distinct geographical isolates (i.e. 3D7 and HB3) and sub-telomeric var gene sequences of a third isolate (IT4) known to adhere in culture condition. Probes in the array have been selected based on their efficiency to detect transcripts through a 244K array experimentation. Array performance for the 15K array, was evaluated and validated using RNA materials from P. falciparum clinical isolates. A large percentage (91%) of the represented transcripts was detected from Indian P. falciparum patient isolates. Replicated probes and multiple probes representing the same gene showed perfect correlation between them suggesting good probe performance. Additional transcripts could be detected due to inclusion of unique probes representing HB3 strain transcripts. Variant surface antigen (VSA) transcripts were detected by optimized probes representing the VSA genes of three geographically distinct strains. The 15K cross strain P. falciparum array has shown good efficiency in detecting transcripts from P. falciparum parasite samples isolated from patients. The low parasite loads and presence of host RNA makes arrays a preferred platform for gene expression studies over RNA-Seq.
Abbreviations: PfENO, enolase (PF10_0155); PfCP, conserved hypothetical protein (PF14_0683); PfSec14, Sec 14 domain containing protein (PF1280w); PfROM3, rhomboid protease 3 (MAL8P1.16); PfGK, glycerol kinase (PF13_0269)
Keywords: Plasmodium falciparum, Microarray, Uncomplicated malaria, Complicated malaria, PfEMP1, RIFIN
1. Introduction
Malaria imposes a significant burden on world health. There were an estimated 149–303 million cases of infections along with 0.48 million deaths reported worldwide in the year 2015 [39]. In the year 2014, 1.6 million confirmed cases were reported from the 10 malaria endemic South-East Asian countries, of which, India registered the highest number of confirmed cases (70%) [39]. Plasmodium falciparum is often considered the most virulent among the 5 Plasmodium species known to infect humans. Malaria elimination programs remain challenged due to the emergence and reemergence of drug resistant parasites and inability to develop an effective vaccine [11], [14].
The failure to develop a successful vaccine and emergence of drug resistant parasites can partly be attributed to the complex life cycle of the parasite. Interestingly, many basic aspects of its biology, while in the host, still need to be explored. This has been partially achieved in recent years due to the advancement in techniques like microarray (DNA microarrays, Protein binding microarrays, SNP microarrays), array-comparative genomic hybridization (aCGH) [19], RNA-seq [26], ChIP-chip [9], Chip-seq [3], mass spectrometry techniques [25], transfection techniques [5], mutagenesis methods [2], gene knock out techniques [8], [20] and imaging techniques. All these techniques have helped in exploring some fundamental questions, which was not possible earlier.
The evolution of P. falciparum DNA microarrays has started few years before the release of its first sequence draft using PCR amplified inserts from a P. falciparum DNA library (shotgun microarray) or cDNAs [12], [21]. Following the release of the first draft of P. falciparum 3D7 genome sequence from Malaria Genome Consortium (The Sanger center Plasmodium falciparum Genome Project, Stanford Genome Technology Malaria Genome Project, TIGR Plasmodium falciparum Genome Database), many oligonucleotide based whole genome microarrays has been designed, Importantly high density short oligonucleotide (25mer) microarray on Affymetrix platform came from Winzeler lab [18] and long oligonucleotide (70mer) spotted microarray came from DeRisi lab [4]. Upon release of subsequent updates of P. falciparum genome database annotation, many new versions of microarrays were evolved and utilized various platforms (To see the comprehensive list of P. falciparum microarrays designed till date, please refer to NCBI's Gene Expression Omnibus website http://www.ncbi.nlm.nih.gov/geo/.
Use of microarrays has provided new avenues for understanding expression and regulation of genes at a genome-wide level. DNA microarrays have been utilized to show the stage specific temporal expression pattern of the parasite during its intra-erythrocytic developmental cycle (IDC) [4], [18], [21]. Other microarray based studies have also helped to explore the transcriptional status of genes in other life cycle stages like gametocyte and sporozoite stages, differences and commonality of transcriptional programming at a cross strain level [19], transcripts possibly involved in gametocytogenesis [32], [41], in vivo transcriptional states of the parasite in infected patients [6], soft-wired transcriptional reprogramming in response to drug perturbations [23], [36] or environmental cues [24] and expression pattern of variant surface antigens particularly in clinical conditions [37]. Although the microarray technique has helped in addressing many fundamental questions, its use has been limited due to its cost and comparatively large amount of starting biological material required which is a limitation in the case of clinical isolates. Most of the arrays for P. falciparum have been designed based on a single reference strain genome (3D7 laboratory strain) [4], [13], [18]. These arrays have not considered the possibilities of genomic variations among strains that could limit transcript detection.
A cross strain P. falciparum custom 15K microarray has been designed in a platform, which provides in many ways the flexibility to design a custom array. This extends from ordering the number of slides, allowing updation of the microarray design as and when required. Further, due to the cRNA amplification strategy adopted, the amount of starting material required makes studies with field derived samples more feasible. The use of ink-jet based in situ synthesized 60mer long oligonucleotide probes in the selected platform enhances its sensitivity and due to outstanding spot uniformity, it reduces the inter-array variability. It has a broad dynamic range of signal intensity and shows low and uniform background signal [16]. Probes in our 15K cross-strain array represent the genomes of two geographically distinct strains i.e. 3D7 and HB3 and the var gene sequences of a third strain (IT4). Probes have been validated using RNA material from Indian P. falciparum clinical isolates in a 244K array format [33] and best probes were chosen for the 15K array design. The pre-probe selection experiment helped in identification of probes for efficient detection of transcript sequences of field isolates. Here, we report, the further validation of our cross strain P. falciparum custom 15K arrays using 13 clinical isolates from patients with uncomplicated/complicated symptoms due to P. falciparum infection.
2. Results and discussion
This study intends to provide validation of data from the custom cross strain microarray. The overall examination of the microarray data is presented as a analysis to validate the strength and accuracy of the array design and procedures.
2.1. Probe selection for 15K Plasmodium falciparum cross strain whole genome GXP microarray
To screen best probes for transcriptome studies of Plasmodium falciparum Indian field isolates for which genome sequence information is limited, a 244K custom whole genome array was designed on an Agilent platform with 60mer oligonucleotide probes (Agilent Microarray Design Identity number (AMADID) 024956). Probes represent P. falciparum 3D7 strain transcript sequences (PlasmoDB v5.3) [1] and NCBI EST sequences (NCBI 2007). Details about the 244K array can be found in our previous report [33].
Probe selection for 15K array was carried out by considering only sense probes representing PlasmoDB transcripts. GE gMedianSignal (green channel) and GE_rMedianSignal intensity (red channel) for each probe was collected. Maximum value of the GE green and red channel intensity of each probe representing a transcript was calculated, ranked and probe with maximum GE_Median signal intensity selected for each transcript. Probes with signal intensity > 96 (~ twice the average background intensity) were taken for consideration (3025 probes). Transcripts for which representing probes did not qualify the criteria of showing intensity > 96, probes were selected based on their proximity to the 3′ end of the representing sequences. In this way, 5629 probes representing 3D7 transcript sequences were collected from the 244K array.
