Abstract
For decades, the fruit fly, Drosophila melanogaster, has been among the premiere genetic model systems for probing fundamental neurobiology, including elucidation of mechanisms responsible for human neurologic disorders. Flies continue to offer virtually unparalleled versatility and speed for genetic manipulation, strong genomic conservation, and a nervous system that recapitulates a range of cellular and network properties relevant to human disease. I focus here on four critical challenges emerging from recent advances in our understanding of the genomic basis of human neurologic disorders where innovative experimental strategies are urgently needed: (1) pinpointing causal genes from associated genomic loci; (2) confirming the functional impact of allelic variants; (3) elucidating nervous system roles for novel or poorly studied genes; and (4) probing network interactions within implicated regulatory pathways. Drosophila genetic approaches are ideally suited to address each of these potential translational roadblocks, and will therefore contribute to mechanistic insights and potential breakthrough therapies for complex genetic disorders in the coming years. Strategic collaboration between neurologists, human geneticists, and the Drosophila research community holds great promise to accelerate progress in the post-genomic era.
Keywords: Drosophila melanogaster, Animal model, Genomics, Genome-wide association study, Exome sequencing, Epistasis, Neurology, Complex genetics, Mendelian disease
While experimentation in the fruit fly Drosophila melanogaster has contributed to many profound insights on heritability and neuroscience over the last hundred years (Bellen et al., 2010), that flies could serve as useful genetic models for human disease is a rather new concept. Based on PubMed search criteria, a 1971 study in Acta Neuropathol was among the first to highlight the potential for direct “modeling” of human nervous system injury in flies, based on observations of age-related changes in the fly brain similar to established human neuropathologic findings (Herman et al., 1971). It is striking that from this earliest conception of Drosophila disease models, applications were specifically imagined in experimental neurology. Indeed, numerous subsequent reports heralded fly models for Huntington’s disease (Jackson et al., 1998), Spinocerebellar Ataxia (Fernandez-Funez et al., 2000), Alzheimer’s disease (Finelli et al., 2004;Wittmann et al., 2001), and Parkinson’s disease (Feany and Bender, 2000), among other applications (Shulman et al., 2003). This rapid progress was enabled by the relative ease of Drosophila transgenesis, the availability of versatile, targeted expression systems (Brand and Perrimon, 1993), and contemporaneous discoveries of human genes responsible for autosomal dominant, familial forms of neurodegeneration with toxic gain-of-function mechanisms. The resulting fly models have contributed enormously to our understanding of neurologic disorders and continue to spur mechanistic insights, as reviewed previously (Bellen et al., 2010; Jaiswal et al., 2012; Lessing and Bonini, 2009; Shulman et al., 2003) and discussed elsewhere in this special issue.
In recent years, however, powerful methods for gene manipulation have become available in mammalian models, including conditional knockout strategies, optogenetics, and genome-editing technology. Further, advances in induced-pluripotent stem (iPS) cell methods now permit modeling of disease biology directly in human patient-derived neurons. Therefore, for many applications in experimental neurology, flies no longer offer all the unique advantages they once did. Importantly, there has also been a paradigm shift from a simple to amore complex genetic framework for understanding common neurologic conditions. In contrast to Mendelian diseases characterized by single-gene etiologies, complex genetic disorders are defined by substantial heterogeneity and polygenicity. Although the precise genomic architectures remain to be fully elucidated, we now appreciate that most common neurologic diseases (e.g., migraine, stroke, epilepsies, multiple sclerosis, and neurodegenerative conditions) are likely influenced by a combination of many common and rare genomic variants with a range of effect sizes. Based on the current rapid rate of progress, we are beginning to have a glimpse of the “post-genomic era,” when the majority of genes or genomic loci responsible for most neurologic conditions are known. While this is an exciting prospect, it also presents a number of unprecedented challenges (Chakravarti et al., 2013) and is creating an urgent need for new experimental models and approaches. Thus, in this altered landscape, what will be the future role of Drosophila in experimental neurology? The primary goal of this review is to address this question, and I will argue that Drosophila is ideally suited to tackle many of the key emerging obstacles. I organize my discussion around four major problems arising from current human genomic studies, drawing on recent examples to illustrate how flies can offer potential solutions. Overcoming each of these roadblocks will be essential for moving from genomic discoveries to clinical applications. At the conclusion, I propose how multi-disciplinary teams including many in the Drosophila research community will ensure sustained momentum for effective translational research in neurogenomics.
1. From susceptibility locus to causal gene
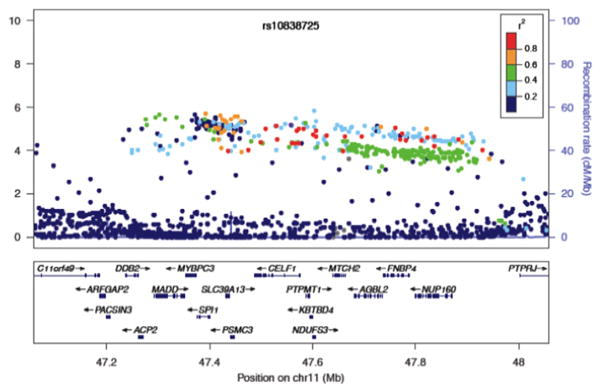
Over the last decade, genome-wide association studies (GWAS) have identified thousands of genomic loci (http://www.genome.gov/gwastudies/) that contribute to common and complex human genetic traits (Welter et al., 2014), including many neurologic and neuropsychiatric disorders. This successful strategy has begun to reveal genetic determinants for many conditions that long were relatively resistant to genetic dissection, including ischemic stroke (Kilarski et al., 2014), migraine (Anttila et al., 2013), Alzheimer’s disease (Lambert et al., 2013), Parkinson’s disease (Nalls et al., 2014), multiple sclerosis (International Multiple Sclerosis Genetics Consortium IMSGC et al., 2013), and schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), among others. While the odds ratios for many such loci are modest, they are estimated to have broad population impact on disease risk since most are defined by common sites of genomic variation [single nucleotide polymorphisms (SNPs) with minor allele frequencies of >1%]. Most signals discovered by GWAS do not unambiguously define the responsible causal genes but rather implicate a broad genomic region tagged by associated SNPs (Fig. 1). Such regions usually contain a number of candidate genes. Thus, experimental confirmation is critical to move beyond an “educated guess” to a validated target for further mechanistic investigation.
Fig. 1.
Multiple gene candidates implicated at an Alzheimer’s disease susceptibility locus. Regional association plot is shown from the GWAS discovery phase, with −log(P-value) for each SNP plotted on the left (Lambert et al., 2013). Following joining with replication phase data (not shown), the rs10838725 SNP exhibited genome-wide significant evidence of association with disease susceptibility; however, numerous genes fall under the association peak, based on regional linkage-disequilibrium (measured in r2). Independently, functional screening in Drosophila highlighted CELF1 as a potential causal gene for this locus (Shulman et al., 2014). Figure adapted by permission from Macmillan Publishers Ltd: Lambert et al. Nature Genetics 45(12):1452–1458 (2013), copyright 2013.
Based on several studies, flies provide an outstanding platform for functional validation of genes from GWAS, and this approach has already shown success for neurologic (Chapuis et al., 2013;Macleod et al., 2013; Shulman et al., 2011, 2014) as well as non-neurologic disorders (Hoed et al., 2013; van der Harst et al., 2012). Given the strong conservation between the human and Drosophila genomes (Fortini et al., 2000; Rubin et al., 2000), the majority of candidate genes at associated loci can usually be tested in flies. In fact, evolutionary conserved genes appear more likely to be implicated in disease (Hu et al., 2011). Further, resources are immediately available to facilitate manipulation of virtually all such candidates (Matthews et al., 2005), including genome-wide RNA-interference (RNAi) transgenic stocks (Dietzl et al., 2007; Ni et al., 2011), classical mutant alleles including transposable element insertions from the gene disruption project (Bellen et al., 2011), and strains that facilitate gene overexpression in many cases (Bischof et al., 2013; Jenett et al., 2012). Coupled with the many established transgenic neurologic disease models (Jaiswal et al., 2012; Shulman et al., 2003), the array of available Drosophila genetic reagents is a potent combination for medium- to high-throughput functional validation of gene candidates nominated by GWAS. The underlying hypothesis for this strategy is that many susceptibility alleles identified by GWAS modulate mechanisms of pathogenesis recapitulated in fly models. In our recent study (Shulman et al., 2014), we examined whether genetic manipulation of fly homologs of candidate genes at susceptibility loci from Alzheimer’s disease GWAS (Hollingworth et al., 2011; Lambert et al., 2013; Naj et al., 2011; Seshadri et al., 2010) interact with the neurotoxicity of human Tau protein, responsible for the characteristic neurofibrillary tangle pathology. RNAi-mediated knockdown of fly homologs of CD2AP, FERMT2, and CELF1 each enhanced retinal degeneration in transgenic models expressing human Tau, and in the case of FERMT2 and CELF1, overexpression reciprocally suppressed Tau toxicity. At the CELF1 locus, for example (Fig. 1), 8 out of 10 gene candidates within the implicated genomic region were conserved, and only manipulation of aret, the fly ortholog of CELF1, demonstrated modifier activity. Besides highlighting the most likely causal gene for GWAS signals, these results also provide important clues to potential mechanisms, suggesting that the implicated susceptibility loci may impact Tau-mediated neurodegeneration. Using similar experimental strategies, two other candidate-based studies recently provided support for functional validation of BIN1 in Alzheimer’s disease (Chapuis et al., 2013) and the RAB7L1 gene at the PARK16 locus in Parkinson’s disease (Macleod et al., 2013) (discussed further below).
