Abstract
Purpose: Helicobacter pylori is one of the most prevalent infectious agents in the world which causes a variety of gastrointestinal diseases including gastritis, peptic ulcer and gastric carcinoma. The objective of this study was to comparatively evaluate invasive (rapid urease test and polymerase chain reaction) and non-invasive (enzyme-linked immunosorbent assay) tests in diagnosis of infection with cytotoxigenic H. pylori.
Methods: Biopsy specimens and sera were collected from 105 patients with gastric disorders. The presence of H. pylori infection in gastric biopsies was evaluated by RUT and PCR methods using chemotaxis signal transduction protein gene (CSTP), Urea C and HP-16srRNA primers. Serum samples were used for the ELISA test. Detection of infection with cag A-positive strains was performed by PCR and cag A-IgG ELISA kit.
Results: Patients with at least two out of three positive results were regarded as infected. The sensitivity, specificity, predictive value and accuracy of the three different methods were evaluated. Of the 105 gastric biopsies, H. pylori were positive in 51 patients (48.57%). The best sensitivity (92.16%) belonged to RUT. The sensitivities of other tests including PCR and ELISA test were 88.24% and 90.20%, respectively. PCR showed the best specificity (94.44%), and the specificities of the other tests including RUT and ELISA test, were 90.74 % and 61.11%, respectively. Furthermore, results of PCR and cag A-IgG ELISA showed high prevalence of cag A-positive strain in the study population.
Conclusion: Based on our findings, serum ELISA is a rapid noninvasive test for screening of H. pylori infection in the absence of endoscopy indication. In addition, considering the high prevalence of cytotoxigenic H. pylori strains, cag A is suggested as a promising target for PCR and non- invasive ELISA tests for detection of infection with toxigenic strains.
Keywords: Helicobacter pylori, Polymerase Chain Reaction, Enzyme-Linked Immunosorbent Assay, Cag A, Rapid Urease Test
Introduction
Helicobacter pylori (H. pylori) is a Gram-negative, microaerophilic bacterium which was identified in 1982 by Marshall and Warren.1,2 H. pylori is one of the most common human-specific pathogens which exclusively inhabits the gastric mucosa.3 Infection with H. pylori is always associated with chronic gastric inflammation, gastritis and peptic ulceration which can lead to gastric cancers such as adenocarcinoma, lymphoma of the stomach or benign mucosal-associated lymphoid tissues (MALT).4,5 H. pylori infection is prevalent throughout the world and more than half of the world population harbors this organism.6 There is a higher incidence of infection in less developed and developing countries.7,8 The prevalence of H. pylori in the Iranian population is around 80% in adults and 50% in children,9 beginning at infancy.10
The appearance of symptoms of H. pylori infection varies depending on the strains of H. pylori and the interaction of both bacterial and host factors. However, most H. pylori-infected persons are asymptomatic due to cofactors shortage of the host or bacteria or colonization by less virulent strains.11,12 The spiral shape, motility and production of urease are important virulence factors of H. pylori which facilitate the colonization of bacterium in the stomach mucosa.11 Furthermore, the bacterium releases several pathogenic proteins such as cytotoxin-associated antigen (Cag A) and vacuolating cytotoxin (Vac A).13
The cytotoxin-producing strains of Helicobacter contains the cag A gene (type I strains) and are frequently isolated from patients with gastric diseases. Hence, the detection of cag A is used for identifying infection with harmful strains.14
A number of methods are currently available for detection of H. pylori infection that divided into two groups of invasive and noninvasive methods according to the necessity of endoscopic biopsy, each having their own merits and demerits. Biopsy-based invasive tests for detection of H. pylori infection includes histological examination, culture, rapid urease test (RUT) and polymerase chain reaction (PCR).15 PCR is the accurate method that is used for detecting the H. pylori DNA by using several gene targets such as urease operon genes, cag A and Hsp60. Although PCR could be performed even with a traces of bacterial DNA, it is mainly considered as an invasive method that needs biopsy.16
On the other hand simple breath tests (UBT), serology and stool antigen test as well as Enzyme-Linked Immunosorbent Assay (ELISA) are known as non-invasive assays which are usually used for patients who are not advised undergoing gastroscopy.17 To date, several commercially available ELISA kits have been used for detection of H. pylori infection which differs in target antigens and antibody preparations. The prevalence of antibody against H. pylori varies according to geographic regions and populations.18,19 The aim of this study was to comparatively evaluate invasive (RUT and PCR) and non-invasive (ELISA) methods for diagnosis of infection with cytotoxigenic H. pylori in northwest of Iran.
