Abstract
Background: S100 calcium-binding protein A9 (S100A9) has previously been identified as a type 2 diabetes (T2D) gene. However, this finding requires independent validation and more in-depth analyses in other populations and ancestries.
Objectives: We aimed to replicate the associations between an S100A9 variant and insulin resistance and T2D and to initiate an investigation of potential interactions with the habitual diet in several independent populations.
Design: We investigated the association of the S100A9 variant rs3014866 with insulin resistance and T2D risk and its interactions with diet in 3 diverse populations as follows: the CORDIOPREV (Coronary Diet Intervention with Olive Oil and Cardiovascular Prevention; n = 711), which consisted of Spanish white adults; the GOLDN (Genetics of Lipids Lowering Drugs and Diet Network; n = 818), which involved North American non-Hispanic white adults; and Hispanic adults who participated in the BPRHS (Boston Puerto Rican Health Study; n = 1155).
Results: Meta-analysis indicated that T carriers presented a lower risk of T2D than CC carriers (pooled OR: 0.714; 95% CI: 0.584, 0.845; P = 0.002). In all 3 populations (CORDIOPREV, GOLDN, and BPRHS), we showed a significant interaction between the rs3014866 single nucleotide polymorphism and dietary SFA:carbohydrate ratio intake for the homeostasis model assessment of insulin resistance (HOMA-IR) (P = 0.028, P = 0.017, and P = 0.026, respectively). CC carriers had a significantly higher HOMA-IR only when SFA:carbohydrate intake was high (P = 0.045 for the CORDIOPREV, P = 0.033 for the GOLDN, and P = 0.046 for the BPRHS) but not when SFA:carbohydrate ratio intake was low.
Conclusions: The minor allele (T) of the S100A9 variant rs3014866 is associated with lower T2D risk in 3 populations of different ancestries. Note that individuals with the high-risk CC genotype may be more likely to benefit from a low SFA:carbohydrate ratio intake to improve insulin resistance as evaluated with the use of the HOMA-IR. These trials were registered at clinicaltrials.gov as NCT00924937 (CORDIOPREV), NCT00083369 (GOLDN), and NCT01231958 (BPRHS).
Keywords: gene-diet interaction, insulin resistance, SFA:carbohydrate ratio; S100A9 gene, type 2 diabetes
INTRODUCTION
Type 2 diabetes (T2D)14 is associated with increased morbidity and mortality, and its growing global prevalence represents a major public health concern (1). The International Diabetes Federation has estimated that 382 million people worldwide are currently presenting with T2D and has projected that this number will rise to 592 million people by 2035 (2).
T2D is a metabolic disease that is characterized by hyperglycemia that results from dysregulation in insulin secretion, insulin action, or both (3). It has been established that the development of T2D is the result of a combination of genetics and the exposome, including dietary intake, exercise, sedentary lifestyle, sleep, and stress (4). A growing number of genetic loci related to T2D have been identified through genome-wide association studies (GWASs), but the cumulative effect of the currently identified loci explains only ∼10% of the estimated T2D heritability (5). Therefore, it has been proposed that interactions between genetic and environmental factors may contribute to the identification of some of the missing heritability. Thus, the identification of these gene-environment interactions could provide a major step in the prevention of this disease.
S100 proteins belong to the EF-hand superfamily of Ca2+-binding proteins and have diverse intracellular and extracellular functions (6). Of them, the S100 calcium-binding protein S100A9 (calgranulin B) and S100A8 (calgranulin A) form a heterocomplex called calprotectin, which has been shown to be involved in chronic inflammation and risk of several other metabolic diseases (7, 8). However, calgranulin monomers have pleiotropic functions, some of which may reduce excessive tissue damage or facilitate the resolution of inflammation (9). Indeed, an S100A9 variant (rs3014866) was associated with protection against T2D development in one European population (10). However, this finding requires independent validation in other populations and ancestries.
Therefore, the aims of the present study were as follows: 1) to examine both the associations between the S100A9 variant rs3014866 and the HOMA-IR as a marker of insulin resistance and T2D and 2) to evaluate possible interactions of this variant with diet in the following 3 populations of different ancestries and geographical locations: the CORDIOPREV (Coronary Diet Intervention with Olive Oil and Cardiovascular Prevention) (clinicaltrials.gov; NCT00924937) population, the GOLDN (Genetics of Lipids Lowering Drugs and Diet Network) (clinicaltrials.gov; NCT00083369) population, and the BPRHS (Boston Puerto Rican Health Study) (clinicaltrials.gov; NCT01231958) population.
METHODS
Study populations
The CORDIOPREV study is a long-term secondary cardiovascular prevention intervention trial. The main objective of the study is to evaluate the efficacy of a Mediterranean diet compared with that of a low-fat diet to prevent clinical events and mortality in patients with a history of cardiovascular disease. The eligibility criteria, design, and methods of the CORDIOPREV clinical trial have been previously reported (11, 12), and the protocol is available at clinicaltrials.gov. CORDIOPREV participants were predominantly of European ancestry and were recruited in only one center in a specific region in the south of Spain (Cordoba, Andalucia, Spain). In the current study, we included 711 participants (596 men and 115 women) who had complete genotype and dietary and biochemical baseline data. The local ethics committees approved the trial protocol and all amendments, which followed the Helsinki Declaration and good clinical practices. The experimental protocol conformed to international ethical standards (13), and written informed consent was obtained from all individuals.
GOLDN participants were predominantly of European ancestry and were recruited from 2 genetically homogeneous centers (Minneapolis, Minnesota, and Salt Lake City, Utah). In the current study, 818 participants (404 men and 414 women) of European ancestry were included in our analyses. The primary aim of the GOLDN was to examine the influence of genetic and dietary factors on the response of individuals to fenofibrate. Baseline data obtained from participants before they entered the intervention were selected for this analysis. The study details and related methodology of the GOLDN have been previously described (14) and are also shown at clinicaltrials.gov. The protocol was approved by the institutional review boards at the University of Alabama, the University of Minnesota, the University of Utah, and Tufts University.
The BPRHS is a longitudinal cohort study of stress, nutrition, health, and aging for which study participants were self-identified as Puerto Rican and living in Boston and its metropolitan area (15). The ancestry composition of the BPRHS population is 57.2% European, 27.4% African, and 15.4% Native American (16). For this study, we included 1155 participants (339 men and 816 women) with complete genotype and dietary data. The study protocol was approved by the institutional review boards at Tufts University, Northeastern University, and the University of Massachusetts Lowell.
Biochemical, anthropometric, physical activity, and dietary intake measurements and T2D diagnosis
Blood samples were drawn after an overnight fast. In the CORDIOPREV, lipid variables and glucose were measured with the use of Architect c-16000 analyzers (Abbott) with the following spectrophotometric techniques (enzymatic colorimetric methods): a hexokinase method for glucose and oxidation-peroxidation for total cholesterol, HDL cholesterol, and triglycerides. LDL cholesterol was calculated with the use of the Friedewald formula (provided that the triglyceride concentration was <400 mg/dL). The fasting insulin concentration was measured with the use of a chemiluminescent microparticle immunoassay and an i-2000 analyzer (Abbott Architect). The plasma concentration of high-sensitivity C-reactive protein was determined with the use of a high-sensitivity ELISA (BioCheck Inc.). In the GOLDN, fasting insulin was obtained with the use of a radioimmunoassay and a commercial kit (Linco Research), and fasting glucose was measured with the use of a hexokinase-mediated reaction in the Hitachi commercial kit (Roche Diagnostics). Measurements of blood lipids, including triglycerides and HDL cholesterol, have been described (17). Plasma high-sensitivity C-reactive protein was measured with the use of the K-Assay CRP calibrator (2), a latex particle–enhanced immunoturbidimetric assay on the Hitachi 911 analyzer (Kamiya Biomedical Co.). In the BPRHS, fasting insulin was measured with the use of an Immulite 1000 Insulin Kit (LKIN1; Siemens Medical Solution Diagnostics) on the Immulite 1000 system (Siemens Medical Solution Diagnostics), and Olympus Au400e with Olympus glucose reagents (Olympus America Inc.) were used to measure fasting glucose. Fasting triglycerides and HDL cholesterol were measured with the use of Olympus HDL cholesterol reagents (OSR6195; Olympus America Inc.) and Olympus triglyceride reagents (OSR6033; Olympus America Inc.). C-reactive protein in serum was analyzed with the use of an immunoturbidimetric reaction in a Cobas Fara II Centrifugal Analyzer (Roche) with a DiaSorin CRP SPQ test-system antibody reagent set II (AM-0039; Atlantic Antibodies).
