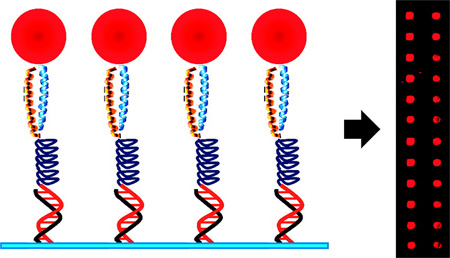
Abstract
A new method for protein surface functionalization was developed that utilizes DNA-conjugated artificial polypeptides to capture recombinant target proteins from the solution phase and direct their deposition onto DNA-functionalized matrices. Protein capture is accomplished through the coiled-coil association of an engineered pair of heterodimeric leucine zippers. Incorporating half of the zipper complex directly into the polypeptides and labeling these polymers with ssDNA enables the polypeptide conjugates to form intermediate linkages that connect the target proteins securely to DNA-functionalized supports. This synthetic route provides an important alternative to conventional DNA-conjugation techniques by allowing proteins to be outfitted site-specifically with ssDNA while minimizing the need for postexpression processing. We demonstrate these attributes by (i) using the capture probes to prepare protein microarrays, (ii) demonstrating control over enzyme activity via deposition of DNA, and, (iii) synthesizing finite-sized, multiprotein complexes that are templated on designed DNA scaffolds in near quantitative yield.
Graphical abstract
Protein coupling and immobilization strategies remain central to the investigation of protein interactions and constitute an important design element for a wide variety of protein detection technologies (1–3). To meet these needs, an array of polypeptide-based affinity tags has been implemented that can mediate the surface anchoring of proteins (4, 5). In addition to being expressed as small protein fusions, these functionalities offer significant advantages over physical adsorption as well as most direct covalent attachment schemes, since they provide control over the orientations of proteins on their supports and, in many circumstances, help preserve the native folded state of proteins (6).
Alternatively, DNA–protein conjugate molecules have been developed to facilitate protein immobilization onto DNA-coated matrices such as DNA microarrays (7–9) and organized molecular scaffolds formed from DNA (10). DNA-directed immobilization of proteins allows oligonucleotide sequences to encode the spatial positioning and composition of multiple proteins on solid supports via the controlled deposition of DNA (7). This approach can simplify the preparation of solid supports for protein immobilization and provide sensitive control over protein deposition, since DNA-surface chemistries are more established and robust than most protein immobilization techniques. DNA-directed protein deposition can also provide routes to tune the quantitative activities of enzymes in ways that can be difficult to achieve using protein affinity tags alone and offer new opportunities to build artificial systems of interacting proteins with complex kinetic properties. Such control has been demonstrated by recent work to “program” the stoichiometric ratios of multiple enzymes immobilized on the surfaces of DNA-coated 96-well plates (11). Similarly, multienzyme complexes have been created using DNA as a molecular scaffold and used to explore how proximity effects influence their coupled-enzyme reaction dynamics (11, 12).
Despite the advantages of using DNA hybridization to direct protein deposition, various technical obstacles still limit the utility of this approach for a variety of applications. Particularly, the production of large numbers of DNA–protein conjugates remains challenging. Although commercially available cross-linking agents can be used to couple amine- or thiol-functionalized DNA strands to surface cysteine and lysine residues of proteins (13, 14), the efficiency of these reactions tends to be low, and postlabeling purification is generally required to isolate the conjugates. Furthermore, site-specific DNA conjugation through this route requires that the majority of surface cysteines and lysines are mutated to nonreacting residues. Even if these or alternative modifications are possible, the direct labeling of proteins with DNA may interfere with protein function (14). These issues can be avoided by coupling DNA to proteins indirectly through protein affinity tags. Protocols that employ intein tags along with expressed protein ligation (15, 16), DNA–streptavidin conjugates and biotin labeling (7), reactions of chemically modified DNA molecules with SNAP-tag fusion (17), and protein farnesyltransferase labeling coupled with click chemistry (18) have all been adopted for this purpose. However, most of these methods still require purification steps to isolate the DNA–protein conjugates. Consequently, a host of technological applications stand to benefit from the development of new synthetic routes to prepare DNA–protein conjugates that emulate the simple and robust processing afforded by commonly used protein affinity tags alone.
