Abstract
The thyroid gland is an important endocrine organ modulating development, growth, and metabolism, mainly by controlling the synthesis and secretion of thyroid hormones (THs). However, little is known about the pig thyroid transcriptome. Long non-coding RNAs (lncRNAs) regulate gene expression and play critical roles in many cellular processes. Yorkshire pigs have a higher growth rate but lower fat deposition than that of Jinhua pigs, and thus, these species are ideal models for studying growth and lipid metabolism. This study revealed higher levels of THs in the serum of Yorkshire pigs than in the serum of Jinhua pigs. By using Ribo-zero RNA sequencing—which can capture both polyA and non-polyA transcripts—the thyroid transcriptome of both breeds were analyzed and 22,435 known mRNAs were found to be expressed in the pig thyroid. In addition, 1189 novel mRNAs and 1018 candidate lncRNA transcripts were detected. Multiple TH-synthesis-related genes were identified among the 455 differentially-expressed known mRNAs, 37 novel mRNAs, and 52 lncRNA transcripts. Bioinformatics analysis revealed that differentially-expressed genes were enriched in the microtubule-based process, which contributes to THs secretion. Moreover, integrating analysis predicted 13 potential lncRNA-mRNA gene pairs. These data expanded the repertoire of porcine lncRNAs and mRNAs and contribute to understanding the possible molecular mechanisms involved in animal growth and lipid metabolism.
Keywords: Ribo-zero RNA sequencing, lncRNA, thyroid, pig
1. Introduction
The thyroid is one of the largest endocrine glands in the body, mainly synthesizing and secreting two vital thyroid hormones (THs), tetraiodothyronine or thyroxine (T4) and triiodothyronine (T3). THs play critical roles in differentiation, growth, metabolism, and reproduction [1]. Increased levels of THs can stimulate almost all aspects of carbohydrate metabolism, increase the basal metabolic rate, and stimulate both lipogenesis and lipolysis—although lipolysis is more influenced than synthesis [2,3]. Thyrocytes—the cells responsible for producing and secreting T4 and T3—integrate complicated extracellular signals from the hypothalamus (thyrotropin releasing hormone, TRH) and the pituitary (thyroid stimulating hormone, TSH), the feedback from serum THs [4], and from other factors, such as iodine and leptin [5,6], which then influence the intracellular signaling pathways modulating THs synthesis and secretion. Thus, many interacting genes determine the regulation of THs synthesis and secretion. Among others, the genes encoding thyroglobulin (Tg), thyroid stimulating hormone receptor (TSHR), solute carrier family 5 member 5 (SLC5A5), iodotyrosine deiodinase (IYD), and dual oxidase 2 (DUOX2), as well as the calcium signaling pathway, glutathione metabolism, lysosome, and tyrosine metabolism signaling pathways, are involved in the synthesis and secretion of THs [7].
Long noncoding RNAs (lncRNAs) are a type of RNA transcript, longer than 200 bases but without evident protein-coding capacity. Accumulating evidence suggests that lncRNAs regulate gene expression during cellular development and homeostasis by recruiting epigenetic complexes or directly affecting the process of transcription [8,9,10]. The expression of lncRNAs is highly regulated, displaying discrete temporal and spatial expression patterns [11,12], and some groups have revealed their crucial role in the normal development and function of several endocrine organs, such as the pancreas, prostate gland, and adipose tissue [13]. However, little is known about the physiological function of lncRNAs in the thyroid gland.
Recent studies suggest that non-polyadenylated (polyA) RNA transcripts (both protein-coding and noncoding) are functionally important [14,15]. Although high-throughput RNA sequencing (RNA-seq) elucidates on global gene expression profiles, the standard method of enriching polyA RNAs with oligo (dT) primers cannot be used to reveal non-polyA transcripts and partially degraded mRNAs. This limitation has been overcome by the Ribo-Zero RNA-seq, a new method that can capture both polyA and non-polyA transcripts [16].
The pig (Sus scrofa) is an economically important food source, corresponding to approximately 40% of all meat consumed worldwide. The Jinhua pig is a Chinese indigenous fat-type pig breed that has low growth rate and body weight, but high fat deposition. The Yorkshire pig, on the contrary, is a lean-type pig breed characterized by high growth rate and body weight, but low fat deposition. Therefore, Jinhua and Yorkshire pigs are ideal models for studying animal growth and fat deposition.
In this study, we compared the transcriptome profiles of the thyroid gland of Jinhua and Yorkshire pigs using Ribo-Zero RNA-seq. To our knowledge, this is the first description of lncRNA expression profiles in the porcine thyroid. This study aimed to extend the transcript catalog (both mRNAs and lncRNAs) in pig species, identifying putative mRNAs, lncRNAs, and pathways that are involved in the regulation of animal growth and fat deposition.
2. Materials and Methods
2.1. Animal Material
All procedures described here were conducted under an experimental license from the Animal Care and Use Committee of Zhejiang University, in agreement with the relevant guidelines and regulations imposed by the Administration of Affairs Concerning Experimental Animals. Three castrated Jinhua pigs and three castrated Yorkshire pigs aged 120 days were humanely killed and their body weights were recorded. Thyroid glands were weighted and thyroid weight indices were calculated as thyroid weight/ body weight ratios (g/kg). Thyroid glands were cut up and preserved in liquid nitrogen within 10 min post-slaughter, and then stored at −80 °C until RNA extraction.
2.2. Serum THs Assays
Blood samples were collected from the anterior vena cava of each animal, kept at 37 °C for 2 h, and then centrifuged at 4 °C for 10 min (3000 × g). Serum was collected and its total thyroxine (TT4), total triiodothyronine (TT3), free thyroxine (FT4), and free triiodothyronine (FT3) concentrations were measured through an enzyme-linked immunosorbent assay (ELISA) provided at the Affiliated Hospital of Hangzhou Normal University.
2.3. RNA Isolation, Library Preparation, and Sequencing
Total RNA was isolated from each individual sample using TRIzol™ reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. RNA was quantified using a Qubit RNA Assay Kit in a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA), and checked for purity and integrity in a Bioanalyzer 2100 system (Agilent Technologies, Palo Alto, CA, USA). RNA integrity number (RIN) was larger than 8.0 in all samples.
