Abstract
Elizabethkingia anophelis is an emerging pathogen involved in human infections and outbreaks in distinct world regions. We investigated the phylogenetic relationships and pathogenesis-associated genomic features of two neonatal meningitis isolates isolated 5 years apart from one hospital in Central African Republic and compared them with Elizabethkingia from other regions and sources. Average nucleotide identity firmly confirmed that E. anophelis, E. meningoseptica and E. miricola represent demarcated genomic species. A core genome multilocus sequence typing scheme, broadly applicable to Elizabethkingia species, was developed and made publicly available (http://bigsdb.pasteur.fr/elizabethkingia). Phylogenetic analysis revealed distinct E. anophelis sublineages and demonstrated high genetic relatedness between the African isolates, compatible with persistence of the strain in the hospital environment. CRISPR spacer variation between the African isolates was mirrored by the presence of a large mobile genetic element. The pan-genome of E. anophelis comprised 6,880 gene families, underlining genomic heterogeneity of this species. African isolates carried unique resistance genes acquired by horizontal transfer. We demonstrated the presence of extensive variation of the capsular polysaccharide synthesis gene cluster in E. anophelis. Our results demonstrate the dynamic evolution of this emerging pathogen and the power of genomic approaches for Elizabethkingia identification, population biology and epidemiology.
Elizabethkingia is a genus of aerobic, non-motile bacteria belonging to the family Flavobacteriaceae in the phylum Bacteroidetes. Elizabethkingia strains are considered ubiquitous in nature and highly prevalent in the gut of Anopheles mosquitoes1,2,3. The genus currently comprises four species. E. meningoseptica is a common cause of neonatal meningitis, especially in premature infants during the first weeks of life, and also causes a wide range of human infections as an opportunistic pathogen4,5,6. E. miricola is rarely associated with sepsis7. E. anophelis was recently described from the midgut of Anophelis gambiae mosquitoes8, while E. endophytica was recently described based on a strain from corn9.
In 2013, some of us described the two first human cases of neonatal meningitis caused by E. anophelis in Bangui, Central African Republic10. A nosocomial origin was suspected for these strains given that patients had a history of intubation and mechanical ventilation at birth. The two meningitis cases occurred in the same hospital five years apart and an epidemiological link between them was not established. E. anophelis was also implicated in an outbreak in an intensive care unit in Singapore11 and in a transmission from mother to infant in Hong Kong12, resulting in late-onset meningitis in the baby. At the time of writing, an unprecedented large outbreak of E. anophelis from a yet unknown source is ongoing in Wisconsin, USA. E. anophelis is therefore considered as an emerging opportunistic pathogen, and its clinical significance was underlined recently, with a majority of Elizabethkingia isolates being in fact identified as E. anophelis, rather than E. meningoseptica13. In all previously reported clinical cases10,11,12,13, E. anophelis isolates were resistant to most antibiotics recommended for empirical therapy, including third-generation cephalosporins, aminoglycosides and carbapenems14, probably accounting for the poor infection outcomes. Consistently, most E. anophelis genomes harbor multiple antimicrobial resistance genes, including the two metallo-betalactamase genes blaGOB and blablaB, associated with resistance to carbapenems15. Currently, the clinical significance and biological distribution of E. anophelis is restricted, as this novel species is not well distinguished from other Elizabethkingia species in routine microbiological identification procedures, and as reference identification of E. anophelis relies mostly on the low-resolution 16S rRNA gene.
Genomic studies of E. anophelis strains from clinical, environmental and mosquito origins have revealed interesting features that may contribute to the adaptation of this bacterium to mosquito and that may be implicated in its pathogenesis1,12,16. Genomic sequencing has also been used to derive epidemiological insights during hospital outbreaks12,16. However, knowledge on the genomic diversity and population structure of E. anophelis is still scant, and there is currently no standard way to compare isolates at local, regional or global scales, which limits our understanding of the epidemiology and global emergence of E. anophelis.
The aims of this study were to determine the phylogenetic origin and genomic features of the two African neonatal meningitis E. anophelis isolates; to compare them with Elizabethkingia isolates from other world regions and sources; to take advantage of the high resolution of whole genome sequencing to distinguish Elizabethkingia species unambiguously, and to develop a publicly-accessible genotyping system enabling a common language on strain types in order to ease future collaborations on biodiversity studies and epidemiological investigations of Elizabethkingia clinical and environmental isolates.
Results and Discussion
Genome sequences of African isolates E27017 and E18064
The two African isolates were sequenced using paired-end Illumina technology. The genome sequence of isolate E18064 was assembled into 213 contigs of total length 4,080,763 bp. It is 35.7% G + C rich and contains 3,648 CDSs. The E27107 genome sequence is 4,059,474 bp long, 35.5% G + C rich and contains 3,674 CDSs distributed over 89 contigs. The genome characteristics of the two African isolates thus appeared similar to those of previously sequenced Elizabethkingia genomes (Table S1).
Genome-based identification of members of the Elizabethkingia genus
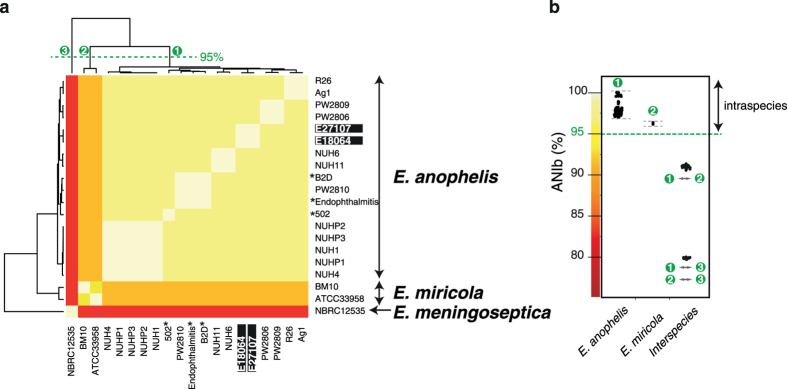
To determine with confidence the species-level identification of the two African and the 18 other Elizabethkingia isolates, we calculated average nucleotide identity (ANI) based on their genomic sequences (Table S2)17,18. The distribution of ANI values among the 20 genomes classified them into three main groups (Fig. 1a) and showed a clear separation of E. anophelis strains from E. meningoseptica and E. miricola strains. These results fully demonstrate that these three taxa represent distinct genomic groups. The two African isolates clearly belong to E. anophelis based on their high ANI with R26T, the type strain of this species, fully confirming their initial identification10. Even though they were initially identified as E. meningoseptica, the three strains 502, Endophthalmitis and B2D also appeared closely related to E. anophelis isolates based on ANI values. Therefore, these three strains may be re-assigned to E. anophelis. Based on their ANI, the E. miricola reference strain ATCC 33958 and the unclassified Elizabethkingia sp. strain BM10 isolated from a termite, clearly belong to a single species. Urea degradation is a differential phenotypic characteristic of E. miricola, and we detected the presence in both ATCC 33958 and BM10 genomes of the urease cluster, whereas it was absent from all other genomes. The ANI values among E. anophelis genomes ranged from 96.97% to 100% (Fig. 1b), thus clearly higher than the classical 95% threshold used for species delineation. This result confirms that E. anophelis isolates belong to a single genomic species. Of note, the highest ANI value computed between E. anophelis and E. meningoseptica genomes was only 79.7%, showing that these species are separated by an evolutionary distance similar to the one observed between Escherichia coli and Salmonella spp. (80.4%), two model species with very distinct ecological lifestyles and virulence features.
