Abstract
Gamma-aminobutyric acid (GABA) is a widely conserved signaling molecule that in animals has been adapted as a neurotransmitter. GABA is synthesized from the amino acid glutamate by the action of glutamate decarboxylases (GADs). Two vertebrate genes, GAD1 and GAD2, encode distinct GAD proteins: GAD67 and GAD65, respectively. We have identified a third vertebrate GAD gene, GAD3. This gene is conserved in fishes as well as tetrapods. We analyzed protein sequence, gene structure, synteny, and phylogenetics to identify GAD3 as a homolog of GAD1 and GAD2. Interestingly, we found that GAD3 was lost in the hominid lineage. Because of the importance of GABA as a neurotransmitter, GAD3 may play important roles in vertebrate nervous systems.
Glutamate decarboxylases (GADs) are essential for the conversion of glutamate to γ-aminobutyric acid (GABA), the predominant inhibitory neurotransmitter in central nervous systems1. GADs are members of the Group II pyridoxal-5′-phosphate-dependent decarboxylases, which includes decarboxylases that operate on several different substrates2. Two GAD proteins found in vertebrate species, GAD67 and GAD65, are encoded by the paralogous genes GAD1 and GAD2, respectively3. While both GADs synthesize GABA and are co-expressed in most vertebrate GABAergic neurons, GAD1 synthesizes cytoplasmic GABA that is used for extrasynaptic and metabolic purposes and GAD2 regulates the vesicular pool for release4,5,6. Nevertheless, GAD1 and GAD2 sequences are highly similar to each other, and they share a common intron-exon organization, indicating a common origin7.
The evolutionary history of GAD genes is long and diverse. Genes with homology to GAD arose before the evolution of eukaryotes8. Genes encoding GAD are found, for example, in Escherichia coli9, Saccharomyces cerevisiae10, Drosophila melanogaster11, and Caenorhabditis elegans12. Furthermore, GABA signaling via membrane receptors elicits hyperpolarization in plants as well as mammals, suggesting conserved or convergent roles for the product of GAD enzymatic activity13. In most vertebrate species, only two GAD genes have been described. Another gene in the GAD family, GAD-like 1 (GADL1) resembles GAD1 and GAD2 in sequence, but is expressed in mouse skeletal muscles and kidney rather than in the brain14. There have also been some hints of greater diversity in vertebrate GAD genes.
In addition to the teleost gad1 and gad2 genes, a third gene, gad3, was found in brain cDNA of the abyssal grenadier (Coryphaenoides (Nematonurus) armatus), a benthic teleost fish15. A similar gad3 sequence was subsequently identified in the brain cDNA of goldfish (Carassius auratus)16. The sequences of goldfish and abyssal grenadier gad3 are clearly related to gad1a, gad1b, and gad2 sequences, but their evolutionary history remained unknown16. Furthermore, no gad3 genes were reported in any species other than grenadier and goldfish. This absence remained an anomaly, since the goldfish (order Cypriniformes), is very distantly related to the abyssal grenadier (order Gadiformes). Recent teleost phylogenies indicate that the Ostariophysians, of which Cypriniformes including goldfish are members, diverged from the Euteleosts, which include the abyssal grenadier, over 250 million years ago17. Thus, the conservation of a gad3 gene in these two divergent species suggested that gad3 was present in an early teleost ancestor. Because the teleost lineage is known to have experienced a whole-genome duplication early in its evolution, one reasonable possibility could therefore have been that gad3 was a teleost-specific gad paralog18,19,20,21.
Since the original identification of gad3 from teleost brain cDNA, many comparative genomic resources have become available. The sequencing of teleost and other vertebrate genomes has been accompanied by the development of databases and software for analyzing the conservation of genes. Sarcopterygii species with sequenced genomes include primitive fishes, e.g. elephant shark22 and coelacanth23, as well as tetrapods, e.g. chicken24, dog25, human26, Tasmanian devil27, Chinese softshelled turtle28, and Xenopus29. Actinopterygii species with sequenced genomes include the spotted gar30 as well as teleosts like fugu19, medaka31, tilapia32, and zebrafish33.
We used recently generated genomic resources to ask whether gad3 is present and expressed in species other than the goldfish and abyssal grenadier. Our results revealed a surprisingly broad conservation of GAD3 in mammals, reptiles, birds, and amphibians, as well as fishes.
Methods
Throughout this paper, we use standard gene nomenclature. For fishes, gene symbols are lowercase and italicized and protein symbols are capitalized. For other vertebrates, human conventions are used: gene symbols in all capitals and italicized, protein symbols in all capitals.
Vertebrate sequence data (Table 1) for GAD1, GAD2, and GAD3 homolog transcripts were downloaded from Ensembl genomes for the following species: chicken (Gallus gallus), coelacanth, fugu, human, medaka, spotted gar, Tasmanian devil, tilapia, Chinese softshell turtle, xenopus, zebrafish34. Transcript DNA sequences for elephant shark were retrieved from the elephant shark Ensembl server. Transcript cDNA sequences for grenadier were retrieved from NCBI15,35. The spotted gar genome shares extensive similarity with both tetrapod and teleost genomes, so we chose to focus on this species for sequence alignment, phylogenetics, and intron/exon structure comparisons30. Additionally, we obtained sequences for fruitfly (Drosophila melanogaster) GAD1: NM_079190, sea urchin (Strongylocentrotus purpuratus) GAD: XM_779763, amphioxus (Branchiostoma floridae) GAD: XP_002592141, and tunicate (Ciona intestinalis) GAD: ENSCINT00000004013.