Materials and Methods
Patients
A total of 105 patients with gastric disorders undergoing endoscopy at Emam Reza Hospital in Tabriz, Iran were participated in this study. The study population consisted of 43 males and 62 females with a mean age of 43 years (ranging 17 to 75 years).
Samples
Two biopsy specimens were obtained from each patient; one was used for RUT and one for PCR. In addition, serum samples from these patients were collected for ELISA tests.
Rapid Urease Test
RUT was performed at the time of endoscopy and by adding the biopsy specimens to 0.5 mL of 10% (w/v) urea in deionized water containing phenol red indicator. A positive result was recorded when the color changed from yellow to pink within two hours.
Enzyme-Linked Immunosorbent Assay
The presence of Anti- H. pylori IgG was determined by ELISA using the H. pylori IgG kit (DIA.PRO). Subsequently, a serological assay for anti-cag A antibody was performed by commercial ELISA kit (DIA.PRO)according to the manufacturer’s instructions. Briefly, a 1/100 dilution of sera in buffer was introduced in H. pylori-coated microtiter wells. After one hour incubation, the wells were washed and incubated with peroxidase-conjugated anti-human IgG. The tetramethylbenzidine substrate was then added and the optical density (OD) was measured at 450 nm and 620 nm. The results were expressed as unit per milliliter according to a calibrator curve.
Polymerase chain reaction
Each sample was examined by five different primers. Three of them for detecting the H. pylori DNA and two for amplification of the cag A gene sequence in order to diagnose infection with harmful strains. Primers used in this study were from chemotaxis signal transduction protein (CSTP)(987 bp), urease C (337 bp) and 16S rRNA (439 bp) gene fragments. The primers sequences and product sizes are listed in Table 1.
Table 1. Primer Sequences Used for Polymerase Chain Reaction Amplifications .
| Primer | Sequence (5'->3') | Tm | Product length |
| CSTP-F | GAAGTCATGGCTGATAGTTTA | 59.81 | 987 bp |
| CSTP-R | TAGTGCTGTATTTTTTCATGCTAA | ||
| Urea C-F | CTAGTGGTGGTGGACAATTTAGG | 58 | 337 bp |
| Urea C-R | CTTGCTTACTTTCTAACACTAACGC | ||
| HP16s- F | CAGCTTGTTGGTAAGGTAATGGC | 56 | 439 bp |
| HP16s- R | GATCTCTACGGATTTTACCCCTACAC | ||
| cag595-F | AACAGGCAAGCTTTTGATGG | 60.25 | 595 bp |
| cag 595-R | GCGGTAAGCCTTGTATGTGAG | ||
| cag 750-F | ACAATGACTAACGAAACTATTGA | 59.78 | 750 bp |
| cag 750-R | ACATCACGCCATCATGTTTTA |
For PCR, genomic DNAs were extracted by standard CTAB/NaCl method.20 Briefly, samples were resuspended overnight at 40°C in TE buffer (Tris 10 mM, EDTA 1 mM, pH=8) together with 10% sodium dodecyl sulfate (SDS) and proteinase K. Then, the DNA was extracted by CTAB/NaCl solution (10% CTAB and 0.7 M NaCl). The cell debris and proteins were removed by two times phenol/chloroform/isoamylalcohol (25:24:1) extraction. DNA was precipitated by isopropanol and washed with ethanol (70%), dried, and then resuspended in TE buffer. One microliter of the extracted DNA was used as the template for PCR.
