Abstract
Matrix-attachment regions (MARs) may function as domain boundaries and partition chromosomes into independently regulated units. We have tested whether MAR sequences from the chicken lysozyme locus, the so-called A-elements, can confer position-independent regulation to a whey acidic protein (WAP) transgene in mammary tissue of mice. In the absence of MARs, expression of WAP transgenes was observed in 50% of the lines, and regulation during pregnancy, during lactation, and upon hormonal induction did not mimic that of the endogenous WAP gene and varied with the integration site. In contrast, all 11 lines in which WAP transgenes were juxtaposed to MAR elements showed expression. Accurate position-independent hormonal and developmental regulation was seen in four out of the five lines analyzed. These results indicate that MARs can establish independent genetic domains in transgenic mice.
Full text
PDF

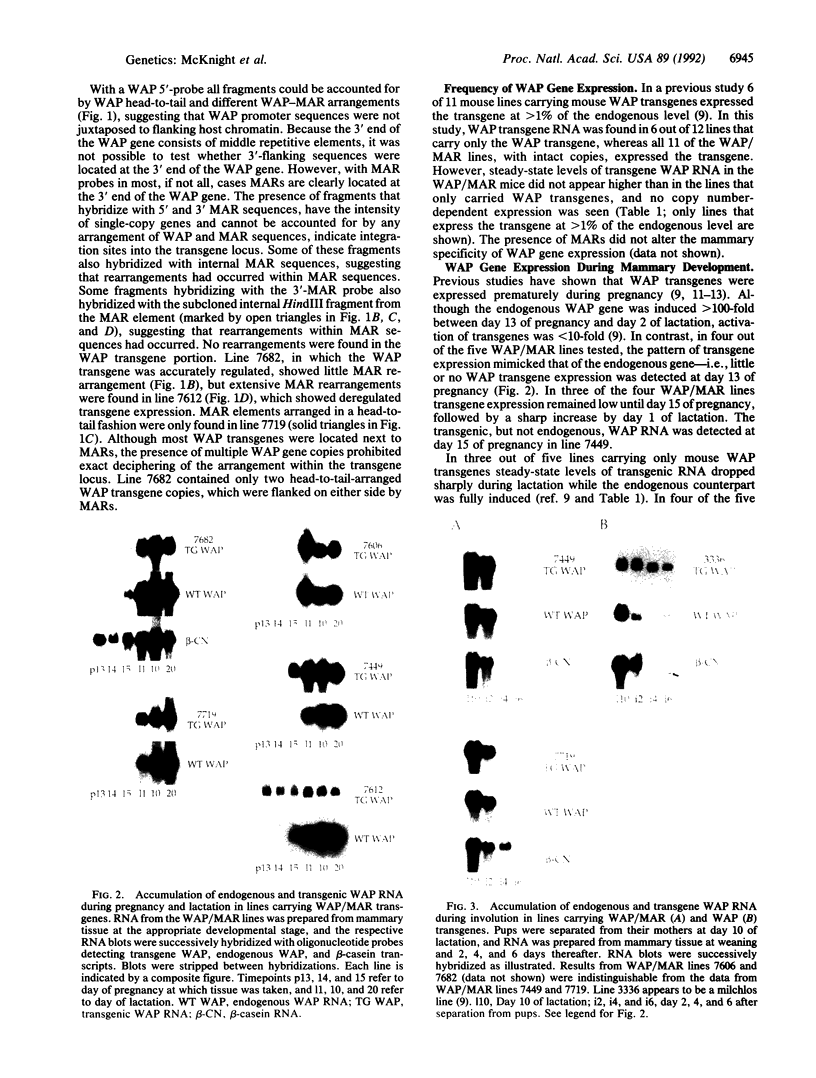


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayna E. M., Rosen J. M. Tissue-specific, high level expression of the rat whey acidic protein gene in transgenic mice. Nucleic Acids Res. 1990 May 25;18(10):2977–2985. doi: 10.1093/nar/18.10.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifer C., Vidal M., Grosveld F., Sippel A. E. Tissue specific and position independent expression of the complete gene domain for chicken lysozyme in transgenic mice. EMBO J. 1990 Sep;9(9):2843–2848. doi: 10.1002/j.1460-2075.1990.tb07473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T., Sankaran L., Wall R. J., Spencer M., Hennighausen L. Expression of a whey acidic protein transgene during mammary development. Evidence for different mechanisms of regulation during pregnancy and lactation. J Biol Chem. 1991 Apr 15;266(11):6909–6914. [PubMed] [Google Scholar]
- Burdon T., Wall R. J., Shamay A., Smith G. H., Hennighausen L. Over-expression of an endogenous milk protein gene in transgenic mice is associated with impaired mammary alveolar development and a milchlos phenotype. Mech Dev. 1991 Dec;36(1-2):67–74. doi: 10.1016/0925-4773(91)90073-f. [DOI] [PubMed] [Google Scholar]
- Campbell S. M., Rosen J. M., Hennighausen L. G., Strech-Jurk U., Sippel A. E. Comparison of the whey acidic protein genes of the rat and mouse. Nucleic Acids Res. 1984 Nov 26;12(22):8685–8697. doi: 10.1093/nar/12.22.8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Dale T. C., Krnacik M. J., Schmidhauser C., Yang C. L., Bissell M. J., Rosen J. M. High-level expression of the rat whey acidic protein gene is mediated by elements in the promoter and 3' untranslated region. Mol Cell Biol. 1992 Mar;12(3):905–914. doi: 10.1128/mcb.12.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Furth P. A., Hennighausen L., Baker C., Beatty B., Woychick R. The variability in activity of the universally expressed human cytomegalovirus immediate early gene 1 enhancer/promoter in transgenic mice. Nucleic Acids Res. 1991 Nov 25;19(22):6205–6208. doi: 10.1093/nar/19.22.