In the CORDIOPREV, subjects also underwent an oral glucose tolerance test at baseline. Therefore, they were considered to have T2D if they had a medical diagnosis of diabetes, were receiving glucose-lowering treatment, or had any of the American Diabetes Association criteria for the diagnosis of T2D (fasting glucose concentration ≥7 mmol/L, 2-h glucose concentration ≥11.1 mmol/L, or glycated hemoglobin ≥6.5%) (18). In the GOLDN and BPRHS, T2D was defined by self-reported diabetes, a fasting glucose concentration ≥7 mmol/L, or the use of a glucose-lowering medication. For the 3 populations, the HOMA-IR, which was calculated as
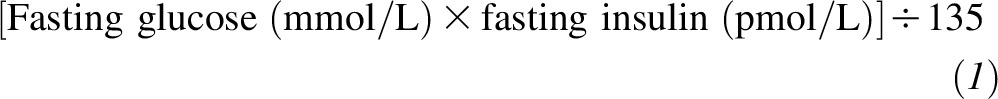 |
was used to represent insulin resistance.
Weight, height, and waist circumference were measured according to standardized protocols in the 3 populations, and BMI (in kg/m2) was calculated as weight divided by height square.
Physical activity in the CORDIOPREV was assessed by the calculation of the validated physical activity factor the leisure time physical activity (19) by multiplying the metabolic equivalent value (20) by the number of hours per week of each reported physical activity and summed across activities to estimate the overall leisure time physical activity. In the GOLDN, a questionnaire was used to estimate the number of hours per day that were dedicated to different activities (21). In the BPRHS, a physical activity score that was based on the Paffenbarger questionnaire of the Harvard Alumni Activity Survey (22) was estimated.
Dietary intake was assessed in the CORDIOPREV with the use of a 137-item semiquantitative food-frequency questionnaire (FFQ) that was validated in Spain (23). Energy and nutrient intakes were calculated with the use of Spanish food-composition tables (24). In the GOLDN, dietary intake was collected with the use of a diet-history questionnaire, which was developed by the National Cancer Institute and was validated in 2 studies (25, 26). The food list and nutrient database used with the diet-history questionnaire were based on US dietary data (27). In the BPRHS, dietary intake was assessed with the use of an FFQ that was designed for and validated in this population (28). Energy and nutrient intakes were calculated with the use of Nutrition Data System for Research software (version 2007; Nutrition Coordinating Center, University of Minnesota). To normalize dietary intake in cohorts, we evaluated protein, carbohydrate, and total and type of fat intakes as percentages of total energy. In addition, we also examined specific SFAs and types of carbohydrates as percentages of total energy.
DNA isolation and S100A9 genotyping
The single nucleotide polymorphism (SNP) rs3014866 (4811713T>C), which is located ∼1.2-kbp upstream of the S100A9 messenger RNA start site, was selected according to previous evidence (6, 10). DNA samples were isolated from blood samples with the use of the salting-out method in the CORDIOPREV, with Gentra Puregene Blood Kits (Gentra Systems) in the GOLDN, and with QIAamp DNA Blood Mini Kits (Qiagen) in the BPRHS.
In the CORDIOPREV, genotyping was performed on an OpenArray SNP Genotyping System of Applied Biosystems (Thermo Fisher Scientific) with the use of assays that were available from the same company. Data were analyzed with the Applied Biosystems TaqManGenotyper Software 2.1 (Thermo Fisher Scientific). For the GOLDN, the S100A9 genotype was imputed with the use of MACH (29) on the basis of the original genome genotype data that was generated from the Genome-Wide Human SNP Array 6.0 (Affymetrix) and the reference panel of CEU-phased haplotypes from HapMap (30); and for the BPRHS, the S100A9 variant was genotyped with the use of Axiom Genome-Wide LAT Array (Affymetrix), which was designed especially for Hispanic populations and contains probe sets to genotype 817,810 SNPs.
Statistical analysis
The statistical analysis was carried out with the use of SPSS software (version 21.0 for Windows; SPSS Inc.), and differences were considered statistically significant at P < 0.05. The normal distribution was evaluated for all measured variables, and skewed variables (triglycerides, fasting insulin, HOMA-IR, and C-reactive protein) were normalized by log10. Nonadjusted data are presented as mean ± SDs for continuous variables and as frequencies for categorical variables except as indicated. The Hardy-Weinberg equilibrium for the S100A9 variant was assessed with the use of chi-square tests in each population. Because of the limited sample size, a dominant genetic model for the rs3014866 (4811713T>C) was considered for the analysis to increase the statistical power after confirmation that the model was valid in all studied populations. Thus, CT and TT participants were grouped and compared with CC participants. We also tested the statistical homogeneity by sex, and men and women were analyzed together. Nutrient intake expressed as the percentage of total energy intake (except as indicated) was dichotomized on the basis of the median intake of each population for the interaction analysis.
We performed a linear regression analysis to examine the associations between SNP rs3014866 and glucose metabolism variables (fasting glucose, fasting insulin, and HOMA-IR). To avoid potential confounding across cohorts, we adjusted the analyses for sex, age, current smoking, alcohol consumption, physical activity, diabetes medication use, and BMI. In addition, we adjusted the model for the study center and family relationships in the GOLDN and for population structure in the BPRHS. In the GOLDN, family relationships within the population were estimated with the use of the SAS statistical software (version 9.1 for Windows; SAS Institute Inc.) GENMOD procedure with the assumption of an exchangeable correlation structure with pedigree (14). In the BPRHS, the population admixture was estimated with the use of the first component of a principal component analysis (16). The principal component analysis was estimated with the use of the SVS program (version 8; Golden Helix Inc.) on the basis of 50,704 SNPs that were selected from GWAS genotype data (see DNA isolation and S100A9 genotyping) according to the following criteria: call rate >97%, minor allele frequency ≥5%, pairwise linkage disequilibrium R2 ≤ 0.1, and a Hardy-Weinberg equilibrium P ≥10−6.
In all populations, logistic regression models were fitted to test the rs3014866 genotypes in determination of the OR of T2D. Study-specific ORs and 95% CIs were estimated for each population. Multivariable adjustments were performed as indicated.
Meta-analyses were conducted with the Open Meta-Analyst program (Brown University; the lastest version is available at http://www.cebm.brown.edu/openmeta/) under fixed-effect models. For continuous outcomes, meta-analyses were conducted by combining z statistics across the 3 populations, which were weighted by sample size. For the binary outcome (T2D presence), we used a meta-analysis to combine effect-size estimates (β coefficients and 95% CIs) from the 3 studies, which were weighted by the inverse of the corresponding SEs. Heterogeneity was tested with the use of Cochran’s Q statistic and was quantified with the use of I2.
Finally, we used univariate general linear regression models to consider gene-diet interactions in relation to glycemic traits. We fitted separate models for each population, including rs3014866 genotypes and the dietary variables (dichotomous) for interaction terms. Multivariable adjustments were performed as indicated.
RESULTS
Characteristics of the populations and S100A9 variant
We included 2755 individuals from 3 independent cohorts; one cohort was from Spain (the CORDIOPREV), and 2 cohorts were from the United States (the GOLDN and the BPRHS). Table 1 shows the demographic, anthropometric, clinical, biochemical, and lifestyle characteristics of the participants according to S100A9 polymorphism (rs3014866) for each population. Genotype frequencies were consistent with Hardy-Weinberg equilibrium in all populations. In the CORDIOPREV and BPRHS, the prevalence of T2D (P = 0.040 and P = 0.031, respectively) and the prevalence of diabetes medication use (P = 0.026 and P = 0.042, respectively) were greater in CC than in T carriers but not in the GOLDN. In the CORDIOPREV, CC subjects also had higher fasting glucose concentrations than T carriers (P = 0.020). No significant differences were shown between carriers of the T allele and CC genotype for the rest of the measured variables in all 3 populations.