Table 1. Vertebrate GAD1, GAD2, and GAD3 transcript sequence IDs.
| Species | Scientific Name | GAD1 | GAD2 | GAD3 |
|---|---|---|---|---|
| Chicken | Gallus gallus | ENSGALT00000043162 | ENSGALT00000012268 | |
| Burton’s mouthbrooder | Astatotilapia burtoni | Gad1a: XM_014332345 Gad1b: XM_014340384 | XM_005932121 | XM_005950266 |
| Coelacanth | Latimeria chalumnae | ENSLACT00000014577 | ENSLACT00000011268 | ENSLACT00000005682 |
| Dog | Canis familiaris | ENSCAFT00000049584 | ENSCAFT00000006929 | ENSCAFT00000000144 |
| Elephant shark | Callorhinchus millii | SINCAMT00000000719 | SINCAMT00000011054 | SINCAMT00000005039 |
| Fugu | Takifugu rubripes | Gad1a:ENSTRUT00000045798 Gad1b:ENSTRUT00000020549 | ENSTRUT00000024751 | ENSTRUT00000021119 |
| Abyssal grenadier | Coryphaenoides armatus | AF043268 | AF043267 | AF043269 |
| Human | Homo sapiens | ENST00000358196 | ENST00000376261 | ENST00000592477* |
| Medaka | Oryzias latipes | Gad1a:ENSORLT00000021605 Gad1b:ENSORLT00000011550 | ENSORLT00000016248 | |
| Spotted Gar | Lepisosteus oculatus | ENSLOCT00000009532 | ENSLOCT00000009370 | ENSLOCT00000015874 |
| Tasmanian Devil | Sarcophilus harrisii | ENSSHAT00000013524 | ENSSHAT00000015741 | ENSSHAT00000004379 |
| Nile tilapia | Oreochromis niloticus | Gad1a:ENSONIT00000011023 Gad1b:ENSONIT00000023558 | ENSONIT00000008095 | ENSONIT00000008040 |
| Chinese softshelled turtle | Pelodiscus sinensis | ENSPSIT00000002371 | ENSPSIT00000019780 | ENSPSIT00000019627 |
| Xenopus | Xenopus tropicalis | ENSXETT00000040862 | ENSXETT00000040531 | ENSXETT00000012900 |
| Zebrafish | Danio rerio | Gad1a:ENSDART00000140425 Gad1b:ENSDART00000003008 | ENSDART00000021609 | ENSDART00000109561 |
*pseudogene transcript sequence.
In addition to the species listed in Table 1, we identified several other tetrapod species with GAD3 genes. These included: Orangutan: ENSPPYG00000009199, Rhesus: ENSMMUG00000001554, Rabbit: ENSOCUG00000022124, Horse: ENSECAG00000009017, Platypus: ENSOANG00000002106, Lizard: ENSACAG00000008555.
Sequence Alignment
Both DNA and amino acid sequences were aligned using MAFFT v7.01736,37 (by translation alignment for CDS sequences); algorithm E-INS-I; scoring matrix: BLOSUM62; gap open penalty: 1.53; offset value: 0.
Model Testing
MEGA 6 software was used to compare 24 DNA evolution models for the aligned GAD CDS sequences38. A generalized time-reversible plus gamma (GTR + G + I) model had the lowest BIC score (Bayesian Information Criterion) and AICc value (Akaike Information Criterion, corrected), so it was used for subsequent phylogenetic analyses. In this model, non-uniformity of evolutionary rates among sites is modeled by estimating a discrete Gamma distribution (+G) of rates and by assuming that certain sites are evolutionarily invariable (+I).
Bayesian Phylogenetic Inference
MrBayes 3.2.639 was used to infer phylogenetic relationships between GAD homologs based on aligned nucleotide CDS sequences, and was accessed via the CIPRES web portal40. In MrBayes, the GTR + G + I model of evolution was used; with default settings except for the following specified parameters: nruns(number of runs) = 2; ngen(number of generations) = 1000000; samplefreq = 500; nchain(number of chains) = 8; temp(chain heating temperature) = 0.1; savebrlens = yes; burninfrac(fraction of initial generations discarded) = 0.25.
Diagnostics of the MCMC sampling were carried out using Tracer v1.6 (http://tree.bio.ed.ac.uk/software/tracer/). The effective sample size (ESS) for each parameter was >300 for each run, allowing adequate sampling of the Markov chain.