Figure 1.
Heat-map of the ANIb for each pairwise comparison (a) and intra and inter-species ANIb variability (b). Green dashed line on the top dendogram marks groups of taxa that are from the same species or have more than 95% ANIb values and therefore might be regarded as part of the same species. Black asterisks indicate strains previously described as E. meningoseptica. Black rectangles indicate the two hospital-acquired isolates from Central African Republic sequenced in this study.
The two African E. anophelis isolates were initially identified based on 16S rRNA sequencing10. Phylogenetic analysis of the 16S rRNA sequences (Figure S1a) was consistent with the clustering into three species. However, four E. anophelis isolates (502, B2D, Endophthalmitis and PW2810) were linked to the other E. anophelis strains in a clade with an atypically long branch. Inspection of the alignment revealed that this long branch was caused by 10 nucleotide differences clustered within a single region of 35 nucleotides (Figure S1b). These results indicate that careful interpretation may be required when classifying Elizabethkingia isolates based on the divergence values estimated from the classical 16S rRNA marker.
Elizabethkingia species are difficult to distinguish based on biochemical tests8,13. Although 16S rDNA sequencing is currently used in practice for species-level identification, MALDI-TOF mass spectrometry may be a method of choice in the future, after appropriate reference spectrum databases have been constructed13. We recommend that molecular approaches such as genome sequence-based ANI or protein-coding gene sequencing should be used for reference taxonomic work17,18. Future use of appropriate identification methods will help defining better the clinical importance of E. anophelis, which has been underreported until now due to frequent misidentification as E. meningoseptica8.
Gene repertoire of Elizabethkingia
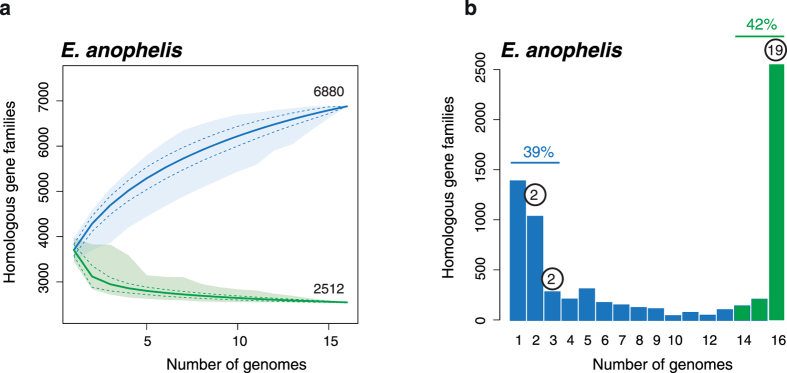
To compare the gene repertoire of the two African isolates with other Elizabethkingia isolates, we first quantified the diversity of Elizabethkingia genes by computing the set of ubiquitous genes (core genome) and the set of different homologous genes families (pan-genome) among the 20 genomes. At the level of the genus Elizabethkingia, the core genome contained 2,221 orthologous protein families, corresponding to 65% of the size of the smallest proteome (strain E. meningoseptica NBRC 12535). The core-genome of the species E. anophelis had 2,512 orthologous protein families, corresponding to 72% of the size of the smallest proteome (strain B2D). Gene rarefaction analyses showed that the E. anophelis core genome varies little with the addition of the last genomes (Fig. 2a), suggesting that this core genome estimate is robust. Both the pan-genomes of the Elizabethkingia genus and of E. anophelis were large with, respectively, 7,801 and 6,880 gene families. Gene rarefaction analyses showed that the addition of future E. anophelis genomes to the analysis will still significantly increase the size of the pan-genome16. The spectrum of gene frequencies for the E. anophelis pan-genome (Fig. 2b) showed that the vast majority of gene families were either encoded in a few genomes (39% in three or less) or in most of them (42% in more than 14 genomes), confirming that the genome sequencing of additional Elizabethkingia strains will uncover multiple novel gene families.
Figure 2.
Core- and pan-genome sizes of Elizabethkingia anophelis (a) and spectrum of frequencies for E. anophelis gene repertoires (b). The pan- and core-genomes were used to perform gene accumulation curves. These curves describe the number of new genes (pan-genome) and genes in common (core-genome) obtained by adding a new genome to a previous set. The procedure was repeated 1,000 times by randomly modifying the order of integration of genomes in the analysis. The spectrum of frequencies represents the number of genomes where the families of the pan-genome can be found, from 1 for strain-specific genes to 16 for core genes. Blue indicates accessory genes and green the genes that are highly persistent in E. anophelis. Numbers in circles above the bars indicate the number of antimicrobial resistance (AR) gene families.
Core genome-based phylogenetic structure of E. anophelis
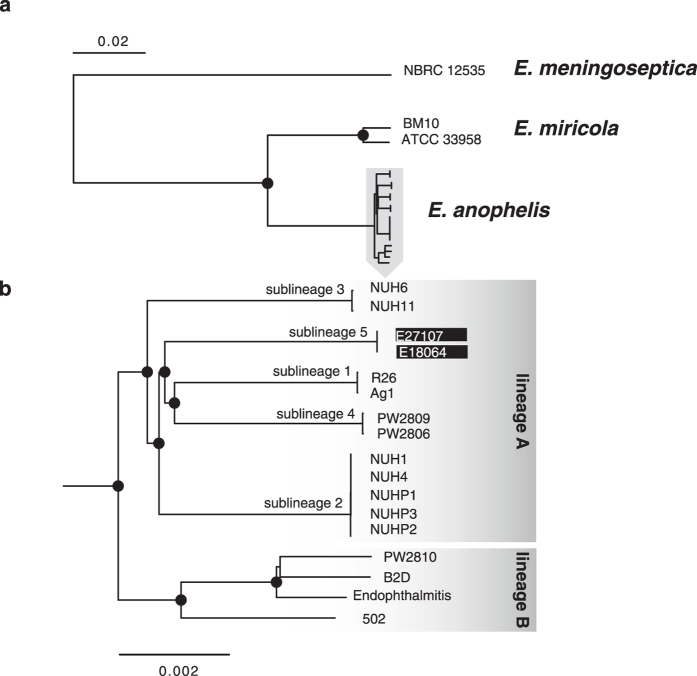
To determine the phylogenetic origin of the African strains with respect to strains from other world regions, we selected a subset of 1,546 genes families with very reliable alignments (few indels) to infer robust phylogenies. Phylogenetic analysis based on concatenated alignments of the 1,546 gene sequences showed that all E. anophelis isolates were clearly separated from closely related species E. miricola (its sister group) and E. meningoseptica (Fig. 3a). These results fully corroborated the classification into three genomic groups based on ANI and 16S rRNA analyses.