Approximately 3 μg RNA per sample (two individuals per breed) were used to construct the complementary (cDNA) library, according to the following procedures: ribosomal RNA (rRNA) was removed from total RNA by Epicentre Ribo-zero™ rRNA Removal Kit (Epicentre, Madison, WI, USA); rRNA-depleted RNA was then used to generate strand-specific RNA-seq libraries using the NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, Ipswich, UK), following manufacturer’s recommendations. Briefly, after RNA fragmentation, double-stranded cDNA was synthesized replacing dTTPs with dUTPs in the reaction buffer used in second strand cDNA synthesis. The resulting double-stranded cDNA was ligated to NEBNext® adaptors, after being end-repaired and A-tailed, and 150–200 bp cDNA fragments were isolated. Single strand cDNA was then obtained using USER Enzyme. Finally, PCR amplification was performed to enrich cDNA libraries.
Sequencing was performed on an Illumina® Hiseq 2500 (Illumina, San Diego, CA, USA) instrument using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina®) to generate 125 bp paired-end reads.
2.4. Mapping to the Reference Genome
Quality control and reads statistics were determined by FASTQC v0.11.2 [17]. The Phred score (Q20, Q30) and GC content of the raw reads were calculated. After discarding the reads containing adapter, reads containing over 10% poly-Ns, and reads of low quality (>50% of bases with Phred scores <5), the remaining clean reads were aligned to the reference pig genome (S. scrofa 10.2) using TopHat v2.0.9 [18] and its underlying aligner Bowtie v2.0.6 [19]. TopHat run parameters were set to default except for “-library-type fr-firststrand”. The number of reads annotated in mRNAs, misc_RNAs, ncRNAs, precursor RNAs, pseudogenes, rRNAs, and tRNAs were counted in HTSeq v0.5.4p5 [20].
2.5. Transcriptome Assembly
Mapped reads belonging to each sample were assembled in Scripture (beta2) [21] using the default parameters, and in Cufflinks v2.1.1 [22] using the default parameters, except for “min-frags-per-transfrag = 0” and “-library-type fr-firststrand”. Transcripts resulting from the two assemblers were pooled, and they were considered reliable if it found for at least two samples or in both assemblers.
2.6. Identification of lncRNA and Novel mRNA
Assembled transcripts were processed as follows:
The reads coverage of every transcript was calculated by Cufflinks, and those with reads coverage less than three were eliminated;
Single exon transcripts and transcripts <200 bp were excluded;
Transcripts blasted to known mRNA and belonged to pseudogenes, pre-microRNA, tRNA, rRNA, and snoRNA were removed;
CNCI v2 (score < 0) [23], CPC-0.9-r2 (score < 0) [24], Pfam-scan v1.3 (E-value < 0.001) [25], and PhyloCSF v20121028 (no sufficiently long ORFs found) [26] were used to evaluate the coding potential of transcripts. Transcripts revealing coding potential in any of the four tools were filtered out.
Transcripts that passed all the filters mentioned above were considered candidate lncRNAs, and those with coding potential were considered novel mRNAs. Candidate lncRNAs were blasted to pig lncRNAs in the ALDB v1.0 database.
2.7. Homology Analysis with lncRNAs in Human and Mouse
Human and mouse lncRNAs were downloaded from NONCODE v5 database [27], which included several public databases (Ensembl [28], RefSeq [29], lncRNAdb [30], and GENCODE [31]). Blastn was used to search for homology between candidate lncRNAs identified in our study and human or mouse lncRNAs in the NONCODE v5 database, considering a 1 × 10−6 E-value threshold.
2.8. Differential Expression Analysis
The expression levels of mRNAs and lncRNAs were calculated in fragments per kilo-base of exon per million mapped fragments (FPKM) using the Cuffdiff tool in Cufflinks v2.1.1 [22]. In addition, Cuffdiff was applied to determine differentially-expressed mRNAs and lncRNAs using a model based on the negative binomial distribution. A q-value < 0.05 (Benjamini and Hochberg’s false discovery rate) was set as the threshold for significantly differential expression. Hierarchical clustering was performed to visualize mRNAs and lncRNAs differential expression patterns among samples.
2.9. Functional Enrichment Analysis
Gene Ontology (GO) analysis of the differentially-expressed mRNAs was performed using the GOseq R package (Release2.12) [32], considering GO terms with corrected p-value < 0.05 as significantly enriched. Pathway analysis, using KO-Based Annotation System (KOBAS) v2.0 [33], allowed statistical evaluation of the differentially-expressed mRNAs in KEGG pathways.
2.10. Target Gene Prediction
Differentially-expressed lncRNAs were selected for target prediction. The cis role of lncRNAs was their acting on neighboring target genes [34]. In this study, mRNAs 100 kb up- and down-stream of the differentially-expressed lncRNAs were evaluated, as they might be regulated by the adjacent lncRNAs. In order to reduce false positives, the differentially-expressed mRNAs were selected as potential target genes.
2.11. Validation of the Sequencing Data by qRT-PCR
Quantitative real-time PCR was performed on an ABI Step One Plus system (Applied Biosystem, Carlsbad, CA, USA) using SYBR Premix Ex Taq kit (TaKaRa, Dalian, China) with specific primers (Table S11). Gene relative-expression levels were quantified based on GAPDH gene expression by using ΔΔCt method [35]. Three independent biological replicates were used. All measurements were performed in triplicate. Statistical differences between two pig breeds were evaluated using Student’s t-test and p < 0.05 as the threshold for significance.
3. Results
3.1. Body Index and THs Concentrations in the Serum of the Two Porcine Breeds
As shown in Table 1, body weight and thyroid gland weight of Yorkshire pigs were significantly higher than in Jinhua pigs (p < 0.05); however, the thyroid index was almost identical for the two pig breeds. Yorkshire pigs had significantly higher levels of TT4 and TT3 than Jinhua pigs (p < 0.05), and higher levels of FT4 and FT3 than Jinhua pigs, although these were not statistically significant (p > 0.05).