The tree file generated using MrBayes was visualized using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Synteny
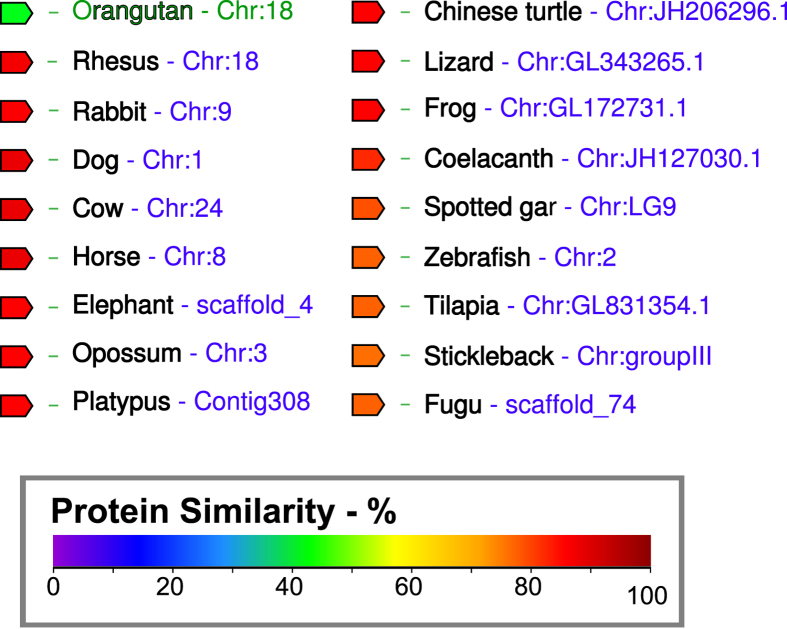
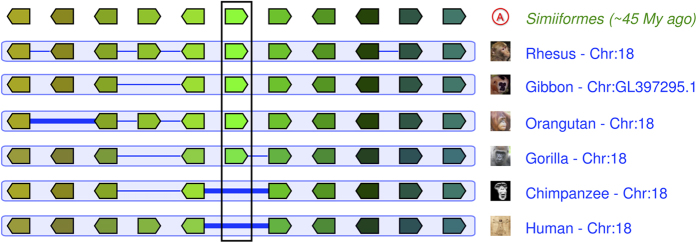
Gene synteny for GAD3 genes was compared to the syntenic region near GAD3 using Genomicus41. Orangutan was used as a reference species for cross-species synteny and protein similarity to highlight conservation of synteny and GAD3 protein sequence in vertebrates despite the absence of GAD3 in some hominids. For comparing primate synteny, simiiformes (last common ancestor of simians) was used as the reference taxon. Synteny data, protein similarity, and species images were downloaded from Genomicus.
Results
We found previously uncharacterized GAD3 genes in many vertebrate genomes, including diverse fishes and tetrapods. A teleost gad3 transcript was found in a transcriptome library generated from testis tissue from Astatotilapia burtoni: (>comp56037_c0_seq1_indA_testis). Zebrafish gad3 has been previously referred to with the identifier zgc:163121. Interestingly, GAD3 had already been annotated in the Xenopus tropicalis genome as GAD1.2. It appears, however, that it has not yet been studied in Xenopus.
Sequence Similarity
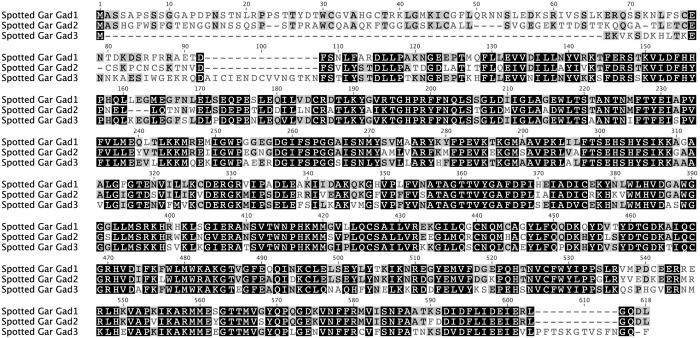
Gad3 predicted protein sequence from spotted gar (Lepisosteus oculatus) is more similar to Gad1 (Pairwise Identity: 60.5%) than to Gad2 (Pairwise Identity: 53.9%) (Fig. 1). Gad1 and Gad2 share 67.1% pairwise identity. The N-terminal domain, which is quite variable between Gad1 and Gad2, is truncated and highly divergent in Gad3. The N-terminal 92 amino acids (aa) of Gad1 align to the N-terminal 84 aa of Gad2. Gad3 has 42 aa aligning in this range, only 27 of which align to Gad1 and Gad2 sequence (with a 15 aa gap). These 27 aa of Gad3 have: 23.1% pairwise identity with Gad1, 11.5% pairwise identity with Gad2.
Figure 1. Alignment of predicted protein sequences translated from spotted gar (Lepisosteus oculatus) GAD genes.
Black: amino acids similar in all three sequences; Gray: similar to one corresponding residue; White: not similar to either corresponding residue. Sequence similarity was calculated using BLOSUM62 matrix with threshold = 1.
Phylogeny
Pyridoxal 5′-phosphate (PLP)-dependent decarboxylase genes include Glutamate Decarboxylase-Like 1 (GADL1), Cysteine Sulfinic Acid Decarboxylase (CSAD), and histidine decarboxylase (HDC), in addition to GADs. Therefore, we tested the phylogenetic relationship of GAD3 to other genes in this group, using HDC as an outgroup for GAD, GADL1, and CSAD genes42. A neighbor-joining tree of aligned predicted amino acid sequences from the spotted gar (Lepisoteus oculatus) place gad3 most closely related to the gad1/gad2 clade (Fig. 2).
Figure 2. GAD3 is closely related to GAD1 and GAD2, and more distantly related to other members of the PLP-dependent decarboxylase gene family.