Despite the successes, there are some notable limitations to the generalizability of the described strategy. Only about 60–70% of candidate human disease genes are conserved, based on cross-species comparisons (Fortini et al., 2000;Hu et al., 2011). At some genomic loci implicated in GWAS, there are no obvious gene candidates to test at all. In other cases, there can potentially be a very large number of candidates, especially if one accounts for the possibility of regulatory variants with long-range impact on gene expression. In addition, the described strategy relies on available fly disease models that can be deployed for modifier screening. Many but certainly not all neurologic disorders have useful transgenic models, and even where such models are available, they only permit probing the potential impact of candidate susceptibility genes on selected aspects of disease biology. Nevertheless, fly genetics shows great promise for accelerating the fine mapping of many susceptibility loci from GWAS, pinpointing likely causal genes and linking them to disease-relevant mechanisms.
2. Confirming the functional impact of genetic variants
Next-generation sequencing technology has revolutionized the discovery of genes responsible for familial neurologic disorders with Mendelian inheritance (Bamshad et al., 2011) and is beginning to successfully define less common and rare variants that contribute to many neurologic diseases with complex genetic inheritance (Pittman and Hardy, 2013). In contrast to GWAS-defined susceptibility loci, sequencing-based approaches are capable of more precisely defining the most likely causal genes and variants when performed in sufficiently large family pedigrees or population samples (Goldstein et al., 2013; MacArthur et al., 2014). The discovery pipeline for sequencing studies incorporates analytic filters to distinguish variants with anticipated deleterious consequences from those that are benign. While some variants are obviously damaging (nonsense mutations, splicing mutations, or insertions/deletions that cause frameshifts), others cause non-synonymous amino acid changes of uncertain functional significance. Numerous bioinformatic algorithms (Goldstein et al., 2013; Liu et al., 2013) facilitate prediction of functional consequences for such variants. However, direct experimentation is critical for confirming the pathogenicity of identified mutations, understanding how such changes disrupt gene functions, and defining the broader role of the encoded protein in the nervous system context. For conserved genes, flies remain an outstanding animal model to explore the functions of genetic variants.
One recent study that illustrates the power of this approach began with an unusual, three-generation family pedigree with an autosomal dominant disorder consisting of variable limb weakness and electrophysiologic evidence of presynaptic neuromuscular junction failure (Herrmann et al., 2014). Whole-exome sequencing of three individuals followed by stringent analytic filters and segregation analysis in additional family members narrowed an initially unwieldy variant list to a missense mutation in the synaptotagmin II gene (SYT2), encoding a key mediator of synaptic vesicle exocytosis. The candidate variant fell within a conserved calcium-binding domain and was predicted to be deleterious to protein function using bioinformatics. To confirm this finding and reveal the potential mechanisms, the corresponding mutation was introduced in the single Drosophila synaptotagmin gene, DSyt1, and expression of the mutant transgene was directed throughout the nervous system. Interestingly, whereas mutant DSyt1 was unable to rescue DSyt1−/− null animals, consistent with reduced function, expression of the mutant protein in a wild-type genetic background also disrupted synaptic transmission. Overall, these data suggest a dominant-negative mechanism for this human disorder and demonstrate how flies can be leveraged to rapidly dissect the functional consequences of missense variants in conserved genes. Indeed, understanding the mechanism of disease-associated mutations is often of critical importance to progress from a disease gene to potential therapies. For example, potential loss-of-function changes that reduce risk of disease (i.e., protective variants) may identify excellent candidates for potential drug targets.
With whole-exome sequencing becoming an increasingly available tool, both in the research (Bamshad et al., 2011) and clinical (Yaping Yang et al., 2013) settings, we can expect exponential growth in the pace of variant discovery underlying diverse neurologic disorders. In fact, large National Institutes of Health (NIH)-funded efforts are now under way with the goal of “solving” as many Mendelian disorders as possible (Bamshad et al., 2012). Drosophila is a powerful platform for accelerating the functional follow-up of promising variants emerging from such studies. As mentioned, genome-wide RNAi transgenic stocks (Dietzl et al., 2007; Ni et al., 2011) and large, publicly available collections of mapped transposon insertion alleles (Bellen et al., 2011) enable rapid genetic analysis of nearly all Drosophila genes. Other resources and technologies, including bacterial artificial chromosome libraries covering the fly genome (Venken et al., 2009) and the more recently developed CRISPR/Cas9 system (Bassett et al., 2013; Gratz et al., 2013; Kondo and Ueda, 2013; Yu et al., 2013), offer limitless flexibility for genomic manipulation in flies (Venken et al., 2011). One generalizable strategy for functional validation of a novel variant linked to human disease begins with characterization of the loss-of-function phenotype for the conserved gene homolog in flies. Importantly, any robust phenotype (e.g., lethality) can potentially serve as a substrate for variant functional analysis. Next, rescue experiments can be implemented comparing the activities of the wild-type gene or versions harboring disease-implicated variants. Such experiments can also be conducted using human cDNAs to demonstrate cross-species functional substitution. Where sufficient sequence conservation allows, corresponding mutations can additionally be engineered into the context of a fly cDNAor genomic rescue construct (or endogenous genomic locus using CRISPR). This cross-species strategy permits efficient characterization of variants of unknown significance, delineating those that alter protein function consistent with damaging, loss-of-function alleles versus other potential mechanisms (e.g., gain-of-function and/or dominant-negative). Such studies would also be a prelude to more detailed functional investigation in the relevant nervous system context.
3. Linking conserved genes to disease-relevant biology
Unbiased human genomic studies, whether GWAS or sequencing, often lead to candidate genes with little or no prior functional studies to support mechanistic hypotheses of disease pathogenesis. In the case of evolutionarily conserved genes, Drosophila remains a premiere system for efficient investigation of the consequences of gene loss-of function. In some cases, phenotypic consequences can reveal remarkable and unexpected parallels with the human disease. One illustrative example comes from a study of the Drosophila dBTBD9 gene (Freeman et al., 2012), homologous to a candidate gene from human GWAS in Restless Legs Syndrome(RLS) (Stefánsson et al., 2007). RLS is a common neurologic condition characterized by bothersome nighttime, sensory symptoms of the lower extremities that disrupt sleep, often forcing affected individuals out of bed to pace about for relief. Besides the strong association of an intronic BTBD9 SNP with RLS in humans, little was previously known to connect this gene to informative, disease biology. Based on public databases, BTBD9 showed fairly widespread tissue expression in mammals and sequence analysis revealed only a conserved, BTB protein interaction domain. To learn more, Freeman et al. (2012) generated a null allele of dBTBD9 in Drosophila through imprecise excision of an available transposon insertion stock, and characterized the resulting loss-of-function phenotype. Remarkably, dBTBD9−/− flies exhibited disrupted and fragmented sleep and a hyperlocomotor phenotype that resembles restlessness. In further studies, the authors found that loss-of-dBTBD9 function is associated with reduced brain dopamine levels and altered iron homeostasis, both of which have been implicated in the pathophysiology of human RLS (e.g., iron supplementation and dopamine agonists are both employed as effective therapies).