PCR amplification was carried out in a final volume of 25 μL containing 2 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate (dATP, dCTP, dTTP and GTP), 0.4 μL of each primer (Forward, Reverse) and 2.5 U of Taq DNA polymerase. The PCR reactions were as follows: an initial denaturation at 94°C for 5 minutes, with 35 cycles of denaturation at 94°C for 30 seconds, annealing at Primer Specific Tm for 30 seconds, extension at 72°C for 30 seconds and final extension at 72°C for 7 minutes. The PCR products were visualized on 1% agarose gel under UV light, after staining with ethidium bromide (Figure 1).
Figure 1.
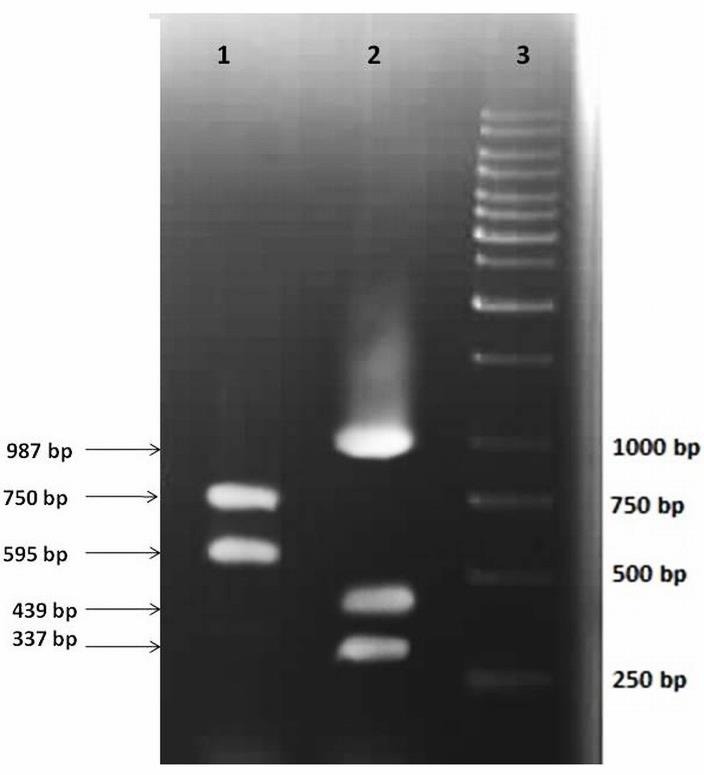
Agarose gel electrophoresis of PCR products related to the amplification of target genes. Lane 1, PCR products of cag 595 and cag 750; lane2, PCR products of Urea C, 16s rRNA and CSTP genes.
For amplification of the cag A gene, 1 μL of the prepared DNA from all biopsy specimens was subjected to PCR. The 750 bp and 595 bp fragments of the cag A gene sequence were amplified using the primers cag 595-F and cag 595-R (595 bp) and cag 750-F and cag 750-R (750 bp) primers, respectively (Table 1).
Results
The amplification of target genes by PCR was visualized on agarose gel, revealing specified bands of about 337 bp for the urease C gene, 439 bp for the HP16s rRNA gene and 987 bp for the CSTPgene.
Patients with at least two out of three positive tests (gold standard) were regarded as infected. According to this definition, of 105 gastric biopsies, 51 (48.57%) were positive for H. pylori, and 54 (51.42%) were diagnosed as uninfected. Out of 105 examined samples, 36(34.28%) were positive and 25 (23.80%) were negative by all diagnostic techniques (Table 2).
Table 2. Sensitivity, specificity, predictive values and accuracy of RUT, PCR and ELISA test for detection of H. pylori infections.
| Assay | Gold standard | Sensitivity | Specificity | Accuracy | PPV | NPV | ||
| Positive | not detected | |||||||
| RUT | Positive | 47 | 5 | 92.16% | 90.74% | 91.42 % | 90.38% | 92.45% |
| Negative | 4 | 49 | ||||||
| PCR | Positive | 45 | 3 | 88.24% | 94.44% | 91.42 % | 93.75% | 89.47% |
| Negative | 6 | 51 | ||||||
| ELISA | Positive | 46 | 21 | 90.20% | 61.11% | 72.23% | 68.66% | 86.84% |
| Negative | 5 | 33 | ||||||
| Total | 51 | 54 | ||||||
Abbreviations: RUT, rapid urease test; PCR, polymerase chain reaction; ELISA, enzyme-linked immunosorbent assay.