2.2. Plasmodium falciparum 15K cross strain whole genome GXP microarray design
We also designed and included probes against sequences of two other geographically distinct sequenced P. falciparum strains i.e. HB3 and IT4 so as to represent diverse sequences of P. falciparum in the array. To design a cross strain P. falciparum array, transcript sequences were collected from PlasmoDB v6.3 for 3D7 strain (5595 transcript sequences), from Broad Institute for HB3 strain (5623 transcript sequences) and from NCBI for IT4 strain (80 var gene transcript sequences) and a unique database was made. Total number of sequences thus collected were 11,298 out of which 9842 were unique sequences (Although sequence similarity was observed between them) and 1427 transcripts sequences of HB3 were identical to 3D7 transcripts sequences (determined from BLAST results). Probes collected from the 244K array experiment were BLASTed against the database and probes having only single significant hit with any of the strain were selected. New probes were designed for the transcripts for which no probes were found, using Agilent eArray tool. In the case of transcripts for which specific probes could not be designed, probes having minimum number of hits were selected (probes with a potential to cross hybridize) and these were flagged. Criterion for significant hit was alignment of 30 bp or more with > 84% identity with the transcript sequence. Although, the criteria for significant hit was as mentioned above, 90% of the probes showed 60 bp alignment with 100% identity to the transcript sequences. All probes, collected and designed were BLASTed against the human transcriptome to check for their cross hybridizing potential with the human transcripts. Potential cross hybridizing probes were removed from the array design. A total of 6362 user defined long oligonucleotide (60mer) probes were designed against the transcript sequences present in the database. Based on the BLAST results, probes were annotated as specific to any one strain (Single significant hit within that strain-3D7 specific/HB3 specific/IT4 specific), common in between any two or more strain (Single significant hit within each strain-3D7 and HB3 or 3D7 and IT4 or HB3 and IT4 or 3D7, HB3 and IT4) and cross hybridizing probes (multiple significant hits within a strain). Array was re-annotated recently using the PlasmoDB v8.2. After the re-annotation, the final design of the P. falciparum cross strain whole genome GXP array (AMADID: 024956) is summarized in Supplementary Table S1 and S2. Out of 6362 probes present in the array, 6120 probes can be used against the transcripts of any one of the strain, 194 probes are cross hybridizing in nature and 48 probes cannot be used after re-annotation. Briefly, 5791 probes representing 5276 3D7 transcripts, 5374 probes representing 5251 HB3 transcripts and 69 probes representing 43 IT4 var gene transcripts are present in the array. Further classification of the probes based on the specificity was performed and there are 691 3D7 specific probes, 224 HB3 specific probes and 41 IT4 specific probes. These probes were designated specific for these strains, as they either did not show alignment, or showed multiple transcript alignments, for the genome of the other strains. 5164 probes are common between strains of which, 5077, 13, 6 and 9 can be used in common between 3D7 and HB3, 3D7 and IT4, HB3 and IT4 and between 3D7, HB3, and IT4 strain respectively. Of the, 5276 3D7 transcripts which are represented by specific probes in the array, 5203 (94%) are protein coding, 14 of them are non protein coding (NPC), 2 of them are snRNA coding, 10 of them are r RNA coding and 47 of them are t RNA coding genes. Because of the dynamic nature of the genome sequence annotation, a few transcripts that were represented by a specific probe in the earlier annotation are not represented by any probes in the new annotation. Probes against these transcripts can be designed and incorporated in the new version of the array as Agilent provides the flexibility in up gradation of the custom array.
The array also contains 22 probes representing 8 human housekeeping genes and 536 Agilent controls. The total number of features that were available in the 8 × 15K array after filling the 536 Agilent control features was 15,208. Each designed probe (6362 P. falciparum probes and 22 human probes) was replicated to fill the features. The remaining 2440 blank features were filled by randomly selected duplicate probes from the set of probes specific to any one strain. All the 60mer oligonucleotide probes were designed and synthesized in situ as per algorithms and methodologies used by Agilent technologies for 60mer in situ oligonucleotide DNA microarrays.
2.3. Performance of probes to detect transcripts of Plasmodium falciparum field isolates
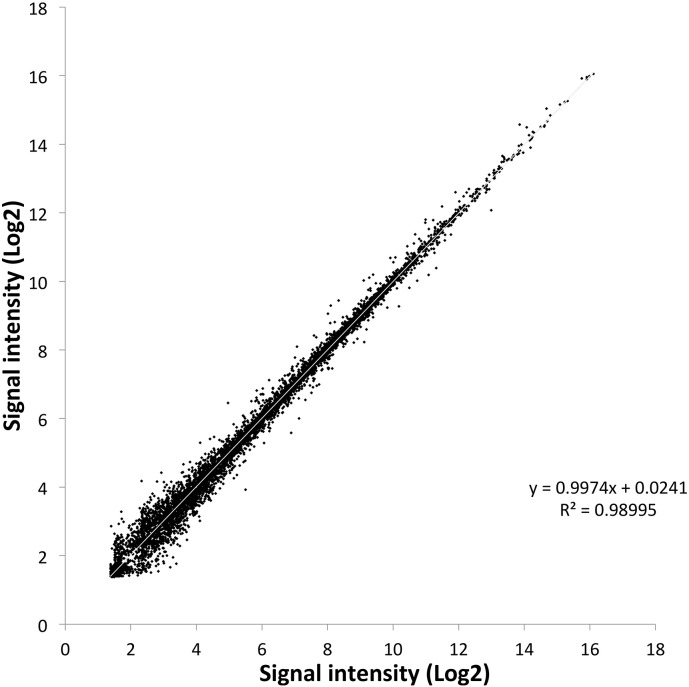
Probe efficiency was evaluated for its ability to detect the transcripts of the Indian P. falciparum field isolates. Parasite RNA samples (n = 13) were hybridized to individual arrays. Based on the normalized signal intensity, 5502 probes (90%) were able to detect 91% of 3D7 and 92% of HB3 transcript sequences in at least one out of thirteen samples analyzed. Transcripts were not detected by 618 (10%) probes and may represent genes which might express in parasite stages not present in the samples analyzed here or might represent variant surface antigen coding genes. This point has been analyzed in detail in the next section. Due to the in situ probe creation process, probes were of high quality and the background signal was quite low. There are 6120 probes representing P. falciparum transcripts sequences that are present as duplicates in the array. All the duplicate probes showed almost perfect correlation (r = 0.99) between them (Fig. 1).
Fig. 1.
Correlation of replicated probes. Signal intensity (log2) from the median of 13 RNA samples of replicated probe pairs (6120 pairs; R2 = 0.99).
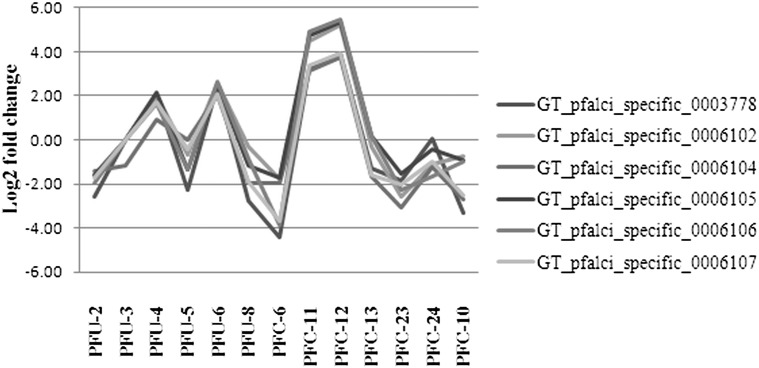
The array contains 10% of 3D7 genes represented by more than one probe (1101 probes representing 522 genes) and accuracy of measurements of these individual probes was verified. Pair wise Pearson correlation of multiple probes representing the same gene was calculated. Seventy two percent of genes represented by multiple probes and detected in at least 4 out of 13 samples showed average Pearson correlation (r) above 0.8 between the probes representing the same gene. Percentage of genes with Pearson correlation between their representative probes > 0.8 was found to increase, if probe detection status increased for number of samples, e.g. increased from 4 samples to 5 or 6 samples (Supplementary Fig. 1). This indicates that the correlation between probes representing the same transcript is high and multiple probes which could detect transcripts in the majority of samples can be taken for further analysis with high confidence level. One of the examples for genes with multiple probes is the gene encoding DBL like protein (PF11_0506). This gene is represented by 9 probes out of which 6 probes were detected in at least 4 samples and showed an average Pearson correlation of 0.986 (Fig. 2). Probes representing a transcript with low Pearson correlation value may bind to sequence regions having variations, where one oligonucleotide may bind to the transcript efficiently in case of a test sample and the others may not. This would provide in-consistent signal intensity across the samples in comparison to the efficient binder. In case of such probes, those showing the highest consistent detection values across samples have been considered.