Figure 3. Phylogenetic tree of the Elizabethkingia genus based on the multiple sequence alignments of the 1,546 cgMLST loci.
Black dots correspond to bootstrap supports higher than 90%.
The phylogenetic structure of E. anophelis revealed two main lineages (A and B, Fig. 3b), each containing clearly demarcated sublineages. Mosquito strains Ag1 and R26T formed a single sublineage (sublineage 1) within lineage A, which comprised four additional sublineages: sublineage 2, which includes three patient isolates and two hospital environment isolates from a Singapore outbreak11; Sublineage 3, which includes two unrelated hospital environment isolates from the above outbreak investigation; Sublineage 4, which includes strains PW2806 and PW2809 from Hong Kong12; and sublineage 5, made of the two African isolates from Bangui10. Within each sublineage, the strains were highly related. These results show that the African strains formed a distinct sublineage, therefore having a clearly distinct evolutionary origin from Asian nosocomial strains and from mosquito strains. The nearly simultaneous recognition of the emergence of E. anophelis infections in Africa and Asia thus reflects independent epidemiological events.
Antimicrobial resistance-associated genomic features of African E. anophelis isolates
All Elizabethkingia genomes contained at least 17 antimicrobial resistance genes (Table S3). Most prominent among these were genes coding for beta-lactamases, including the metallo-beta-lactamase genes blablaB and blaGOB15,19, and for efflux systems, which may contribute largely to the phenotypic resistance of Elizabethkingia isolates to most antibiotics. These findings are consistent with previous descriptions of multiple resistance genes in Elizabethkingia genomes1,12,16,20. Most resistance genes belonged to persistent families, i.e., they were part of the core genome of Elizabethkingia. This was the case for genes blablaB and blaGOB, which were previously described as intrinsic and chromosome-borne in E. meningoseptica15. We observed that both blablaB and blaGOB were located in highly syntenic regions in all members of the Elizabethkingia genus, but were absent from the closely related genera Flavobacterium, Riemerella and Chryseobacterium (not shown). These results show that multiple potential resistance genes are ancestral in the genus Elizabethkingia and suggest acquisition by horizontal gene transfer into this genus before the evolutionary separation of the three Elizabethkingia species.
Interestingly, several additional resistance genes were detected in the African isolates (24 genes in total) as compared to other Elizabethkingia genomes (range: 17–20). Whereas all Elizabethkingia genomes contained one conserved chloramphenicol acetyltransferase (CAT) gene, the two African isolates contained an additional CAT gene that had 80.7% amino-acid identity with the conserved copy. The phylogenetic relationships of the conserved copy (Figure S2) were concordant with the classification of the strains into E. meningoseptica, E. miricola and E. anophelis, indicating vertical evolution of the conserved CAT copy from an ancestral Elizabethkingia gene. In contrast, the additional CAT copy from African isolates was closely related to Riemerella sequences (Figure S2), suggesting acquisition by horizontal gene transfer from this genus.
Each of the African isolates also carried two additional metallo-beta-lactamases of the B1 (NDM-CcrA) subclass. In addition, an aminoglycoside acetyltransferase (AAC3-I) was found only in the two African isolates and in one isolate (NUH6) from Singapore. Finally, a tet(X) gene coding for a tetracycline inactivating enzyme 99.7% identical to that of Bacteroides fragilis21 was found in the African isolates and in the Singapore isolate NUH4. Interestingly, the additional CAT gene, tet(X) and one subclass B1 beta-lactamase gene were clustered in a genomic region that also contained a putative class D beta-lactamase gene, a mercuric reductase gene and a lincosamide nucleotidyltransferase (LinF) gene associated with resistance to lincosamides22. This region had high similarity to plasmid pRA0511 of Riemella anatipestifer23. Furthermore, the other subclass B1 beta-lactamase gene and the AAC3-I gene were adjacent to each other and to a transposase gene. These observations indicate that the additional resistance genes of the African E. anophelis isolates were acquired through the transfer of mobile genetic elements.
Identification and diversity of capsular polysaccharide synthesis clusters
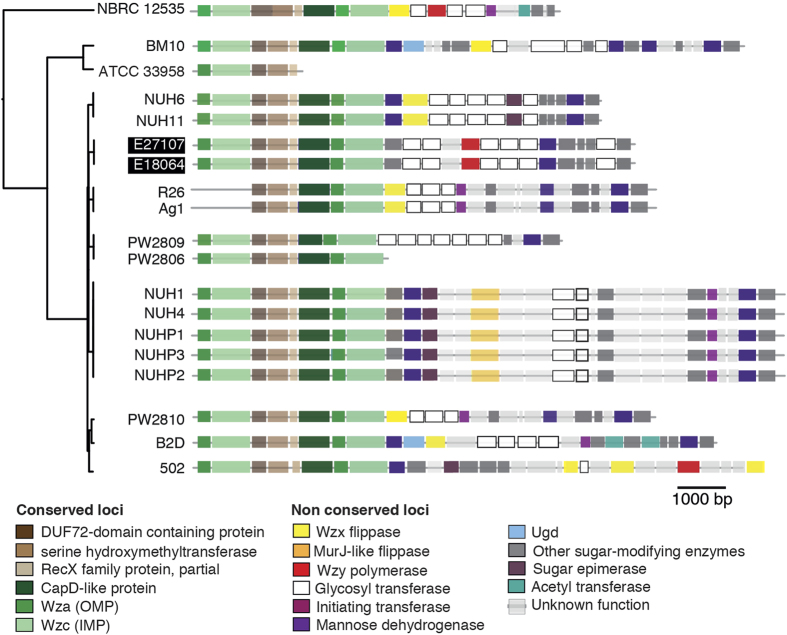
Currently, the pathophysiology of Elizabethkingia infections is poorly understood2. We searched for virulence-associated features in the African E. anophelis and other Elizabethkingia genomes by using the VFDB database (Table S4). A number of coding sequences showed homologs to iron and heme acquisition, to hemolysins and to oxidative stress resistance proteins, consistent with previous reports1,12,16,20. Among the hits found in the VFDB database, several corresponded to proteins putatively involved in capsule synthesis (Table S4). Further inspection of genome sequences revealed a Wzy-dependent cps gene cluster24 comprising 27 co-oriented coding sequences in the two African isolates and in most other Elizabethkingia strains (Fig. 4). The upstream (3′) part of the gene cluster was highly conserved (Fig. 4) and included the two well-characterized genes, wzc and wza. The former codes for an inner membrane protein necessary for capsular polymerization and translocation across the inner membrane. The latter encodes a channel that allows translocation of the polysaccharide across the outer membrane. Interestingly, these two genes were found in two copies in most strains separated by three other conserved genes (a RecX family transcription factor, and two other genes of unknown function).