The sensitivity, specificity, predictive values and accuracy of three different methods including RUT, PCR and ELISA were determined to identify the most appropriate test for the diagnosis of infection with H. pylori (Table 2).
Detection of the cag A
H. pylori was detected in 67 (70%) of 105 cases using HP- IgG ELISA Kit, whereas anti-cag A ELISA was positive in 44 (43%) patients. Of 71 positive cases, 40 (56%) samples were positive for both ELISA tests, 27 (38%) were positive only by HP- IgG ELISA test and 4 (5.6%) were positive only by cag A-IgG ELISA test.
For detection of the cag A gene, 105 samples were examined by PCR using two pairs of specific primer yielded products of 750 bp and 595 bp portions of the cag A gene sequence. Of 105 patients, 25 samples were positive by cag-595 primers and 22 were positive by cag-750. Of the positive samples, 13 were positive by both primers, 9 only by cag-595 and 12 only by cag-750. Out of 48 PCR-positive samples, 47 were infected with H. pylori cag A-positive strains.
Discussion
Although numerous methods for the presence of H. pylori have been developed, the gold standard for the detection of H. pylori infection is controversial. None of the diagnostic methods is entirely failsafe or suitable for all situations and each has its own drawbacks. Although there is a need for rapid, cost-effective and highly accurate test in clinical settings, there is no single appropriate test for diagnosis of H. pylori infection yet.16,21
Invasive tests such as the rapid urease test (RUT) and histology have been considered as the gold standard in several studies owing to their high sensitivity (above 80%-100%) and specificity (ranging from 97%-99%).22-24 In our study, RUT test presented the best sensitivity of 92.16 %, but specificity of 90.74% which was lower than the PCR method. Only four false positive and four false negative results were observed in this study which were in line with those reported by other authors.25,26
Several factors affect the result of RUT including the biopsy condition as well as the type of disease. The accuracy of RUT is dependent on site, number, size and bacterial density of biopsy specimen.23 Biopsies from both antrum and corpus and combining them prior to RUT increase the sensitivity of the test. In contrast, it was shown that the sensitivity of RUT decreased in patients with bleeding peptic ulcers.15,21 Compared to conventional methods, molecular tests such as PCR are faster, more accurate and sensitive. The need for limited quantity of bacteria enables PCR to recognize infection when other tests are negative due to low bacterial density.15 Additionally, this method is used not only for detection of antibiotic resistance and related mutations but also for characterization of pathogenic genes and virulence determinants, which give this modality an advantage over others techniques.16
Various genes have been used as targets for PCR analysis of Helicobacter infection. These target genes could be classified into two major groups. The conserved genes for detection of H. pylori include urease operon genes, ureC gene (glmM), 16S rRNA gene, 23S rRNA gene, hsp60 gene, a 26-kDa species-specific antigen gene (SSA) and pathogenic genes for characterization of virulent strains such as cag A gene, babA2 gene, oip A gene and vac A.27,28
To the best of our knowledge application of single primer pair is not sufficient for detecting H. pylori infection because none of the primers show 100% sensitivity or specificity.29 Our results indicate that the combination of primers can significantly improve the detection of H. pylori infection.