Fig. 2.
Correlation of 6 probes representing antigen 332 encoding gene (PF11_0506). PF11_0506 is represented by 9 probes out of which 6 probes were detected in at least 4 samples. Pair wise Pearson correlation was calculated for detected probes. Average Pearson correlation of all the 6 detected probes was 0.986. Figure shows the expression patterns of all the 6 detected probes in the 13 RNA samples as 13 data points.
The frequency of signal distribution of RNA/transcripts showed that many features were with low signal intensity, when probes irrespective of the filtering criteria (detected and not detected probe) were taken into consideration. After applying the filtering criteria and considering probes that were detected in at least one sample, many probes with low signal intensity were removed and the graph showed a bell shaped appearance adhering to the well accepted frequency distribution of signal intensities for whole genome expression profile (Supplementary Fig. 2). The low intensities for many features may be due to low expression of the genes in the blood stages of the parasite that are normally present in the peripheral blood of the infected humans from where the parasite material was isolated. The parasite exclusively expresses many of the genes that are stage specific (maximum expression at one point/stage of its lifecycle and express in low abundant or do not express in other stages) [4], [18].
2.4. Stage specific expression pattern of detected and undetected transcripts
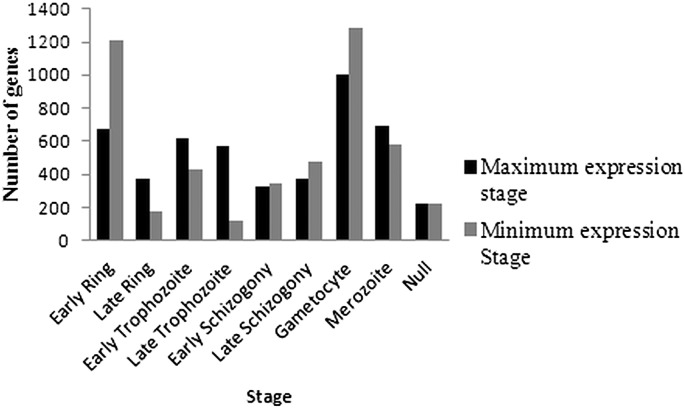
Developmental stage specific gene expression profiles of this parasite from culture condition have been reported and maximum and minimum expression stages for most of the genes have been determined [1], [4], [18]. Transcripts, which were not detected and detected in this study, were analyzed to find out the blood stages in which they express maximum and minimum [1], [4], [18]. Transcripts were detected from 4846 genes (91% of the genes represented in the array) in at least 1 sample out of the 13 samples analyzed (Supplementary Table S3). Majority of these have been reported to express maximum in G (21%), ER (14%), M (14%), LR (7%) and ET (13%) stages. Stages in which these genes expressed minimum were G (27%), ER (25%), M (12%) and LS (10%) stages (Fig. 3). There were 4.1% genes for which expression data is not available from the previous studies using microarray (PlasmoDB v8.2) [1], [18]. This indicates that the probes present in the array were able to detect most of the represented transcripts if expressed in a detectable range.
Fig. 3.
Maximum and minimum expression stages of detected transcripts in different blood stages of the P. falciparum. Blood stages at which each detected transcript was reported to be expressed maximum and minimum were retrieved from PlasmoDB v8.2.
In this study, 6.9% transcripts of 3D7 strain represented by specific probes were not detected (Supplementary Table S4). Investigation of their expression in the reported data revealed that 29% of these (108 transcripts) expressed maximum in stages like early ring (ER)—5% (n = 20), late ring (LR)—13% (49) and Gametocyte (G)—11% (39) which are normally found in peripheral blood of infected patients (Supplementary Fig. 3). We have investigated the probable cause for not detecting these transcripts although they were represented by specific probes. We observed that many of these, which expressed maximum in early ring and late ring stages in culture condition constitutes variant parasite encoded erythrocyte surface antigens like Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) (n = 5), repetitive interspersed family protein (RIFIN) (n = 14), sub-telomeric variable open reading frame (STEVOR) (n = 5) and Plasmodium falciparum Maurer's cleft-2 trans membrane domain containing protein (PfMC-2TM) (n = 4). There are 29 genes in the list for which expression data is not available in the PlasmoDB v9.2 (transcripts have not been detected in culture condition using microarray) [1], [18]. Many genes which express maximum in merozoite (M) stage might also express in ring stage due to possible involvement in linked processes like erythrocyte invasion and infected erythrocyte remodeling, so we have scanned the list of genes which express maximum in this stage too. In the merozoite stage, 112 genes from the not detected list have been reported to express maximum, out of which again rifin (n = 54) predominates in the list. Other genes that are present in the list are genes encoding for STEVOR (n = 8), PfEMP1 (n = 3), PHISTa (n = 2) and hypothetical proteins (n = 24).
Members of variant surface antigens (PfEMP1, RIFIN and STEVOR) thought to be expressed in a mutually exclusive pattern where one member at a time expresses and others shut down. This has been well established in case of var genes encoding PfEMP1 antigens and might be the cause for un-detection of these transcripts. This might also hold true for the other erythrocyte surface antigens (i.e. RIFINs and STEVORs). Members of rifin and stevor gene family were also reported to transcribe at a later stage of the intra erythrocytic developmental cycle. Plasmodium helical interspersed sub-telomeric (PHIST) family genes containing PHIST domains have been clustered into three groups (i.e. PHISTa, PHISTb and PHISTc). Microarray data from P. falciparum 3D7 strain has shown that PHISTa genes are generally not detectably transcribed [30]. Genes which expressed in other developmental stages (early trophozoite (ET), Late trophozoite (LT), early schizont (ES), late schizont (LS), liver and mosquito stages) and are not found in the peripheral blood of the infected patients may produce low amounts of transcripts under certain in-vivo conditions, and are also found in the not detected transcripts list.
2.5. Probes specific to HB3 strain and transcripts detection
The array contains 288 HB3 strain specific probes (only can be used for HB3 strain) representing 282 HB3 transcripts. Out of these 288 probes, 147 probes are specific to HB3 strain because they did not show similarity with any of the 3D7 transcript sequences (based on BLAST results) and 141 probes were assigned as specific to HB3 strain because they showed multiple hits against 3D7 transcripts (cross hybridizing probes for 3D7 strain) but showed single hit to any one of the HB3 strain transcript (Supplementary Table S5). As many as 202 probes representing 196 transcripts were detected in at least one sample. Of the 147 probes, that are specific to HB3 (no hits found against the 3D7 transcripts), 106 probes representing 103 genes were able to detect transcripts (Supplementary Table S5). Most of these transcripts code for hypothetical proteins except for some PfEMP1 and RIFINs. This suggests that variations in the transcript sequences between strains and transcript of genes that are additional to a strain can be captured by the array. This would give information about the expression status of the transcripts that are expressed, but could not be earlier detected due to strain specific sequence variability, as most of the arrays relied on the genome sequence of P. falciparum 3D7 reference strain. This could be true for studies investigating the in vivo transcriptome status [6].