The BTBD9 story exemplifies the potential power of “reverse genetic analyses” in flies—the targeted characterization of loss-of-function phenotypes for genes initially implicated by other studies (i.e., in humans). The diversity of available reagents and methods for targeted gene knockdown makes this an attractive strategy for probing functions of genes implicated in disease. In at least some cases, such manipulations unexpectedly lead to phenotypes that recapitulate key features of disease, as in RLS. Moreover, the reduced genetic redundancy within Drosophila, that is the number of gene paralogs that can potentially functionally substitute for one another, often accelerates investigation of gene functions in this system. Therefore, a single genetic knockdown can frequently provide answers where more time-consuming double- or triple-knockdown would be required in vertebrate models. Finally, a variety of experimental strategies are available to facilitate tissue-specific and/or conditional knockdown. Indeed, many genes with adult nervous system functions are expected to have earlier, developmental roles or to mediate similar essential functions in non-neuronal tissues. In such cases, fly experimental approaches enable the genetic analysis of nervous system phenotypes, including within the adult, aging animal, which is the most relevant context for many adult-onset, progressive neurologic disorders. Available strategies include classical approaches such as the generation of genetic mosaic tissues, consisting of labeled homozygous mutant cell clones—including in the brain (Lee and Luo, 2001)—in an otherwise heterozygous animal. Besides RNAi transgenic lines, newer strategies targeting GFP-tagged transcripts (Neumüller et al., 2012; Pastor-Pareja and Xu, 2011) or proteins (Caussinus et al., 2011) allow precise spatial and temporal control as well as reversible knockdown. Importantly, the Drosophila gene disruption project (Jaiswal et al., in press) will generate GFP-tagged stocks for thousands of genes in the coming years, including prioritization of human disease gene homologs.
Despite the ease and potential power of reverse genetic approaches, Drosophila has historically excelled at “forward genetic” investigations (St Johnston, 2002; Venken and Bellen, 2014)—the identification of genes based on unbiased screening for selected mutant phenotypes. Indeed, genetic screens in flies have generated innumerable insights with relevance to diverse neurologic disorders (Bellen et al., 2010; Lessing and Bonini, 2009). In the post-genomic era, there is great potential for forward genetics to even more directly inform the discovery and functional elucidation of genes responsible for human disease. For example, based on the results of a large-scale, chemical mutagenesis screen, Yamamoto et al. (2014) recently reported on 165 essential genes with secondary requirements for neuronal development, function, and/or maintenance, many of which had never before been examined. Importantly, these genes were highly enriched for evolutionarily conserved loci, a third of which were previously linked to human disease in the Online Mendelian Inheritance in Man database (http://omim.org/). In order to determine if the screen may have also identified novel disease genes, the investigators teamed with the Baylor-Hopkins Centers for Mendelian Genomics, a large-scale effort to use next-generation sequencing technology to identify unsolved Mendelian genetic disorders (Bamshad et al., 2012). In an innovative cross-species analysis, the nervous system functional annotation of genes from the Drosophila screen were integrated with data from whole-exome sequencing in nearly 2,000 human subjects. Remarkably, the results of this joint analysis helped define likely causal genes/variants in several families (Yamamoto et al., 2014). For example, three cases of bull’s eye maculopathy, a late-onset, progressive retinal disorder, were associated with dominant mutations in the human CRX gene, based in part on the complementary discovery that mutations in the fly CRX homolog, ocelliless, led to age-dependent disruptions in the electroretinogram. In another compelling case, loss-of-function of a previously uncharacterized fly gene, l(1)G0222 (dANKLE2), disrupted development of thoracic sensory bristles, and mutations in the human homolog, ANKLE2, were associated with a recessively-inherited microcephaly syndrome. Subsequent detailed phenotypic characterization further demonstrated that loss-of-dANKLE2 function leads to diminished neuroblast numbers and concomitantly decreased larval brain size. In prior related work (Bayat et al., 2012; Neely et al., 2010), Drosophila forward genetic screening identified mutant phenotypes (straightjacket and Aats-met) prompting follow-up human genetic studies that highlighted conserved roles of the homologous loci (MARS2 and CACNA2D3) in neuronal maintenance and nociception, respectively.
These studies highlight the urgent need for comprehensive and systematic functional annotation of evolutionary conserved genes for possible roles in the nervous system, perhaps using a combination of reverse and forward genetic strategies in Drosophila. With such a valuable community resource, any gene implicated from a human genomic study could be instantly cross-referenced in a convenient online database, such as FlyBase (St Pierre et al., 2014). Such functional annotation might provide critical clues to support fine mapping of an associated locus from GWAS (From Susceptibility Locus to Causal Gene section) or to enhance confidence in a gene with potential loss-of-function variants from a family or population-based sequencing study (Confirming the Functional Impact of Genetic Variants section). But what phenotypes should such large-scale screening efforts focus on? One potential strategy is to generate flies with nervous system phenotypes that mimic human neurologic disorders. While reflexively satisfying, this approach is based on a potentially misguided assumption that most genes, including those highly conserved throughout evolution, will necessarily generate similar phenotypes when disrupted in flies as in humans. With this problem in mind, Marcotte and colleagues proposed the powerful concept of “phenologs,” or homologous phenotypes (McGary et al., 2010; Woods et al., 2013). Using bioinformatic approaches incorporating available model organism data, these investigators mapped human disease-associated orthologous gene sets with less obvious, homologous phenotypes in other species. For example, a statistically significant overlap was discovered between genes causing breast and ovarian cancer in humans and the high incidence of male progeny in the nematode, Caenorhabditis elegans, among many other similar and striking comparisons. While this study did not consider Drosophila, an extension of this successful approach has the potential to redefine the nature of fly disease models in the future. Rather than emphasizing the particular clinical and pathologic manifestations characteristic of human disease, this provocative work teaches us to embrace the species-specific consequences emerging from dysfunction in conserved molecular modules. Besides providing a potential path to new “disease models,” the phenolog conceptual framework provides an attractive approach to implicate new disease genes. Specifically, once the phenolog for a given human disease is defined, one can mount screens for mutants causing similar phenotypes, thereby identifying additional genes that may participate in common functional pathways.
4. From genes to pathways and networks
Many complex genetic disorders are believed to be polygenic, potentially arising from gene–gene (and gene–environment) interactions among a large number of susceptibility loci. This model contrasts with the single-gene etiologies underlying many simple Mendelian disorders. Grouping genes into functional pathways and networks, and probing the logic of their interactions, is therefore a major goal for understanding the pathophysiology of many common neurologic diseases with complex genetic architectures (Chakravarti et al., 2013). A major advantage of Drosophila is the capability to efficiently map epistatic, additive, synergistic, or antagonistic interactions by examining the phenotypic consequences of multiple gene manipulations in combination. Such pairwise genetic interaction experiments facilitate the reconstruction of linear models for serial gene action. In one notable example relevant to autosomal recessive juvenile-onset parkinsonism, studies of fly genes homologous to human PARK2/parkin and PARK6/pink1 demonstrated that the encoded proteins function in a sequential regulatory pathway impinging on mitochondrial dynamics (Clark et al., 2006; Deng et al., 2008; Greene et al., 2003; Park et al., 2006; Poole et al., 2008; Yufeng Yang et al., 2008). In more recent work also related to Parkinson’s disease, MacLeod et al. (2013) used flies to validate interactions between LRRK2 and the PARK16 locus, initially suggested from analyses of large-scale human genomic and transcriptomic data sets. In Drosophila, the expression of the Parkinson’s disease-associated LRRK2G2019S mutant protein led to reduced survival and loss of dopaminergic neurons, and both phenotypes were suppressed by co-expression of a constitutively active form of RAB7L1 protein, encoded by a candidate gene at the PARK16 locus identified in GWAS (Nalls et al., 2014). Loss-of-function mutations in lightoid, the fly homologue of RAB7L1, also caused reduced dopaminergic neuron counts. Interestingly, additional experiments further suggested that the LRRK2-RAB7L1 pathway impinges on another Parkinson’s disease gene product, VPS35 (Vilariño-Güell et al., 2011; Zimprich et al., 2011), which regulates the sorting of lysosomal substrates. In flies, MacLeod et al. found that knockdown or overexpression of the VPS35 homolog reciprocally enhanced or suppressed mutant LRRK2-induced dopaminergic neuron toxicity, respectively. These elegant experiments integrate human genomic data and studies in Drosophila to interlink susceptibility genes from GWAS (PARK16/RAB7L1) and sequencing studies (LRRK2, VPS35) into a unified functional pathway.