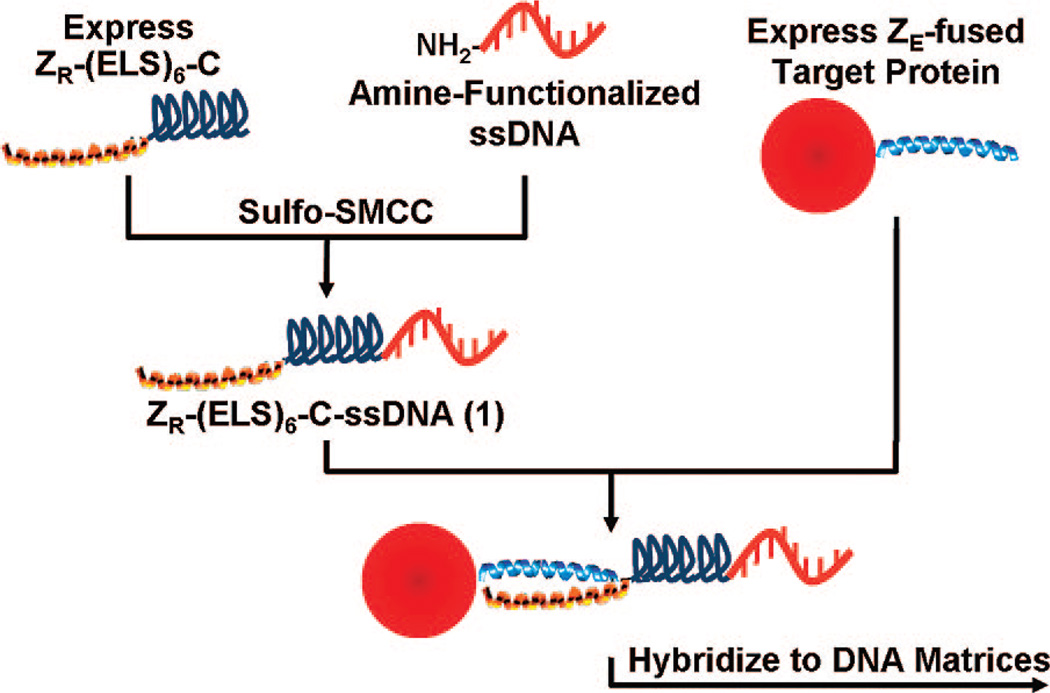
In this report, we describe a new synthetic strategy to couple proteins to DNA matrices that involves the production of DNA-conjugated polypeptide polymers 1 that function as capture probes by associating with recombinant affinity-tagged proteins in solution and then directing their deposition onto DNA-coated supports. These polymers are based on artificial proteins that some of us have previously used to control the surface immobilization of proteins (19) and to build finite-sized multiprotein complexes (20). With this system, protein capture is achieved through the coiled-coil association of an engineered parallel pair of heterodimeric leucine zippers, designated ZE and ZR. The zipper sequences are derived from polypeptides developed by Vinson et al. (21) and form exceptionally strong heterodimeric complexes (KD ~ 10−15 M) with much weaker homodimeric complexes (KD ~ 10−3 to 10−6 M). Half of the zipper complex is fused to a target protein as an affinity tag, while the other component is incorporated into the polymer as a genetic fusion. The polymers also contain a mechanically flexible and repetitive domain based on the elastomeric poly(VPGVαG) structural motif of the protein elastin (EL). Substitution of amino acids at the Vα position of this domain provides control over the hydrophilicity of the polymer and can be used to either direct or minimize physical adhesion of the polymers to functionalized surfaces (19). Here, we demonstrate that a DNA-conjugated version of these artificial proteins can be used to direct the self-assembly of target proteins onto DNA supports by forming a monovalent and stable intermediate linkage between immobilized ssDNA and the target proteins (Scheme 1).