2.6. Variant surface antigens expression pattern
This array is supplemented with probes representing the parasite genes encoding variant surface antigens of three geographically distinct strains. Probes in the array represent the var genes of 3D7, HB3 and IT4 strains and rifin and stevor genes of 3D7 and HB3 strains. Transcript detection capability of these probes was ascertained.
Based on the 5′ upstream sequence and chromosomal location, the PfEMP1 proteins in the P. falciparum have been divided into five groups (A, B, C, B/A and B/C) [29]. Arrays used in this study contain specific probes representing var genes of the three strains (40 var genes of 3D7 strain, 30 of HB3 strain and 34 of IT4 strain) (Supplementary Table S6). Probes representing var genes were either specific for one strain or could be common between strains. All the full var genes were considered for this analysis and var pseudo genes, var gene fragments and var like genes were excluded from the analysis, although many of these are represented by specific probes. Many var genes that are not represented by specific probes are represented by cross hybridizing probes, where a single probe represents multiple var genes of a strain. Usually these represent genes from the same var group. Because of the high sequence similarity between var genes, it is difficult to design 3′ biased specific probes for all the members of the var gene family. Transcripts for 35, 3D7 var genes, 25 HB3 var genes and 23 IT4 var genes could be detected in at least 1 sample by representing probes (Supplementary Table S6). Taking the 3D7 strain as an example, transcripts were detected from members of all the var group represented by probes but was dominated by var group B sub-family genes.
Joannin et al. [15] grouped 134, 3D7 and 59, HB3 RIFIN protein sequences into two groups (97 RIFINs in group A and 37 RIFINSs in group B for 3D7 and 50 RIFINs in group A and 9 RIFINs in group B for HB3 strain respectively) [15]. Array contains specific probes for 80 group A and 19 group B rifin genes of 3D7 strain and 43 group A and 9 group B rifin genes of HB3 strain. Transcripts were detected from 40, 3D7 rifin genes (34 rifin group A and 6 rifin group B genes) and 27 HB3 rifin genes (22 rifin group A and 5 rifin group B genes) by probes in at least 1 sample (Supplementary Table S7).
Array contains probes for 26 stevor genes out of 28 genes present in the P. falciparum 3D7 reference strain [10]. Transcripts were detected from 14 stevor genes (Supplementary Table S8). Out of 28 stevor genes reported to be present in the HB3 strain [15], 23 are represented by specific probes. Transcripts were detected from 13 of these genes (Supplementary Table S8). The expression of rifins were reported to peak at 12–27 h (ring and trophozoite stages) post invasion whereas stevors was reported to peak at 22–32 h (trophozoite and schizont stages) post invasion of merozoites but expression of these genes was not only restricted to these stages and might express in other stages too [31]. Probes representing these VSA gene transcripts could efficiently detect the transcripts although some of them expressed at low levels. It is essential to detect these transcripts in vivo as their expression might determine their possible interaction with the host molecules and henceforth the pathological outcome [22].
2.7. Gene expression profiling of clinical isolates
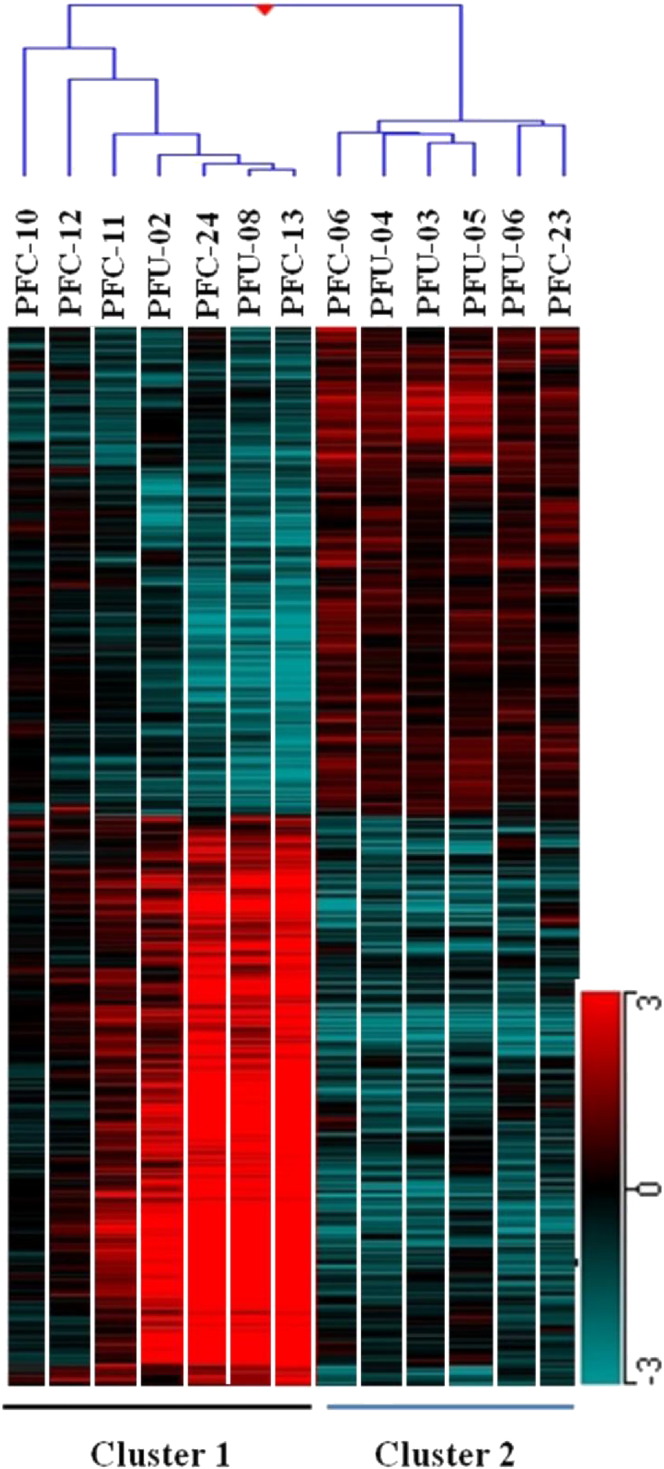
Hierarchical clustering (similarity measure used Pearson centered, linkage rule used average) of the parasite whole genome expression profiles of 13 samples, segregated parasite samples into two broad clusters (i.e. cluster 1 and cluster 2). Cluster 1 contains 7 samples and cluster 2 contains 6 samples (Fig. 4). Only probes representing the 3D7 strain were considered for this analysis. Comparison between cluster 1 and cluster 2 was performed to investigate genes, which differed between the two clusters. After allowing multiple testing corrections (Benjamini Hochberg False Discovery Rate) by applying a cut-off of ≥ 2.0 fold changes based on unpaired statistical t-test, the number of probes that differed between cluster 1 and cluster 2 were 1260 representing 1214 genes. Of the 1214 genes declared as significant, 53% (649) were up-regulated and 47% (564) were down-regulated in cluster 1 compared to cluster 2 (Supplementary Table S9).