In the current study, we utilized ureaC, 16S rRNA and CSTP as conserved genes for detection of infection by different strains of H. pylori. Multiplex PCR yielded the best specificity of 94.44% and sensitivity of 88.24%, respectively. 16srRNA displayed the highest sensitivity followed by CSTP and UreaC genes. These findings were consistent with results of previously reported studies.30,31 Low Sensitivity of PCR is possibly due the presence of inhibitors of the polymerase enzyme which adversely impact the outcomes.16 Besides, as a passive test, distinction between live and dead organisms is not possible via PCR and it might results in false positive.15,32
Although PCR has been reported as a highly sensitive and specific test in several studies:25,33 a disadvantage of this method is that in contrast to non-invasive tests such as ELISA, patients must undergo oral endoscopy. Not only generalized use of endoscopy is impractical but also some patients cannot tolerate this procedure. In this light, patients could be screened non-invasively for H. pylori infection based on clinical goal or “test-and-treat” strategy.26 ELISA is safe, not influenced by sampling errors and less of a burden for patients.34
In our study, ELISA yielded 90.20 % sensitivity and 61.11% specificity, respectively. Compared to PCR, ELISA presented higher sensitivity and lower specificity. Sensitivity is an important parameter where the test is used to identify a serious but treatable disorder.35 Therefore, despite lower specificity, ELISA could be considered as a first-line method for detection of H. pylori infection. To accurately diagnose disorders, it is recommended to subject the initially positive patients with "high sensitivity/ low specificity" tests to a second line-test with "low sensitivity/high specificity". In this way, the majority of false positives will be identified as disease negative.35
Nowadays, the geographical differences of H. pylori strains and high prevalence of virulent strains particularly in Asian countries necessitated the cag A screening of clinical samples. It has shown that the pathogenicity of H. pylori strains is significantly higher in cag A-positive strains.12,36 Hence, all the samples were screened for the presence of cag A gene by PCR using two pairs of specific primers related to C-terminus and N-terminus of the cag A protein. The results indicated that all but one patient were infected with cag A-positive strains. The C-terminus of this gene is polymorphic and bears different motifs.37 Our findings showed that the sensitivity of N-terminus primers was higher than the C-terminus primers. The detection of cag A protein was performed by cag A-IgG ELISA kit and demonstrated the similar results with cag A-PCR. The results indicated that cag A is a promising target not only for PCR but also for non- invasive ELISA test in the Iranian population.
Conclusion
The results of the present study indicated that noninvasive ELlSA is highly sensitive test for first-line detection of H. pylori infection. PCR, RUT or UBT could be considered for determination of H. pylori eradication in patients subjected to antimicrobial treatments.
Acknowledgments
The present study was financially supported by a grant from Tabriz University of medical science and Biotechnology Research Center, Tabriz, Iran.
Ethical Issues
The project was approved by the ethics committee of Tabriz university of Medical Sciences.
Conflict of Interest
The authors report no conflict of interest.
References
- 1.Ahmed N. 23 years of the discovery of Helicobacter pylori: Is the debate over? Ann Clin Microbiol Antimicrob. 2005;4:17. doi: 10.1186/1476-0711-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishaq S, Nunn L. Helicobacter pylori and Gastric Cancer: A State of the Art Review. Gastroenterol Hepatol Bed Bench. 2015;8(Suppl 1):S6–S14. [PMC free article] [PubMed] [Google Scholar]