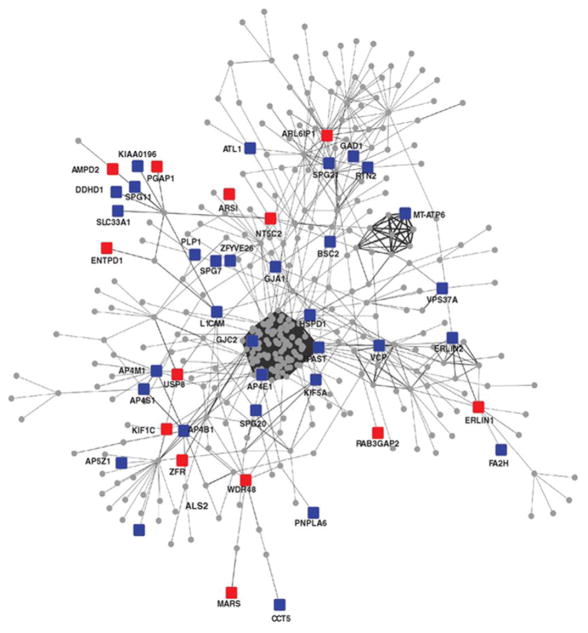
Despite the power of relatively simple pathway models for informing hypothesis-based investigation, such a conceptual framework may be ill-equipped to fully explain the pathophysiologic mechanisms underlying most complex genetic diseases, where widespread gene and allelic heterogeneity, incomplete penetrance, and polygenicity predominate (Chakravarti et al., 2013). The potential challenge is illustrated by a recent study focused on Hereditary Spastic Paraplegias (HSPs), a heterogeneous group of disorders characterized in part by corticospinal tract dysfunction and progressive lower limb weakness. Although mutations in greater than 20 distinct genes have been implicated to cause HSP, the majority of cases remain unexplained. Novarino et al. (2014) performed whole-exome sequencing on more than 90 individuals from 60 families with autosomal recessive HSP, identifying candidate mutations in 15 new genes. For confirmation, many of the genes were replicated in an independent HSP cohort, and functional validation of several other loci was pursued using zebrafish models. The investigators then developed a protein interaction network based on all previously established and newly identified HSP genes (Fig. 2). This “interactome” model identifies several highly conserved subnetworks in HSP pathogenesis, including proteins involved in endoplasmic reticulum biology, endosomal/membrane trafficking, and purine metabolism. Similar large-scale sequencing projects are now in progress for many other neurologic disorders, including Alzheimer’s disease, Parkinson’s disease, and epilepsies, among others. Due to the large number of new candidate genes likely to arise from these studies, there is a pressing need for new experimental strategies to test the predictions emerging from the resulting disease networks. Even more intricate models are on the horizon, as systems biology approaches (Civelek and Lusis, 2014) are applied toward integration of large-scale genomic data sets with complementary transcriptomic, proteomic, metabolomic, and epigenomic surveys conducted in neurologic patients. Unlike genomic variation, changes in other “-omic” data (e.g., the transcriptome) can be either a proximate cause of disease or a more distal, secondary effect (e.g., a biomarker). Fly models may be useful for pinpointing network features representing primary causes and decoding whether observed changes are pathogenic or rather compensatory, and potentially protective.
Fig. 2.
A hereditary spastic paraplegia (HSP) protein interaction network. In combination with proteins previously known to harbor mutations in HSP (blue), newly discovered gene candidates from whole-exome sequencing (red) populate a complex network of protein interactions with evidence of significant connectivity. From Novarino et al. Science 346 (6170): 506–511. Reprinted with permission from AAAS.
Currently prevailing approaches in flies and most other experimental systems predominantly investigate the consequences of isolated gene manipulations, ignoring the likelihood that majority of genetic variants probably act in combinations. In budding yeast, for example, a systematic study of more than 5 million pairwise interactions among ~1,700 genes identified ~170,000 interactions affecting cellular fitness (Costanzo et al., 2010). This study highlights the pervasive role that genetic interactions play in the heritability and expressivity of complex phenotypes. While many of the Drosophila experimental methodologies introduced earlier may be adaptable for probing gene–gene interactions, further innovations will also be needed to enable similar genome-wide combinatorial analyses that are currently feasible only in yeast or cell-based models. Even experimental strategies for pairwise testing of gene interactions within implicated functional pathways constitute an oversimplification. The phenotypic consequences of many genetic variants are probably influenced by higher-order interactions within large-scale disease regulatory networks comprising the “genetic background.” One innovative approach that may lead to advances in functional dissection of large-scale gene and protein interaction networks comes from studies of the “Drosophila Genetic Reference Panel” (DGRP) (Huang et al., 2012; Mackay et al., 2012). This remarkable resource includes 205 distinct inbred Drosophila strains that have been comprehensively sequenced to generate a reference map of naturally occurring genomic variation. There is already evidence to support a shared genomic architecture between flies and humans for selected phenotypes; for example, an analysis of fly sleep using the DGRP identified gene candidates homologous to loci implicated in human sleep disorders (Harbison et al., 2013). In the future, it is possible that similar population genetic experimental approaches might elucidate large-scale genomic regulatory networks with pleiotropic effects on nervous system traits and potentially conserved roles from flies to humans.
5. Conclusions
In sum, there is tremendous potential for Drosophila experimentation to confront challenges in the post-genomic era. However, no single experimental model species is likely to provide the complete solution. Indeed, there are some dimensions to neurologic disease, where cross-species genomic and biologic conservation is more limited. For example, besides autoimmune disorders, such as multiple sclerosis, there is now increasing evidence that immune and inflammatory mechanisms play important roles in primary neurodegenerative conditions, including Alzheimer’s and Parkinson’s disease. Because Drosophila lack adaptive immunity, modeling T- or B-cell related processes is not possible. However, flies do possess an innate immune system with rich conservation of many immune-related signaling systems, including the Toll-NFkB pathway that was originally elucidated in flies (Lemaitre, 2004). Moreover, recent studies of fly brain glial subpopulations have defined a type of ensheathing glia that can mediate phagocytic responses to neuronal injury similar to mammalian microglia (Doherty et al., 2009). Another potential limitation of fly models relate to diseases or selected disease manifestations that arise from brain network properties that are found only in higher vertebrates. For example, many of the defining features of movement disorders, including basal ganglionic (tremor, dyskinesia) or cerebellar (ataxia) dysfunction, relate to emergent properties of neural circuits that are simply not present in flies. In fact, many neurologic and neuropsychiatric disorders remain “functional disorders,” in which the neuroanatomic and pathologic substrates remain poorly defined. Examples include dystonia, essential tremor, autism spectrum disorders, major depression, and schizophrenia. As discussed earlier however, many of the implicated genes and functional pathways underlying such conditions may in fact be evolutionary conserved; therefore mapping the corresponding homologous loss-of-function phenotypes in flies may be a successful strategy for probing relevant mechanisms [see earlier discussion of “phenologs” (McGary et al., 2010; Woods et al., 2013)].
Given the scope of the challenges outlined above, a successful functional genomics strategy with broad applicability to diverse neurologic disorders will likely require contributions from numerous complementary experimental models. For first-pass screening of large lists of candidate genes/variants, truly high-throughput approaches, including in silico, in vitro, or cell-based genetic systems (e.g., yeast, iPS cells, etc.), are attractive. Importantly, while human iPS cell strategies show remarkable promise for disease modeling, many critical aspects of neurologic disorders, including properties of neural networks, interactions between neurons and glia, systemic influences on nervous system function (e.g., from the endocrine or cardiovascular system), as well as the essential role of aging, may be difficult if not impossible to recapitulate in cell-based systems. Thus, Drosophila along with the nematode, C. elegans, and zebrafish offer an intermediate platform, bridging in vitro and ex vivo systems to more time-intensive mammalian genetic models, such as mouse, rat, or primates. Ultimately, the confirmation and functional elucidation of genes and genomic variants responsible for neurologic disease, including dissection of the gene–gene interaction networks, will require unprecedented cooperation among investigators with varied skills, including neurologists, computational/systems biologists, and both human and model organism geneticists, among others. Many of the cross-disciplinary studies discussed above point to examples for how such collaborations can be leveraged effectively. In recent years, large human genomics consortia have formed to successfully pursue gene discovery in neurology, and it stands to reason that such organizations might naturally evolve next to grapple with the urgently needed functional investigations. Funders, including the NIH, will likely also serve an important organizing and enabling role. Compared with the deep investment in technology and resources for gene discovery, the field still needs to be effectively mobilized for the essential subsequent translational research. Reason for hope comes from the Undiagnosed Disease Network (http://commonfund.nih.gov/Diseases/), which began as an NIH intramural initiative to help patients with unknown diseases, and was recently expanded to include partnerships among several academic medical centers. This effort draws together diverse clinical and genomics expertise, given the expectation that many rare/undiagnosed conditions will have genetic underpinnings. Importantly, a model organism screening component for functional follow-up studies was also recently announced, including a pivotal role for flies. Similar national and worldwide efforts, including deep engagement of the Drosophila research community, will likely be needed to sustain the current momentum in our progress toward understanding the mechanisms responsible for neurologic disorders in the post-genomic era.