TABLE 1.
General characteristics by S100A9 polymorphism (rs3014866) genotypes in the studied populations1
| CORDIOPREV |
GOLDN |
BPRHS |
||||
| TT+CT | CC | TT+CT | CC | TT+CT | CC | |
| n (%)2 | 510 (71.7) | 201 (28.3) | 684 (83.6) | 134 (16.4) | 787 (68.1) | 368 (31.9) |
| Men/women, n | 431/78 | 165/37 | 337/347 | 67/67 | 228/559 | 111/257 |
| Age, y | 59.3 ± 9.23 | 60.0 ± 7.95 | 48.9 ± 16.3 | 48.7 ± 14.5 | 57.1 ± 7.7 | 57.0 ± 7.6 |
| Type 2 diabetes, n (%) | 187 (36.7) | 91 (45.3)* | 49 (7.2) | 11 (8.2) | 312 (39.6) | 168 (45.7)* |
| BMI, kg/m2 | 31.0 ± 4.2 | 31.1 ± 4.8 | 28.4 ± 5.8 | 28.7 ± 5.5 | 31.9 ± 6.7 | 31.8 ± 6.9 |
| Waist circumference, cm | 105.1 ± 11.5 | 105.1 ± 11.6 | 96.7 ± 15.7 | 99.5 ± 19.1 | 101.0 ± 14.4 | 102.0 ± 15.6 |
| Total cholesterol, mmol/L | 4.11 ± 0.78 | 4.09 ± 0.83 | 4.93 ± 1.02 | 5.05 ± 1.06 | 4.73 ± 1.12 | 4.69 ± 1.09 |
| HDL cholesterol, mmol/L | 1.09 ± 0.23 | 1.07 ± 0.27 | 1.20 ± 0.34 | 1.18 ± 0.31 | 1.20 ± 0.50 | 1.18 ± 0.46 |
| LDL cholesterol, mmol/L | 2.31 ± 0.63 | 2.25 ± 0.66 | 3.16 ± 0.82 | 3.31 ± 0.85 | 2.79 ± 0.90 | 2.69 ± 0.91 |
| Serum triglycerides, mmol/L | 1.46 ± 0.69 | 1.56 ± 0.85 | 1.59 ± 1.14 | 1.52 ± 0.95 | 1.66 ± 0.74 | 1.72 ± 1.28 |
| C-reactive protein,4 mmol/L | 22.9 ± 18.9 | 24.6 ± 21.1 | 2.38 ± 4.67 | 2.19 ± 3.71 | 33.5 ± 24.9 | 32.6 ± 24.8 |
| Fasting glucose (mmol/L) | 6.27 ± 2.11 | 6.68 ± 2.54* | 5.64 ± 1.03 | 5.79 ± 1.41 | 6.52 ± 2.58 | 6.78 ± 2.64 |
| Fasting insulin (pmol/L) | 78.0 ± 57.1 | 83.7 ± 61.6 | 83.8 ± 49.6 | 87.5 ± 49.4 | 101.7 ± 85.7 | 98.1 ± 86.0 |
| HOMA-IR | 3.83 ± 4.04 | 4.56 ± 6.98 | 3.61 ± 2.60 | 3.92 ± 3.16 | 5.46 ± 6.65 | 5.77 ± 8.89 |
| Physical activity5 | 22.4 ± 5.4 | 24.3 ± 8.8 | 33.8 ± 6.0 | 34.8 ± 7.5 | 29.1 ± 9.2 | 29.7 ± 9.0 |
| Current smoking, n (%) | 48 (9.4) | 17 (8.4) | 55 (8.0) | 11 (8.2) | 201 (25.5) | 82 (22.3) |
| Current drinking, n (%) | 301 (59.0) | 116 (57.7) | 338 (49.4) | 68 (50.7) | 552 (70.1) | 251 (68.2) |
| Diabetes medication use, n (%) | 167 (32.7) | 84 (41.8)* | 33 (4.8) | 8 (6.0) | 258 (32.8) | 139 (37.8)* |
| Lipid medication use, n (%) | 435 (85.3) | 175 (87.1) | 114 (16.7) | 22 (16.4) | 318 (40.4) | 147 (39.9) |
Genotype frequencies in the CORDIOPREV were as follows: TT, n = 158; CT, n = 352; and CC, n = 201. Genotype frequencies in the GOLDN were as follows: TT, n = 266; CT, n = 418; and CC, n = 134. Genotype frequencies in the BPRHS were as follows: TT, n = 217; CT, n = 570; CC, n = 368. *P < 0.05 between carriers of the T allele and CC genotype for the corresponding variable in each population (univariate general linear model). BPRHS, Boston Puerto Rican Health Study; CORDIOPREV, Coronary Diet Intervention with Olive Oil and Cardiovascular Prevention; GOLDN, Genetics of Lipids Lowering Drugs and Diet Network; S100A9, S100 calcium-binding protein A9 gene.
For all categorical variables.
Mean ± SD (all such values for continuous variables).
Measured with the use of different techniques in the CORDIOPREV, GOLDN, and BPRHS as described in Methods.
Measured with the use of different physical activity scores in the CORDIOPREV, GOLDN, and BPRHS as described in Methods.
Table 2 shows dietary intakes of the populations. In the CORDIOPREV, no differences were observed between CC and TT+CT genotypes. In the GOLDN, compared with CC carriers, T carriers reported higher intakes (percentages of total energy intake) of total and complex carbohydrates and lower intakes of total fat, saturated fat (SFA), and myristic, palmitic, and stearic fatty acids and a lower SFA-to-carbohydrate (SFA:carbohydrate) ratio (P < 0.01). In contrast, BPRHS participants with the TT+CT genotype reported higher intakes of SFA and myristic and palmitic fatty acids and a higher SFA:carbohydrate ratio (P < 0.05) than did participants with the CC genotype.
TABLE 2.