Figure 4. Genetic organization of the putative capsular polysaccharide synthesis (cps) gene clusters in Elizabethkingia genomes.
Open reading frames (ORFs) are shown as rectangles and are drawn to scale (see scale bar). Intergenic spaces are not to scale. Three highly conserved genes, including recX, are depicted in brown. Green indicates highly conserved genes involved in capsule translocation (capD, wza, wzc). Yellow and red indicate genes wzx and wzy, respectively.
The downstream region of the cps gene cluster was highly variable and may be responsible for the Wzy-dependent synthesis of polysaccharides with distinctive chemical compositions and which would possibly correspond to distinct capsular serotypes as observed in other capsulated species25,26. This region comprised the flippase (wzx) and the polymerase (wzy) and other genes coding for glycosyl transferases, acetyl transferases and other sugar-modifying enzymes. In some strains, the essential genes involved in polymerization–wzx and wzy–could not be confidently identified. These genes are known to be poorly conserved even among phylogenetically closely related isolates, rendering their detection difficult27.
Interestingly, the genetic organization was strongly conserved among strains belonging to the same phylogenetic sublineage (as defined above; Fig. 4). These results suggest that capsular types are conserved within E. anophelis sublineages, but not between them. For example, the groups of African isolates, and mosquito isolates (R26T and Ag1) showed few intra-group and many inter-group differences in the downstream regions of the capsular cluster locus. More specifically, the two African genomes seem to code for a unique and distinctive capsule, relative to other Elizabethkingia strains. Capsular type variation in Elizabethkingia may restrict immune cross-reactions and could be associated with distinct pathogenic properties, as observed in other capsulated bacterial groups28,29,30. Using the India ink method, we did not observe capsular material in the two African strains, as tested during stationary phase in BHI and in LB medium. A capsule was observed previously for some Elizabethkingia strains, but capsule production was variable across in-vitro and in-vivo conditions31. Given that bacterial capsules represent one of the main bacterial virulence factors, particularly in pathogens that may be responsible for meningitis32, these results warrant future studies into the pathophysiological and epidemiological implications of capsular variation in Elizabethkingia.
Secretion systems
Bacterial secretion systems are associated with important functions such as detoxification, antibiotic resistance and scavenging. We used MacSyFinder33, together with TXSScan profiles34, to identify the secretion systems encoded in the Elizabethkingia genomes (Table S5). While T9SS (PorSS) is present in many members of the phylum Bacteroidetes35, we found none in the Elizabethkingia genomes. We also found no evidence for the presence of Type 2, 3, 5 protein secretion systems, which fits previous suggestions that these systems are absent from Bacteroidetes34. In contrast, we found two families of T1SS in the genomes of Elizabethkingia, one of which was present in both African strains.
A third variant of the type VI secretion system (T6SSiii) was recently uncovered in Bacteroidetes36. T6SS can be involved in bacterial competition36 or pathogenicity37. Interestingly, one locus encoding a T6SSiii was found in every strain of our dataset. This system was highly conserved, except for the gene family tssI that showed extensive copy number variability. This gene encodes the cell-puncturing device and it can be fused with a variety of toxic domains, thus encoding a toxin38. These systems might provide Elizabethkingia strains a way to antagonize competing bacteria in the complex environments such as the mosquito gut.
Type 4 secretion systems (T4SS) implicated in conjugative DNA transfer were previously described in E. anophelis1,20. We identified such systems in nearly all the genomes. Some strains encoded up to six conjugative systems (e.g., strain NUHP3), which is among the highest values observed among bacteria39. Expectedly, all these systems were of the MPFB type, the one specific to Bacteroidetes. All the genomes encoding a T4SS also encoded one or several relaxases, suggesting that these systems are involved in conjugation of mobile genetic elements and not in protein secretion. The abundance of conjugative mobile genetic elements suggests that they might drive much of the genetic diversification observed in Elizabethkingia.
Standardized strain typing method based on genome sequencing for Elizabethkingia epidemiology
In order to explore the epidemiological links between E. anophelis isolates, we translated whole genome sequences into high-resolution genotyping data. For this purpose, we followed the cgMLST strategy, which allows creating universal genotype nomenclatures and comparing isolates globally40,41,42. A cgMLST scheme was developed based on 1,546 genes that could be aligned with high confidence among Elizabethkingia genomes. We then tested this scheme by scanning the 20 isolates for allelic variation at these loci. The identity of the 1,546 alleles was recorded for each isolate. The resulting allelic profiles were then clustered to obtain groups of isolates. These groups coincided with the sublineages from the phylogenetic analysis of the core gene sequences (Figure S4).
To validate our cgMLST approach, we took advantage of the availability within the dataset of public genomes, of groups of isolates that were epidemiologically related, i.e., which were isolated during single outbreaks or were associated with direct mother-to-child transmission11,12. First, the five E. anophelis strains NUHP1, NUHP2, NUHP3, NUH1 and NUH4 from a single outbreak from Singapore were compared11. We found that four of these strains were almost identical (2 to 6 cgMLST allelic mismatches), whereas NUH4 was slightly more distant (32 or more allelic mismatches with the others). This is highly consistent with the whole genome single nucleotide polymorphism (SNP) analysis by Teo and colleagues16, who found 24 to 38 SNPs among the four closely related isolates, whereas at least 176 SNPs separated these from isolate NUH4. In a second study12, two isolates (HKU37 and HKU38) from a mother and her neonate were considered to be directly related based on their genomic sequences, which showed no difference out of 2,000 genes. Based on our cgMLST approach, these two isolates were also identical, as none of the 1,546 loci was variable between them. In contrast, 1,433 cgMLST loci were distinct between this pair of isolates and a third independent isolate (HKU36)12. These results demonstrate the extremely high efficiency of the cgMLST approach to distinguish epidemiologically related isolates from unrelated ones. The core-genome MLST approach and attached genomic sequences were made publicly available through the Institut Pasteur server (http://bigsdb.pasteur.fr).