- 3.Logan RP, Walker MM. ABC of the upper gastrointestinal tract: Epidemiology and diagnosis of Helicobacter pylori infection. BMJ. 2001;323(7318):920–2. doi: 10.1136/bmj.323.7318.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thung I, Aramin H, Vavinskaya V, Gupta S, Park JY, Crowe SE. et al. Review article: the global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol Ther. 2016;43(4):514–33. doi: 10.1111/apt.13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redeen S, Petersson F, Tornkrantz E, Levander H, Mardh E, Borch K. Reliability of diagnostic tests for Helicobacter pylori infection. Gastroenterol Res Pract. 2011;2011:940650. doi: 10.1155/2011/940650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conteduca V, Sansonno D, Lauletta G, Russi S, Ingravallo G, Dammacco F. H. pylori infection and gastric cancer: state of the art (review) Int J Oncol. 2013;42(1):5–18. doi: 10.3892/ijo.2012.1701. [DOI] [PubMed] [Google Scholar]
- 7.Goddard AF, Logan RPH. Diagnostic methods for Helicobacter pylori detection and eradication. Br J Clin Pharmacol. 2003;56(3):273–83. doi: 10.1046/j.1365-2125.2003.01941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Doorn LJ, Henskens Y, Nouhan N, Verschuuren A, Vreede R, Herbink P. et al. The Efficacy of Laboratory Diagnosis of Helicobacter pylori Infections in Gastric Biopsy Specimens Is Related to Bacterial Density and vacA, cagA, and iceA Genotypes. J Clin Microbiol. 2000;38(1):13–7. doi: 10.1128/jcm.38.1.13-17.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falsafi T, Lavasani P, Basardeh I, Massarrat S, Landarani Z. Evaluation of an Iranian Home-made Helicobacter pylori Stool Antigen ELISA Kit. Jundishapur J Microbiol. 2014;7(6):e10629. doi: 10.5812/jjm.10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alborzi A, Soltani J, Pourabbas B, Oboodi B, Haghighat M, Hayati M. et al. Prevalence of Helicobacter pylori infection in children (south of Iran) Diagn Microbiol Infect Dis. 2006;54(4):259–61. doi: 10.1016/j.diagmicrobio.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Dubois A. Spiral bacteria in the human stomach: the gastric helicobacters. Emerg Infect Dis. 1995;1(3):79–85. doi: 10.3201/eid0103.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7(11):629–41. doi: 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones KR, Whitmire JM, Merrell DS. A tale of two toxins: Helicobacter pylori CagA and VacA modulate host pathways that impact disease. Front Microbiol. 2010;1:115. doi: 10.3389/fmicb.2010.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreson H, Loivukene K, Sillakivi T, Maaroos HI, Ustav M, Peetsalu A. et al. Association of cagA and vacA genotypes of Helicobacter pylori with gastric diseases in Estonia. J Clin Microbiol. 2002;40(1):298–300. doi: 10.1128/JCM.40.1.298-300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garza-Gonzalez E, Perez-Perez GI, Maldonado-Garza HJ, Bosques-Padilla FJ. A review of Helicobacter pylori diagnosis, treatment, and methods to detect eradication. World J Gastroenterol. 2014;20(6):1438–49. doi: 10.3748/wjg.v20.i6.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel SK, Pratap CB, Jain AK, Gulati AK, Nath G. Diagnosis of Helicobacter pylori: What should be the gold standard? World J Gastroenterol. 2014;20(36):12847–59. doi: 10.3748/wjg.v20.i36.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Testerman TL, Morris J. Beyond the stomach: An updated view of Helicobacter pylori pathogenesis, diagnosis, and treatment. World J Gastroenterol. 2014;20(36):12781–808. doi: 10.3748/wjg.v20.i36.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laheij RJ, Straatman H, Jansen JB, Verbeek AL. Evaluation of commercially available Helicobacter pylori serology kits: a review. J Clin Microbiol. 1998;36(10):2803–9. doi: 10.1128/jcm.36.10.2803-2809.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren Z, Borody T, Pang G, Dunkley M, Clancy R, Xia HH. et al. Evaluation of anti-Helicobacter pylori IgG2 antibody for the diagnosis of Helicobacter pylori infection in western and Chinese populations. Aliment Pharmacol Ther. 2005;21(1):83–9. doi: 10.1111/j.1365-2036.2004.02293.x. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Russell DW. The condensed protocols from molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]