Fig. 4.
Heat map showing combined hierarchical clustering of differentially regulated probes in 13 samples. Clustering applied on both samples and probes. Pearson centered distance matrix and average linkage rule was used. 1260 differentially regulated probes were included to generate the tree. Hierarchical clustering demarcated total samples under investigation into two clusters.
2.8. Real time PCR based validation of few differentially regulated genes
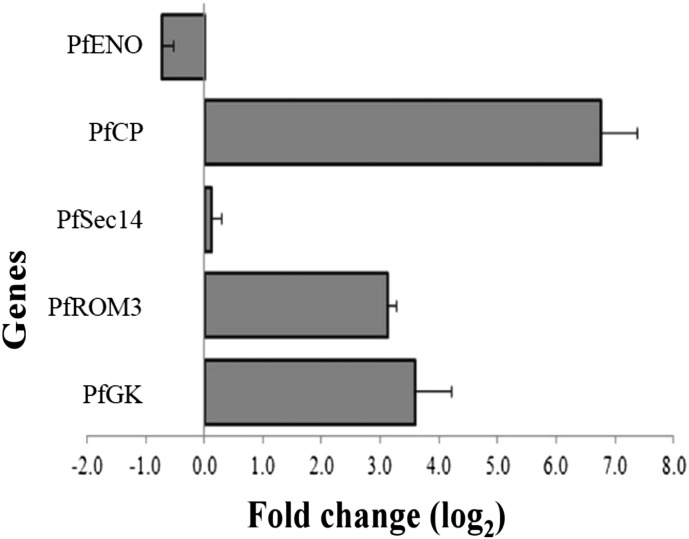
To confirm the microarray results, we examined five genes by real-time qPCR. Either Northern blot or qPCR can be performed as an independent gene expression profiling method to confirm microarray data. We have chosen qPCR over northern blot because it requires less amount of input RNA (in our case this is important because the amount of RNA material we get from clinical samples is very less), very good detection sensitivity, sequence specificity, large dynamic range as well as high precision and reproducible quantitation. Five randomly selected differentially regulated genes were selected for validation. Real time qPCR results confirmed up-regulation of three genes out of four up-regulated genes from the microarray analysis (> 1 Log2 fold change) and one gene showed Log2 fold change expression of 0.13 (this did not qualify our criterion considered for designating up-regulated transcript expression) (Fig. 5). In case of down-regulated gene, a Log2 fold change expression of − 0.70 was observed from qPCR analysis (Fig. 5). Real time qPCR validated overall expression pattern of selected genes as seen in the microarray data (Supplementary Fig. 4) (Spearman's rank correlation = 0.99).
Fig. 5.
Quantitative real-time PCR based validation of 5 differentially expressed genes in cluster 1 compared to cluster 2. Log2 fold change expression was calculated using 2− ΔΔCt method. Seryl-tRNA synthetase was used as a reference gene. Y-axis represent genes and X-axis represent normalized fold change expression(mean ± corrected S.D.; n = 2).
3. Significance, utility of the array and concluding remarks
A P. falciparum array designed on Agilent platform and its validation using RNA material from 3D7 strain has also been reported [16]. Although the array mentioned in this manuscript has utilized the Agilent platform, it has differences from the already reported Agilent array. The probes in the latter one were designed against one reference strain (3D7). In contrast, our array contains probes designed against three different strains (3D7, HB3, IT4) wherever possible. The objective of the array being detailed here was to develop the same for use with field material. We have used standard, certified cRNA amplification, labeling and hybridization protocols of Agilent whereas the previous publication has used their own protocols. This is very important as our array can be utilized by malaria community by out sourcing the microarray service from any Agilent certified microarray service provider. The uniqueness of our array lies in the fact that multiple probes, representing the sequences from 3 P. falciparum laboratory strains of differing geographical origins, were first tested for its efficiency to detect transcripts from field isolates. Further, bioinformatics analysis was also performed to remove probes showing significant hit with the human genome. This step is imperative for arrays, which are being designed for use with parasite samples extracted from patients. It is a matter of fact that the already published array has also been validated for its efficiency and has successfully been utilized in many studies for transcriptome analysis of laboratory strains; our array is a good alternate for those handling clinical isolates with uncharacterized genomes.
In conclusion, a cross-strain P. falciparum array has been designed for in vivo studies utilizing parasite material taken directly from patients. This array has been used for detection and analysis of the “sense” transcriptome of patient derived parasite material from Western India. Analysis of the array signals has established the strength of the array design and procedures adopted. This array has recently been used successfully by our group in two studies where we have reported the expression pattern of specific sub-groups of var and rifin genes in patients with severe malaria [34] and identified P. falciparum disease specific network modules by generating a P. falciparum specific weighted co-expression network [35]. As more sequence information of Indian P. falciparum strains emerge, additional oligonucleotide probes will be added to this flexible and evolving cross strain array. In addition to probes detecting “sense” transcriptome, probes detecting “antisense” transcriptome could also be included for the documentation of pervasive antisense transcription in this parasite. Emerging evidence also suggests the transcription of non-coding RNAs and probes against these may also be included as this platform provides flexibility in probes inclusion and updating array.
4. Materials and methods
4.1. 15K cross-strain P. falciparum microarray
P. falciparum transcript sequences were downloaded from PlasmoDB v5.3 and NCBI (2007). Eight to ten probes per sequence were designed in both sense and antisense direction which constituted 2,41,399 probes and additional 2105 Agilent control probes were added to the total probe list to design a 244K array. P. falciparum RNA samples were hybridized onto it in an Agilent two color format where pooled uncomplicated and complicated malaria RNA samples were labeled with Cy3 and Cy5 respectively. Best probes were chosen for 15K array based on the signal intensities (both Cy3 and Cy5 signal intensities were considered). Probes representing the same transcript and showing signal intensity more than twice the back ground were ranked and the top ranked probe/s was chosen. In some cases, probes showing intensity slightly lesser than twice the background were not excluded and retained in the array. These were probes representing mostly the 3′ end of the transcript. A total of 5629 probes representing the 3D7 transcript sequences were collected from the 244K array.
Array was designed using Genotypic Right Design Technology that enables the user to design the best probes based on GC content, melting temperature and checking cross hybridization potential across transcriptome as well as genome of a species to create custom microarray designs. For the P. falciparum cross strain 15K custom array design, the shortlisted probes from the 244K content were BLASTed against transcript sequences from PlasmoDB v5.3 (5595) [1], [10] for 3D7 strain, from the Broad Institute for HB3 strain (5623) and from NCBI for IT4 strain (80 var transcript sequences). Probes having single significant hit within any one of the strain's transcript sequence were selected in addition to the probes shortlisted by hybridization intensity as mentioned above. Additionally, probes were included for transcripts unique to the PlasmoDB v5.3, HB3 and IT4 strains using Agilent eArray tool (http://earray.chem.agilent.com). In case of transcripts for which specific probes could not be designed, probes having minimum hits were selected. Criteria for significant hit were alignment of 30 bp or more with > 84% identity with the transcript sequence. Although, the criteria for significant hit was as mentioned above, there are 90% of probes in the array with 60 bp alignment and 100% identity. Probes were BLASTed against Human transcript sequences and probes cross hybridizing with the human transcript sequences were removed from the list. Strain specific specificity of probes was determined by BLAST analysis and annotated accordingly: probes specific to only one strain, common to two or all the three represented strains, cross hybridizing to one strain but specific to the other strain or cross hybridizing probes. A total of 6362 probes were selected and included in the final array design. All the probes were replicated twice and randomly distributed across the array and empty features were filled with randomly selected duplicate probes.