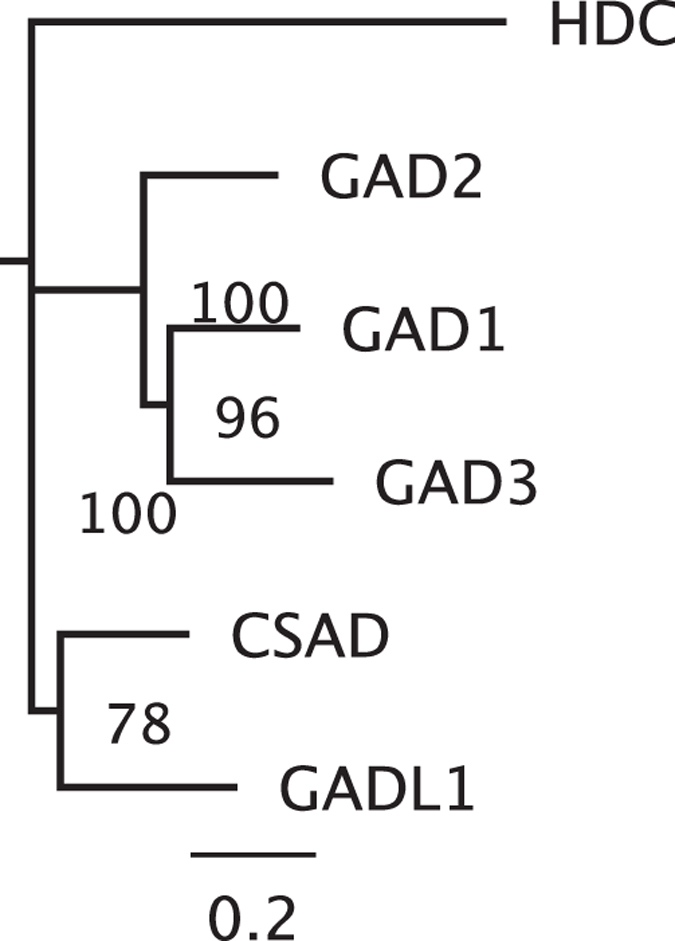
A phylogenetic tree of aligned PLP-dependent decarboxylase amino acid sequences from the spotted gar was generated using neighbor-joining and 2000 bootstrap iterations. Percent bootstrap support for nodes are shown. The scale bar (bottom) indicates substitutions per site.
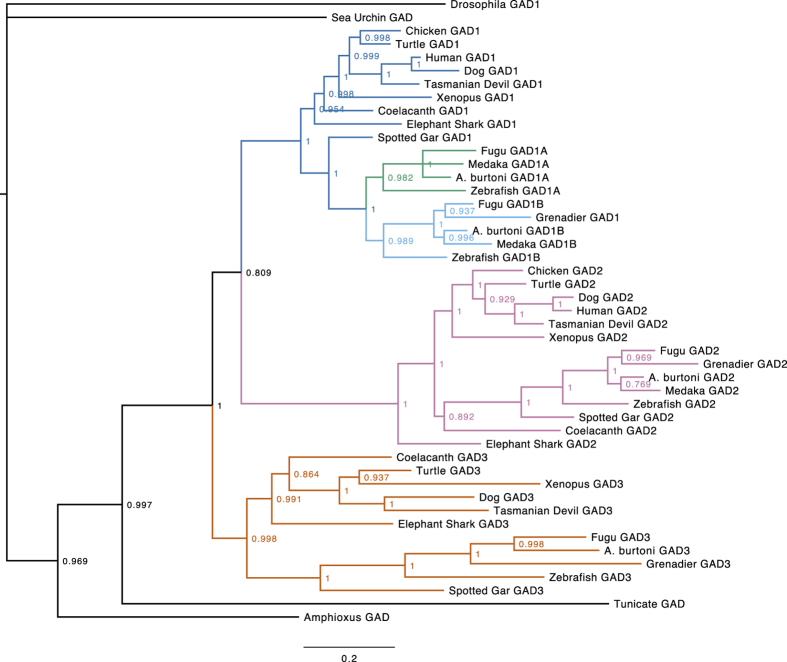
A phylogenetic tree of vertebrate GAD1, GAD2, and GAD3 nucleotide coding sequences was generated using MrBayes (Fig. 3). In insects, GAD1 is the single homolog of vertebrate GAD genes (insect GAD2 is homologous to vertebrate CSAD and GADL1). Therefore we chose Drosophila melanogaster GAD1 as the outgroup for the vertebrate and deuterostome GAD genes.
Figure 3. Phylogenetic tree of GAD1, GAD2, and GAD3 nucleotide sequences.
This consensus tree was generated using MrBayes with Drosophila melanogaster GAD1 as the outgroup. Nodes are labeled with posterior probabilities. Distinct gene lineages are indicated by colors. The scale bar (bottom) indicates substitutions per site.