To our surprise, the two African E. anophelis isolates, which were isolated 5 years apart in the same hospital in Bangui10, differed at only 4 alleles. This level of variation is similar to the variation observed among related isolates of the Singapore outbreak and thus strongly suggests to us that the two African hospital isolates may also be epidemiologically related. We therefore hypothesize that a single E. anophelis strain caused the two infections five years apart, due to its persistence in a hospital environmental source. Alternately, it is conceivable that an epidemiological reservoir, such as the mosquito gut, comprises an E. anophelis population with an extremely reduced genetic diversity. Although we favor the hospital persistence hypothesis, more data on the diversity of E. anophelis in various environments is needed to interpret genotyping data for epidemiological questions.
Clustered regularly interspaced short palindromic repeats (CRISPR)
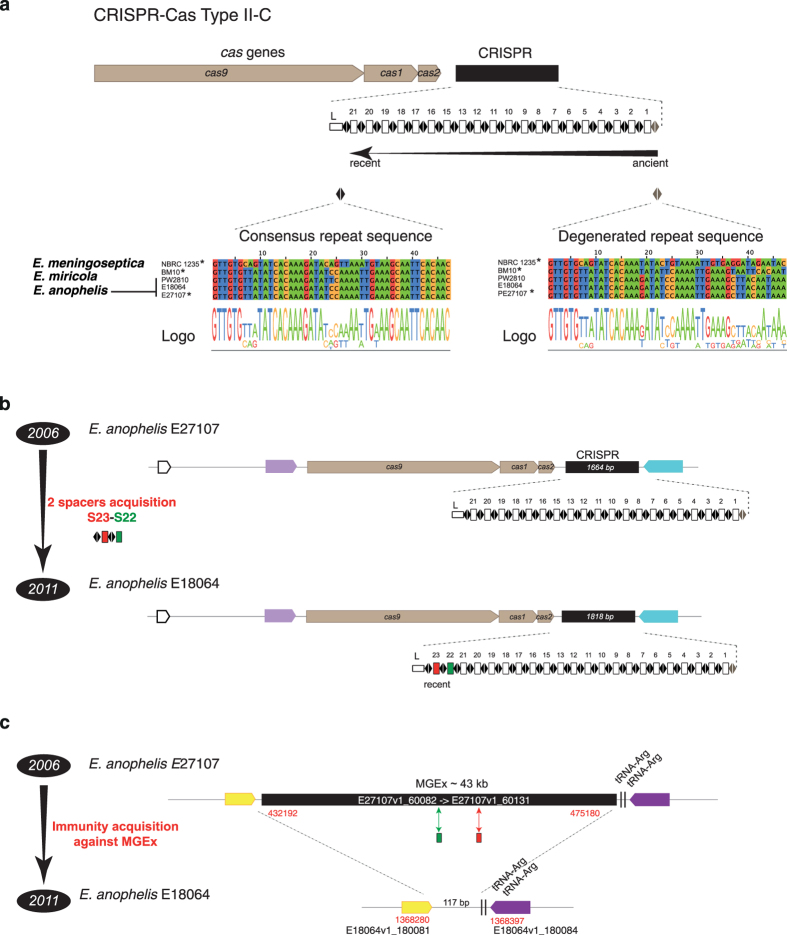
CRISPR loci exhibit spacer diversity that reflects a history of prior invasions by different phages and plasmids43. In addition, the high variability of CRISPR loci in some bacterial species implies that they represent useful markers for epidemiological investigations44,45. To extend our understanding of genomic variation between the African isolates and among the other Elizabethkingia isolates, we investigated CRISPR-Cas systems and analyzed spacer content in the 20 strains. All three species contained subtype II-C CRISPR-Cas systems comprising of Cas1, Cas2, and Cas9 proteins, while no homolog of Cas4 and Csn2 were identified46. The loci were characterized by unusually long repeats (that is, 47 bp in size, Fig. 5a), as previously observed in bacteria belonging to the Bacteroidetes phylum46. However, the CRISPR locus was only present in a few Elizabethkingia strains (Figure S3). These results demonstrate the occurrence of CRISPR systems in Elizabethkingia and the variable presence of these systems among phylogenetic sublineages of E. anophelis.
Figure 5. CRISPR-Cas type II-C organization and diversity.
(a) System organization is depicted on the top, with cas genes in brown, and the repeat-spacer array in black. Below the gene scheme, the repeat and spacer (captured MGEs) content is detailed as black diamonds and white rectangles, respectively. L corresponds to the leader sequence. Bottom line, consensus repeat sequence and degenerated repeat sequence in the 5 genomes containing such system. (b) Diversity of spacer content between the two Central African Republic hospital isolates. (c) Presence of a large MGE containing perfect matches with the two additional spacers S22 and S23.
Analysis of the spacer diversity among Elizabethkingia CRISPR arrays revealed no shared spacer between or within species, with the exception of the two hospital acquired African isolates, which showed a strong CRISPR conservation (Fig. 5b). This finding suggests a high rate of gain and loss of complete CRISPR-Cas system during E. anophelis evolution. CRISPR arrays of the two African isolates were almost identical, with the exception that E18064, which was isolated 5 years later (in 2011), contained two additional spacers at the 5′ end of the array (called S22 and S23, Fig. 5b). Importantly, according to the current spacer acquisition model, these two spacers are the most recent acquired spacers (see Methods). This clear genetic difference between CRISPR arrays of the African isolates was confirmed by PCR. CRISPR-based typing may thus offer the high level of discrimination needed for strain subtyping in a local epidemiological context.
Surprisingly, while the S22 and S23 spacers were clearly absent in the CRISPR array of the earlier African isolate E27107 (isolated in 2006), we found perfect matches (i.e. protospacers) in one E27107 specific genomic region of 43 kb (not including the CRISPR array) (Fig. 5c). This particular genomic island was totally absent in other strains and was characterized by the presence of a putative integrase gene located near a tRNA-Arg gene, three genes encoding putative phage tail and lysis proteins, with the remaining genes having unknown functions. Further, this region contained only co-oriented genes. Thus, we hypothesize that this putative mobile genetic element (MGEx) encodes a prophage. In sum, whereas isolate E27107 contained a shorter CRISPR array devoid of S22 and S23 and a putative prophage harboring S22 and S23 protospacers, isolate E18064 harbored the two spacers in its CRISPR array and lacked the corresponding prophage. Based on these observations, we speculate that the presence of these two spacers provides resistance against this particular MGE. The CRISPR and phage dynamics uncovered here emphasize the rapid diversification of MGE-related genomic features in Elizabethkingia genomes, even at the short evolutionary timescale of 5 years that separates the two African hospital infections.