Table 1.
Body Index and serum concentration of thyroid hormones (THs) in the Jinhua and Yorkshire pig breeds.
| Traits | Jinhua | Yorkshire | p-Value |
|---|---|---|---|
| Body weight (kg) | 29.33 ± 0.88 | 47.07 ± 1.93 | 0.001 |
| Thyroid gland weight (g) | 3.20 ± 0.06 | 4.93 ± 0.38 | 0.011 |
| Thyroid index (g/kg) | 0.109 ± 0.004 | 0.105 ± 0.009 | 0.702 |
| Serum total thyroxine level (nmol/L) | 30.36 ± 0.76 | 37.85 ± 1.73 | 0.017 |
| Serum total triiodothyronine level (nmol/L) | 0.53 ± 0.02 | 1.00 ± 0.12 | 0.020 |
| Serum free thyroxine level (pmol/L) | 9.34 ± 0.20 | 11.31 ± 0.73 | 0.058 |
| Serum free triiodothyronine level (pmol/L) | 1.84 ± 0.09 | 2.58 ± 0.33 | 0.096 |
All data are expressed as mean ± SEM; and a p-value < 0.05 reveals significant differences.
3.2. Characterization of the Thyroid Gland Transcriptome
The four Ribo-zero RNA sequencing libraries (two Jinhua and two Yorkshire) were sequenced on the Illumina® HiSeq 2500 platform. Sequencing yielded about 351 million 125-bp paired-end reads. Approximately 83% of these reads were mapped to the pig reference genome (S. scrofa 10.2) using Tophat, and about 72% aligned with unique loci (Table 2). HTSeq was used to calculate the proportion of reads aligning to mRNAs, misc_RNAs, ncRNAs, precursor RNAs, pseudogenes, rRNAs, and tRNAs (Table S1). Most reads were matched mRNAs (51%–58%). However, 33%–39% of the reads were mapped outside of annotated loci.
Table 2.
Summary of the sequencing reads alignment to the reference genome.
| Sample | Total Reads | Total Mapped % | Uniquely Mapped % |
|---|---|---|---|
| Jinhua-1 | 89460072 | 81.83 | 70.63 |
| Jinhua-2 | 90102440 | 81.50 | 72.83 |
| Yorkshire-1 | 84160098 | 83.63 | 72.62 |
| Yorkshire-2 | 87285462 | 84.37 | 73.80 |
3.3. Known mRNA Profiling in Pig Thyroid
There are 38,222 known mRNAs so far annotated in the pig genome, according to the RefSeq database. 22,435 mRNAs were expressed in the pig thyroid, which represents ~59% of the total annotated pig mRNAs (Table S2). The majority of these identified mRNAs (18,464) were expressed in both Jinhua and Yorkshire pigs, while 1,492 were Yorkshire-specific and 2,479 were Jinhua-specific. Most of the breed-specific mRNAs were expressed at low FPKM, suggesting these transcripts were generally expressed at low levels in the thyroid gland, being hard to discover. Tg mRNA exhibited the highest abundance in both pig breeds, accounting for 13.37% and 16.12% of all thyroid transcripts expression in Yorkshire and Jinhua pigs, respectively, which was consistent with previous studies [36]. TH-production-related transcripts (i.e., HSPA5, IYD, DUOX2, SLC5A5, LRP2, and TSHR) were also highly expressed in the pig thyroid, reaching more than 1000 FPKM. KEGG analysis showed that the transcripts whose expression levels were more than 1000 FPKM were enriched in the ribosome, focal adhesion, proteasome, fatty acid metabolism, spliceosome, and regulation of actin cytoskeleton pathways, suggesting that these mRNAs might be essential for maintaining the basic functions of the thyroid gland.
3.4. Identification of lncRNA
A series of filtering criteria was applied to define a set of putative lncRNAs in the porcine thyroid transcriptome (Figure 1). After assembling, 265,605 transcript isoforms were assembled by Cufflinks and Scripture. Putative lncRNAs were distinguished from the numerous lowly-expressed, single-exon, unreliable fragments, employing a read coverage threshold [12] and focusing only on multiexonic transcripts [21] with at least 200 bp. These procedures filtered 36,691 transcripts. Finally, annotated non-lncRNA transcripts (e.g., annotated protein-coding genes, pre-microRNA, tRNAs, rRNA, and pseudogenes) and transcripts with coding potential were eliminated, and a rigorous lncRNA set including 1018 transcripts was obtained (Table S3). Among these, 659 (64.7%) candidate lncRNA transcripts were included in the pig ALDB database (Table S4).
Figure 1.
Schematic diagram of the pipeline used in the identification of long non-coding RNAs (lncRNAs) and novel mRNAs. This pipeline took the reads that mapped to the pig reference genome as input data (top). The four criteria required for the identification of lncRNAs are indicated in red. LncRNAs are defined as reliably expressed, long, multi-exonic, and noncoding transcripts. Transcripts that predicted with positive coding potential by any of the four tools (CNCI, CPC, Pfam-scan and PhyloCSF) were considered as novel mRNAs.
Blasting candidate lncRNAs to human and mouse lncRNA revealed 473 and 237 lncRNAs showed significant sequence similarity with human and mouse lncRNA, respectively (Table S5). Furthermore, 198 candidate lncRNAs showed sequence similarity with known lncRNAs in both human and mouse.
3.5. Identification of Novel mRNAs
Transcripts that predicted with coding potential by any of the four tools (CNCI, CPC, Pfam-scan, and PhyloCSF) were considered as novel mRNAs (Figure 1). We detected 1189 novel mRNAs not linked to any annotated genes in the NCBI database (Table S6).