4.2. Patients, blood sample collection and processing
Venous blood samples were collected (approximately 5 ml) from 13 P. falciparum infected adult patients admitted to S.P. Medical college, Bikaner, India during two transmission seasons, August to October in 2008 and 2009. The patients exhibited symptoms categorized as uncomplicated (n = 6) and complicated malaria (n = 7) as shown in Supplementary Table S10. Criteria for determination of complicated disease were based on World Health Organization (WHO) guidelines [38]. Infection with P. falciparum was confirmed by slide microscopy and RDTs (OptiMal test; Diamed AG, Cressier sur Morat, Switzerland, Falcivax test; Zephyr Biomedical System, Goa, India) in the hospital. All relevant laboratory investigations to rule out any possible cause for symptoms exhibited (Complicated P. falciparum cases) were done as described by Kochar et al. [17]. The samples were collected on informed consent and approval for sample collection was as per Approval No. F (Acad) SPMC/2003/2395 of the Hospital Ethics Committee.
Blood was immediately (Within 15–20 min of collection) subjected to density gradient-based separation (Histopaque 1077, Sigma Aldrich, USA) to separate the peripheral blood mononuclear cells (PBMCs) from the infected and uninfected erythrocytes following manufacturer's instructions. Both fractions were washed twice with phosphate buffered saline and lysed using 4 volumes of Tri-Reagent (Sigma Aldrich, USA) and stored at − 80 °C. Subsequently, samples were transported in cold chain to the research center at BITS, Pilani, processed and evaluated by 18S rRNA based multiplex PCR and 28S rRNA based nested PCR, to rule out any possibility of P. vivax co-infection [7], [27], [28].
4.3. RNA sample preparation
Total RNA and DNA were isolated from samples using the manufacturer's protocol (Tri Reagent, Sigma Aldrich, USA). All RNA samples were processed on denaturing agarose gel and found to be intact. Total RNA integrity was also assessed using RNA 6000 Nano Lab Chip on the 2100 Bioanalyzer (Agilent, Palo Alto, CA) following the manufacturer's protocol. Total RNA purity was assessed by the NanoDrop® ND-1000 UV–Vis Spectrophotometer (Nanodrop Technologies, Rockland, USA).
4.4. RNA sample labeling, hybridization and data acquisition
The total RNA samples for Gene expression were labeled using Agilent Quick-Amp labeling Kit (p/n5190-0442). Five hundred nanograms from each of the samples were incubated with reverse transcription mix at 40 °C and converted to double stranded cDNA primed by oligodT with a T7 polymerase promoter. The cleaned up double stranded cDNA was used as template for cRNA generation. cRNA was generated by in vitro transcription and the dye Cy3 CTP(Agilent) was incorporated during this step. The cDNA synthesis and in vitro transcription steps were carried out at 40 °C.
The labeled cRNA samples were hybridized on to a custom P. falciparum 8 × 15K Array (AMADID: 24956) designed with the assistance of Genotypic Technology Pvt. Ltd., Bangalore, India. 600 ng of Cy3 labeled cRNA samples were fragmented and hybridized. Fragmentation of labeled cRNA and hybridization were done using the Gene Expression Hybridization kit of Agilent (Part Number 5188-5242). Hybridization was carried out in Agilent's Surehyb Chambers at 65 °C for 16 h. The hybridized slides were washed using Agilent Gene Expression wash buffers (Part No: 5188-5327) and scanned using the Agilent Microarray Scanner G2505C at 5 μm resolution.
4.5. Pre-processing, data normalization and analysis
4.5.1. Feature extraction
Data extraction from Images, within array normalization, background subtracted signal intensity calculation and flagging of any outlier spots either due to saturation or non-uniformity was done using Feature Extraction software v 10.5.1.1 of Agilent. Processed signals from the feature extraction software were used for the analysis. Further details can be obtained from the feature extraction user guide http://www.chem.agilent.com.
4.5.2. Data analysis
Feature extracted data was analyzed using GeneSpring GX Version 11.5 software from Agilent. For finding out differentially regulated probes, data was filtered by considering all those probes detected in at least 4 samples out of thirteen samples analyzed. Filtered data was subjected to between array normalization using “quantile” method and was baseline transformed using median signal intensity of all samples. Agilent Feature extraction software uses the spot information in the data to flag the features. Spot information that can be used to flag a feature as present, marginal or as absent are features that are positive and significant, above background, not uniform, saturated and population outlier. Features which were positive and significant above background (IsPosAndSignif) established via a 2-sided t-test and well above background (IsWellAboveBG) were flagged as detected. IsWellAboveBG is a flagging option which first determines if the feature is IsPosAndSignif and additionally calculates if the background subtracted signal is > 2.6 times the background subtracted standard deviation for that feature. Differentially regulated genes were identified based on a volcano plot. Genes were declared as deferentially regulated if they showed fold change of ≥ 2 with corrected p value ≤ 0.05 (after Benjamini Hochberg based False Discovery Rate correction).
4.6. Data validation by quantitative real-time PCR validation
Total RNA from each sample was treated with DNaseI (Fermentas) for 30 min at 37 °C. Inactivation of DNase I was performed by adding 1 μl of 50 Mm EDTA per unit of DNase I used and incubated for 10 min at 65 °C. The absence of DNA in RNA samples was confirmed by no detection of DNA band in 2% agarose gel after 40 cycles of PCR with seryl-tRNAsynthetase primers. DNase I treated total RNA samples from cluster 1 and cluster 2 samples were pooled in equi-molar amount separately. First strand cDNA synthesis was carried out using iScript cDNA synthesis kit (BIO-RAD) in a total volume of 20 μl according to the manufacturer's recommendations. Quantitative real-time PCR using IqSYBR green PCR supermix (BIO-RAD) was performed on an IQ5 real-time PCR cycler (BIO-RAD) using the seryl-tRNAsynthetase gene as an endogenous control. Fold change expression was calculated using 2− ΔΔCt method. The cycling parameters used were 1 cycle of 94 °C for 4 min, 55 °C for 2 min and 72 °C for 2.5 min, 35 cycles of 94 °C for 1.5 min, 55 °C for 1.5 min, and 72 °C for 2 min. All the primers were designed using NCBI Primer BLAST (Supplementary Table S11) [40].
The following are the supplementary data related to this article.
Supporting information contains supplementary figures S1-S4 and supplementary tables S1 and S10-S12.
Agilent 15K cross strain P. falciparum gene expression microarray probe annotation.
3D7 transcripts detected in this study.
3D7 Transcripts not detected in this study although represented by specific probes.
Transcripts detected in this study represented by probes specific only to HB3 strain.
var gene transcripts detected in this study. var genes transcripts detected for all the 3 representing strain is included in three separate excel sheets. Details about the var genes represented by specific probe in the array and detection status of each var gene in the three strains are included.
rifins gene transcripts detected in this study. Details about the rifin genes of the two strains i.e. 3D7 and HB3 represented by specific probe in the array and detection status of each rifin genes in the two strains are included.
stevor gene transcripts detected in this study. Details about the stevor genes of the two strains i.e. 3D7 and HB3 represented by specific probe in the array and detection status of each stevor gene in the two strains are included.
List of differentially regulated genes between two clusters.