Acknowledgments
I would like to thank Michael Wangler, Shinya Yamamoto, Hugo Bellen, and Zongqi Xia for helpful discussion and critical feedback on the manuscript. Funding was provided by the NIH (U01AG046161, R01AG033193, R01NS078009, R21NS089854, R56NS089674), the Alzheimer’s Association, the American Federation of Aging Research, and a Career Award for Medical Scientists from the Burroughs Wellcome Fund.
References
- Anttila V, Winsvold BS, Gormley P, Kurth T, Bettella F, McMahon G, Kallela M, Malik R, de Vries B, Terwindt G, Medland SE, Todt U, Mcardle WL, Quaye L, Koiranen M, Ikram MA, Lehtimäki T, Stam AH, Ligthart L, Wedenoja J, Dunham I, Neale BM, Palta P, Hamalainen E, Schürks M, Rose LM, Buring JE, Ridker PM, Steinberg S, Stefánsson H, Jakobsson F, Lawlor DA, Evans DM, Ring SM, Färkkilä M, Artto V, Kaunisto MA, Freilinger T, Schoenen J, Frants RR, Pelzer N, Weller CM, Zielman R, Heath AC, Madden PAF, Montgomery GW, Martin NG, Borck G, Göbel H, Heinze A, Heinze-Kuhn K, Williams FMK, Hartikainen AL, Pouta A, van den Ende J, Uitterlinden AG, Hofman A, Amin N, Hottenga JJ, Vink JM, Heikkilä K, Alexander M, Müller-Myhsok B, Schreiber S, Meitinger T, Wichmann HE, Aromaa A, Eriksson JG, Traynor BJ, Trabzuni D, Rossin E, Lage K, Jacobs SBR, Gibbs JR, Birney E, Kaprio J, Penninx BW, Boomsma DI, van Duijn C, Raitakari O, Jarvelin MR, Zwart JA, Cherkas L, Strachan DP, Kubisch C, Ferrari MD, van den Maagdenberg AMJM, Dichgans M, Wessman M, Smith GD, Stefánsson K, Daly MJ, Nyholt DR, Chasman DI, Palotie A North American Brain Expression Consortium, UK Brain Expression Consortium, International Headache Genetics Consortium. Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat Genet. 2013;45:912–917. doi: 10.1038/ng.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- Bamshad MJ, Shendure JA, Valle D, Hamosh A, Lupski JR, Gibbs RA, Boerwinkle E, Lifton RP, Gerstein M, Gunel M, Mane S, Nickerson DA Centers for Mendelian Genomics. The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am J Med Genet A. 2012;158A:1523–1525. doi: 10.1002/ajmg.a.35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayat V, Thiffault I, Jaiswal M, Tétreault M, Donti T, Sasarman F, Bernard G, Demers-Lamarche J, Dicaire MJ, Mathieu J, Vanasse M, Bouchard JP, Rioux MF, Lourenco CM, Li Z, Haueter C, Shoubridge EA, Graham BH, Brais B, Bellen HJ. Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol. 2012;10:e1001288. doi: 10.1371/journal.pbio.1001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Tong C, Tsuda H. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat Rev Neurosci. 2010;11:514–522. doi: 10.1038/nrn2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E, Kim J, Metaxakis A, Savakis C, Schulze KL, Hoskins RA, Spradling AC. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics. 2011;188:731–743. doi: 10.1534/genetics.111.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Björklund M, Furger E, Schertel C, Taipale J, Basler K. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development. 2013;140:2434–2442. doi: 10.1242/dev.088757. [DOI] [PubMed] [Google Scholar]
- Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Caussinus E, Kanca O, Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol. 2011;19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Clark AG, Mootha VK. Distilling pathophysiology from complex disease genetics. Cell. 2013;155:21–26. doi: 10.1016/j.cell.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis J, Hansmannel F, Gistelinck M, Mounier A, Van Cauwenberghe C, Kolen KV, Geller F, Sottejeau Y, Harold D, Dourlen P, Grenier-Boley B, Kamatani Y, Delepine B, Demiautte F, Zelenika D, Zommer N, Hamdane M, Bellenguez C, Dartigues JF, Hauw JJ, Letronne F, Ayral AM, Sleegers K, Schellens A, Broeck LV, Engelborghs S, De Deyn PP, Vandenberghe R, O’Donovan M, Owen M, Epelbaum J, Mercken M, Karran E, Bantscheff M, Drewes G, Joberty G, Campion D, Octave JN, Berr C, Lathrop M, Callaerts P, Mann D, Williams J, Buee L, Dewachter I, Van Broeckhoven C, Amouyel P, Moechars D, Dermaut B, Lambert JC. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry. 2013;18:1225–1234. doi: 10.1038/mp.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AHY, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pal C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Taşdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- Fernandez-Funez P, Nino-Rosales M, de Gouyon B, She W, Luchak J, Martinez P, Turiegano E, Benito J, Capovilla M, Skinner P, McCall A, Canal I, Orr H, Zoghbi H, Botas J. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature. 2000;408:101–106. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- Finelli A, Kelkar A, Song H, Yang H, Konsolaki M. A model for studying Alzheimer’s Abeta42-induced toxicity in Drosophila melanogaster. Mol Cell Neurosci. 2004;26:365–375. doi: 10.1016/j.mcn.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Fortini M, Skupski M, Boguski M, Hariharan I. A survey of human disease gene counterparts in the Drosophila genome. J Cell Biol. 2000;150:F23–F30. doi: 10.1083/jcb.150.2.f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A, Pranski E, Miller RD, Radmard S, Bernhard D, Jinnah HA, Betarbet R, Rye DB, Sanyal S. Sleep fragmentation and motor restlessness in a Drosophila model of restless legs syndrome. Curr Biol. 2012;22:1142–1148. doi: 10.1016/j.cub.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB, Allen A, Keebler J, Margulies EH, Petrou S, Petrovski S, Sunyaev S. Sequencing studies in human genetics: design and interpretation. Nat Rev Genet. 2013;14:460–470. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM. Genome engineering of drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison ST, McCoy LJ, Mackay TFC. Genome-wide association study of sleep in Drosophila melanogaster. BMC Genomics. 2013;14:281. doi: 10.1186/1471-2164-14-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MM, Miquel J, Johnson M. Insect brain as a model for the study of aging. Age-related changes in drosophila melanogaster. Acta Neuropathol. 1971;19:167–183. doi: 10.1007/BF00684595. [DOI] [PubMed] [Google Scholar]