Conclusions
Elizabethkingia anophelis was recently recognized as a cause of nosocomial infections, and more recently, of community-acquired infections. In this work, the genome sequences of two hospital meningitis African isolates were established and compared with available genomes from other regions and sources. Our comparative phylogenetic analyses led us to refine the identification of Elizabethkingia strains and to demonstrate firmly the phylogenetic distinctness of the three established species of the genus. Comparative genomics analysis revealed specific features of the African isolates including additional resistance genes, a unique CRISPR locus and a specific putative capsular synthesis (cps) cluster. Demonstration of the capsulated nature of the African E. anophelis isolates, and the presence of variable cps clusters suggests an important determinant of the pathogenicity mechanisms and virulence heterogeneity among Elizabethkingia strains. Analysis of the phylogenetic structure of E. anophelis revealed several well-demarcated sublineages and demonstrated a distinct evolutionary origin of African clinical isolates, excluding an epidemiological link between recent African and Asian hospital infections. The core-genome MLST approach defined here and made publicly available through the Institut Pasteur bacterial genome database and analysis platform represents a standardized high-resolution genotyping tool. It will enable to share a common language on E. anophelis isolates sublineages, which will facilitate collaborative work on the population biology, epidemiology and pathophysiology of this newly recognized bacterial species. Here, the analysis of nosocomial infection isolates using this genotyping approach suggested that the hospital environment, rather than mosquitoes, acts as a reservoir for E. anophelis, and that the high persistence of the pathogen in this environment, if uncontrolled, allows for long-term transmissions.
Materials and Methods
Isolates for genome sequencing
The two clinical strains E18064 (alias: V0378064) and E27107 (alias: Po0527107) isolated in Central African Republic were sequenced with a 100 base pair (bp) paired-end protocol in Illumina HiSeq-2000. Libraries were constructed with the Nextera DNA Sample Prep Kit (Illumina). Genome assembly was performed with the CLC Assembly Cell analysis package v.3.2.2. The average number of contigs and the N50 statistic values (i.e., the length for which half of the bases of a draft genome are situated in contigs of that length or longer) observed from the two assembled genomes were similar to those of the publicly available draft genomes (Table S1). The draft genome sequences of strains E18064 and E27107 were annotated using the MicroScope/MaGe platform47 and were deposited in the European Nucleotide Archive under accession numbers CCAB000000000 and CCAC000000000, respectively.
Genomes from sequence database
The eighteen Elizabethkingia genomic sequences available on April 1st, 2015 were downloaded from the NCBI genome sequence repository (www.ncbi.nlm.nih.gov/genome). The sequences comprised one complete genome and 17 whole-genome shotgun sequences available as scaffolds or contigs. The strains were initially identified as E. anophelis (12 isolates), E. meningoseptica (n = 4), E. miricola (n = 1), and Elizabethkingia sp. (n = 1) (Table S1).
Average nucleotide identity and 16S sequence analysis
Average nucleotide identity values were estimated from the genomic assemblies using JSpecies v.1.2.1 with the BLASTN option17. 16S rRNA sequences were gathered from genomic sequences by BLASTN similarity search using the 16S sequence of the type strain R26T (GenBank accession number EF426425) as a query.
Core genome and pan-genome
Core genomes (the set of genes present in all isolates) were built either for the genus or for E. anophelis species. Orthologs were identified as bidirectional best hits, using end-gap free global alignment, between the proteome of E. anophelis NUHP1 as a pivot and each of the other proteomes (18 for the genus and 15 for the species). Strain Endophthalmitis contained a large fraction of pseudogenes (likely due to low quality of the sequence) and was therefore excluded from this analysis. Hits with less than 60% (genus) or 80% (species) amino acid sequence similarity or more than 20% difference in sequence length were discarded. Genomes from the same species typically show low levels of genome rearrangements and this information can be used to identify orthologs more accurately48,49. Therefore, the core-genome of the species was defined as the intersection of pairwise lists of strict positional orthologs (as in ref. 50). The core-genomes consist in the genes present in all genomes of the two sets. They were defined as the intersection of the lists of orthologs between pairs of genomes.
Pan-genomes were built by clustering homologous protein-coding sequences (CDS) into families. We determined the lists of putative homologs between pairs of genomes with BLASTP used the E-values (<10−4) to perform single-linkage clustering with SiLiX v.1.251. A CDS is thus included in a family if it shares a relation of homology to at least one CDS already belonging to the family. SiLiX parameters were set to consider two CDS as homologs if the aligned part showed at least 60% (Elizabethkingia genus) or 80% (E. anophelis) identity and represented more than 80% of the smallest CDS length. The pan-genomes thus represent the full complements of genes in the genus and in the species, respectively. The pan-genomes of the genus Elizabethkingia and of the species E. anophelis were determined independently.
Core genome multilocus sequence typing (MLST)
To obtain a subset of CDSs that are highly reliable as genotypic markers, a subset of the core genome was selected based on length conservation criterion leading to the selection of 1,546 CDSs. These loci together constitute a cgMLST scheme useful for genotyping of E. meningoseptica, E. anophelis, and E. miricola isolates. The scheme comprises fewer loci than the core genome as expected from our more stringent selection criteria. The genome sequences were then scanned for allelic variation using the BIGSdb tool41 using the allele sequences from reference strain R26T as the initial query sequences.
Characterization of gene associated with drug resistance and horizontal gene transfer mechanisms
Acquired antimicrobial resistance (AR) genes were detected using HMMER3 (v.3.1b1)52 against the ResFams (Core v.1.2), a curated database of AR protein families and associated profile hidden Markov models, with the tblout and cut_ga options53 (Table S3). Virulence-associated genes were searched on the VFDB 201654 using BLASTP (minimum 40% identity with E-value <10−5), as in ref. 20 (Table S4).
Comparison of genomes
Local genomic alignments visualization was performed using software BioNumerics v.7.5 (Applied-Maths, Sint-Martens Latem, Belgium) with default seed matching and stretch extension parameters.
Phylogenetic analyses
For each of the 1,546 cgMLST loci, a multiple amino acid sequence alignment was performed with MAFFT v.7.205 (default options)55 that was back-translated in order to obtain a codon-level alignment. The concatenation of these 1,546 multiple sequence alignments was used to estimate the p-distance between each pair of isolates. These estimated evolutionary distances were used to infer a genome-level phylogenetic tree with FastME v.2.07 (Balanced Minimum Evolution criterion, SPR-based BME tree search)56.
Phylogenetic tree analyses from single markers (i.e. 16S rRNA and CAT) were performed from well-suited characters selected by BMGE v.1.12 (up to 50% allowed gap proportion with models PAM1 and BLOSUM62, respectively)57 from the multiple sequence alignments generated by MAFFT v.7.205 (default options). Tree inferences from 16S rRNA and CAT sequences were performed by PhyML v.2013101658 (SPR-based ML tree search with evolutionary models GTR + I and LG + Γ4 + I, respectively).