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Grosveld F., van Assendelft G. B., Greaves D. R., Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987 Dec 24;51(6):975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Gyurkovics H., Gausz J., Kummer J., Karch F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 1990 Aug;9(8):2579–2585. doi: 10.1002/j.1460-2075.1990.tb07439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günzburg W. H., Salmons B., Zimmermann B., Müller M., Erfle V., Brem G. A mammary-specific promoter directs expression of growth hormone not only to the mammary gland, but also to Bergman glia cells in transgenic mice. Mol Endocrinol. 1991 Jan;5(1):123–133. doi: 10.1210/mend-5-1-123. [DOI] [PubMed] [Google Scholar]
- Kellum R., Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991 Mar 8;64(5):941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- Loc P. V., Strätling W. H. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988 Mar;7(3):655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phi-Van L., von Kries J. P., Ostertag W., Strätling W. H. The chicken lysozyme 5' matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol Cell Biol. 1990 May;10(5):2302–2307. doi: 10.1128/mcb.10.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piletz J. E., Heinlen M., Ganschow R. E. Biochemical characterization of a novel whey protein from murine milk. J Biol Chem. 1981 Nov 25;256(22):11509–11516. [PubMed] [Google Scholar]
- Pittius C. W., Sankaran L., Topper Y. J., Hennighausen L. Comparison of the regulation of the whey acidic protein gene with that of a hybrid gene containing the whey acidic protein gene promoter in transgenic mice. Mol Endocrinol. 1988 Nov;2(11):1027–1032. doi: 10.1210/mend-2-11-1027. [DOI] [PubMed] [Google Scholar]
- Pursel V. G., Bolt D. J., Miller K. F., Pinkert C. A., Hammer R. E., Palmiter R. D., Brinster R. L. Expression and performance in transgenic pigs. J Reprod Fertil Suppl. 1990;40:235–245. [PubMed] [Google Scholar]
- Rexroad C. E., Jr, Hammer R. E., Behringer R. R., Palmiter R. D., Brinster R. L. Insertion, expression and physiology of growth-regulating genes in ruminants. J Reprod Fertil Suppl. 1990;41:119–124. [PubMed] [Google Scholar]
- Stief A., Winter D. M., Strätling W. H., Sippel A. E. A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature. 1989 Sep 28;341(6240):343–345. doi: 10.1038/341343a0. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Maine E., Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985 Sep 20;185(2):341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- Webb C. F., Das C., Eneff K. L., Tucker P. W. Identification of a matrix-associated region 5' of an immunoglobulin heavy chain variable region gene. Mol Cell Biol. 1991 Oct;11(10):5206–5211. doi: 10.1128/mcb.11.10.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kries J. P., Buhrmester H., Strätling W. H. A matrix/scaffold attachment region binding protein: identification, purification, and mode of binding. Cell. 1991 Jan 11;64(1):123–135. doi: 10.1016/0092-8674(91)90214-j. [DOI] [PubMed] [Google Scholar]