3.6. Differential Expression Analysis
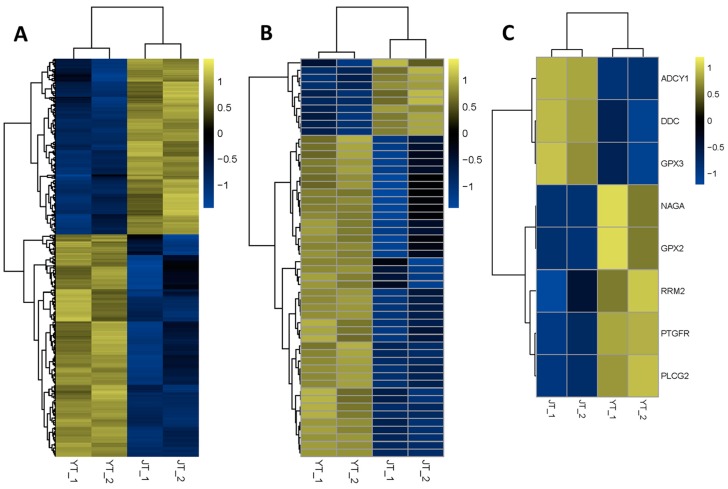
Differentially-expressed mRNAs and lncRNAs in the thyroid glands of Yorkshire and Jinhua pigs were obtained by Cuffdiff. In Yorkshire pigs, 246 known mRNAs and 29 novel mRNAs were upregulated, whereas 209 known mRNAs and eight novel mRNAs were downregulated (q-value < 0.05) (Table S7). From the lncRNA expression data, 42 lncRNA transcripts were upregulated and 10 lncRNAs were down-regulated (q-value < 0.05) in the Yorkshire pigs (Table S8). The heat maps displayed mRNA (Figure 2A) and lncRNA (Figure 2B) significantly differentially-expressed between the two pig breeds.
Figure 2.
Heat maps of the distinguishable expression profiles in the thyroid gland of Jinhua and Yorkshire pig breeds. (A) Hierarchical clustering of the differentially-expressed mRNAs; (B) Differentially-expressed lncRNA transcripts; and (C) TH-regulation-related genes is performed. Yellow indicates relatively high expression and blue denotes relatively low expression.
Due to the importance of THs in pig growth and lipid metabolism, we focused on TH-regulation-related genes identified in previous studies (Figure 2C) [7]. The expression levels of transcripts in the calcium signaling pathway (PTGFR, PLCG2), lysosome (NAGA), tyrosine metabolism (DDC), and glutathione metabolism (RRM2) differed between Yorkshire pigs and Jinhua pigs. In addition, the expression of the TH-synthesis-related genes ADCY1, GPX2, and GPX3 differed significantly between the two breeds.
3.7. Functional Enrichment of Differentially-Expressed mRNAs
The GO enrichment analysis performed to explore the functions of the differentially-expressed mRNAs between two pig breeds revealed that only the microtubule-based process (ontology: biological process) was significantly enriched (p-adjust < 0.05). In addition, the majority of mRNAs (10/14) assigned to this GO term were significantly upregulated in Yorkshire pigs (Table S9).
The KEGG pathway analysis performed to uncover biological functions of these genes indicated that the only significantly enriched network was “Cell cycle” (p-adjust < 0.05). Proliferating cell nuclear antigen (PCNA) was assigned to this pathway, and had a much higher expression level in Yorkshire pigs than Jinhua.
3.8. Functional Prediction of lncRNAs
To investigate the function of lncRNAs, we predicted the potential targets of lncRNAs in cis, using 100 kb as the cutoff. Thirteen differently expressed protein-coding genes were found close to 10 differentially-expressed lncRNA genes (Table S10). Seven of these 10 lncRNA genes were predicted to regulate only one target gene, while three were predicted to regulate two target genes. In summary, nine of the lncRNA-mRNA gene pairs were regulated in the same direction (up-up) and four pairs in the opposite direction (down-up). The expression levels of two target genes (RAB27B and Novel000261) for XLOC_250893, which were regulated in the same direction (Figure 3A), and of two targets (IFIT1 and IFIT3) for XLOC_1411136, which were regulated in the opposite direction (Figure 3B), were validated by qRT-PCR.
Figure 3.
Validation of the RNA-seq data by qRT-PCR. (A) The expression patterns of XLOC_250893 and its target genes (RAB27B and Novel000261); and (B) for XLOC_1411136 and its target genes (IFIT1 and IFIT3) were verified by qRT-PCR; (C) The relative expression levels of six randomly selected genes were expressed as the mean ± SEM. * p < 0.05, ** p < 0.01; (D) Fold-change of the Yorkshire pigs versus the Jinhua pigs were verified by qPCR.
3.9. Quantitative Real-Time PCR Validation
To validate RNA-seq results, three differentially-expressed mRNAs (SCPEP1, CALCB, and PDK4) and three differentially-expressed lncRNAs (XLOC_3395075, XLOC_3395082, and XLOC_3395055) were subjected to qRT-PCR, with three biological replicates for each breed (Figure 3C). As shown in Figure 3D, the relative fold changes in expression detected by qRT-PCR were consistent with RNA-seq data, suggesting our transcripts identification and abundance estimation were highly reliable.
3.10. Genomic Features of lncRNAs
The candidate lncRNAs identified in this study were shorter in transcript length and contained fewer exons than mRNAs (Figure 4), which was consistent with previous studies [22,37].
Figure 4.
Genomic features of candidate lncRNAs. (A) Length distribution of the 38,222 coding transcripts (white) and 1,018 predicted lncRNAs (black); (B) Exon number distribution of coding transcripts and lncRNAs.
4. Discussion
Although animal growth and fat deposition are complex processes regulated by many factors, the thyroid gland is one of the most important. It synthesizes, stores, and secretes THs, which are controlled by thousands of molecules, but until now, little is known about the porcine thyroid transcriptome. In this study, we conducted a preliminary investigation of mRNA and lncRNA expression profiles in the thyroid gland of Yorkshire and Jinhua pigs to assess the potential regulators of porcine growth and fat deposition related to thyroid functions.
Dramatic differences in growth performance and fat deposition were observed between Yorkshire and Jinhua pigs. Yorkshire pigs had significantly higher levels of TT3 and TT4 than Jinhua pigs. Serum levels of FT4 and FT3 in the Yorkshire pig also tended to be higher than in the Jinhua pig. These results are in agreement with those of a previous study reporting that serum FT3 and FT4 levels in Jinhua pigs were significantly lower than those in Landrace pigs, which is another lean-type pig breed [38].