Scheme 1.
Synthetic Route to Employ Artificial Protein–DNA Conjugates for Recombinant Protein Capture
The polypeptide polymers employed here contain the basic portion of the zipper complex (ZR), a polymerized elastic “midblock domain” or ELS fragment (VPGVG VPGSG VPGVG VPGSG VPGVG), and a C-terminal cysteine, yielding ZR-(ELS)6-C. After the in vivo expression and purification of these polymers, sulfosuccinimidyl 4-[N-maleimidomethyl]cyclohexane-1-carboxylate (sulfo-SMCC) was used as a heterobifunctional cross-linker to couple an amine-functionalized oligonucleotide to the C-terminal cysteine of the polymers (13). The large shift in the isoelectric point of the polymers after DNA labeling allowed the conjugates to be purified to homogeneity using fast protein liquid chromatography (FPLC). Since the artificial proteins can sustain, without loss of function, lyophilization, resuspension in denaturing buffers, and extended dialysis, the labeling reactions can be scaled up, and this route allows concentrated stock solutions of ZR-(ELS)6-C-ssDNA to be prepared despite the intrinsic inefficiency of the sulfo-SMCC cross-linking reaction.
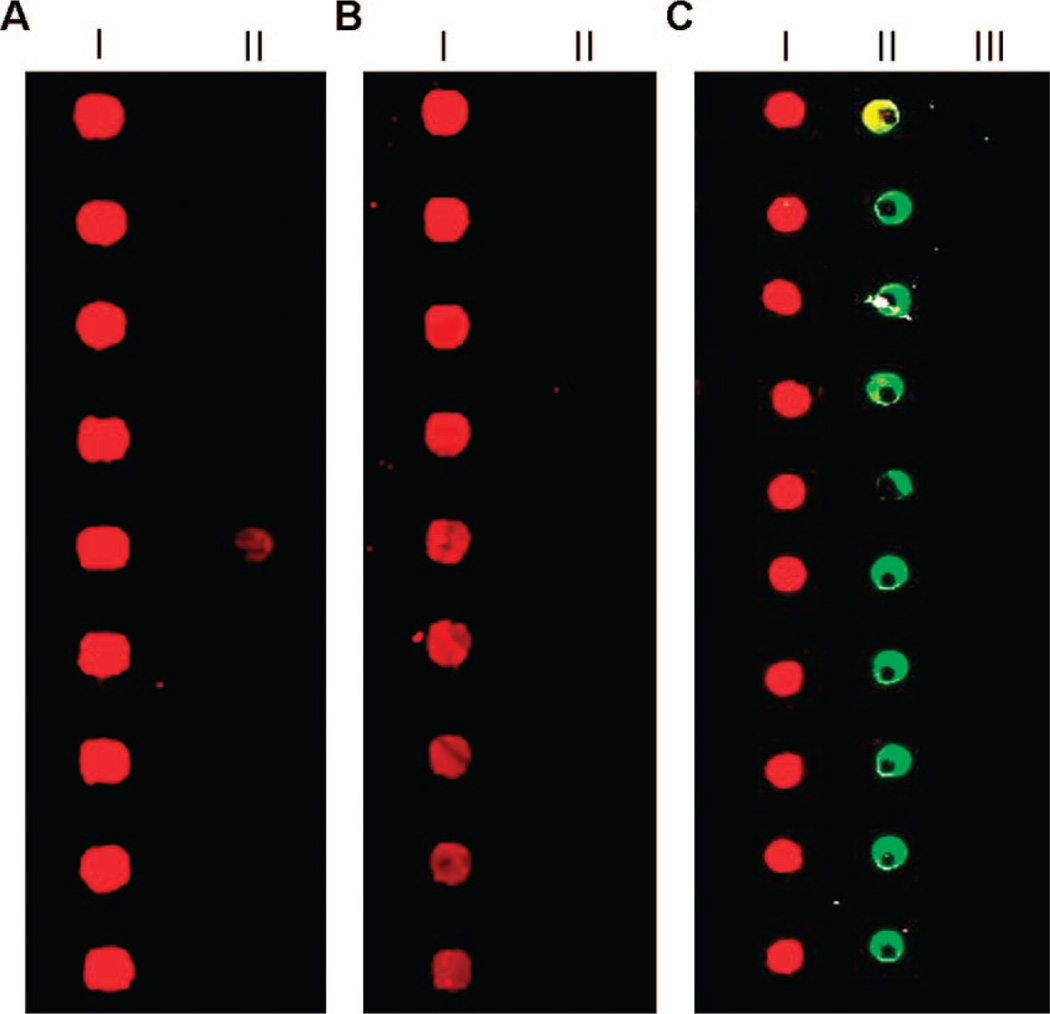
To test the ability of 1 to control the deposition of protein on DNA matrices, we labeled two samples of ZR-(ELS)6-C with different oligonucleotides containing non-complementary sequences. These polymers were then used as solution-phase capture probes that encode the spatial deposition of recombinant proteins onto DNA-printed microarrays (Figure 1). In this procedure, green fluorescent protein (GFP) and glutathione-S-transferase (GST) were chosen as target proteins and were expressed in vivo with C-terminal ZE fusions. Protein capture was achieved by mixing samples of either Ni2+-NTA purified target proteins (~1 mg/mL) or expressed proteins in unpurified cell lysates with a single conjugated version of 1 at a 3:1 molar ratio of target protein to polymer. After a brief incubation period (<30 min), these solutions were incubated for ~12 h over