Dietary intake by S100A9 polymorphism (rs3014866) genotype in the studied populations1
| CORDIOPREV |
GOLDN |
BPRHS |
||||
| TT+CT | CC | TT+CT | CC | TT+CT | CC | |
| n (%) | 510 (71.7) | 201 (28.3) | 684 (83.6) | 134 (16.4) | 787 (68.1) | 368 (31.9) |
| Total energy, kcal | 2283 ± 5292 | 2214 ± 462 | 2129 ± 1248 | 2193 ± 1310 | 2271 ± 1140 | 2197 ± 1194 |
| Protein, E% | 18.5 ± 2.7 | 18.4 ± 2.7 | 15.8 ± 2.8 | 15.8 ± 2.6 | 17.4 ± 3.5 | 17.1 ± 3.4 |
| Carbohydrate, E% | 41.8 ± 6.8 | 41.4 ± 6.1 | 49.2 ± 8.6 | 47.6 ± 7.7* | 51.3 ± 7.4 | 52.0 ± 7.3 |
| Simple sugars, E% | 15.9 ± 4.4 | 16.0 ± 4.8 | 11.7 ± 6.4 | 11.7 ± 5.7 | 20.2 ± 7.9 | 20.7 ± 8.3 |
| Complex carbohydrate, E% | 23.1 ± 5.7 | 22.7 ± 5.7 | 34.4 ± 7.0 | 32.9 ± 6.0* | 28.0 ± 5.5 | 28.2 ± 5.8 |
| Total fiber, g/d | 25.2 ± 7.7 | 25.1 ± 7.8 | 19.2 ± 10.4 | 18.6 ± 10.5 | 19.1 ± 9.2 | 18.2 ± 9.0 |
| Total fat, E% | 36.9 ± 6.0 | 37.4 ± 5.6 | 35.2 ± 6.8 | 36.9 ± 6.6* | 31.3 ± 5.2 | 30.9 ± 5.3 |
| MUFA, E% | 18.0 ± 3.6 | 18.2 ± 3.3 | 13.2 ± 2.8 | 13.7 ± 2.7 | 11.4 ± 2.1 | 11.3 ± 2.0 |
| PUFA, E% | 6.3 ± 1.7 | 6.5 ± 1.8 | 7.6 ± 2.2 | 7.8 ± 2.2 | 7.7 ± 1.7 | 7.8 ± 1.8 |
| SFA, E% | 9.0 ± 1.9 | 9.0 ± 1.8 | 11.7 ± 2.6 | 12.6 ± 2.6* | 9.6 ± 2.2 | 9.3 ± 2.2* |
| Myristic acid (14:0), E% | 0.7 ± 0.4 | 0.7 ± 0.3 | 1.0 ± 0.4 | 1.2 ± 0.4* | 0.8 ± 0.3 | 0.7 ± 0.3* |
| Palmitic acid (16:0), E% | 4.1 ± 1.2 | 4.1 ± 1.1 | 6.3 ± 1.3 | 6.7 ± 1.2* | 5.5 ± 1.1 | 5.3 ± 1.1* |
| Stearic acid (18:0), E% | 1.6 ± 0.5 | 1.6 ± 0.5 | 2.9 ± 0.6 | 3.1 ± 0.6* | 2.3 ± 0.6 | 2.2 ± 0.6 |
| SFA:carbohydrate ratio | 0.23 ± 0.10 | 0.23 ± 0.07 | 0.25 ± 0.10 | 0.28 ± 0.09* | 0.20 ± 0.07 | 0.19 ± 0.05* |
*P < 0.05 between carriers of the T allele and CC genotype for the corresponding variable in each population (univariate general linear model). BPRHS, Boston Puerto Rican Health Study; CORDIOPREV, Coronary Diet Intervention with Olive Oil and Cardiovascular Prevention; E%, percentage of energy; GOLDN, Genetics of Lipids Lowering Drugs and Diet Network; S100A9, S100 calcium-binding protein A9 gene.
Mean ± SD (all such values for continuous variables).
Meta-analysis of S100A9 variant with glucose metabolism variables and T2D
Meta-analyzed T carriers had significantly lower fasting glucose (β = −2.58; 95% CI: −4.94, −0.23; P = 0.03) than CC carriers. No differences were shown with respect to fasting insulin values between genotypes, but participants with the TT+CT genotype tended to have a lower HOMA-IR than participants with the CC genotype (β = −0.31; 95% CI: −0.67, 0.04; P = 0.08). No significant heterogeneity was observed for both variables (Q = 0.72, I2 = 0%, and P-heterogeneity = 0.695 for fasting glucose; Q = 0.90, I2= 0%, and P-heterogeneity = 0.634 for HOMA-IR) (Supplemental Table 1).
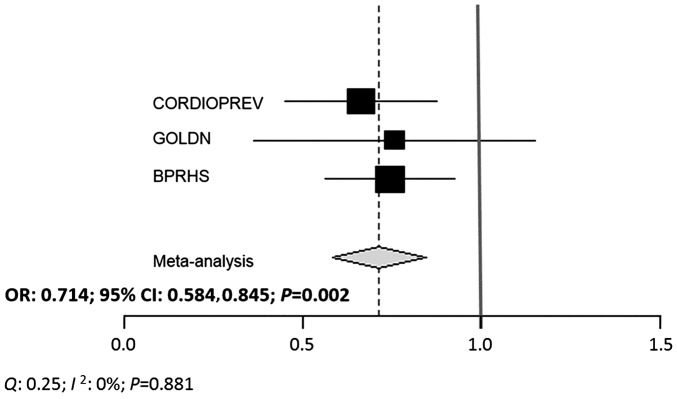
When the CORDIOPREV and BPRHS populations were analyzed separately, subjects with the TT+CT genotype showed a lower OR of T2D than participants with the CC genotype (OR for CORDIOPREV: 0.662; 95% CI: 0.450, 0.974; P = 0.036 and OR for BPRHS: 0.744; 95% CI: 0.562, 0.986; P = 0.039) Although the tendency was the same, we did not find significant results in the GOLDN population (Table 3). Nevertheless, the overall meta-analysis for all 3 cohorts showed that T carriers presented lower risk of T2D than CC carriers (pooled OR: 0.714; 95% CI: 0.584, 0.845; P = 0.002). No significant heterogeneity was observed (Q = 0.25, I2 = 0%, and P-heterogeneity = 0.881) (Figure 1).
TABLE 3.
Association of S100A9 variant (rs3014866) with T2D1
| T2D subjects, n (%) |
||||
| No | Yes | OR (95% CI) | P | |
| CORDIOPREV | 0.036 | |||
| TT+CT | 323 (63.3) | 187 (36.7) | 0.662 (0.450, 0.974) | |
| CC | 110 (54.7) | 91 (45.3) | 1 (reference) | |
| GOLDN | 0.456 | |||
| TT+CT | 635 (92.8) | 49 (7.2) | 0.757 (0.364, 1.574) | |
| CC | 123 (91.8) | 11 (8.2) | 1 (reference) | |
| BPRHS | 0.039 | |||
| TT+CT | 475 (60.4) | 312 (39.6) | 0.744 (0.562, 0.986) | |
| CC | 200 (54.3) | 168 (45.7) | 1 (reference) | |
ORs (95% CIs) of T2D risk were estimated by linear regression analysis. P values were adjusted for age, sex, BMI, smoking status, alcohol use, and physical activity. In addition, we adjusted the model for the study center and family relationships in the GOLDN and for the population structure in the BPRHS. BPRHS, Boston Puerto Rican Health Study; CORDIOPREV, Coronary Diet Intervention with Olive Oil and Cardiovascular Prevention; GOLDN, Genetics of Lipids Lowering Drugs and Diet Network; S100A9, S100 calcium-binding protein A9 gene; T2D, type 2 diabetes.
FIGURE 1.
Meta-analysis of associations between rs3014866 and T2D risk in the 3 populations. A fixed-effect model was used to get the overall effect estimate. Black squares indicated βs, square sizes reflect corresponding weights, and horizontal bars denote 95% CIs. Heterogeneity was tested by Cochran’s Q statistic and quantified with the use of I2. BPRHS, Boston Puerto Rican Health Study; CORDIOPREV, Coronary Diet Intervention with Olive Oil and Cardiovascular Prevention; GOLDN, Genetics of Lipids Lowering Drugs and Diet Network.