To our knowledge, this is the first application of Ribo-zero RNA sequencing to study the porcine thyroid transcriptome. This method allows non-polyA transcripts to be obtained, as well as fragmented RNAs, which is advantageous compared to polyA sequencing [39]. Most of the 351 million reads obtained (83%) were mapped to the current S. scrofa genome assembly, which was higher than the percentage obtained for testis (77.4%–78.37%) [40] and muscle (79.31%–82.21%) transcriptomes [41]. The extensive sequencing depth yielded high sequence read coverage on the pig reference genome. With the higher sequencing depth and higher mapping ratio, we obtained more transcripts. However, a large proportion of clean reads (33%–39%) were mapped to unannotated loci, reinforcing the need to improve current pig genome annotation.
A total of 22,435 known and 1189 novel mRNAs were expressed in this study, and almost all novel mRNAs had low expression, which suggested that their roles in the thyroid need to be further confirmed. The 492 differentially-expressed mRNAs between Jinhua and Yorkshire pigs might explain the different levels of THs found between the two pig breeds, as many important differentially-expressed genes participate in TH synthesis and secretion. It is well established that glutathione metabolism, lysosome, tyrosine metabolism, and calcium signaling pathway are involved in the synthesis and secretion of THs [7], and any change in these pathways might affect the levels of serum THs. The present study results evidenced differentially-expressed genes in all these pathways. More importantly, the majority of the differentially-expressed genes assigned to these pathways were significantly upregulated in Yorkshire pigs, which might improve the effect of the upstream regulator on TH production. Notably, our study also showed that the expression of TH-synthesis-related genes was significantly different between Yorkshire and Jinhua pigs. For example, adenylate cyclase 1 (ADCY1) is activated by TSHR and involved in the process of adopting and organifying iodide [42]. Glutathione peroxidase 2 (GPX2) and Glutathione peroxidase 3 (GPX3)—which are induced by TSH in vitro [43]—participate in the degradation of thyroglobulin and in the liberation of T4 and T3 [44,45]. Anoctamin 1 (ANO1) is a calcium-activated anion channel with a preferential affinity for iodide over chloride, and may contribute to the rapid delivery of iodide into the follicular lumen for the synthesis of THs [46,47], and improve the TH-secretion response to extracellular stimulus (such as TSH, iodine). Secretogranin III (SCG3) was also reported as upregulated in thyroid regeneration [48]. In addition, a group of differentially-expressed genes were reported to be related to thyroid diseases, such as caspase 8 (CASP8) [49], dual specificity phosphatase 4 (DUSP4) [50], BTG family, member 2 (BTG2) [51], potassium channel, voltage gated Shal related subfamily D, member 3 (KCND3) [52], interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) [53], chloride channel, voltage-sensitive 5 (CLC-5) [54] and connective tissue growth factor (CCN2) [55]. In this study, 14 genes assigned to the GO term "microtubule-based process" were differentially regulated between the two pig breeds, and previous studies suggested a role for microtubules in thyroid secretion through endocytosis [56,57]. The currently accepted mechanism for TH secretion is that TG molecules are first taken up by polarized thyrocytes through vesicle-mediated endocytosis, and then transported to the lysosome for proteolytic cleavage, releasing T4 and T3. Thus, the first step is the limiting point in the TH secretory pathway. Taken together, differential expression of these genes may lead to a difference in thyroid function and serum THs concentration, which may partly explain the different growth rate and fat deposition between Yorkshire and Jinhua pigs, although their exact mechanism needs further investigation.
Given their low expression level and few annotated functions, lncRNAs were thought to be transcriptional noise. With the development of high-throughput sequencing technologies for large-scale expression profile analysis, the discovery and characterization of lncRNAs became feasible [58,59]. A total of 1018 candidate lncRNA transcripts were identified in this study. This number is much lower than the pig lncRNAs in the ALDB database (12,103) [60]. One possible reason for this difference is that lncRNAs are expressed in a tissue-specific pattern [61], and many of the lncRNAs in the ALDB are exclusively expressed in other tissues. This also partly explains why only 64.7% of the candidate lncRNAs identified in this study were included in the ALDB database. Similar to the lncRNAs in other species [31,62], the candidate lncRNAs identified in pig thyroid are shorter, have fewer exons, and lower expression levels than mRNAs.
Analyzing conservation of lncRNAs across species might help understand lncRNA evolution and function [63]. The comparative analysis found that 473 (46.5%) and 237 (23.3%) candidate lncRNAs showed significant sequence similarities with human and mouse lncRNAs, respectively. Furthermore, 198 (19.4%) lncRNAs showed significant sequence similarities with both human and mouse lncRNAs. These observations are consistent with the results of Zhou et al., who reported a 40% similarity between pig and human long intergenic non-coding RNAs [64], suggesting that pig might be a good model to analyze lncRNAs function. Compared to the more than 84% sequence similarity between pig and human mRNAs [65], the homology between pig and human lncRNAs is small, supporting the hypothesis that lncRNA evolved more rapidly than mRNA [31,63].