a custom-fabricated DNA microarray. Each array was washed and probed with fluorescently labeled antibodies (either Alexa647-anti-GFP or both Alexa647-anti-GFP and Cy3-anti-GST at 1 µg/mL) and scanned using a GenePix 4000B microarray scanner (Molecular Devices). DNA-directed self-assembly of both GFP-ZE and GST-ZE was found to be highly selective. As expected, arrays formed using either purified proteins or cell lysates demonstrated the oligonucleotide sequence-specific targeting of the proteins. Microarray spots containing the appropriate complementary ssDNA sequence possessed average signal-tonoise ratios (SNR) with a range of 20 to 60. Proteins appearing in the Cy3 channel tended toward the higher end of this range. On the other hand, average SNR values were significantly lower for spots containing non-complementary strands, an SNR range of 1.6 to 5. Furthermore, when precaptured protein solutions were combined for 1, 2, 4, or 8 h prior to their incubation on the microarray, we did not observe evidence of exchange between proteins displaying the zipper fusion.
Figure 1.
The use of artificial polypeptide conjugates to create protein microarrays encoded by DNA. Arrays probed with Alexa647-anti-GFP were prepared using purified GFP-ZE (A) and cell lysates containing expressed GFP-ZE (B). In both cases, column I denotes spots containing the complementary sequence for GFP-ZE/polymer complex while a non-complementary control strand was spotted in column II. Images in A and B were cropped and placed together for clarity. The pitch between spots is 250 µm. (C) Two-component arrays were fabricated through a single incubation of precaptured GFP-ZE and GST-ZE proteins. Column I contains a complementary strand for GFP-ZE/polymer complex, column II contains a complementary sequence for a second GST-ZE /polymer complex, and column III contains spots of a non-complementary control strand. Multiprotein arrays were probed simultaneously with a mixture of Alexa647-anti-GFP and Cy3-anti-GST. The pitch between spots is 500 µm.