Interaction of S100A9 variant with dietary intake for glucose metabolism variables
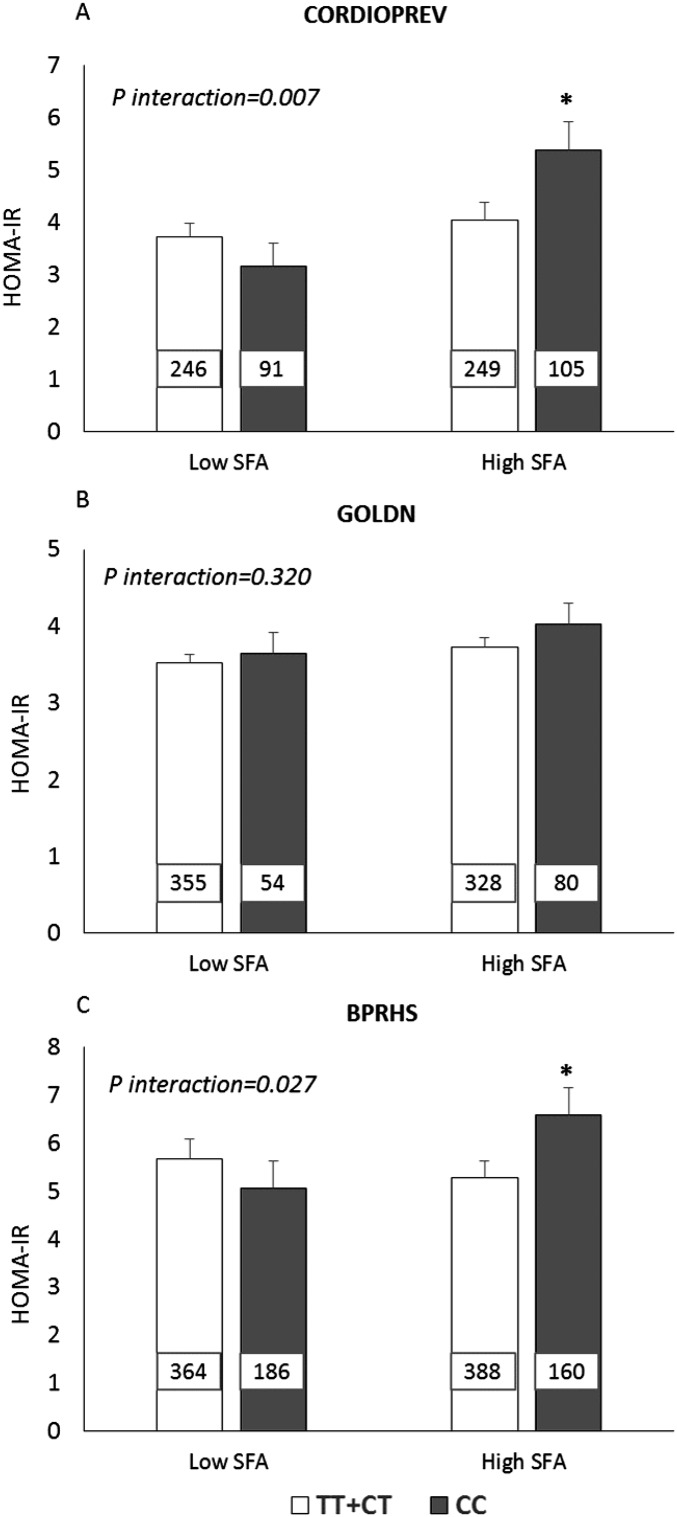
To perform the SNP-by-diet interaction tests, dietary factors were dichotomized at median intake in each population. In the CORDIOPREV and BPRHS, rs3014866 interacted significantly with dietary SFA intake (P = 0.007 and P = 0.027, respectively) for the HOMA-IR. In these 2 populations, CC carriers, compared with T carriers, had a significantly higher HOMA-IR when they had high SFA intake (P = 0.029 in the CORDIOPREV and P = 0.048 in the BPRHS) but not when they had low SFA intake (Figure 2).
FIGURE 2.
Estimated mean ± SEM interactions between the S100A9 variant (rs3014866) and SFA intake (percentage of total energy) on HOMA-IR. (A) For the CORDIOPREV, low and high SFA intakes indicate median intakes (≤8.8% and >8.8%). (B) For the GOLDN, low and high SFA intakes indicate median intakes (≤11.8% and >11.8%). (C) In the BPRHS, low SFA and high SFA indicate SFA median intakes (≤9.3% and >9.3%). The number inside each bar indicates the number of subjects in that group with data available for HOMA-IR. Analyses were adjusted for age, sex, BMI, smoking status, alcohol use, diabetes medication use, and physical activity. In addition, we adjusted the model for the study center and family relationships in the GOLDN and for the population structure in the BPRHS. P-interactions between SFA intake (as dichotomous) and the S100A9 variant in each population were obtained with the use of a univariate model analysis. *P < 0.05 for mean comparisons of HOMA-IR values between CC and TT+CT genotypes with high SFA intake (univariate model analysis). BPRHS, Boston Puerto Rican Health Study; CORDIOPREV, Coronary Diet Intervention with Olive Oil and Cardiovascular Prevention; GOLDN, Genetics of Lipids Lowering Drugs and Diet Network; S100A9, S100 calcium-binding protein A9 gene.
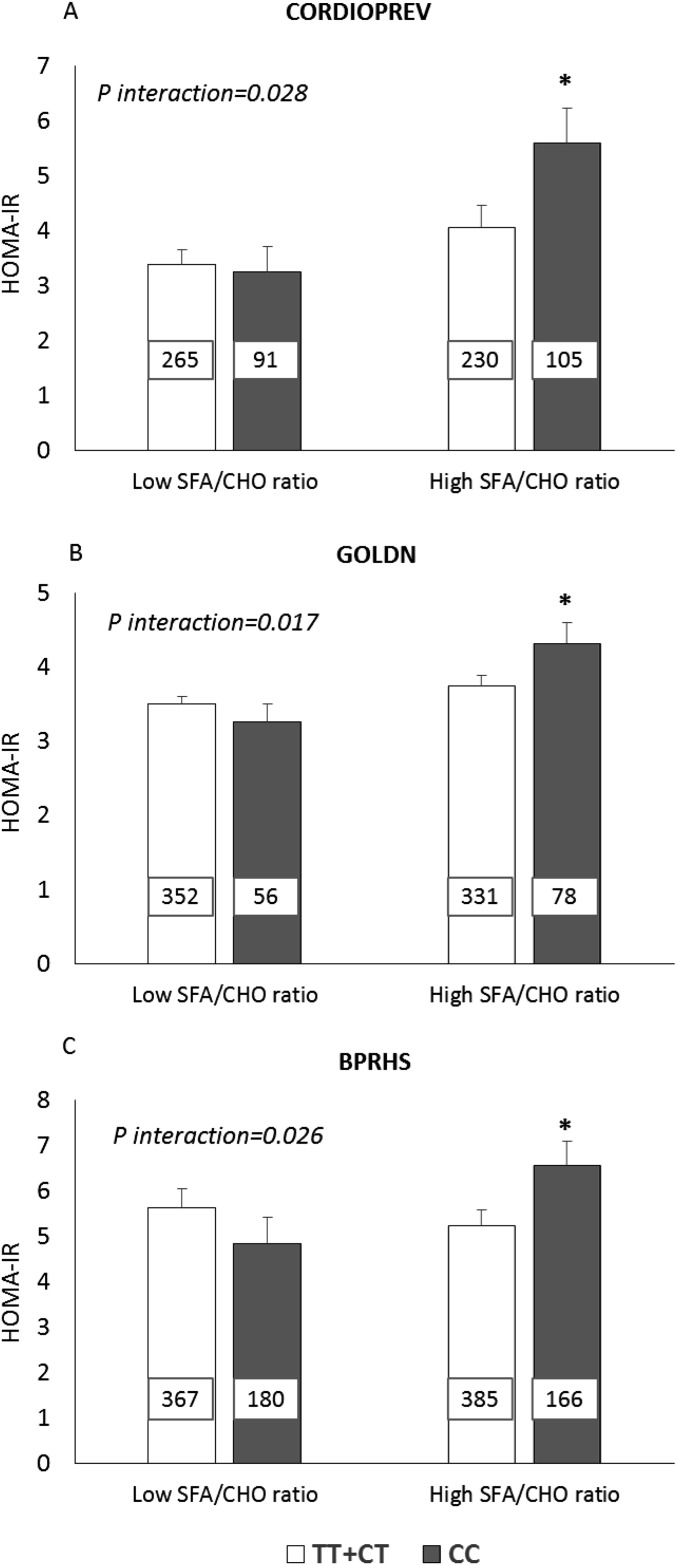
In each of all 3 populations (in the CORDIOPREV, GOLDN, and BPRHS), we showed a significant interaction between the rs3014866 SNP and dietary SFA:carbohydrate intake for the HOMA-IR (P = 0.028, P = 0.017, and P = 0.026, respectively). CC carriers, compared with T carriers, had a significantly higher HOMA-IR when their SFA:carbohydrate intake was high (P = 0.045 in the CORDIOPREV, P = 0.033 in the GOLDN, and P = 0.046 in the BPRHS) but not when their SFA:carbohydrate intake was low (Figure 3).
FIGURE 3.