Fifty-two lncRNA transcripts were significantly differentially expressed in the thyroid gland of Yorkshire and Jinhua pigs, and they might play critical roles in porcine thyroid function. The roles of some lncRNAs in the development and function of some endocrine organs (pancreatic β cells, white and brown adipose tissue, adrenal gland, and mammary gland) have been reported in previous studies [13]. The function of lncRNAs was not inferred from their sequences and structures. In addition, recent studies have demonstrated that some lncRNAs can regulate the expression levels of their chromosomal neighboring genes [66], and this has been used to predict the function of lncRNAs [12,62,67]. In the present study, we searched for potential target genes 100 kb up- and downstream of lncRNAs. To minimize the false positive rates to predict the target genes of lncRNAs, only differentially-expressed lncRNAs and mRNAs were used in this analysis, which found 13 potential lncRNA-mRNA gene pairs. These were either co- or inversely-regulated, in agreement with what has been reported in previous studies [68,69]. Co-regulation of lncRNA-mRNA pairs suggested they might be under the same transcriptional control or that lncRNAs activated the expression of their target genes. Ørom et al. reported some lncRNAs display an enhancer-like function on their neighboring mRNAs [70]. Inverse expression patterns between lncRNAs and their neighboring protein-coding genes indicate that lncRNAs might silence or inhibit the expression of their target genes. For example, the Xist lncRNA is expressed from one of the two X chromosomes in female mammals, and induces silencing of the whole chromosome [71]. We also found that some lncRNAs might exert their function in the pig thyroid through their potential target mRNAs. For example, lncRNA XLOC_250893 was predicted to act on Rab27b, a member of the Rab family of small GTPase proteins. Rab GTPases regulate almost all membrane trafficking processes, including vesicle formation, movement, and fusion [72]. Rab27b has been reported to be widely expressed in secretory organs such as the pituitary gland, pancreas, and thyroid gland [73], and is up-regulated in papillary thyroid carcinoma [74]. LncRNA XLOC_1411136 was predicted to act on the potential garget gene IFIT1. Kuang et al. reported that IFIT1 might play an important role in the pathology of Graves’ disease [53], an autoimmune disorder of the thyroid gland characterized by hyperthyroidism. These findings suggest that lncRNAs might play regulatory roles in pig thyroid function and health. However, these predicted lncRNA-mRNA pairs require further experimental verification.
5. Conclusions
This study represented the first description of mRNA and lncRNA profiles in the porcine thyroid employing Ribo-Zero RNA-seq technology. In addition, the 1189 novel mRNAs and 1018 candidate lncRNA transcripts detected, 22,435 known mRNAs were expressed in the porcine thyroid. Among the 455 known mRNAs, 37 novel mRNAs, and 52 lncRNA transcripts differentially-expressed in Yorkshire and Jinhua pigs, several were known to regulate the synthesis and secretion of THs, according to previous studies. Bioinformatics analysis suggested that differentially-expressed lncRNAs might also play a role in regulating the synthesis and secretion of THs by governing their potential target genes. Considering the known important role of THs in metabolism, the mRNAs and lncRNAs revealed in this study would contribute to the understanding of the possible mechanisms involved in animal growth and fat deposition, although more detailed studies are required.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No.31272424) and a grant from Transgenic Organisms Breeding Major Projects (No. 2014ZX08006-003).
Abbreviations
The following abbreviations are used in this manuscript:
| T3 | Triiodothyronine |
| T4 | Tetraiodothyronine or Thyroxine |
| THs | Thyroid Hormones |
| PCR | Polymerase Chain Reaction |
| FPKM | Fragments per kilo-base of exon per million mapped fragments |
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/7/7/34/s1, Table S1: Categorization of reads annotation, Table S2: Known mRNAs identified in porcine thyroid, Table S3: Candidate lncRNA identified in porcine thyroid, Table S4: Candidate lncRNAs included in pig ALDB database, Table S5: Candidate lncRNAs with sequence similarities with human or mouse, Table S6: Novel mRNA identified in porcine thyroid, Table S7: Differentially-expressed mRNAs, Table S8: Differentially-expressed lncRNAs, Table S9: Genes involved in microtubule-based process, Table S10: LncRNA-mRNA pairs, Table S11: Primers for qRT-PCR.
Author Contributions
Yifei Shen performed the main experiment and wrote the paper; Zhaowei Cai and Ying Wang assisted with the experimental design; Haiguang Mao, Lixing Chen and Minjie Huang were involved in executing the study; Jiucheng Chen was involved in data analysis; Ningying Xu guided the execution of the study and revised the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Yen P.M. Physiological and molecular basis of thyroid hormone action. Physiol. Rev. 2001;81:1097–1142. doi: 10.1152/physrev.2001.81.3.1097. [DOI] [PubMed] [Google Scholar]
- 2.Zhu X.G., Cheng S.Y. New insights into regulation of lipid metabolism by thyroid hormone. Curr. Opin. Endocrinol. Diabetes Obes. 2010;17:408–413. doi: 10.1097/MED.0b013e32833d6d46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullur R., Liu Y.-Y., Brent G.A. Thyroid hormone regulation of metabolism. Physiol. Rev. 2014;94:355–382. doi: 10.1152/physrev.00030.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiamolera M.I., Wondisford F.E. Thyrotropin-releasing hormone and the thyroid hormone feedback mechanism. Endocrinology. 2009;150:1091–1096. doi: 10.1210/en.2008-1795. [DOI] [PubMed] [Google Scholar]
- 5.Schomburg L., Köhrle J. On the importance of selenium and iodine metabolism for thyroid hormone biosynthesis and human health. Mol. Nutr. Food Res. 2008;52:1235–1246. doi: 10.1002/mnfr.200700465. [DOI] [PubMed] [Google Scholar]
- 6.Ortiga-Carvalho T., Oliveira K., Soares B., Pazos-Moura C. The role of leptin in the regulation of TSH secretion in the fed state: In vivo and in vitro studies. J. Endocrinol. 2002;174:121–125. doi: 10.1677/joe.0.1740121. [DOI] [PubMed] [Google Scholar]