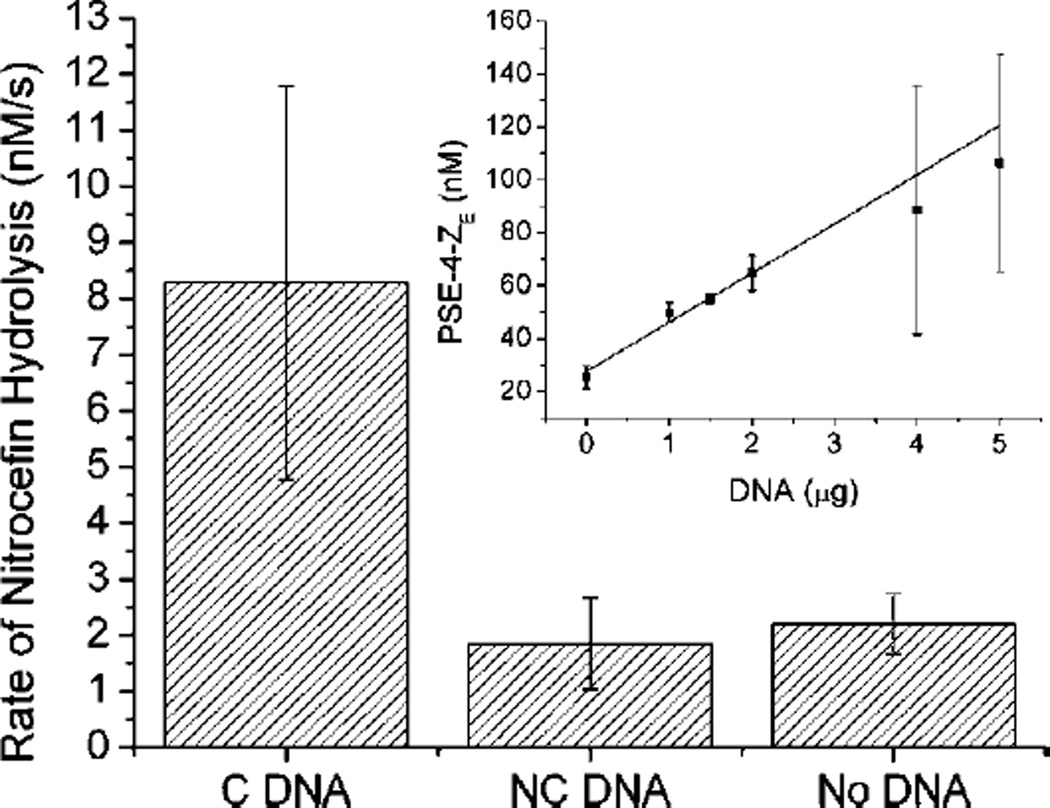
We also examined the effect our DNA-directed immobilization strategy had on enzymatic activity. As a test case, we immobilized a β-lactamase enzyme (PSE-4) onto the surface of 96-well plates that were functionalized with streptavidin and then biotin-labeled oligonucleotides. DNA-directed deposition was performed using a similar procedure to that developed for the microarrays except that the PSE-4-ZE fusion protein was prepared using standard periplasmic expression and lysis methods. Nitrocefin was used as a substrate for PSE-4, since the enzymatic opening of its β-lactam bond produced a detectable change in absorption at 485 nm using a Tecan Infinite 200 microplate reader. Measurements of nitrocefin hydrolysis rates again verified ability of 1 to selectively control enzyme deposition of target proteins (Figure 2B). Wells containing 5 µg of a complementary oligonucleotide to 1 showed a 4-fold increase in activity over wells that contained non-complementary sequences or where the ssDNA was omitted (Figure 2). Consistent with previous reports that utilize DNA-directed immobilization of enzymes (11), levels of PSE-4 activity were found to be linearly dependent on the amount of DNA that is immobilized in the wells (Figure 2 inset). Using soluble PSE-4-ZE as a calibration standard, the hydrolysis rate was converted to PSE-4-ZE concentration, yielding a slope of 18.6 nM PSE-4-ZE per microgram of DNA.
Figure 2.
DNA-directed control over immobilized enzyme activity. A 4-fold increase in PSE-4-ZE activity over background nitrocefin hydrolysis is seen in wells that were incubated with 5 µg of cDNA strands (C DNA) when compared to wells containing the same amount of a non-cDNA strand (NC DNA). Control wells where the DNA-functionalization step was omitted (No DNA) are also shown for comparison. The inset shows the linear dependence of PSE-4-ZE activity on the amount of cDNA deposited in the wells.