Microarray data accession number
The array data are deposited in the Gene Expression Omnibus (GEO) under accession number GSE54253.
Acknowledgements
We thank all the patients and technical workers for their participation in and support of this project. A.K.S. acknowledges Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR), New Delhi, India and Project Assistantship from Department of Biotechnology (DBT), New Delhi, India. P.A.B. acknowledges Basic Scientific Research fellowship from University Grant Commission, New Delhi, India and Project Assistantship from Department of Biotechnology (DBT), New Delhi, India. A.K.D., S.K.K. and D.K.K. acknowledges Department of Biotechnology (DBT), New Delhi, India for the financial support through the grant BT/PR7520/BRB/10/481/2006 and Birla Institute of Technology and Science, Pilani, India and S.P. Medical College, Bikaner, India for providing the required infrastructural facilities during this study. We thank the PlasmoDB team for making available the whole genome expression data for P. falciparum. We also thank Genotypic Technology Pvt. Ltd., Bangalore, India for the microarray hybridization service provided by them. We acknowledge National Institute of Allergy and Infectious Disease (NIAID), National Institute of Health (NIH) funded Genome Sequencing Center for Infectious Diseases at the Broad Institute for making available the HB3 genome sequence and annotation.
Contributor Information
Amit Kumar Subudhi, Email: amit4help@gmail.com.
P.A. Boopathi, Email: boopathiarunachalama@gmail.com.
Sheetal Middha, Email: sheetumig21@gmail.com.
Jyoti Acharya, Email: jyotiacharya2@gmail.com.
Sudha Narayana Rao, Email: Sudha.rao@genotupic.co.in.
Raja C. Mugasimangalam, Email: raja@genotypic.co.in.
Paramendra Sirohi, Email: drpsirohi@gmail.com.
Sanjay K. Kochar, Email: drskkochar@rediffmail.com.
Dhanpat K. Kochar, Email: drdkkochar@yahoo.com.
Ashis Das, Email: ashisd28@gmail.com, adas@pilani.bits-pilani.ac.in.
References
- 1.Aurrecoechea C., Brestelli J., Brunk B.P., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O.S., Heiges M., Innamorato F., Iodice J., Kissinger J.C., Kraemer E., Li W., Miller J.A., Nayak V., Pennington C., Pinney D.F., Roos D.S., Ross C., Stoeckert C.J., Treatman C., Wang H. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balu B., Shoue D.A., Fraser M.J., Jr., Adams J.H. High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16391–16396. doi: 10.1073/pnas.0504679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartfai R., Hoeijmakers W.A., Salcedo-Amaya A.M., Smits A.H., Janssen-Megens E., Kaan A., Treeck M., Gilberger T.W., Francoijs K.J., Stunnenberg H.G. H2A.Z demarcates intergenic regions of the Plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bozdech Z., Llinas M., Pulliam B.L., Wong E.D., Zhu J., DeRisi J.L. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1 doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caro F., Miller M.G., DeRisi J.L. Plate-based transfection and culturing technique for genetic manipulation of Plasmodium falciparum. Malar. J. 2012;11:22. doi: 10.1186/1475-2875-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daily J.P., Scanfeld D., Pochet N., Le Roch K., Plouffe D., Kamal M., Sarr O., Mboup S., Ndir O., Wypij D., Levasseur K., Thomas E., Tamayo P., Dong C., Zhou Y., Lander E.S., Ndiaye D., Wirth D., Winzeler E.A., Mesirov J.P., Regev A. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature. 2007;450:1091–1095. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- 7.Das A., Holloway B., Collins W.E., Shama V.P., Ghosh S.K., Sinha S., Hasnain S.E., Talwar G.P., Lal A.A. Species-specific 18S rRNA gene amplification for the detection of Plasmodium falciparum and Plasmodium vivax malaria parasites. Mol. Cell. Probes. 1995;9:161–165. doi: 10.1006/mcpr.1995.0025. [DOI] [PubMed] [Google Scholar]
- 8.Duraisingh M.T., Voss T.S., Marty A.J., Duffy M.F., Good R.T., Thompson J.K., Freitas-Junior L.H., Scherf A., Crabb B.S., Cowman A.F. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 9.Flueck C., Bartfai R., Volz J., Niederwieser I., Salcedo-Amaya A.M., Alako B.T., Ehlgen F., Ralph S.A., Cowman A.F., Bozdech Z., Stunnenberg H.G., Voss T.S. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., Paulsen I.T., James K., Eisen J.A., Rutherford K., Salzberg S.L., Craig A., Kyes S., Chan M.S., Nene V., Shallom S.J., Suh B., Peterson J., Angiuoli S., Pertea M., Allen J., Selengut J., Haft D., Mather M.W., Vaidya A.B., Martin D.M., Fairlamb A.H., Fraunholz M.J., Roos D.S., Ralph S.A., McFadden G.I., Cummings L.M., Subramanian G.M., Mungall C., Venter J.C., Carucci D.J., Hoffman S.L., Newbold C., Davis R.W., Fraser C.M., Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay S.I., Okiro E.A., Gething P.W., Patil A.P., Tatem A.J., Guerra C.A., Snow R.W. Estimating the global clinical burden of Plasmodium falciparum malaria in 2007. PLoS Med. 2010;7 doi: 10.1371/journal.pmed.1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayward R.E., Derisi J.L., Alfadhli S., Kaslow D.C., Brown P.O., Rathod P.K. Shotgun DNA microarrays and stage-specific gene expression in Plasmodium falciparum malaria. Mol. Microbiol. 2000;35:6–14. doi: 10.1046/j.1365-2958.2000.01730.x. [DOI] [PubMed] [Google Scholar]
- 13.Hu G., Cabrera A., Kono M., Mok S., Chaal B.K., Haase S., Engelberg K., Cheemadan S., Spielmann T., Preiser P.R., Gilberger T.W., Bozdech Z. Transcriptional profiling of growth perturbations of the human malaria parasite Plasmodium falciparum. Nat. Biotechnol. 2010;28:91–98. doi: 10.1038/nbt.1597. [DOI] [PubMed] [Google Scholar]
- 14.Imwong M., Dondorp A.M., Nosten F., Yi P., Mungthin M., Hanchana S., Das D., Phyo A.P., Lwin K.M., Pukrittayakamee S., Lee S.J., Saisung S., Koecharoen K., Nguon C., Day N.P., Socheat D., White N.J. Exploring the contribution of candidate genes to artemisinin resistance in Plasmodium falciparum. Antimicrob. Agents Chemother. 2010;54:2886–2892. doi: 10.1128/AAC.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joannin N., Abhiman S., Sonnhammer E.L., Wahlgren M. Sub-grouping and sub-functionalization of the RIFIN multi-copy protein family. BMC Genomics. 2008;9:19. doi: 10.1186/1471-2164-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kafsack B.F., Painter H.J., Llinas M. New Agilent platform DNA microarrays for transcriptome analysis of Plasmodium falciparum and Plasmodium berghei for the malaria research community. Malar. J. 2012;11:187. doi: 10.1186/1475-2875-11-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kochar D.K., Tanwar G.S., Khatri P.C., Kochar S.K., Sengar G.S., Gupta A., Kochar A., Middha S., Acharya J., Saxena V., Pakalapati D., Garg S., Das A. Clinical features of children hospitalized with malaria—a study from Bikaner, northwest India. Am.J.Trop. Med. Hyg. 2010;83:981–989. doi: 10.4269/ajtmh.2010.09-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Roch K.G., Zhou Y., Blair P.L., Grainger M., Moch J.K., Haynes J.D., De La Vega P., Holder A.A., Batalov S., Carucci D.J., Winzeler E.A. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 19.Mackinnon M.J., Li J., Mok S., Kortok M.M., Marsh K., Preiser P.R., Bozdech Z. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maier A.G., Rug M., O'Neill M.T., Brown M., Chakravorty S., Szestak T., Chesson J., Wu Y., Hughes K., Coppel R.L., Newbold C., Beeson J.G., Craig A., Crabb B.S., Cowman A.F. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell. 2008;134:48–61. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mamoun C.B., Goldberg D.E. Plasmodium protein phosphatase 2C dephosphorylates translation elongation factor 1beta and inhibits its PKC-mediated nucleotide exchange activity in vitro. Mol. Microbiol. 2001;39:973–981. doi: 10.1046/j.1365-2958.2001.02289.x. [DOI] [PubMed] [Google Scholar]