Estimated mean ± SEM interactions between the S100A9 variant (rs3014866) and SFA/CHO ratio intake on HOMA-IR. (A) For the CORDIOPREV, low and high SFA/CHO ratios indicate median intakes (≤0.21 and >0.22). (B) For the GOLDN, low and high SFA/CHO ratios indicate median intakes (≤0.24 and >0.25). (C) In the BPRHS, low and high SFA/CHO ratios indicate median intakes (≤0.19 and >0.20). The number inside each bar indicates the number of subjects in that group with data available for HOMA-IR. Analyses were adjusted for age, sex, BMI, smoking status, alcohol use, diabetes medication use, and physical activity. In addition, we adjusted the model for the study center and family relationships in the GOLDN and for the population structure in the BPRHS. P-interactions between the SFA/CHO ratio (as dichotomous) and the S100A9 variant in each population were obtained with the use of a univariate model analysis. *P < 0.05 for mean comparisons of HOMA-IR values between CC and TT+CT genotypes with high SFA/CHO ratio intake (univariate model analysis). BPRHS, Boston Puerto Rican Health Study; CHO, carbohydrate; CORDIOPREV, Coronary Diet Intervention with Olive Oil and Cardiovascular Prevention; GOLDN, Genetics of Lipids Lowering Drugs and Diet Network; S100A9, S100 calcium-binding protein A9 gene.
We did not observe significant interactions between the rs3014866 genotype and SFA and SFA:carbohydrate intakes for fasting glucose or fasting insulin. Likewise, interactions between the genetic variant and carbohydrate intake for fasting glucose, fasting insulin, and the HOMA-IR were not identified (Supplemental Table 2). Moreover, no interactions reached significance with specific SFAs (myristic, palmitic, and stearic fatty acids) or with simple sugars, complex carbohydrates, or dietary fiber (Supplemental Tables 3 and 4).
DISCUSSION
The current study showed that the minor allele of the S100A9 rs3014866 variant was associated with lower T2D risk in 3 independent populations. This finding is consistent with that of a previous study in a European population (10). In addition, to our knowledge, we have provided novel evidence that showed an interaction between rs3014866 and saturated fat and SFA-to-carbohydrate ratio intakes that appeared to influence the HOMA-IR in these populations. This result suggests that CC homozygous individuals may benefit from low SFA:carbohydrate ratio intake to decrease insulin resistance. These findings were robust because we were able to replicate them in populations of different ancestries and diverse clinical, demographic, and lifestyle characteristics.
S100A9 is a T2D gene that was identified in a GWAS. The gene is located at 1q21, which is a known susceptibility region for T2D (31), and codes for S100A9 protein, also known as calgranulin B. The rs3014866 SNP is located in the 5′-upstream region of the S100A9 gene, and it has been reported to increase gene expression and S100A9 concentrations in humans (10). The protein S100A9 has been associated with a range of acute and chronic inflammatory responses (32), latter-involved in the progression of insulin resistance and the development of T2D (33). However, we did not observe differences in values of C-reactive protein between genotypes in any of the studied populations. Therefore, our results support the hypothesis that S100A9 is not merely a marker of inflammation but, in addition, may be related to increased insulin-stimulated glucose uptake in some tissues (10). The calprotectin protein (S100A8/S100A9 complex) has been described as a transporter that shuttles arachidonic acid into target cells (34), and there has been evidence that this fatty acid could stimulate glucose uptake in adipocytes by increasing the glucose transporter 1 and the glucose transporter 4 concentrations (35). Moreover, rs3014866 has been associated with the expression of several genes in adipose and lymphoblastic tissues in humans. Specifically, the rs3014866 T allele was associated with significantly decreased expression of interleukin enhancer binding factor 2 in both tissues (36). Interleukin enhancer binding factor 2 is a nuclear factor that modulates IL-2, and it is overexpressed in the peripheral blood of T2D patients (37). Additional studies are needed to clarify the mechanism for the association between this S100A9 variant and T2D risk.
In contrast, rs3014866 was associated with fasting glucose but not with fasting insulin or the HOMA-IR. Similarly, the Meta-Analyses of Glucose and Insulin-related traits Consortium showed no relation between this SNP and glycemic variables (38). This lack of an association could be explained by the gene-diet interactions that we have reported here. These novel and consistent interactions indicated that rs3014866 CC carriers showed a less HOMA-IR when their SFA and SFA:carbohydrate intakes were low, which suggested a protective effect of a diet that was low in saturated fat in relation to carbohydrate in these participants. Dietary SFA are thought to compromise insulin sensitivity by affecting cell-membrane fluidity and functions, including insulin receptor binding and affinity (39). In addition, some studies have suggested that lipolytic products of SFAs may exert unfavorable effects on the expression of genes encoding lipogenic and lipid-transport proteins (40). Despite these reports, the relation between total SFA intake and T2D risk remains unclear (41), and it has been suggested that the SFA-chain length could be associated with different biological effects (42). However, we did not find significant interactions of the genotype with myristic, palmitic, and stearic fatty acids on measures of the HOMA-IR.
Current dietary recommendations that are aimed to prevent diabetes agree on the importance of reducing dietary saturated fat intake (18, 43) although debate remains as to which macronutrient should optimally replace the saturated fat and what dietary pattern should be recommended (41). MUFAs, PUFAs and complex carbohydrates have been shown to have beneficial effects in glycemic control and insulin sensitivity when substituted for SFAs in the diet (44, 45). In this context, gene-diet interactions may play a role, and several studies have shown that various dietary approaches could provide differing effects on glucose metabolism depending on the genetic background of the participants (46). Therefore, our findings reinforce the hypothesis that a single dietary model may not be optimal for all individuals (47), and dietary advice that takes the genotype into account may be the key for a more effective prevention of T2D.
The potential mechanisms for the interactions observed should be explored in future studies. Nevertheless, an in silico analysis (http://snpinfo.niehs.nih.gov/) of the S100A9 variant rs3014866 suggested that the major allele (C) might create a specific binding site for the transcription factor peroxisome proliferator-activated receptor γ (PPARG), which might be eliminated by the presence of the minor allele (T). This transcription factor is known to regulate adipocyte differentiation and has been implicated in lipid and glucose metabolism (48). A reduction in PPARG activity has been shown to be associated with insulin sensitivity (49). Several studies have suggested that PPARG may play a role in the development of insulin resistance in response to a Western-type high-fat diet (50, 51). Therefore, these findings provide support for the observed interaction by which rs3014866 CC carriers have a lower HOMA-IR with lower SFA:carbohydrate ratio intake.
Some limitations of the current study should be mentioned. The CORDIOPREV, GOLDN, and BPRHS populations represent individuals of different ancestries, lifestyles, and clinical and baseline characteristics. For example, GOLDN participants are younger with lower BMI and a lower prevalence of T2D, which could explain the lack of a direct association between the S100A9 variant and T2D risk in this population. Dietary intakes also differ significantly in the 3 populations, and it was not possible to find a cutoff for SFA or SFA:carbohydrate ratio intakes that was common to all subjects for the interaction with rs3014866 on insulin resistance. Moreover, different FFQs and methodologies were used to evaluate dietary data. In addition, although the HOMA-IR is a validated index for estimating insulin resistance in human studies (52) and was the only method available in the 3 populations, we recognize that the use of this single measure was a limitation. Despite these shortcomings, we showed consistent trends across the 3 populations in terms of the main genetic associations and gene-diet interactions. In addition, moderate sample sizes of the studied populations may have limited the statistical power. Nevertheless, to our knowledge, this is the first study to replicate the association between rs3014866 and T2D risk, and novel interactions between this SNP and dietary factors to modulate insulin resistance were identified.
In conclusion, the minor allele (T) of the S100A9 variant rs3014866 is associated with decreased T2D risk in 3 populations of different ancestries and diverse clinical and demographic characteristics. Note that individuals who lack the protective (T) allele (CC carriers) may benefit from low SFA:carbohydrate ratio intake to improve insulin sensitivity. Our study adds support to accumulating evidence that T2D arises from the complex interplay between genetic and environmental factors and, if replicated, may contribute to the development of effective individualized dietary recommendations to prevent T2D.