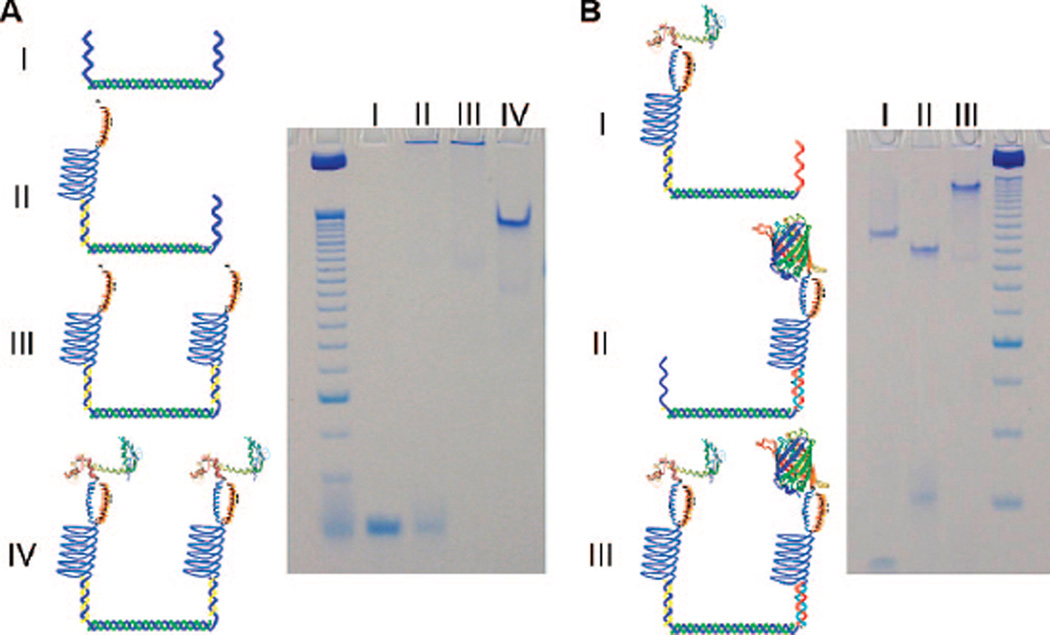
Finally, the two different conjugated versions of 1 were synthesized and used to facilitate the self-assembly of multiprotein complexes onto molecular-scale scaffolds formed from DNA. Complexes containing either two GFP-ZE proteins or a GFP-ZE and a calmodulin protein (CaM-ZE) were prepared (Figure 3). The DNA scaffold consisted of a 10-nm-long double helix (34 bases) flanked by unique single-strand “overhangs” comprising 20 bases on either end. The “overhangs” possessed either the same or orthogonal oligonucleotide sequences depending on whether single or multicomponent complexes were being synthesized. Multiprotein assemblies were synthesized by preincubating 1 with GFP-ZE or CaM-ZE, and then adding the DNA scaffold. In each case, the protein, polymer, and “overhang” concentrations were held at a 1:1:1 stoichiometry. Complex formation was examined using non-denaturing polyacrylamide gel electorphoresis (PAGE) and by staining for DNA and protein using Stains-All (Sigma). Clear separation between the scaffold and both partially and fully formed assemblies was observed. For each complex, gel intensity analysis indicated that the multiprotein complexes formed in high yield (>95%) using these conditions, proving that the association of the zipper complex and DNA-hybridization provided stable and selective linkages to drive the assembly process near quantitative yield, and that this process does not require the addition of large excesses of individual assembly components to form complete complexes.
Figure 3.
Non-denaturing PAGE-gels confirming the synthesis of multiprotein complexes. (A) Native PAGE-gel showing the successful formation of the complex containing two CaM-ZE proteins assembled on a single DNA double helix. The dominant species observed in the individual lanes of the gel are indicated by Roman numerals. The labeling scheme is as follows: I. DNA scaffold alone; II. assemblies prepared at a 1 to 0.5 ratio of ssDNA scaffold to conjugates; III. a “half” complex composed of the DNA scaffolds and two artificial polypeptides; IV. complete assemblies formed at a 1:1:1 ratio of all assembly components. (B) Gel demonstrating the assembly of multiprotein complexes containing both a GFP-ZE and a CaM-ZE (III), templated on a DNA scaffold possessing two different “overhang” sequences. Partial complexes formed using only one type of protein and corresponding to either CaM-ZE (I), or GFP-ZE (II) are also shown.