Detection of CRISPR-Cas systems
Clusters of cas genes were identified and classified using MacSyFinder33 with Cas-Finder profiles33. To identify cas pseudogenes, all Cas protein sequences previously detected were searched in all the Elizabethkingia draft genomes using TBLASTN v.2.0 (E-value < 10−3). CRISPR arrays were identified following a previously published methodology59. In short, they were identified using CRT (CRISPR Recognition Tool) with the option -maxRL = 6060, in all the Elizabethkingia draft genomes. For each array, the repeats were extracted and were aligned using MAFFT v.7.205 (default options). Then, we used cons (www.bioinformatics.nl/cgi-bin/emboss/help/cons) to obtain consensus sequences from these multiple sequence alignments of each array. In all cases, the consensus sequence corresponds to the most frequent sequence within a particular array. We used the consensus sequence of the repeats as patterns to identify additional, smaller and/or degenerate repeat clusters in all draft genomes with fuzznuc (www.bioinformatics.nl/cgi-bin/emboss/help/fuzznuc). This step has also confirmed the lack of any repeat in some draft genomes devoid of detectable CRISPR array and cas genes.
Spacer diversity
To identify additional spacers (i.e., located at contig extremities) or to verify the lack of a particular spacer in a given strain, all spacer sequences previously detected (i.e., flanked by repeats at both sides) were searched in all the Elizabethkingia draft genomes using BLASTN. Only hits showing at least 90% identity with the query, less than 20% difference in sequence length and flanked by sequences similar to repeats were considered as additional spacers. We then compared the spacer content within and between strains and species. Therefore, two spacers were considered as similar (within strain) or common (between strains) if they had less than 20% difference in sequence length and at least 90% identity.
CRISPR locus orientation for polarized spacer acquisition
During the adaptation phase, spacer acquisition occurs in a polarized fashion: new spacers are typically integrated at the 5′ (leader) end of the CRISPR array, which involves the duplication of the first repeat of the array, and older spacers and repeats decay at the 3′ end. Consistent with this spacer acquisition model, we found that the most degenerated repeat was located at one end of each CRISPR arrays. Hence, such particular repeat was considered as the signature of the 3′ end of the array, which corresponds to the oldest part of the locus. Such positional information represents a timeline of spacer acquisition events.
CRISPR locus PCR
To confirm the genomic differences observed between CRISPR arrays of the two African isolates based on Illumina sequence data, three primer pairs were designed for PCR amplification and sequencing of the CRISPR locus in three overlapping fragments: (i) L1F-CGTAAACGTCTGTTAGATGATGG and sp15R-AAACCATTCTACGGAGAAC; (ii) sp17F-GATGTAATAAGAGTTGTTGCG and sp5R-TCGGATTTATGAGGTGATCCC; and (iii) sp7F-CATAGATCACACATACAGGGC and L1R-TGAGCGCCCATGTTGTCTCCG. PCR conditions for all amplification reactions were as follows: initial denaturation at 94 °C for 5 min; 30 cycles at 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 30s; and final extension at 72 °C for 5 min. PCR products were purified by ultrafiltration (Millipore), and nucleotide sequences were obtained using the PCR primers and BigDye Terminator v1.1 chemistry (Applied Biosystems, Foster City, CA) on an ABI 3730XL apparatus (Applied Biosystems, Foster City, CA). Sequence traces were edited and assembled using BioNumerics.
Detection of capsular gene clusters
To identify capsular gene clusters, we performed a keyword search of the Pfam database (pfam.xfam.org) for protein profiles involved in capsular polysaccharide production such as glycosyl transferases, ABC transporters, wzx flippase and wzy polymerase (Table S6). We then performed a search of these profiles in Elizabethkingia genomes using HMMER3 (v.3.1b1)52, with the tbl_out option. This allowed us to clearly identify two capsule clusters in each of the strains 502 and NBRC 12535. These clusters were both located in the same genomic region, which included a highly conserved RecX family transcriptional regulator. We then searched for recX in the other genomes and reconstructed the putative capsular polysaccharide synthesis (cps) cluster in the recX neighborhood using the aforementioned protein profiles.
Additional Information
How to cite this article: Breurec, S. et al. Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis. Sci. Rep. 6, 30379; doi: 10.1038/srep30379 (2016).
Supplementary Material
Acknowledgments
We would like to express our most sincere thanks to the director of Institut Pasteur in Bangui, Dr. Mirdad Kazanji, who made this work possible. This study was supported by the Institut Pasteur (Programme Transversal de Recherche CEVACAR No. 385), by the CNRS and the European Research Council (Grant EVOMOBILOME, number 281605), and by the French government’s Investissement d’Avenir program Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (grant ANR-10-LABX-62-IBEID). O.R. is supported by an FRM fellowship (grant number ARF20150934077).
Footnotes
Author Contributions S. Breurec, M.T. and S. Brisse conceived the study. V.C. and S. Brisse designed the experiments. L.D. and V.P. performed the experiments. S. Breurec, A.C., L.D., O.R., M.V., E.P.C.R., M.T. and S. Brisse analyzed the data. S. Breurec, A.C., L.D., O.R., E.P.C.R., M.T. and S. Brisse wrote the paper. All authors commented on the manuscript.