Acknowledgments
The authors’ responsibilities were as follows—RB-R: designed the study hypothesis, analyzed the data, and wrote the manuscript; RB-R and JMO: had primary responsibility for the final content of the manuscript; JD-L, PP-M, BH, JFA-D, FG-D, LDP, and KLT: contributed to the editing of the manuscript; Y-CL, C-QL, OR-Z, and CES: conducted the research; DKA, KLT, and JL-M: provided essential data; JD-L, JL-M, and JMO: designed the research and provided study oversight; and all authors: read and approved the final manuscript. None of the authors reported a conflict of interest related to the study.
Footnotes
Abbreviations used: BPRHS, Boston Puerto Rican Health Study; CORDIOPREV, Coronary Diet Intervention with Olive Oil and Cardiovascular Prevention; FFQ, food-frequency questionnaire; GOLDN, Genetics of Lipids Lowering Drugs and Diet Network; GWAS, genome-wide association study; PPARG, peroxisome proliferator-activated receptor γ SNP, single nucleotide polymorphism; S100A9, S100 calcium-binding protein A9; T2D, type 2 diabetes.
REFERENCES
- 1.World Health Organization. Global status report on noncommunicable diseases 2014. Geneva (Switzerland): WHO; 2014. [Google Scholar]
- 2.Guariguata L, Whiting DR, Hambleton I, Beagley J, Linnenkamp U, Shaw JE. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract 2014;103:137–49. [DOI] [PubMed] [Google Scholar]
- 3.American Diabetes Assocation. Diagnosis and classification of diabetes mellitus. Diabetes Care 2014;37(Suppl 1):S81–90. [DOI] [PubMed] [Google Scholar]
- 4.Bi Y, Wang T, Xu M, Xu Y, Li M, Lu J, Zhu X, Ning G. Advanced research on risk factors of type 2 diabetes. Diabetes Metab Res Rev 2012;28(Suppl 2):32–9. [DOI] [PubMed] [Google Scholar]
- 5.Marullo L, El-Sayed Moustafa JS, Prokopenko I. Insights into the genetic susceptibility to type 2 diabetes from genome-wide association studies of glycaemic traits. Curr Diab Rep 2014;14:551. [DOI] [PubMed] [Google Scholar]
- 6.Lim SY, Raftery MJ, Geczy CL. Oxidative modifications of DAMPs suppress inflammation: the case for S100A8 and S100A9. Antioxid Redox Signal 2011;15:2235–48. [DOI] [PubMed] [Google Scholar]
- 7.Catalán V, Gomez-Ambrosi J, Rodriguez A, Ramirez B, Rotellar F, Valenti V, Silva C, Gil MJ, Fernandez-Real JM, Salvador J, et al. Increased levels of calprotectin in obesity are related to macrophage content: impact on inflammation and effect of weight loss. Mol Med 2011;17:1157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortega FJ, Sabater M, Moreno-Navarrete JM, Pueyo N, Botas P, Delgado E, Ricart W, Fruhbeck G, Fernandez-Real JM. Serum and urinary concentrations of calprotectin as markers of insulin resistance and type 2 diabetes. Eur J Endocrinol 2012;167:569–78. [DOI] [PubMed] [Google Scholar]
- 9.Geczy CL, Chung YM, Hiroshima Y. Calgranulins may contribute vascular protection in atherogenesis. Circ J 2014;78:271–80. [DOI] [PubMed] [Google Scholar]
- 10.Ortega FJ, Mercader JM, Moreno-Navarrete JM, Sabater M, Pueyo N, Valdes S, Ruiz B, Luche E, Serino M, Naon D, et al. Targeting the association of calgranulin B (S100A9) with insulin resistance and type 2 diabetes. J Mol Med (Berl) 2013;91:523–34. Erratum in: J Mol Med (Berl) 2013;91:535 [DOI] [PubMed] [Google Scholar]
- 11.Delgado-Lista J, Perez-Martinez P, Garcia-Rios A, Alcala-Diaz JF, Perez-Caballero AI, Gomez-Delgado F, Fuentes F, Quintana-Navarro G, Lopez-Segura F, Ortiz-Morales AM, et al. CORonary Diet Intervention with Olive oil and cardiovascular PREVention study (the CORDIOPREV study): rationale, methods, and baseline characteristics: a clinical trial comparing the efficacy of a Mediterranean diet rich in olive oil versus a low-fat diet on cardiovascular disease in coronary patients. Am Heart J 2016;177:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Martinez P, Alcala-Diaz JF, Delgado-Lista J, Garcia-Rios A, Gomez-Delgado F, Marin-Hinojosa C, Rodriguez-Cantalejo F, Delgado-Casado N, Perez-Caballero AI, Fuentes-Jimenez FJ, et al. Metabolic phenotypes of obesity influence triglyceride and inflammation homoeostasis. Eur J Clin Invest 2014;44:1053–64. [DOI] [PubMed] [Google Scholar]
- 13.Portaluppi F, Smolensky MH, Touitou Y. Ethics and methods for biological rhythm research on animals and human beings. Chronobiol Int 2010;27:1911–29. [DOI] [PubMed] [Google Scholar]
- 14.Corella D, Arnett DK, Tsai MY, Kabagambe EK, Peacock JM, Hixson JE, Straka RJ, Province M, Lai CQ, Parnell LD, et al. The -256T>C polymorphism in the apolipoprotein A-II gene promoter is associated with body mass index and food intake in the genetics of lipid lowering drugs and diet network study. Clin Chem 2007;53:1144–52. [DOI] [PubMed] [Google Scholar]
- 15.Tucker KL, Mattei J, Noel SE, Collado BM, Mendez J, Nelson J, Griffith J, Ordovas JM, Falcon LM. The Boston Puerto Rican Health Study, a longitudinal cohort study on health disparities in Puerto Rican adults: challenges and opportunities. BMC Public Health 2010;10:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai CQ, Tucker KL, Choudhry S, Parnell LD, Mattei J, Garcia-Bailo B, Beckman K, Burchard EG, Ordovas JM. Population admixture associated with disease prevalence in the Boston Puerto Rican health study. Hum Genet 2009;125:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai MY, Hanson NQ, Straka RJ, Hoke TR, Ordovas JM, Peacock JM, Arends VL, Arnett DK. Effect of influenza vaccine on markers of inflammation and lipid profile. J Lab Clin Med 2005;145:323–7. [DOI] [PubMed] [Google Scholar]
- 18.American Diabetes Association. Standards of medical care in diabetes–2014. Diabetes Care 2014;37(Suppl 1):S14–80. [DOI] [PubMed] [Google Scholar]
- 19.Arem H, Moore SC, Patel A, Hartge P, Berrington de Gonzalez A, Visvanathan K, Campbell PT, Freedman M, Weiderpass E, Adami HO, et al. Leisure time physical activity and mortality: a detailed pooled analysis of the dose-response relationship. JAMA Intern Med 2015;175:959–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ainsworth BE, Haskell WL, Herrmann SD, Meckes N, Bassett DR Jr, Tudor-Locke C, Greer JL, Vezina J, Whitt-Glover MC, Leon AS. 2011 Compendium of Physical Activities: a second update of codes and MET values. Med Sci Sports Exerc 2011;43:1575–81. [DOI] [PubMed] [Google Scholar]
- 21.Smith CE, Arnett DK, Tsai MY, Lai CQ, Parnell LD, Shen J, Laclaustra M, Junyent M, Ordovas JM. Physical inactivity interacts with an endothelial lipase polymorphism to modulate high density lipoprotein cholesterol in the GOLDN study. Atherosclerosis 2009;206:500–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee IM, Paffenbarger RS Jr. Physical activity and stroke incidence: the Harvard Alumni Health Study. Stroke 1998;29:2049–54. [DOI] [PubMed] [Google Scholar]