- 7.Mansourian A. Metabolic pathways of tetraidothyronine and triidothyronine production by thyroid gland: A review of articles. Pak. J. Biol. Sci. 2011 doi: 10.3923/pjbs.2011.1.12. [DOI] [PubMed] [Google Scholar]
- 8.Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallory A.C., Shkumatava A. LncRNAs in vertebrates: Advances and challenges. Biochimie. 2015;117:3–14. doi: 10.1016/j.biochi.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kretz M., Siprashvili Z., Chu C., Webster D.E., Zehnder A., Qu K., Lee C.S., Flockhart R.J., Groff A.F., Chow J. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponjavic J., Ponting C.P., Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoll M., Lodish H.F., Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat. Rev. Endocrinol. 2015;11:151–160. doi: 10.1038/nrendo.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du Z., Fei T., Verhaak R.G., Su Z., Zhang Y., Brown M., Chen Y., Liu X.S. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fatica A., Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 16.Adiconis X., Borges-Rivera D., Satija R., DeLuca D.S., Busby M.A., Berlin A.M., Sivachenko A., Thompson D.A., Wysoker A., Fennell T. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat. Methods. 2013;10:623–629. doi: 10.1038/nmeth.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fastqc. A quality control tool for high throughput sequence data. [(accessed on 8 July 2016)]. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 18.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013 doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B., Salzberg S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S., Pyl P.T., Huber W. HTSeq—A python framework to work with high-throughput sequencing data. Bioinformatics. 2014 doi: 10.1101/002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M., Garber M., Levin J.Z., Donaghey J., Robinson J., Adiconis X., Fan L., Koziol M.J., Gnirke A., Nusbaum C. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., Liu Y., Chen R., Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong L., Zhang Y., Ye Z.-Q., Liu X.-Q., Zhao S.-Q., Wei L., Gao G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J. The Pfam protein families database. Nucleic Acids Res. 2011;28:263–266. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin M.F., Jungreis I., Kellis M. Phylocsf: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y., Li H., Fang S., Kang Y., Hao Y., Li Z., Bu D., Sun N., Zhang M.Q., Chen R. Noncode 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2015;44:D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M. Refseq: An update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quek X.C., Thomson D.W., Maag J.L., Bartonicek N., Signal B., Clark M.B., Gloss B.S., Dinger M.E. LncRNAdb v2. 0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2014;43:D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. The gencode v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Method gene ontology analysis for RNA-Seq: Accounting for selection bias. Genome Biol. 2010 doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao X., Cai T., Olyarchuk J.G., Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 34.Guil S., Esteller M. Cis-acting noncoding RNAs: Friends and foes. Nat. Struct. Mol. Biol. 2012;19:1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 35.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-δδct method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Vilain C., Libert F., Venet D., Costagliola S., Vassart G. Small amplified RNA-sage: An alternative approach to study transcriptome from limiting amount of mRNA. Nucleic Acids Res. 2003;31:e24–e24. doi: 10.1093/nar/gng024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alvarez-Dominguez J.R., Bai Z., Xu D., Yuan B., Lo K.A., Yoon M.J., Lim Y.C., Knoll M., Slavov N., Chen S. De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab. 2015;21:764–776. doi: 10.1016/j.cmet.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao Z.-G., Wang L.-J., Xu Z.-R., Huang J.-F., Wang Y.-R. Developmental patterns in hormone and lipid metabolism of growing jinhua and landrace gilts. Can. J. Anim. Sci. 2008;88:601–607. doi: 10.4141/CJAS08037. [DOI] [Google Scholar]
- 39.Zhao W., He X., Hoadley K.A., Parker J.S., Hayes D.N., Perou C.M. Comparison of RNA-Seq by poly (a) capture, ribosomal RNA depletion, and DNA microarray for expression profiling. BMC Genomics. 2014 doi: 10.1186/1471-2164-15-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ran M., Chen B., Wu M., Liu X., He C., Yang A., Li Z., Xiang Y., Li Z., Zhang S. Integrated analysis of miRNA and mRNA expression profiles in development of porcine testes. RSC Adv. 2015;5:63439–63449. doi: 10.1039/C5RA07488F. [DOI] [Google Scholar]
- 41.Wang Z., Li Q., Chamba Y., Zhang B., Shang P., Zhang H., Wu C. Identification of genes related to growth and lipid deposition from transcriptome profiles of pig muscle tissue. PLoS ONE. 2015;10:34. doi: 10.1371/journal.pone.0141138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marians R., Ng L., Blair H., Unger P., Graves P., Davies T. Defining thyrotropin-dependent and-independent steps of thyroid hormone synthesis by using thyrotropin receptor-null mice. Proc. Natl. Acad. Sci. USA. 2002;99:15776–15781. doi: 10.1073/pnas.242322099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweizer U., Chiu J., Köhrle J. Peroxides and peroxide-degrading enzymes in the thyroid. Antioxid. Redox Signal. 2008;10:1577–1592. doi: 10.1089/ars.2008.2054. [DOI] [PubMed] [Google Scholar]
- 44.Drutel A., Archambeaud F., Caron P. Selenium and the thyroid gland: More good news for clinicians. Clin. Endocrinol. 2013;78:155–164. doi: 10.1111/cen.12066. [DOI] [PubMed] [Google Scholar]
- 45.Howie A.F., Walker S.W., Akesson B., Arthur J.R., Beckett G.J. Thyroidal extracellular glutathione peroxidase: A potential regulator of thyroid-hormone synthesis. Biochem. J. 1995;308:713–717. doi: 10.1042/bj3080713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iosco C., Cosentino C., Sirna L., Romano R., Cursano S., Mongia A., Pompeo G., di Bernardo J., Ceccarelli C., Tallini G. Anoctamin 1 is apically expressed on thyroid follicular cells and contributes to ATP-and calcium-activated iodide efflux. Cell. Physiol. Biochem. 2014;34:966–980. doi: 10.1159/000366313. [DOI] [PubMed] [Google Scholar]