Exon-intron Structure
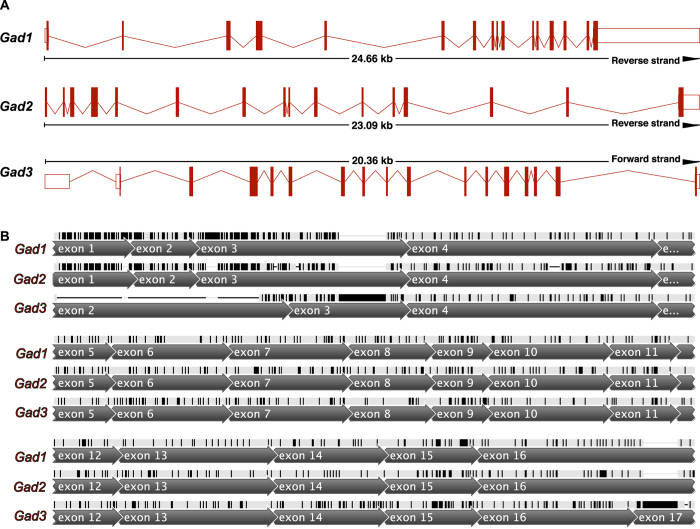
Spotted gar gad1 and gad2 each have 16 exons (Fig. 4). Spotted gar gad3 has 17 exons. While both gad1 and gad2 have coding sequence beginning in exon 1, gad3 coding sequence (CDS) begins in the second exon (exon 2). The predicted coding sequence of gad3 has a gap (does not align) with the 5′ CDS sequence found in gad1 and gad2 exon 1, exon 2, and part of exon 3. The 3′ portion of the gad3 CDS is included on exon 17, while gad1 and gad2 stop codons are found in exon 16.
Figure 4. Shared exon-intron structure of GAD genes from spotted gar (Lepisosteus oculatus).
(A) Maps of the three gad genes showing 16 exons in gad1 and gad2, and 17 exons in gad3. The direction of transcription is from left to right. Genomic distance spanned and strand on which the gene is located are indicated for each gene. (B) Aligned spotted gar gad1, gad2, and gad3 CDS regions annotated with position of exons. Bases colored black indicate disagreements with the consensus sequence of the three gad genes.
Aside from these differences, gad3 exon structure is largely similar to gad1 and gad2. All of the exon junctions from exon 3 to exon 16 are in identical locations for all three gad genes. In our alignment of the three gad genes, the only gaps introduced in gad3 are located in exon 2 and exon 17.
Synteny
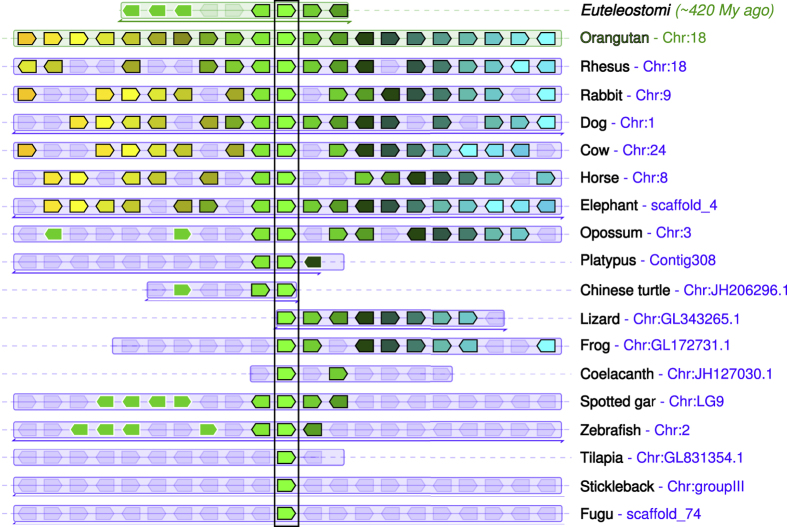
In spotted gar, gad1 and gad2 are located adjacent to the myosin genes myo3b and myo3a, respectively. Similarly, in humans GAD1 is located near MYO3B on chromosome 2, and GAD2 is located adjacent to MYO3A on chromosome 10. On the other hand, gad3 is not located near a myosin gene in the genome of spotted gar. The genes located adjacent to spotted gar gad3 are mc4r and cdh20. This syntenic block of genes is conserved across many vertebrate species, and represents the inferred ancestral state of the bony vertebrates (euteleostomi) (Fig. 5).
Figure 5. GAD3 genes are found in a conserved syntenic region in vertebrates.
Compared to orangutan GAD3 for reference, other species including representatives of mammals, other tetrapods, and Euteleostomi (bony vertebrates) in general have similar chromosomal positions of GAD3 genes. The central green pentagons (surrounded by a vertical rectangle) represent the GAD3 genes. For each species, GAD3 and 10 flanking genes on each side are represented by colored pentagons. The pentagons point in the direction of transcription, and each color identifies a set of orthologous genes. To the right of the figure, both species and chromosome are indicated for each GAD3 ortholog. Figure modified from Genomicus PhyloView output41.
GAD3 Conservation and Gene Loss
GAD3 predicted protein sequence is highly conserved in diverse vertebrate genomes (Fig. 6). Yet primates appear to have experienced varying degrees of gene loss at the GAD3 locus (Fig. 7). We identified a human transcript (ENST00000592477) with homology to GAD3 (Table 1), derived from a pseudogene located in the human genome in the conserved GAD3 syntenic position between MC4R and CDH20 (Fig. 7). Similarly, gorilla (Gorilla gorilla) GAD3 is annotated as a pseudogene in Ensembl (ENSGGOG00000027455). Although macaque (Macaca mulatta) and orangutan (Pongo pygmaeus) predicted GAD3 protein sequences share relatively high pairwise identity (84.5%), the GAD3 genes in these two species appear to have large insertions (or deletions) in their predicted coding sequence.