In summary, we have demonstrated that DNA-conjugated artificial polypeptides can be used as protein capture probes that facilitate the oligonucleotide sequence-dependent deposition onto DNA-functionalized matrices. Furthermore, we have shown that these polymers allow the stoichiometries of enzymes to be deterministically controlled by forming a stable intermediate linkage between DNA-coated supports and target proteins. Importantly, our modular strategy separates the production of recombinant target proteins from DNA-conjugation procedures. Here, the synthesis of a single set of polymer allows diverse combinations of ZE-tagged proteins to be immobilized for different applications. Furthermore, the selectivity and stability of the heterodimeric association of the leucine zipper system employed here permits target protein capture and pull down from unpurified cellular lysates and is compatible with a variety of protein expression systems and techniques. In this way, our strategy presents a flexible and versatile alternative to existing DNA-conjugation methods that is advantageous for applications where the ability to conjugate proteins with DNA is limiting, and in circumstances that require minimal postexpression processing of target proteins.
Supplementary Material
Acknowledgments
This research was generously supported by grants from the NSF (MCB-0645832) and the Welch Foundation (C-1625). P.E.C. was supported by a training fellowship awarded by the Program of the W.M. Keck Center for Interdisciplinary Bioscience (NIH grant No. T90 DK70121-03 and 5R90DK71504-3). We are also grateful for support from the NSF sponsored Center of Biological and Environmental Nanotechnology (CBEN) at Rice University.
Footnotes
Supporting Information Available: A detailed description of materials and methods, a listing of the amino acid and DNA sequences for all reagents, MALDI-TOF MS spectrum of the ZR-(ELS)6-C protein, FPLC chromatogram of the ZR-(ELS)6-C-ssDNA with the corresponding SDS-PAGE gel, Mircoarray from time-dependent incubations, and the full non-denaturing PAGE gel for the multiprotein complex. This material is available free of charge via the Internet at http://pubs.acs.org.
LITERATURE CITED
- 1.Konig T, Skerra A. Use of an albumin-binding domain for the selective immobilisation of recombinant capture antibody fragments on ELISA plates. J. Immunol. Methods. 1998;218:73–83. doi: 10.1016/s0022-1759(98)00112-4. [DOI] [PubMed] [Google Scholar]
- 2.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 3.Wu G, Datar RH, Hansen KM, Thundat T, Cote RJ, Majumdar A. Bioassay of prostate-specific antigen (PSA) using microcantilevers. Nat. Biotechnol. 2001;19:856–860. doi: 10.1038/nbt0901-856. [DOI] [PubMed] [Google Scholar]
- 4.Camarero JA. Recent developments in the site-specific immobilization of proteins onto solid supports. Biopolymers. 2008;90:450–458. doi: 10.1002/bip.20803. [DOI] [PubMed] [Google Scholar]
- 5.Rusmini F, Zhong Z, Feijen J. Protein immobilization strategies for protein biochips. Biomacromolecules. 2007;8:1775–1789. doi: 10.1021/bm061197b. [DOI] [PubMed] [Google Scholar]
- 6.Zhu H, Snyder M. Protein chip technology. Curr. Opin. Chem. Biol. 2003;7:55–63. doi: 10.1016/s1367-5931(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 7.Niemeyer CM, Boldt L, Ceyhan B, Blohm D. DNA-Directed immobilization: efficient, reversible, and site-selective surface binding of proteins by means of covalent DNA-streptavidin conjugates. Anal. Biochem. 1999;268:54–63. doi: 10.1006/abio.1998.3017. [DOI] [PubMed] [Google Scholar]