- 47.Viitanen T.M., Sukumaran P., Löf C., Törnquist K. Functional coupling of TRPC2 cation channels and the calcium-activated anion channels in rat thyroid cells: Implications for iodide homeostasis. J. Cell Physiol. 2013;228:814–823. doi: 10.1002/jcp.24230. [DOI] [PubMed] [Google Scholar]
- 48.Ozaki T., Matsubara T., Seo D., Okamoto M., Nagashima K., Sasaki Y., Hayase S., Murata T., Liao X.-H., Hanson J. Thyroid regeneration: Characterization of clear cells after partial thyroidectomy. Endocrinology. 2012;153:2514–2525. doi: 10.1210/en.2011-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stephen J.K., Chitale D., Narra V., Chen K.M., Sawhney R., Worsham M.J. DNA methylation in thyroid tumorigenesis. Cancers. 2011;3:1732–1743. doi: 10.3390/cancers3021732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee E.K., Chung K.-W., Yang S.K., Park M.J., Min H.S., Kim S.W., Kang H.S. DNA methylation of MAPK signal-inhibiting genes in papillary thyroid carcinoma. Anticancer Res. 2013;33:4833–4839. [PubMed] [Google Scholar]
- 51.Planck T., Shahida B., Sjögren M., Groop L., Hallengren B., Lantz M. Association of BTG2, CYR61, ZFP36, and SCD gene polymorphisms with Graves’ Disease and ophthalmopathy. Thyroid. 2014;24:1156–1161. doi: 10.1089/thy.2013.0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ueno A., Yamamoto T., Sato N., Tanaka K. Ventricular fibrillation associated with early repolarization in a patient with thyroid storm. J. Interv. Card. Electrophysiol. 2010;29:93–96. doi: 10.1007/s10840-010-9507-3. [DOI] [PubMed] [Google Scholar]
- 53.Kuang M., Wang S., Wu M., Ning G., Yao Z., Li L. Expression of IFNα-inducible genes and modulation of HLA-DR and thyroid stimulating hormone receptors in Graves’ disease. Mol. Cell. Endocrinol. 2010;319:23–29. doi: 10.1016/j.mce.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 54.Van den Hove M.-F., Croizet-Berger K., Jouret F., Guggino S.E., Guggino W.B., Devuyst O., Courtoy P.J. The loss of the chloride channel, CLC-5, delays apical iodide efflux and induces a euthyroid goiter in the mouse thyroid gland. Endocrinology. 2006;147:1287–1296. doi: 10.1210/en.2005-1149. [DOI] [PubMed] [Google Scholar]
- 55.Cui L., Zhang Q., Mao Z., Chen J., Wang X., Qu J., Zhang J., Jin D. CTGF is overexpressed in papillary thyroid carcinoma and promotes the growth of papillary thyroid cancer cells. Tumor Biol. 2011;32:721–728. doi: 10.1007/s13277-011-0173-6. [DOI] [PubMed] [Google Scholar]
- 56.Neve P., Willems C., Dumont J.E. Involvement of the microtubule-microfilament system in thyroid secretion. Exp. Cell Res. 1970;63:457–460. doi: 10.1016/0014-4827(70)90238-7. [DOI] [PubMed] [Google Scholar]
- 57.Apodaca G. Endocytic traffic in polarized epithelial cells: Role of the actin and microtubule cytoskeleton. Traffic. 2001;2:149–159. doi: 10.1034/j.1600-0854.2001.020301.x. [DOI] [PubMed] [Google Scholar]
- 58.Weikard R., Hadlich F., Kuehn C. Identification of novel transcripts and noncoding RNAs in bovine skin by deep next generation sequencing. BMC Genomics. 2013 doi: 10.1186/1471-2164-14-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., Xue S., Liu X., Liu H., Hu T., Qiu X., Zhang J., Lei M. Analyses of long non-coding RNA and mRNA profiling using RNA sequencing during the pre-implantation phases in pig endometrium. Sci. Rep. UK. 2016 doi: 10.1038/srep20238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li A., Zhang J., Zhou Z., Wang L., Liu Y., Liu Y. ALDB: A domestic-animal long noncoding RNA database. PLoS ONE. 2015;10:34. doi: 10.1371/journal.pone.0124003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Washietl S., Kellis M., Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014;24:616–628. doi: 10.1101/gr.165035.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pauli A., Valen E., Lin M.F., Garber M., Vastenhouw N.L., Levin J.Z., Fan L., Sandelin A., Rinn J.L., Regev A. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ulitsky I., Bartel D.P. LincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou Z.-Y., Li A.-M., Adeola A.C., Liu Y.-H., Irwin D.M., Xie H.-B., Zhang Y.-P. Genome-wide identification of long intergenic noncoding RNA genes and their potential association with domestication in pigs. Genome Biol. Evol. 2014;6:1387–1392. doi: 10.1093/gbe/evu113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fang X., Mou Y., Huang Z., Li Y., Han L., Zhang Y., Feng Y., Chen Y., Jiang X., Zhao W. The sequence and analysis of a Chinese pig genome. GigaScience. 2012 doi: 10.1186/2047-217X-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pennisi E. Long noncoding RNAs may alter chromosome’s 3D structure. Science. 2013;340:910–910. doi: 10.1126/science.340.6135.910-a. [DOI] [PubMed] [Google Scholar]
- 67.Ran M., Chen B., Li Z., Wu M., Liu X., He C., Zhang S., Li Z. Systematic identification of long non-coding RNAs in immature and mature porcine testes. Biol. Reprod. Biolreprod. 2016 doi: 10.1095/biolreprod.115.136911. [DOI] [PubMed] [Google Scholar]
- 68.Bao J., Wu J., Schuster A.S., Hennig G.W., Yan W. Expression profiling reveals developmentally regulated lncRNA repertoire in the mouse male germline. Biol. Reprod. 2013 doi: 10.1095/biolreprod.113.113308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han L., Zhang K., Shi Z., Zhang J., Zhu J., Zhu S., Zhang A., Jia Z., Wang G., Yu S. LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int. J. Oncol. 2012;40:2004–2012. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- 70.Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wutz A. Gene silencing in x-chromosome inactivation: Advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 72.Stenmark H. Rab gtpases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 73.Gomi H., Mori K., Itohara S., Izumi T. Rab27b is expressed in a wide range of exocytic cells and involved in the delivery of secretory granules near the plasma membrane. Mol. Biol. Cell. 2007;18:4377–4386. doi: 10.1091/mbc.E07-05-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nikolova D.N., Zembutsu H., Sechanov T., Vidinov K., Kee L.S., Ivanova R., Becheva E., Kocova M., Toncheva D., Nakamura Y. Genome-wide gene expression profiles of thyroid carcinoma: Identification of molecular targets for treatment of thyroid carcinoma. Oncol. Rep. 2008;20:105–121. doi: 10.3892/or.20.1.105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.