Figure 6. GAD3 protein sequence is highly conserved across Euteleostomi.
Predicted protein sequences of GAD3 orthologs were compared using Genomicus41. The degree of similarity to the reference sequence (orangutan, in green) is indicated by the color of the block, according to the scale shown at bottom.
Figure 7. GAD3 is lost in hominids. The central green pentagons (surrounded by a vertical rectangle) represent the GAD3 genes.
Human and chimpanzee genomes have no functional gene at the location corresponding to GAD3 in other primates. Humans have a pseudogene at this location. A thick blue line between two genes indicates a “gap”, i.e. a gene lost relative to the ancestral Simiiformes genome. A thin blue line between two genes indicates a “break” in the continuity of the alignment, i.e. a gene added or lost relative to the ancestral Simiiformes genome. Figure modified from Genomicus AlignView41, which relies on data and images from Ensembl34.
Discussion
We identified a novel glutamate decarboxylase homolog, GAD3, found in many vertebrate genomes. We provide phylogenetic and intron/exon structural evidence that GAD3 is an ancient paralog of GAD1 and GAD2. The conserved chromosomal synteny of GAD3 in vertebrates supports an ancient origin for this gene. Surprisingly, GAD3 was lost in the hominid lineage. Taken together, the phylogenetic analyses, comparisons of gene structure, and synteny data suggest that GAD3 arose via gene duplication of a protovertebrate GAD homolog, likely before the duplication of another paralog which gave rise to GAD1 and GAD2.
GAD3 Evolution
Although our data do not rule out the possibility of a local duplication that gave rise to GAD3, they are consistent with an origin of GAD3 in an early vertebrate via whole-genome duplication. Whole-genome duplication is thought to have played a major role in early vertebrate evolution43,44,45. Following genome duplication, these duplicated gene pairs (ohnologs) experienced a range of outcomes including non-functionalization, sub-functionalization, and neo-functionalization46,47. Sub-functionalization may happen via protein changes48 or via regulatory element loss49 in which ancestral expression domains are differentially lost in different genes50. For example, recent evidence indicates that duplication of a corticotropin-releasing hormone (CRH) gene in an early vertebrate led to a broadly expressed CRH1 and a CRH2 with expression restricted to a single hindbrain nucleus51. Like our recent analyses of CRH genes, the discovery of GAD3 as a conserved vertebrate gene relied on freely available genomic resources, pointing to the likelihood that many gene families have unannotated homologs remaining to be found in sequenced genomes51,52.
GAD3 Function
GAD3 is phylogenetically closer to GAD1 and GAD2 than to GADL1, but nonetheless it is possible that its enzymatic functions differ from those of GAD1 and GAD2. The absence of much of the N-terminal region, which regulates intracellular localization of GAD1 and GAD2 proteins, suggests that GAD3 protein may have different localization5.
Little is known regarding the function of GADL1 enzyme, though polymorphisms are linked to differential response to lithium treatment for bipolar disorder53. Mammalian GADL1 does not appear to have glutamate decarboxylase activity, despite its name. Instead, it catalyzes the decarboxylation of aspartate, cysteine sulfinic acid, and cysteic acid to β-alanine, hypotaurine, and taurine, respectively14. Recently, GADL1 and CSAD were found to have preference for cysteine sulfinic acid as a substrate42. Future studies of GAD3 biochemical substrates will be necessary to address the possibility of substrates other than glutamate.
Intriguingly, zebrafish gad3 (referred to as zgc163121) mRNA expression was significantly downregulated by treatment with dexamethasone, a glucocorticoid agonist, in 25hpf larval zebrafish, as measured by microarray and qPCR54. In the deep-sea fish in which gad3 was first described, the armed grenadier, Coryphaenoides (Nematonurus) armatus, gad2 mRNA levels were found to be expressed in the brain in a sexually dimorphic manner, i.e. higher in male hypothalamus than in female, but no differences were found in gad3 levels35. In the goldfish, however, gad3 mRNA levels in the telencephalon were highest in sexually mature fish of both sexes during the breeding period55. Since the specific role of gad3 is unknown in any taxa, the full range of factors that regulate gad3 expression in the brain, and potentially elsewhere, awaits further investigation.
Loss in hominids
The loss of GAD3 in both chimpanzees and humans appears to have been preceded by changes to GAD3 sequences in other hominids. Predicted gorilla, orangutan, and gibbon GAD3 transcripts appear to be truncated relative to fish gad3 sequence, but it may be that not all the exons in these sequences are fully annotated in Ensembl. Glutamate metabolic pathways appear to have been under positive selection in hominids, as seen for example in the origin of glutamate dehydrogenase 2 (GLUD2) by retroposition of GLUD156.
Gene losses have played major roles in human evolution57. For example, loss of L-gulonolactone oxidase (GULO) makes humans and other Haplorhini susceptible to scurvy, a vitamin C deficiency. Despite conferring this disadvantage, GULO gene loss has occurred in multiple mammalian lineages, including guinea pigs58 and some bats59.
Hominids are not the only lineage that has lost GAD3. Rodent genomes, including mice, rats, and squirrels also appear to be missing GAD3 homologs. The absence of GAD3 in both humans and mice likely explains why this gene was not discovered sooner, since many investigators choose to focus on these two species.