- 22.Miller L.H., Baruch D.I., Marsh K., Doumbo O.K. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 23.Natalang O., Bischoff E., Deplaine G., Proux C., Dillies M.A., Sismeiro O., Guigon G., Bonnefoy S., Patarapotikul J., Mercereau-Puijalon O., Coppee J.Y., David P.H. Dynamic RNA profiling in Plasmodium falciparum synchronized blood stages exposed to lethal doses of artesunate. BMC Genomics. 2008;9:388. doi: 10.1186/1471-2164-9-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oakley M.S., Kumar S., Anantharaman V., Zheng H., Mahajan B., Haynes J.D., Moch J.K., Fairhurst R., McCutchan T.F., Aravind L. Molecular factors and biochemical pathways induced by febrile temperature in intraerythrocytic Plasmodium falciparum parasites. Infect. Immun. 2007;75:2012–2025. doi: 10.1128/IAI.01236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olszewski K.L., Morrisey J.M., Wilinski D., Burns J.M., Vaidya A.B., Rabinowitz J.D., Llinas M. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe. 2009;5:191–199. doi: 10.1016/j.chom.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto T.D., Wilinski D., Assefa S., Keane T.M., Sarry L.R., Bohme U., Lemieux J., Barrell B., Pain A., Berriman M., Newbold C., Llinas M. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pakalapati D., Garg S., Middha S., Acharya J., Subudhi A.K., Boopathi A.P., Saxena V., Kochar S.K., Kochar D.K., Das A. Development and evaluation of a 28S rRNA gene-based nested PCR assay for Plasmodium falciparum and Plasmodium vivax. Pathog. Glob. Health. 2013;107:180–188. doi: 10.1179/2047773213Y.0000000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pakalapati D., Garg S., Middha S., Kochar A., Subudhi A.K., Arunachalam B.P., Kochar S.K., Saxena V., Pareek R.P., Acharya J., Kochar D.K., Das A. Comparative evaluation of microscopy, OptiMAL((R)) and 18S rRNA gene based multiplex PCR for detection of Plasmodium falciparum & Plasmodium vivax from field isolates of Bikaner, India. Asian Pac J Trop Med. 2013;6:346–351. doi: 10.1016/S1995-7645(13)60037-1. [DOI] [PubMed] [Google Scholar]
- 29.Petter M., Haeggstrom M., Khattab A., Fernandez V., Klinkert M.Q., Wahlgren M. Variant proteins of the Plasmodium falciparum RIFIN family show distinct subcellular localization and developmental expression patterns. Mol. Biochem. Parasitol. 2007;156:51–61. doi: 10.1016/j.molbiopara.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Sargeant T.J., Marti M., Caler E., Carlton J.M., Simpson K., Speed T.P., Cowman A.F. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 2006;7:R12. doi: 10.1186/gb-2006-7-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scherf A., Lopez-Rubio J.J., Riviere L. Antigenic variation in Plasmodium falciparum. Annu. Rev. Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 32.Silvestrini F., Bozdech Z., Lanfrancotti A., Di Giulio E., Bultrini E., Picci L., Derisi J.L., Pizzi E., Alano P. Genome-wide identification of genes upregulated at the onset of gametocytogenesis in plasmodium falciparum. Mol. Biochem. Parasitol. 2005;143:100–110. doi: 10.1016/j.molbiopara.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 33.Subudhi A.K., Boopathi P.A., Garg S., Middha S., Acharya J., Pakalapati D., Saxena V., Aiyaz M., Orekondy H.B., Mugasimangalam R.C., Sirohi P., Kochar S.K., Kochar D.K., Das A. Natural antisense transcripts in Plasmodium falciparum isolates from patients with complicated malaria. Exp. Parasitol. 2014;141C:39–54. doi: 10.1016/j.exppara.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Subudhi A.K., Boopathi P.A., Pandey I., Kaur R., Middha S., Acharya J., Kochar S.K., Kochar D.K., Das A. Disease specific modules and hub genes for intervention strategies: a co-expression network based approach for Plasmodium falciparum clinical isolates. Infect. Genet. Evol. 2015;35:96–108. doi: 10.1016/j.meegid.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Subudhi A.K., Boopathi P.A., Pandey I., Kohli R., Karwa R., Middha S., Acharya J., Kochar S.K., Kochar D.K., Das A. Plasmodium falciparum complicated malaria: modulation and connectivity between exportome and variant surface antigen gene families. Mol. Biochem. Parasitol. 2015;201:31–46. doi: 10.1016/j.molbiopara.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Tamez P.A., Bhattacharjee S., van Ooij C., Hiller N.L., Llinas M., Balu B., Adams J.H., Haldar K. An erythrocyte vesicle protein exported by the malaria parasite promotes tubovesicular lipid import from the host cell surface. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuikue Ndam N., Bischoff E., Proux C., Lavstsen T., Salanti A., Guitard J., Nielsen M.A., Coppee J.Y., Gaye A., Theander T., David P.H., Deloron P. Plasmodium falciparum transcriptome analysis reveals pregnancy malaria associated gene expression. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health, O. Severe falciparum malaria. Trans. R. Soc. Trop. Med. Hyg. 2000;94:1–90. [Google Scholar]
- 39.WHO Press; Geneva: 2015. World Malaria Report. [Google Scholar]
- 40.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young J.A., Fivelman Q.L., Blair P.L., de la Vega P., Le Roch K.G., Zhou Y., Carucci D.J., Baker D.A., Winzeler E.A. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol. Biochem. Parasitol. 2005;143:67–79. doi: 10.1016/j.molbiopara.2005.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information contains supplementary figures S1-S4 and supplementary tables S1 and S10-S12.
Agilent 15K cross strain P. falciparum gene expression microarray probe annotation.
3D7 transcripts detected in this study.
3D7 Transcripts not detected in this study although represented by specific probes.
Transcripts detected in this study represented by probes specific only to HB3 strain.
var gene transcripts detected in this study. var genes transcripts detected for all the 3 representing strain is included in three separate excel sheets. Details about the var genes represented by specific probe in the array and detection status of each var gene in the three strains are included.
rifins gene transcripts detected in this study. Details about the rifin genes of the two strains i.e. 3D7 and HB3 represented by specific probe in the array and detection status of each rifin genes in the two strains are included.
stevor gene transcripts detected in this study. Details about the stevor genes of the two strains i.e. 3D7 and HB3 represented by specific probe in the array and detection status of each stevor gene in the two strains are included.
List of differentially regulated genes between two clusters.