- 8.Wacker R, Niemeyer CM. DDI-µFIA - A readily configurable microarray-fluorescence immunoassay based on DNA-directed immobilization of proteins. ChemBioChem. 2004;5:453–459. doi: 10.1002/cbic.200300788. [DOI] [PubMed] [Google Scholar]
- 9.Wacker R, Schroder H, Niemeyer CM. Performance of antibody microarrays fabricated by either DNA-directed immobilization, direct spotting, or streptavidin-biotin attachment: a comparative study. Anal. Biochem. 2004;330:281–287. doi: 10.1016/j.ab.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Erben CM, Goodman RP, Turberfield AJ. Single-molecule protein encapsulation in a rigid DNA cage. Angew. Chem., Int. Ed. 2006;45:7414–7417. doi: 10.1002/anie.200603392. [DOI] [PubMed] [Google Scholar]
- 11.Jung GY, Stephanopoulos G. A functional protein chip for pathway optimization and in vitro metabolic engineering. Science. 2004;304:428–431. doi: 10.1126/science.1096920. [DOI] [PubMed] [Google Scholar]
- 12.Niemeyer CM, Koehler J, Wuerdemann C. DNA-directed assembly of bienzymic complexes from in vivo biotinylated NAD(P)H:FMN oxidoreductase and luciferase. ChemBioChem. 2002;3:242–245. doi: 10.1002/1439-7633(20020301)3:2/3<242::AID-CBIC242>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 13.Kukolka F, Niemeyer CM. Synthesis of fluorescent oligonucleotide-EYFP conjugate: towards supramolecular construction of semisynthetic biomolecular antennae. Org. Biomol. Chem. 2004;2:2203–2206. doi: 10.1039/b406492e. [DOI] [PubMed] [Google Scholar]
- 14.Niemeyer CM, Sano T, Smith CL, Cantor CR. Oligonucleotide-directed self-assembly of proteins: semisynthetic DNA-streptavidin hybrid molecules as connectors for the generation of macroscopic arrays and the construction of supramolecular bioconjugates. Nucleic Acids Res. 1994;22:5530–5539. doi: 10.1093/nar/22.25.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon Y, Coleman MA, Camarero JA. Selective immobilization of proteins onto solid supports through split-intein-mediated protein trans-splicing. Angew. Chem., Int. Ed. 2006;45:1726–1729. doi: 10.1002/anie.200503475. [DOI] [PubMed] [Google Scholar]
- 16.Lovrinovic M, Seidel R, Wacker R, Schroeder H, Seitz O, Engelhard M, Goody RS, Niemeyer CM. Synthesis of protein-nucleic acid conjugates by expressed protein ligation. Chem. Commun. 2003;7:822–823. doi: 10.1039/b212294d. [DOI] [PubMed] [Google Scholar]
- 17.Jongsma MA, Litjens RH. Self-assembling protein arrays on DNA chips by auto-labeling fusion proteins with a single DNA address. Proteomics. 2006;6:2650–2655. doi: 10.1002/pmic.200500654. [DOI] [PubMed] [Google Scholar]
- 18.Duckworth BP, Chen Y, Wollack JW, Sham Y, Mueller JD, Taton TA, Distefano MD. A universal method for the preparation of covalent protein-DNA conjugates for use in creating protein nanostructures. Angew. Chem., Int. Ed. 2007;46:8819–8822. doi: 10.1002/anie.200701942. [DOI] [PubMed] [Google Scholar]
- 19.Zhang K, Diehl MR, Tirrell DA. Artificial polypeptide scaffold for protein immobilization. J. Am. Chem. Soc. 2005;127:10136–10137. doi: 10.1021/ja051457h. [DOI] [PubMed] [Google Scholar]
- 20.Diehl MR, Zhang K, Lee HJ, Tirrell DA. Engineering cooperativity in biomotor-protein assemblies. Science. 2006;311:1468–1471. doi: 10.1126/science.1122125. [DOI] [PubMed] [Google Scholar]
- 21.Moll JR, Ruvinov SB, Pastan I, Vinson C. Designed heterodimerizing leucine zippers with a range of pIs and stabilities up to 10−15 M. Protein Sci. 2001;10:649–655. doi: 10.1110/ps.39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.