Additional Information
How to cite this article: Grone, B. P. and Maruska, K. P. Three Distinct Glutamate Decarboxylase Genes in Vertebrates. Sci. Rep. 6, 30507; doi: 10.1038/srep30507 (2016).
Acknowledgments
K.P.M. was supported by startup funds from the College of Science and Department of Biological Sciences at Louisiana State University, a Ralph E. Powe Faculty Enhancement Award from Oak Ridge Associated Universities, and a Louisiana Board of Regents Research Competitiveness Subprogram Grant.
Footnotes
Author Contributions Both authors had full access to all of the data in this study and take responsibility for its collection and analysis. Study concept and design: B.P.G. and K.P.M. Acquisition of data: B.P.G. Analysis and interpretation of data: B.P.G. and K.P.M. Drafting of the manuscript: B.P.G. Critical revision of the manuscript for important intellectual content: K.P.M. Obtained funding: K.P.M. Administrative, technical, and material support: K.P.M.
References
- Roberts E. Gamma-aminobutyric acid. Scholarpedia 2, doi: 10.4249/scholarpedia.3356 (2007). [DOI] [Google Scholar]
- Sandmeier E., Hale T. I. & Christen P. Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. European journal of biochemistry/FEBS 221 (1994). [DOI] [PubMed] [Google Scholar]
- Erlander M. G., Tillakaratne N. J., Feldblum S., Patel N. & Tobin A. J. Two genes encode distinct glutamate decarboxylases. Neuron 7 (1991). [DOI] [PubMed] [Google Scholar]
- Kaufman D. L., Houser C. R. & Tobin A. J. Two forms of the gamma-aminobutyric acid synthetic enzyme glutamate decarboxylase have distinct intraneuronal distributions and cofactor interactions. J Neurochem 56 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soghomonian J. J. & Martin D. L. Two isoforms of glutamate decarboxylase: why? Trends Pharmacol Sci 19, 500–505 (1998). [DOI] [PubMed] [Google Scholar]
- Tian N. et al. The role of the synthetic enzyme GAD65 in the control of neuronal gamma-aminobutyric acid release. Proc Natl Acad Sci USA 96 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu D. F. & Tobin A. J. The exon-intron organization of the genes (GAD1 and GAD2) encoding two human glutamate decarboxylases (GAD67 and GAD65) suggests that they derive from a common ancestral GAD. Genomics 21, doi: 10.1006/geno.1994.1246 (1994). [DOI] [PubMed] [Google Scholar]
- Jackson F. R. Prokaryotic and eukaryotic pyridoxal-dependent decarboxylases are homologous. Journal of molecular evolution 31 (1990). [DOI] [PubMed] [Google Scholar]
- Smith D. K., Kassam T., Singh B. & Elliott J. F. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J Bacteriol 174 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman S. T., Fang T. K., Rovinsky S. A., Turano F. J. & Moye-Rowley W. S. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J Biol Chem 276, doi: 10.1074/jbc.M007103200 (2001). [DOI] [PubMed] [Google Scholar]
- Jackson F. R., Newby L. M. & Kulkarni S. J. Drosophila GABAergic systems: sequence and expression of glutamic acid decarboxylase. Journal of Neurochemistry 54 (1990). [DOI] [PubMed] [Google Scholar]
- Jin Y., Jorgensen E., Hartwieg E. & Horvitz H. R. The Caenorhabditis elegans gene unc-25 encodes glutamic acid decarboxylase and is required for synaptic transmission but not synaptic development. The Journal of neuroscience: the official journal of the Society for Neuroscience 19 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh S. A. et al. GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nat Commun 6, doi: 10.1038/ncomms8879 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. et al. Role of glutamate decarboxylase-like protein 1 (GADL1) in taurine biosynthesis. The Journal of biological chemistry 287, doi: 10.1074/jbc.M112.393728 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma P. T. et al. Multiplicity of glutamic acid decarboxylases (GAD) in vertebrates: molecular phylogeny and evidence for a new GAD paralog. Molecular biology and evolution 16 (1999). [DOI] [PubMed] [Google Scholar]
- Lariviere K. et al. GAD(65) and GAD(67) isoforms of the glutamic acid decarboxylase gene originated before the divergence of cartilaginous fishes. Molecular biology and evolution 19 (2002). [DOI] [PubMed] [Google Scholar]
- Near T. J. et al. Resolution of ray-finned fish phylogeny and timing of diversification. P Natl Acad Sci USA 109, doi: 10.1073/pnas.1206625109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A., Catchen J., Ferrara A., Fontenot Q. & Postlethwait J. H. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics 188, doi: 10.1534/genetics.111.127324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels A. et al. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol Biol Evol 21, doi: 10.1093/molbev/msh114 (2004). [DOI] [PubMed] [Google Scholar]
- Hoegg S., Brinkmann H., Taylor J. S. & Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol 59, doi: 10.1007/s00239-004-2613-z (2004). [DOI] [PubMed] [Google Scholar]
- Jaillon O. et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, doi: nature03025 (2004). [DOI] [PubMed] [Google Scholar]
- Venkatesh B. et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature 505, doi: 10.1038/nature12826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya C. T. et al. The African coelacanth genome provides insights into tetrapod evolution. Nature 496, doi: 10.1038/nature12027 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432, doi: 10.1038/nature03154 (2004). [DOI] [PubMed]
- Kirkness E. F. et al. The dog genome: survey sequencing and comparative analysis. Science 301, doi: 10.1126/science.1086432 (2003). [DOI] [PubMed] [Google Scholar]
- Venter J. C. et al. The sequence of the human genome. Science 291, doi: 10.1126/science.1058040 (2001). [DOI] [Google Scholar]
- Murchison E. P. et al. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell 148, doi: 10.1016/j.cell.2011.11.065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nature genetics 45, doi: 10.1038/ng.2615 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U. et al. The genome of the Western clawed frog Xenopus tropicalis. Science 328, doi: 10.1126/science.1183670 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I. et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet 48, doi: 10.1038/ng.3526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M. et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 447, doi: 10.1038/nature05846 (2007). [DOI] [PubMed] [Google Scholar]
- Brawand D. et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513, doi: 10.1038/nature13726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K. et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, doi: 10.1038/nature12111 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P. et al. Ensembl 2014. Nucleic acids research 42, doi: 10.1093/nar/gkt1196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau V. L., Bosma P. T., Collins M., Priede I. G. & Docherty K. Sexually dimorphic expression of glutamate decarboxylase mRNA in the hypothalamus of the deep sea armed grenadier, Coryphaenoides (Nematonurus) armatus. Brain, behavior and evolution 56, doi: 47210 (2000). [DOI] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H. & Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33, doi: 10.1093/nar/gki198 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K. & Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30, doi: 10.1093/molbev/mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61, doi: 10.1093/sysbio/sys029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W. & Schwartz T. In Proceedings of the Gateway Computing Environments Workshop (GCE) (2010).