- 23.Fernández-Ballart JD, Pinol JL, Zazpe I, Corella D, Carrasco P, Toledo E, Perez-Bauer M, Martinez-Gonzalez MA, Salas-Salvado J, Martin-Moreno JM. Relative validity of a semi-quantitative food-frequency questionnaire in an elderly Mediterranean population of Spain. Br J Nutr 2010;103:1808–16. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Rios A, Gomez-Delgado FJ, Garaulet M, Alcala-Diaz JF, Delgado-Lista FJ, Marin C, Rangel-Zuniga OA, Rodriguez-Cantalejo F, Gomez-Luna P, Ordovas JM, et al. Beneficial effect of CLOCK gene polymorphism rs1801260 in combination with low-fat diet on insulin metabolism in the patients with metabolic syndrome. Chronobiol Int 2014;31:401–8. [DOI] [PubMed] [Google Scholar]
- 25.Subar AF, Thompson FE, Kipnis V, Midthune D, Hurwitz P, McNutt S, McIntosh A, Rosenfeld S. Comparative validation of the Block, Willett, and National Cancer Institute food frequency questionnaires: the Eating at America’s Table Study. Am J Epidemiol 2001;154:1089–99. [DOI] [PubMed] [Google Scholar]
- 26.Thompson FE, Subar AF, Brown CC, Smith AF, Sharbaugh CO, Jobe JB, Mittl B, Gibson JT, Ziegler RG. Cognitive research enhances accuracy of food frequency questionnaire reports: results of an experimental validation study. J Am Diet Assoc 2002;102:212–25. [DOI] [PubMed] [Google Scholar]
- 27.National Technical Information Service. 1994 continuing survey of food intakes by individuals and the 1994 diet and health knowledge survey. Springfield (VA): US Department of Agriculture Agricultural Research Service; 1996. [Google Scholar]
- 28.Tucker KL, Bianchi LA, Maras J, Bermudez OI. Adaptation of a food frequency questionnaire to assess diets of Puerto Rican and non-Hispanic adults. Am J Epidemiol 1998;148:507–18. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Abecasis GR. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet 2006;S79:2290. [Google Scholar]
- 30.Aslibekyan S, Kabagambe EK, Irvin MR, Straka RJ, Borecki IB, Tiwari HK, Tsai MY, Hopkins PN, Shen J, Lai CQ, et al. A genome-wide association study of inflammatory biomarker changes in response to fenofibrate treatment in the Genetics of Lipid Lowering Drug and Diet Network. Pharmacogenet Genomics 2012;22:191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elbein SC, Hoffman MD, Teng K, Leppert MF, Hasstedt SJ. A genome-wide search for type 2 diabetes susceptibility genes in Utah Caucasians. Diabetes 1999;48:1175–82. [DOI] [PubMed] [Google Scholar]
- 32.Cotoi OS, Duner P, Ko N, Hedblad B, Nilsson J, Bjorkbacka H, Schiopu A. Plasma S100A8/A9 correlates with blood neutrophil counts, traditional risk factors, and cardiovascular disease in middle-aged healthy individuals. Arterioscler Thromb Vasc Biol 2014;34:202–10. [DOI] [PubMed] [Google Scholar]
- 33.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest 2006;116:1793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrali M, Signorini C, Ciccoli L, Bambagioni S, Rossi V, Pompella A, Comporti M. Protection of erythrocytes against oxidative damage and autologous immunoglobulin G (IgG) binding by iron chelator fluor-benzoil-pyridoxal hydrazone. Biochem Pharmacol 2000;59:1365–73. [DOI] [PubMed] [Google Scholar]
- 35.Nugent C, Prins JB, Whitehead JP, Wentworth JM, Chatterjee VK, O’Rahilly S. Arachidonic acid stimulates glucose uptake in 3T3-L1 adipocytes by increasing GLUT1 and GLUT4 levels at the plasma membrane. Evidence for involvement of lipoxygenase metabolites and peroxisome proliferator-activated receptor gamma. J Biol Chem 2001;276:9149–57. [DOI] [PubMed] [Google Scholar]
- 36.Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S, Bell JT, Yang TP, Meduri E, Barrett A, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet 2012;44:1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grayson BL, Wang L, Aune TM. Peripheral blood gene expression profiles in metabolic syndrome, coronary artery disease and type 2 diabetes. Genes Immun 2011;12:341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, Magi R, Strawbridge RJ, Rehnberg E, Gustafsson S, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 2012;44:991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Risérus U. Fatty acids and insulin sensitivity. Curr Opin Clin Nutr Metab Care 2008;11:100–5. [DOI] [PubMed] [Google Scholar]
- 40.Savage DB, Petersen KF, Shulman GI. Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol Rev 2007;87:507–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salas-Salvadó J, Martinez-Gonzalez MA, Bullo M, Ros E. The role of diet in the prevention of type 2 diabetes. Nutrition, metabolism, and cardiovascular diseases. Nutr Metab Cardiovasc Dis 2011;21(Suppl 2):B32–48. [DOI] [PubMed] [Google Scholar]
- 42.Mozaffarian D. Saturated fatty acids and type 2 diabetes: more evidence to re-invent dietary guidelines. Lancet Diabetes Endocrinol 2014;2:770–2. [DOI] [PubMed] [Google Scholar]
- 43.Mann JI, De Leeuw I, Hermansen K, Karamanos B, Karlstrom B, Katsilambros N, Riccardi G, Rivellese AA, Rizkalla S, Slama G, et al. Evidence-based nutritional approaches to the treatment and prevention of diabetes mellitus. Nutrition, metabolism, and cardiovascular diseases. Nutr Metab Cardiovasc Dis 2004;14:373–94. [DOI] [PubMed] [Google Scholar]
- 44.Deer J, Koska J, Ozias M, Reaven P. Dietary models of insulin resistance. Metabolism 2015;64:163–71. [DOI] [PubMed] [Google Scholar]
- 45.Pérez-Jiménez F, Lopez-Miranda J, Pinillos MD, Gomez P, Paz-Rojas E, Montilla P, Marin C, Velasco MJ, Blanco-Molina A, Jimenez Pereperez JA, et al. A Mediterranean and a high-carbohydrate diet improve glucose metabolism in healthy young persons. Diabetologia 2001;44:2038–43. [DOI] [PubMed] [Google Scholar]
- 46.Parnell LD, Blokker BA, Dashti HS, Nesbeth PD, Cooper BE, Ma Y, Lee YC, Hou R, Lai CQ, Richardson K, et al. CardioGxE, a catalog of gene-environment interactions for cardiometabolic traits. BioData Min 2014;7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanco-Rojo R, Alcala-Diaz JF, Wopereis S, Perez-Martinez P, Quintana-Navarro GM, Marin C, Ordovas JM, van Ommen B, Perez-Jimenez F, Delgado-Lista J, et al. The insulin resistance phenotype (muscle or liver) interacts with the type of diet to determine changes in disposition index after 2 years of intervention: the CORDIOPREV-DIAB randomised clinical trial. Diabetologia 2016;59:67–76. [DOI] [PubMed] [Google Scholar]
- 48.Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. J Med Chem 2000;43:527–50. [DOI] [PubMed] [Google Scholar]
- 49.Deeb SS, Fajas L, Nemoto M, Pihlajamaki J, Mykkanen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx JA. Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet 1998;20:284–7. [DOI] [PubMed] [Google Scholar]
- 50.Guerre-Millo M, Rouault C, Poulain P, Andre J, Poitout V, Peters JM, Gonzalez FJ, Fruchart JC, Reach G, Staels B. PPAR-alpha-null mice are protected from high-fat diet-induced insulin resistance. Diabetes 2001;50:2809–14. [DOI] [PubMed] [Google Scholar]
- 51.Robitaille J, Despres JP, Perusse L, Vohl MC. The PPAR-gamma P12A polymorphism modulates the relationship between dietary fat intake and components of the metabolic syndrome: results from the Quebec Family Study. Clin Genet 2003;63:109–16. [DOI] [PubMed] [Google Scholar]
- 52.Wallace TM, Levy JC, Matthews DR. Use and abuse of HOMA modeling. Diabetes Care 2004;27:1487–95. [DOI] [PubMed] [Google Scholar]