- Herrmann DN, Horvath R, Sowden JE, Gonzales M, Sanchez-Mejias A, Guan Z, Whittaker RG, Almodovar JL, Lane M, Bansagi B, Pyle A, Boczonadi V, Lochmüller H, Griffin H, Chinnery PF, Lloyd TE, Littleton JT, Züchner S. Synaptotagmin 2 mutations cause an autosomal-dominant form of lambert-eaton myasthenic syndrome and nonprogressive motor neuropathy. Am J Hum Genet. 2014;95:332–339. doi: 10.1016/j.ajhg.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoed den M, Eijgelsheim M, Esko T, Brundel BJJM, Peal DS, Evans DM, Nolte IM, Segrè AV, Holm H, Handsaker RE, Westra HJ, Johnson T, Isaacs A, Yang J, Lundby A, Zhao JH, Kim YJ, Go MJ, Almgren P, Bochud M, Boucher G, Cornelis MC, Gudbjartsson D, Hadley D, van der Harst P, Hayward C, Heijer den M, Igl W, Jackson AU, Kutalik Z, Luan J, Kemp JP, Kristiansson K, Ladenvall C, Lorentzon M, Montasser ME, Njajou OT, O’Reilly PF, Padmanabhan S, St Pourcain B, Rankinen T, Salo P, Tanaka T, Timpson NJ, Vitart V, Waite L, Wheeler W, Zhang W, Draisma HHM, Feitosa MF, Kerr KF, Lind PA, Mihailov E, Onland-Moret NC, Song C, Weedon MN, Xie W, Yengo L, Absher D, Albert CM, Alonso A, Arking DE, De Bakker PIW, Balkau B, Barlassina C, Benaglio P, Bis JC, Bouatia-Naji N, Brage S, Chanock SJ, Chines PS, Chung M, Darbar D, Dina C, Dörr M, Elliott P, Felix SB, Fischer K, Fuchsberger C, de Geus EJC, Goyette P, Gudnason V, Harris TB, Hartikainen A-L, Havulinna AS, Heckbert SR, Hicks AA, Hofman A, Holewijn S, Hoogstra-Berends F, Hottenga J-J, Jensen MK, Johansson A, Junttila J, Kääb S, Kanon B, Ketkar S, Khaw K-T, Knowles JW, Kooner AS, Kors JA, Kumari M, Milani L, Laiho P, Lakatta EG, Langenberg C, Leusink M, Liu Y, Luben RN, Lunetta KL, Lynch SN, Markus MRP, Marques-Vidal P, Mateo Leach I, Mcardle WL, Mccarroll SA, Medland SE, Miller KA, Montgomery GW, Morrison AC, Müller-Nurasyid M, Navarro P, Nelis M, O’Connell JR, O’Connell CJ, Ong KK, Newman AB, Peters A, Polasek O, Pouta A, Pramstaller PP, Psaty BM, Rao DC, Ring SM, Rossin EJ, Rudan D, Sanna S, Scott RA, Sehmi JS, Sharp S, Shin JT, Singleton AB, Smith AV, Soranzo N, Spector TD, Stewart C, Stringham HM, Tarasov KV, Uitterlinden AG, Vandenput L, Hwang S-J, Whitfield JB, Wijmenga C, Wild SH, Willemsen G, Wilson JF, Witteman JCM, Wong A, Wong Q, Jamshidi Y, Zitting P, Boer JMA, Boomsma DI, Borecki IB, van Duijn CM, Ekelund U, Forouhi NG, Froguel P, Hingorani A, Ingelsson E, Kivimaki M, Kronmal RA, Kuh D, Lind L, Martin NG, Oostra BA, Pedersen NL, Quertermous T, Rotter JI, van der Schouw YT, Verschuren WMM, Walker M, Albanes D, Arnar DO, Assimes TL, Bandinelli S, Boehnke M, de Boer RA, Bouchard C, Caulfield WLM, Chambers JC, Curhan G, Cusi D, Eriksson J, Ferrucci L, van Gilst WH, Glorioso N, de Graaf J, Groop L, Gyllensten U, Hsueh W-C, Hu FB, Huikuri HV, Hunter DJ, Iribarren C, Isomaa B, Jarvelin M-R, Jula A, Kähönen M, Kiemeney LA, van der Klauw MM, Kooner JS, Kraft P, Iacoviello L, Lehtimäki T, Lokki M-LL, Mitchell BD, Navis G, Nieminen MS, Ohlsson C, Poulter NR, Qi L, Raitakari OT, Rimm EB, Rioux JD, Rizzi F, Rudan I, Salomaa V, Sever PS, Shields DC, Shuldiner AR, Sinisalo J, Stanton AV, Stolk RP, Strachan DP, Tardif J-C, Thorsteinsdottir U, Tuomilehto J, van Veldhuisen DJ, Virtamo J, Viikari J, Vollenweider P, Waeber G, Widen E, Cho YS, Olsen JV, Visscher PM, Willer C, Franke L, Erdmann J, Thompson JR, Pfeufer A, Sotoodehnia N, Newton-Cheh C, Ellinor PT, Stricker BHC, Metspalu A, Perola M, Beckmann JS, Smith GD, Stefánsson K, Wareham NJ, Munroe PB, Sibon OCM, Milan DJ, Snieder H, Samani NJ, Loos RJF Global BPgen Consortium, CARDIoGRAM Consortium, PR GWAS Consortium, QRS GWAS Consortium, QT-IGC Consortium, CHARGE-AF Consortium. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013;45:621–631. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ERLC, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Rüther E, Schürmann B, Heun R, Kölsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Gallacher J, Hüll M, Rujescu D, Giegling I, Goate AM, Kauwe JSK, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MMB, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alpérovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefánsson H, Stefánsson K, Snædal J, Björnsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossù P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jónsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J Alzheimer’s Disease Neuroimaging Initiative, C.H.A.R.G.E. Consortium, EADI1 Consortium. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinf. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Richards S, Carbone MA, Zhu D, Anholt RRH, Ayroles JF, Duncan L, Jordan KW, Lawrence F, Magwire MM, Warner CB, Blankenburg K, Han Y, Javaid M, Jayaseelan J, Jhangiani SN, Muzny D, Ongeri F, Perales L, Wu YQ, Zhang Y, Zou X, Stone EA, Gibbs RA, Mackay TFC. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc Natl Acad Sci. 2012;109:15553–15559. doi: 10.1073/pnas.1213423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, Shah TS, Spencer C, Booth D, Goris A, Oturai A, Saarela J, Fontaine B, Hemmer B, Martin C, Zipp F, D’alfonso S, Martinelli Boneschi F, Taylor B, Harbo HF, Kockum I, Hillert J, Olsson T, Ban M, Oksenberg JR, Hintzen R, Barcellos LF, Agliardi C, Alfredsson L, Alizadeh M, Anderson C, Andrews R, Søndergaard HB, Baker A, Band G, Baranzini SE, Barizzone N, Barrett J, Bellenguez C, Bergamaschi L, Bernardinelli L, Berthele A, Biberacher V, Binder TMC, Blackburn H, Bomfim IL, Brambilla P, Broadley S, Brochet B, Brundin L, Buck D, Butzkueven H, Caillier SJ, Camu W, Carpentier W, Cavalla P, Celius EG, Coman I, Comi G, Corrado L, Cosemans L, Cournu-Rebeix I, Cree BAC, Cusi D, Damotte V, Defer G, Delgado SR, Deloukas P, di Sapio A, Dilthey AT, Donnelly P, Dubois B, Duddy M, Edkins S, Elovaara I, Esposito F, Evangelou N, Fiddes B, Field J, Franke A, Freeman C, Frohlich IY, Galimberti D, Gieger C, Gourraud P-A, Graetz C, Graham A, Grummel V, Guaschino C, Hadjixenofontos A, Hakonarson H, Halfpenny C, Hall G, Hall P, Hamsten A, Harley J, Harrower T, Hawkins C, Hellenthal G, Hillier C, Hobart J, Hoshi M, Hunt SE, Jagodic M, Jelčić I, Jochim A, Kendall B, Kermode A, Kilpatrick T, Koivisto K, Konidari I, Korn T, Kronsbein H, Langford C, Larsson M, Lathrop M, Lebrun-Frenay C, Lechner-Scott J, Lee MH, Leone MA, Leppä V, Liberatore G, Lie BA, Lill CM, Lindén M, Link J, Luessi F, Lycke J, Macciardi F, Männistö S, Manrique CP, Martin R, Martinelli V, Mason D, Mazibrada G, McCabe C, Mero I-L, Mescheriakova J, Moutsianas L, Myhr K-M, Nagels G, Nicholas R, Nilsson P, Piehl F, Pirinen M, Price SE, Quach H, Reunanen M, Robberecht W, Robertson NP, Rodegher M, Rog D, Salvetti M, Schnetz-Boutaud NC, Sellebjerg F, Selter RC, Schaefer C, Shaunak S, Shen L, Shields S, Siffrin V, Slee M, Sørensen PS, Sorosina M, Sospedra M, Spurkland A, Strange A, Sundqvist E, Thijs V, Thorpe J, Ticca A, Tienari P, van Duijn C, Visser EM, Vucic S, Westerlind H, Wiley JS, Wilkins A, Wilson JF, Winkelmann J, Zajicek J, Zindler E, Haines JL, Pericak-Vance MA, Ivinson AJ, Stewart G, Hafler D, Hauser SL, Compston A, McVean G, De Jager P, Sawcer SJ, McCauley JL International Multiple Sclerosis Genetics Consortium IMSGC, Wellcome Trust Case Control Consortium 2 WTCCC2 International IBD Genetics Consortium (IIBDGC) Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson G, Salecker I, Dong X, Yao X, Arnheim N, Faber P, MacDonald M, Zipursky S. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- Jaiswal M, Sandoval H, Zhang K, Bayat V, Bellen HJ. Probing mechanisms that underlie human neurodegenerative disease in Drosophila. Annu Rev Genet. 2012;46:371–396. doi: 10.1146/annurev-genet-110711-155456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal NS, Lee PT, Campbell ME, Chen K, Anguiano-Zarate S, Cantu Gutierrez M, Busby T, Lin W-W, He Y, Schulze KL, Booth BW, Evans-Holm M, Venken KJT, Levis RW, Spradling AC, Hoskins RA, Bellen HJ. A library of MiMICs allows tagging of genes and reversible spatial and temporal knockdown of proteins in Drosophila. eLife. 2015 doi: 10.7554/eLife.05338. (in press) http://dx.doi.org/10.7554/eLife.05338. [DOI] [PMC free article] [PubMed]