- Louis A., Muffato M. & Roest Crollius H. Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic acids research 41, doi: 10.1093/nar/gks1156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge I. et al. Mammalian CSAD and GADL1 have distinct biochemical properties and patterns of brain expression. Neurochem Int 90, doi: 10.1016/j.neuint.2015.08.013 (2015). [DOI] [PubMed] [Google Scholar]
- Abi-Rached L., Gilles A., Shiina T., Pontarotti P. & Inoko H. Evidence of en bloc duplication in vertebrate genomes. Nature genetics 31, doi: 10.1038/ng855 (2002). [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. (Springer-Verlag, 1970). [Google Scholar]
- Dehal P. & Boore J. L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS biology 3, doi: 10.1371/journal.pbio.0030314 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet F. G. et al. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Molecular biology and evolution 23, doi: 10.1093/molbev/msl049 (2006). [DOI] [PubMed] [Google Scholar]
- Kassahn K. S., Dang V. T., Wilkins S. J., Perkins A. C. & Ragan M. A. Evolution of gene function and regulatory control after whole-genome duplication: comparative analyses in vertebrates. Genome research 19, doi: 10.1101/gr.086827.108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. L. The evolution of functionally novel proteins after gene duplication. Proceedings. Biological sciences / The Royal Society 256, doi: 10.1098/rspb.1994.0058 (1994). [DOI] [PubMed] [Google Scholar]
- Force A. et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. & Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics 154 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grone B. P. & Maruska K. P. A second corticotropin-releasing hormone gene (CRH2) is conserved across vertebrate classes and expressed in the hindbrain of a basal Neopterygian fish, the spotted gar (Lepisosteus oculatus). The Journal of comparative neurology 523, 1125–1143, doi: 10.1002/cne.23729 (2015). [DOI] [PubMed] [Google Scholar]
- Grone B. P. & Maruska K. P. Divergent evolution of two corticotropin-releasing hormone (CRH) genes in teleost fishes. Frontiers in Neuroscience 9, doi: 10.3389/fnins.2015.00365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H. et al. Variant GADL1 and response to lithium therapy in bipolar I disorder. The New England journal of medicine 370, 119–128, doi: 10.1056/NEJMoa1212444 (2014). [DOI] [PubMed] [Google Scholar]
- Chatzopoulou A. Unraveling the glucocorticoid receptor pathway in zebrafish PhD thesis, Leiden University, (2012). [Google Scholar]
- Lariviere K., Samia M., Lister A., Van Der Kraak G. & Trudeau V. L. Sex steroid regulation of brain glutamic acid decarboxylase (GAD) mRNA is season-dependent and sexually dimorphic in the goldfish Carassius auratus. Brain Res Mol Brain Res 141, 1–9, doi: 10.1016/j.molbrainres.2005.06.005 (2005). [DOI] [PubMed] [Google Scholar]
- Burki F. & Kaessmann H. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nature Genetics 36, 1061–1063, doi: 10.1038/ng1431 (2004). [DOI] [PubMed] [Google Scholar]
- Zhu J. et al. Comparative genomics search for losses of long-established genes on the human lineage. PLoS computational biology 3, e247, doi: 10.1371/journal.pcbi.0030247 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikimi M., Kawai T. & Yagi K. Guinea pigs possess a highly mutated gene for L-gulono-gamma-lactone oxidase, the key enzyme for L-ascorbic acid biosynthesis missing in this species. The Journal of biological chemistry 267, 21967–21972 (1992). [PubMed] [Google Scholar]
- Cui J., Pan Y. H., Zhang Y., Jones G. & Zhang S. Progressive pseudogenization: vitamin C synthesis and its loss in bats. Molecular biology and evolution 28, 1025–1031, doi: 10.1093/molbev/msq286 (2011). [DOI] [PubMed] [Google Scholar]