- 21.Wang YK, Kuo FC, Liu CJ, Wu MC, Shih HY, Wang SS. et al. Diagnosis of helicobacter pylori infection: Current options and developments. World J Gastroenterol. 2015;21(40):11221–35. doi: 10.3748/wjg.v21.i40.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo JH, Park JS, Rhee KH, Youn HS. Limitations of urease test in diagnosis of pediatric Helicobacter pylori infection. World J Clin Pediatr. 2015;4(4):143–7. doi: 10.5409/wjcp.v4.i4.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uotani T, Graham DY. Diagnosis of Helicobacter pylori using the rapid urease test. Ann Transl Med. 2015;3(1):9. doi: 10.3978/j.issn.2305-5839.2014.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramis IB, de Moraes EP, Fernandes MS, Mendoza-Sassi R, Rodrigues O, Juliano CR. et al. Evaluation of diagnostic methods for the detection of Helicobacter pylori in gastric biopsy specimens of dyspeptic patients. Braz J Microbiol. 2012;43(3):903–8. doi: 10.1590/S1517-83822012000300008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khalifehgholi M, Shamsipour F, Ajhdarkosh H, Ebrahimi Daryani N, Pourmand MR, Hosseini M. et al. Comparison of five diagnostic methods for Helicobacter pylori. Iran J Microbiol. 2013;5(4):396–401. [PMC free article] [PubMed] [Google Scholar]
- 26.Ables AZ, Simon I, Melton ER. Update on Helicobacter pylori treatment. Am Fam Physician. 2007;75(3):351–8. [PubMed] [Google Scholar]
- 27.Zambon CF, Navaglia F, Basso D, Rugge M, Plebani M. Helicobacter pylori babA2, cagA, and s1 vacA genes work synergistically in causing intestinal metaplasia. J Clin Pathol. 2003;56(4):287–91. doi: 10.1136/jcp.56.4.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oktem-Okullu S, Tiftikci A, Saruc M, Cicek B, Vardareli E, Tozun N. et al. Multiplex-PCR-Based Screening and Computational Modeling of Virulence Factors and T-Cell Mediated Immunity in Helicobacter pylori Infections for Accurate Clinical Diagnosis. PloS One. 2015;10(8):e0136212. doi: 10.1371/journal.pone.0136212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugimoto M, Wu JY, Abudayyeh S, Hoffman J, Brahem H, Al-Khatib K. et al. Unreliability of results of PCR detection of Helicobacter pylori in clinical or environmental samples. J Clin Microbiol. 2009;47(3):738–42. doi: 10.1128/JCM.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorkiewicz G, Feierl G, Schober C, Dieber F, Kofer J, Zechner R. et al. Species-specific identification of campylobacters by partial 16S rRNA gene sequencing. J Clin Microbiol. 2003;41(6):2537–46. doi: 10.1128/JCM.41.6.2537-2546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H, Rahman A, Semino-Mora C, Doi SQ, Dubois A. Specific and sensitive detection of H. pylori in biological specimens by real-time RT-PCR and in situ hybridization. PLoS One. 2008;3(7):e2689. doi: 10.1371/journal.pone.0002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ricci C, Holton J, Vaira D. Diagnosis of Helicobacter pylori: invasive and non-invasive tests. Best Pract Res Clin Gastroenterol. 2007;21(2):299–313. doi: 10.1016/j.bpg.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Lu JJ, Perng CL, Shyu RY, Chen CH, Lou Q, Chong SK. et al. Comparison of five PCR methods for detection of Helicobacter pylori DNA in gastric tissues. J Clin Microbiol. 1999;37(3):772–4. doi: 10.1128/jcm.37.3.772-774.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahman SH, Azam MG, Rahman MA, Arfin MS, Alam MM, Bhuiyan TM. et al. Non-invasive diagnosis of H pylori infection: evaluation of serological tests with and without current infection marker CIM. World J Gastroenterol. 2008;14(8):1231–6. doi: 10.3748/wjg.14.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lalkhen AG, McCluskey A. Clinical tests: sensitivity and specificity. Contin Educ Anaesth Crit Care Pain. 2008;8(6):221–3. doi: 10.1093/bjaceaccp/mkn041. [DOI] [Google Scholar]
- 36.Ghotaslou R, Milani M, Akhi MT, Hejazi MS, Nahaei MR, Hasani A. et al. Relationship Between Drug Resistance and cagA Gene in Helicobacter pylori. Jundishapur J Microbiol. 2013;6(10):e8480. doi: 10.5812/jjm.8480. [DOI] [Google Scholar]
- 37.Sicinschi LA, Correa P, Peek RM, Camargo MC, Piazuelo MB, Romero-Gallo J. et al. CagA C-terminal variations in Helicobacter pylori strains from Colombian patients with gastric precancerous lesions. Clin Microbiol Infect. 2010;16(4):369–78. doi: 10.1111/j.1469-0691.2009.02811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