- Jenett A, Rubin GM, Ngo TTB, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, Iyer N, Fetter D, Hausenfluck JH, Peng H, Trautman ET, Svirskas RR, Myers EW, Iwinski ZR, Aso Y, DePasquale GM, Enos A, Hulamm P, Lam SCB, Li HH, Laverty TR, Long F, Qu L, Murphy SD, Rokicki K, Safford T, Shaw K, Simpson JH, Sowell A, Tae S, Yu Y, Zugates CT. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilarski LL, Achterberg S, Devan WJ, Traylor M, Malik R, Lindgren A, Pare G, Sharma P, Slowik A, Thijs V, Walters M, Worrall BB, Sale MM, Algra A, Kappelle LJ, Wijmenga C, Norrving B, Sandling JK, Rönnblom L, Goris A, Franke A, Sudlow C, Rothwell PM, Levi C, Holliday EG, Fornage M, Psaty B, Gretarsdottir S, Thorsteinsdottir U, Seshadri S, Mitchell BD, Kittner S, Clarke R, Hopewell JC, Bis JC, Boncoraglio GB, Meschia J, Ikram MA, Hansen BM, Montaner J, Thorleifsson G, Stefanson K, Rosand J, De Bakker PIW, Farrall M, Dichgans M, Markus HS, Bevan S GARNET Collaborative Research Group, Wellcome Trust Case Control Consortium 2, Australian Stroke Genetic Collaborative, the METASTROKE Consortium, the International Stroke Genetics Consortium. Meta-analysis in more than 17,900 cases of ischemic stroke reveals a novel association at 12q24.12. Neurology. 2014;83:678–685. doi: 10.1212/WNL.0000000000000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195:715–721. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, DeStefano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thornton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Hollingworth P, Ramirez A, Hanon O, Fitzpatrick AL, Buxbaum JD, Campion D, Crane PK, Baldwin C, Becker T, Gudnason V, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MJ, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuinness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimón J, Lleó A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossù P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Naranjo MCD, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannfelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RFAG, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JSK, Boerwinkle E, Riemenschneider M, Boada M, Hiltunen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Lemaitre B. The road to Toll. Nat Rev Immunol. 2004;4:521–527. doi: 10.1038/nri1390. [DOI] [PubMed] [Google Scholar]
- Lessing D, Bonini NM. Maintaining the brain: insight into human neurodegeneration from Drosophila melanogaster mutants. Nat Rev Genet. 2009;10:359–370. doi: 10.1038/nrg2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, Conrad DF, Cooper GM, Cox NJ, Daly MJ, Gerstein MB, Goldstein DB, Hirschhorn JN, Leal SM, Pennacchio LA, Stamatoyannopoulos JA, Sunyaev SR, Valle D, Voight BF, Winckler W, Gunter C. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, Richardson MF, Anholt RRH, Barrón M, Bess C, Blankenburg KP, Carbone MA, Castellano D, Chaboub L, Duncan L, Harris Z, Javaid M, Jayaseelan JC, Jhangiani SN, Jordan KW, Lara F, Lawrence F, Lee SL, Librado P, Linheiro RS, Lyman RF, Mackey AJ, Munidasa M, Muzny DM, Nazareth L, Newsham I, Perales L, Pu LL, Qu C, Ràmia M, Reid JG, Rollmann SM, Rozas J, Saada N, Turlapati L, Worley KC, Wu YQ, Yamamoto A, Zhu Y, Bergman CM, Thornton KR, Mittelman D, Gibbs RA. The Drosophila melanogaster genetic reference panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod DA, Rhinn H, Kuwahara T, Zolin A, Di Paolo G, Maccabe BD, Marder KS, Honig LS, Clark LN, Small SA, Abeliovich A. RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson’s disease risk. Neuron. 2013;77:425–439. doi: 10.1016/j.neuron.2012.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K, Kaufman T, Gelbart W. Research resources for Drosophila: the expanding universe. Nat Rev Genet. 2005;6:179–193. doi: 10.1038/nrg1554. [DOI] [PubMed] [Google Scholar]
- McGary KL, Park TJ, Woods JO, Cha HJ, Wallingford JB, Marcotte EM. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc Natl Acad Sci. 2010;107:6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JSK, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, Decarli C, Dekosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, Mccormick WC, Mccurry SM, Mcdavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, Schulte C, Keller MF, Arepalli S, Letson C, Edsall C, Stefánsson H, Liu X, Pliner H, Lee JH, Cheng R, Ikram MA, Ioannidis JPA, Hadjigeorgiou GM, Bis JC, Martinez M, Perlmutter JS, Goate A, Marder K, Fiske B, Sutherland M, Xiromerisiou G, Myers RH, Clark LN, Stefánsson K, Hardy JA, Heutink P, Chen H, Wood NW, Houlden H, Payami H, Brice A, Scott WK, Gasser T, Bertram L, Eriksson N, Foroud T, Singleton AB. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, Griffin RS, Belfer I, Dai F, Smith SB, Diatchenko L, Gupta V, Xia CP, Amann S, Kreitz S, Heindl-Erdmann C, Wolz S, Ly CV, Arora S, Sarangi R, Dan D, Novatchkova M, Rosenzweig M, Gibson DG, Truong D, Schramek D, Zoranovic T, Cronin SJF, Angjeli B, Brune K, Dietzl G, Maixner W, Meixner A, Thomas W, Pospisilik JA, Alenius M, Kress M, Subramaniam S, Garrity PA, Bellen HJ, Woolf CJ, Penninger JM. A genome-wide Drosophila screen for heat nociception identifies α2δ3 as an evolutionarily conserved pain gene. Cell. 2010;143:628–638. doi: 10.1016/j.cell.2010.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller RA, Wirtz-Peitz F, Lee S, Kwon Y, Buckner M, Hoskins RA, Venken KJT, Bellen HJ, Mohr SE, Perrimon N. Stringent analysis of gene function and protein-protein interactions using fluorescently tagged genes. Genetics. 2012;190:931–940. doi: 10.1534/genetics.111.136465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. http://dx.doi.org/10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, Abdellateef M, Rosti B, Scott E, Mansour L, Masri A, Kayserili H, Al-Aama JY, Abdel-Salam GMH, Karminejad A, Kara M, Kara B, Bozorgmehri B, Ben-Omran T, Mojahedi F, Mahmoud IGED, Bouslam N, Bouhouche A, Benomar A, Hanein S, Raymond L, Forlani S, Mascaro M, Selim L, Shehata N, Al-Allawi N, Bindu PS, Azam M, Gunel M, Caglayan A, Bilguvar K, Tolun A, Issa MY, Schroth J, Spencer EG, Rosti RO, Akizu N, Vaux KK, Johansen A, Koh AA, Megahed H, Durr A, Brice A, Stevanin G, Gabriel SB, Ideker T, Gleeson JG. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343:506–511. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Pastor-Pareja JC, Xu T. Shaping cells and organs in Drosophila by opposing roles of fat body-secreted Collagen IV and perlecan. Dev Cell. 2011;21:245–256. doi: 10.1016/j.devcel.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman A, Hardy J. Genetic analysis in neurology. JAMA Neurology. 2013;70:696. doi: 10.1001/jamaneurol.2013.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G, Yandell M, Wortman J, Gabor Miklos G, Nelson C, Hariharan I, Fortini M, Li P, Apweiler R, Fleischmann W, Cherry J, Henikoff S, Skupski M, Misra S, Ashburner M, Birney E, Boguski M, Brody T, Brokstein P, Celniker S, Chervitz S, Coates D, Cravchik A, Gabrielian A, Galle R, Gelbart W, George R, Goldstein L, Gong F, Guan P, Harris N, Hay B, Hoskins R, Li J, Li Z, Hynes R, Jones S, Kuehl P, Lemaitre B, Littleton J, Morrison D, Mungall C, O’Farrell P, Pickeral O, Shue C, Vosshall L, Zhang J, Zhao Q, Zheng X, Lewis S. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EMC, Ramirez-Lorca R, Debette S, Longstreth WT, Janssens ACJW, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MMB CHARGE Consortium, GERAD1 Consortium, EADI1 Consortium. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman JM, Shulman LM, Weiner WJ, Feany MB. From fruit fly to bedside: translating lessons from Drosophila models of neurodegenerative disease. Curr Opin Neurol. 2003;16:443–449. doi: 10.1097/01.wco.0000084220.82329.60. [DOI] [PubMed] [Google Scholar]