References
- Kukutla P. et al. Insights from the genome annotation of Elizabethkingia anophelis from the malaria vector Anopheles gambiae. PLoS One 9, e97715 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Bagdasarian M. & Walker E. D. Elizabethkingia anophelis: molecular manipulation and interactions with mosquito hosts. Appl Environ Microbiol 81, 2233–2243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindh J. M., Borg-Karlson A. K. & Faye I. Transstadial and horizontal transfer of bacteria within a colony of Anopheles gambiae and oviposition response to bacteria-containing water. Acta Trop 107, 242–250 (2008). [DOI] [PubMed] [Google Scholar]
- Bloch K. C., Nadarajah R. & Jacobs R. Chryseobacterium meningosepticum: an emerging pathogen among immunocompromised adults. Report of 6 cases and literature review. Medicine 76, 30–41 (1997). [DOI] [PubMed] [Google Scholar]
- Güngör S., Ozen M., Akinci A. & Durmaz R. A. Chryseobacterium meningosepticum outbreak in a neonatal ward. Infect Control Hosp Epidemiol 24, 613–617 (2003). [DOI] [PubMed] [Google Scholar]
- Adachi A. et al. Chryseobacterium meningosepticum septicemia in a recipient of allogeneic cord blood transplantation. Scand J Infect Dis. 36, 539–540 (2004). [DOI] [PubMed] [Google Scholar]
- Green O., Murray P. & Gea-Banacloche J. C. Sepsis caused by Elizabethkingia miricola successfully treated with tigecycline and levofloxacin. Diagn Microbiol Infect Dis. 62, 430–432 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämpfer P. et al. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int. J. Syst. Evol. Microbiol 61, 2670–2675 (2011). [DOI] [PubMed] [Google Scholar]
- Kämpfer P., Busse H. J., McInroy J. A. & Glaeser S. P. Elizabethkingia endophytica sp. nov., isolated from Zea mays and emended description of Elizabethkingia anophelis. Int. J. Syst. Evol. Microbiol 65, 2187–2193 (2015). [DOI] [PubMed] [Google Scholar]
- Frank T. et al. First case of Elizabethkingia anophelis meningitis in the Central African Republic. Lancet 381, 1876 (2013). [DOI] [PubMed] [Google Scholar]
- Teo J. et al. First case of Elizabethkingia anophelis outbreak in an intensive-care unit. Lancet 382, 855–856 (2013). [DOI] [PubMed] [Google Scholar]
- Lau S. K. et al. Evidence for Elizabethkingia anophelis transmission from mother to infant, Hong Kong. Emerg Infect Dis. 21, 232–241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S. K. et al. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci. Rep. 6, 26045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Llorens X. & McCracken G. H. Jr. Bacterial meningitis in children. Lancet 361, 2139–2148 (2003). [DOI] [PubMed] [Google Scholar]
- Gonzalez L. J. & Vila A. J. Carbapenem resistance in Elizabethkingia meningoseptica is mediated by metallo-beta-lactamase BlaB. Antimicrob Agents Chemother 56, 1686–1692 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo J. et al. Comparative genomic analysis of malaria mosquito vector-associated novel pathogen Elizabethkingia anophelis. Genome Biol Evol 6, 1158–1165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M. & Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad Sci. USA 106, 19126–19131 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K. T. & Tiedje J. M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad Sci. USA 102, 2567–2572 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellais S., Aubert D., Naas T. & Nordmann P. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing beta-lactamases in Chryseobacterium meningosepticum. Antimicrob Agents Chemother 44, 1878–1886 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Complete Genome Sequence and Transcriptomic Analysis of the Novel Pathogen Elizabethkingia anophelis in Response to Oxidative Stress. Genome Biol. Evol. 7, 1676–1685 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer B. S., Bedzyk L. & Salyers A. A. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J. Bacteriol 173, 176–183 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heir E., Lindstedt B. A., Leegaard T. M., Gjernes E. & Kapperud G. Prevalence and characterization of integrons in blood culture Enterobacteriaceae and gastrointestinal Escherichia coli in Norway and reporting of a novel class 1 integron-located lincosamide resistance gene. Ann. Clin. Microbiol Antimicrob 3, 12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. P., Tsao M. Y., Lee S. H., Chou C. H. & Tsai H. J. Prevalence and molecular characterization of chloramphenicol resistance in Riemerella anatipestifer isolated from ducks and geese in Taiwan. Avian Pathol 39, 333–338 (2010). [DOI] [PubMed] [Google Scholar]
- Whitfield C. & Paiment A. Biosynthesis and assembly of Group 1 capsular polysaccharides in Escherichia coli and related extracellular polysaccharides in other bacteria. Carbohydr Res. 338, 2491–2502 (2003). [DOI] [PubMed] [Google Scholar]
- Li B., Zhao Y., Liu C., Chen Z. & Zhou D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol 9, 1071–1081 (2014). [DOI] [PubMed] [Google Scholar]
- Geno K. A. et al. Pneumococcal Capsules and Their Types: Past, Present, and Future. Clin Microbiol Rev. 28, 871–899 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y. J. et al. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci. Rep. 5, 15573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J. et al. Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek I. et al. Genetic exchange of determinants for capsular polysaccharide biosynthesis between Klebsiella pneumoniae strains expressing serotypes K2 and K21a. Infect Immun 61, 4208–4216 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon E. R. Molecular basis of invasive Haemophilus influenzae type b disease. J. Infect Dis. 165, S77–S81 (1992). [DOI] [PubMed] [Google Scholar]
- King E. O. Studies on a group of previously classified bacteria associated with meningitis in infants. Am. J. Clin. Pathol 31, 241–247 (1959). [DOI] [PubMed] [Google Scholar]
- Moxon E. R. & Kroll J. S. The role of bacterial polysaccharide capsules as virulence factors. Curr. Top. Microbiol. Immunol. 150, 65–85 (1990). [DOI] [PubMed] [Google Scholar]
- Abby S. S., Neron B., Menager H., Touchon M. & Rocha E. P. MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One 9, e110726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abby S. S. et al. Identification of protein secretion systems in bacterial genomes. Sci. Rep. 6, 23080 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride M. J. & Nakane D. Flavobacterium gliding motility and the type IX secretion system. Curr. Opin. Microbiol 28, 72–77 (2015). [DOI] [PubMed] [Google Scholar]
- Russell A. B. et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 16, 227–236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S., Hood R. D. & Mougous J. D. What is type VI secretion doing in all those bugs? Trends Microbiol. 18, 531–537 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet A. & Nassif X. New players in the toxin field: polymorphic toxin systems in bacteria. Mbio. 6, e00285–00215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J., Quintais L., Garcillan-Barcia M. P., de la Cruz F. & Rocha E. P. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7, e1002222 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek-Davenet S. et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect. Dis. 20, 1812–1820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley K. A. & Maiden M. C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden M. C. et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 11, 728–736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Lawrence C. M. & Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82, 237–266 (2013). [DOI] [PubMed] [Google Scholar]
- Shariat N. & Dudley E. G. CRISPRs: molecular signatures used for pathogen subtyping. Appl. Environ. Microbiol. 80, 430–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre L. et al. CRISPR typing and subtyping for improved laboratory surveillance of Salmonella infections. PLoS One 7, e36995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylinski K., Makarova K. S., Charpentier E. & Koonin E. V. Classification and evolution of type II CRISPR-Cas systems. Nucleic. Acids. Res. 42, 6091–6105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D. et al. MicroScope–an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic acids research 41, D636–D647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar T., Snel B., Huynen M. & Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 23, 324–328 (1998). [DOI] [PubMed] [Google Scholar]
- Rocha E. P. Inference and analysis of the relative stability of bacterial chromosomes. Mol. Biol. Evol. 23, 513–522 (2006). [DOI] [PubMed] [Google Scholar]
- Touchon M. et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS genetics 5, e1000344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele V., Penel S. & Duret L. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics 12, 116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M. K., Forsberg K. J. & Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 9, 207–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiong Z., Sun L., Yang J. & Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 40, D641–D645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K. & Standley D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desper R. & Gascuel O. Theoretical foundation of the balanced minimum evolution method of phylogenetic inference and its relationship to weighted least-squares tree fitting. Mol. Biol. Evol. 21, 587–598 (2004). [DOI] [PubMed] [Google Scholar]
- Criscuolo A. & Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10, 210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. & Gascuel O. Efficient biased estimation of evolutionary distances when substitution rates vary across sites. Mol. Biol. Evol. 19, 534–543 (2002). [DOI] [PubMed] [Google Scholar]
- Touchon M. & Rocha E. P. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One 5, e11126 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland C. et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8, 209 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.