- Shulman JM, Chipendo P, Chibnik LB, Aubin C, Tran D, Keenan BT, Kramer PL, Schneider JA, Bennett DA, Feany MB, De Jager PL. Functional screening of Alzheimer pathology genome-wide association signals in Drosophila. Am J Hum Genet. 2011;88:232–238. doi: 10.1016/j.ajhg.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman JM, Imboywa S, giagtzoglou N, Powers MP, Hu Y, Devenport D, Chipendo P, Chibnik LB, Diamond A, Perrimon N, Brown NH, De Jager PL, Feany MB. Functional screening in Drosophila identifies Alzheimer’s disease susceptibility genes and implicates Tau-mediated mechanisms. Hum Mol Genet. 2014;23:870–877. doi: 10.1093/hmg/ddt478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3:176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- St Pierre SE, Ponting L, Stefancsik R, McQuilton P Consortium FlyBase. FlyBase 102—advanced approaches to interrogating FlyBase. Nucleic Acids Res. 2014;42:D780–D788. doi: 10.1093/nar/gkt1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefánsson H, Rye DB, Hicks A, Pétursson H, Ingason A, Thorgeirsson TE, Palsson S, Sigmundsson T, Sigurdsson AP, Eiriksdottir I, Soebech E, Bliwise D, Beck JM, Rosen A, Waddy S, Trotti LM, Iranzo A, Thambisetty M, Hardarson GA, Kristjansson K, Gudmundsson LJ, Thorsteinsdottir U, Kong A, Gulcher JR, Gudbjartsson D, Stefánsson K. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357:639–647. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- van der Harst P, Zhang W, Mateo Leach I, Rendon A, Verweij N, Sehmi J, Paul DS, Elling U, Allayee H, Li X, Radhakrishnan A, Tan ST, Voss K, Weichenberger CX, Albers CA, Al-Hussani A, Asselbergs FW, Ciullo M, Danjou F, Dina C, Esko T, Evans DM, Franke L, Gögele M, Hartiala J, Hersch M, Holm H, Hottenga JJ, Kanoni S, Kleber ME, Lagou V, Langenberg C, Lopez LM, Lyytikäinen LP, Melander O, Murgia F, Nolte IM, O’Reilly PF, Padmanabhan S, Parsa A, Pirastu N, Porcu E, Portas L, Prokopenko I, Ried JS, Shin SY, Tang CS, Teumer A, Traglia M, Ulivi S, Westra HJ, Yang J, Zhao JH, Anni F, Abdellaoui A, Attwood A, Balkau B, Bandinelli S, Bastardot F, Benyamin B, Boehm BO, Cookson WO, Das D, De Bakker PIW, de Boer RA, de Geus EJC, de Moor MH, Dimitriou M, Domingues FS, Döring A, Engström G, Eyjolfsson GI, Ferrucci L, Fischer K, Galanello R, Garner SF, Genser B, Gibson QD, Girotto G, Gudbjartsson DF, Harris SE, Hartikainen AL, Hastie CE, Hedblad B, Illig T, Jolley J, Kähönen M, Kema IP, Kemp JP, Liang L, Lloyd-Jones H, Loos RJF, Meacham S, Medland SE, Meisinger C, Memari Y, Mihailov E, Miller K, Moffatt MF, Nauck M, Novatchkova M, Nutile T, Olafsson I, Onundarson PT, Parracciani D, Penninx BW, Perseu L, Piga A, Pistis G, Pouta A, Puc U, Raitakari O, Ring SM, Robino A, Ruggiero D, Ruokonen A, Saint-Pierre A, Sala C, Salumets A, Sambrook J, Schepers H, Schmidt CO, Silljé HHW, Sladek R, Smit JH, Starr JM, Stephens J, Sulem P, Tanaka T, Thorsteinsdottir U, Tragante V, van Gilst WH, van Pelt LJ, van Veldhuisen DJ, Völker U, Whitfield JB, Willemsen G, Winkelmann BR, Wirnsberger G, Algra A, Cucca F, d’Adamo AP, Danesh J, Deary IJ, Dominiczak AF, Elliott P, Fortina P, Froguel P, Gasparini P, Greinacher A, Hazen SL, Jarvelin MR, Khaw KT, Lehtimäki T, Maerz W, Martin NG, Metspalu A, Mitchell BD, Montgomery GW, Moore C, Navis G, Pirastu M, Pramstaller PP, Ramirez-Solis R, Schadt E, Scott J, Shuldiner AR, Smith GD, Smith JG, Snieder H, Sorice R, Spector TD, Stefánsson K, Stumvoll M, Tang WHW, Toniolo D, Tönjes A, Visscher PM, Vollenweider P, Wareham NJ, Wolffenbuttel BHR, Boomsma DI, Beckmann JS, Dedoussis GV, Deloukas P, Ferreira MA, Sanna S, Uda M, Hicks AA, Penninger JM, Gieger C, Kooner JS, Ouwehand WH, Soranzo N, Chambers JC. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Bellen HJ. Chemical mutagens, transposons, and transgenes to interrogate gene function in Drosophila melanogaster. Methods. 2014;68:15–28. doi: 10.1016/j.ymeth.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, Bellen HJ, Hoskins RA. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilariño-Güell C, Wider C, Ross OA, Dächsel JC, Kachergus JM, Lincoln SJ, Soto-Ortolaza AI, Cobb SA, Wilhoite GJ, Bacon JA, Behrouz B, Melrose HL, Hentati E, Puschmann A, Evans DM, Conibear E, Wasserman WW, Aasly JO, Burkhard PR, Djaldetti R, Ghika J, Hentati F, Krygowska-Wajs A, Lynch T, Melamed E, Rajput A, Rajput AH, Solida A, Wu RM, Uitti RJ, Wszolek ZK, Vingerhoets F, Farrer MJ. VPS35 mutations in Parkinson disease. Am J Hum Genet. 2011;89:162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann CW, Wszolek MF, Shulman JM, Salvaterra PM, Lewis J, Hutton M, Feany MB. Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science. 2001;293:711–714. doi: 10.1126/science.1062382. [DOI] [PubMed] [Google Scholar]
- Woods JO, Singh-Blom UM, Laurent JM, McGary KL, Marcotte EM. Prediction of gene–phenotype associations in humans, mice, and plants using phenologs. BMC Bioinf. 2013;14:203. doi: 10.1186/1471-2105-14-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman NA, Xiong B, Zhang K, Bayat V, David G, Li T, Chen K, Gala U, Harel T, Pehlivan D, Penney S, Vissers LELM, de Ligt J, Jhangiani SN, Xie Y, Tsang SH, Parman Y, Sivaci M, Battaloglu E, Muzny D, Wan YW, Liu Z, Lin-Moore AT, Clark RD, Curry CJ, Link N, Schulze KL, Boerwinkle E, Dobyns WB, Allikmets R, Gibbs RA, Chen R, Lupski JR, Wangler MF, Bellen HJ. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159:200–214. doi: 10.1016/j.cell.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Yufeng, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Yaping, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, Hardison M, Person R, Bekheirnia MR, Leduc MS, Kirby A, Pham P, Scull J, Wang M, Ding Y, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, Eng CM. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013 doi: 10.1056/NEJMoa1306555. 131002140031007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, Gao G. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013;195:289–291. doi: 10.1534/genetics.113.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Benet-Pagès A, Struhal W, Graf E, Eck SH, Offman MN, Haubenberger D, Spielberger S, Schulte EC, Lichtner P, Rossle SC, Klopp N, Wolf E, Seppi K, Pirker W, Presslauer S, Mollenhauer B, Katzenschlager R, Foki T, Hotzy C, Reinthaler E, Harutyunyan A, Kralovics R, Peters A, Zimprich F, Brücke T, Poewe W, Auff E, Trenkwalder C, Rost B, Ransmayr G, Winkelmann J, Meitinger T, Strom TM. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet. 2011;89:168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]