Keywords: nerve regeneration, sciatic nerve injury, miR-148b-3p, Schwann cells, migration, Cand1, gene expression, microarray, peripheral nerve injury, mechanisms, neural regeneration
Abstract
MicroRNAs (miRNAs) are small, non-coding RNAs that negatively adjust gene expression in multifarious biological processes. However, the regulatory effects of miRNAs on Schwann cells remain poorly understood. Previous microarray analysis results have shown that miRNA expression is altered following sciatic nerve transaction, thereby affecting proliferation and migration of Schwann cells. This study investigated whether miR-148b-3p could regulate migration of Schwann cells by directly targeting cullin-associated and neddylation-dissociated 1 (Cand1). Up-regulated expression of miR-148b-3p promoted Schwann cell migration, whereas silencing of miR-148b-3p inhibited Schwann cell migration in vitro. Further experiments confirmed that Cand1 was a direct target of miR-148b-3p, and Cand1 knockdown reversed suppression of the miR-148b-3p inhibitor on Schwann cell migration. These results suggested that miR-148b-3p promoted migration of Schwann cells by directly targeting Cand1 in vitro.
Introduction
Recovery of injuried central and peripheral nerves remains problematic and difficult (Navarro et al., 2007). This is primarily due to the inability for intrinsic growth and the existence of a regeneration barrier (Zou et al., 2009). Schwann cells (SCs) play a very important role in removing growth obstacles. Following sciatic nerve injury, mature SCs differentiate, proliferate, and migrate, thereby forming a path to guide the growth of new axons (Kury et al., 2001). SCs also contribute to the construction of a microenvironment for nerve regeneration by excreting multiple neurotrophic factors and adhesion molecules (Ngeow, 2010). However, the particular mechanisms that regulate SC proliferation and migration remain unknown. For successful regeneration, it is necessary to explore the molecular mechanisms of SCs.
microRNAs (miRNAs) are endogenous molecules that are approximately 22 nucleotides of non-coding RNA molecules (Bartel, 2009). miRNAs come from either miRNA genes or as a part of intron-encoded proteins; they are further maturated by the endoribonuclease Dicer (Wu and Murashov, 2013). Mature miRNA can play a negative role in the degradation or silencing of mRNA by combining the 3′-untranslated region (UTR) (Filipowicz et al., 2008; Carthew and Sontheimer, 2009). Knocking out the key Dicer not only inhibits differentiation, but also promotes apoptosis and cell death (De Pietri Tonelli et al., 2008). In SCs, Dicer deletion increases proliferation, but blocks myelination (Bremer et al., 2010; Pereira et al., 2010; Verrier et al., 2010). Taken together, these studies suggest that miRNAs play a critical role in cell development.
The role of miRNA has also been studied in a variety of diseases. For example, decreased miR-485-5p promotes BACE1, which stimulates the development of Alzheimer's disease (Faghihi et al., 2010). miR-433 and miR-7 regulate expression of α-synuclein, which is associated with cytotoxicity in Parkinson's disease (Wang et al., 2008; Junn et al., 2009). Previous studies have shown that miR-160b, 30b, and 181b are significantly up-regulated in the frontal cortex of schizophrenia patients (Kim et al., 2010; Santarelli et al., 2011), and miR-148b-3p increases proliferation of breast cancer cell lines (Jiang et al., 2015). Nevertheless, very little is understood about the role that miRNAs play in nerve regeneration (Lu et al., 2014).
Results from microarray analyses and extensive function screening have revealed that expression of many miRNAs, such as miR-221/222 and miR-182, changes after sciatic nerve injury and affects proliferation and migration of SCs (Yu et al., 2011, 2012a, b). The present study investigated whether miR-148b-3p could regulate SC migration by directly targeting cullin-associated and neddylation-dissociated 1 (Cand1), a negative regulator in the proliferation (Murata et al., 2010).
Materials and Methods
Primary Schwann cell culture and transfection with oligonucleotide
Primary SCs were obtained from sciatic nerves of 1-day-old Sprague-Dawley rats of either sex. The SCs were cultured for 2 days with 10 μM Ara-C (Sigma, St Louis, MO, USA) to eliminate fibroblasts. The SCs were then further cultured with 50 ng/mL recombinant glial growth factor 2 (R&D Systems, Minneapolis, MN, USA) and 2 μM forskolin (R&D Systems) for 3 days, and then were purified by incubating with anti-Thy1.1 antibody diluted in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (1:1,000; Sigma) for 1.5 hours on ice (Mantuano et al., 2008). SCs were used when the purity reached 98%, as determined by immunoreaction with S100β. Primary SC cultures were cultured in DMEM containing 10% fetal bovine serum at 37°C and in a humidified 5% CO2 incubator. miR-148b-3p mimics (20 mM), mimic control (20 mM), miR-148b-3p inhibitors (100 mM), inhibitor control (100 mM) or siRNAs (100 mM), and negative control (100 mM) (Ribobio, Guangzhou, Guangdong Province, China) were separately transfected into the SCs using Lipofectamine RNAiMAX transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions (Yu et al., 2012a). Assays were performed three times in triplicate wells. All experimental procedures involving animals were conducted in accordance with institutional animal care guidelines and were ethically approved by the Administration Committee of Experimental Animals (SYXK (Su) 2015-0016), Jiangsu Province, China.
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
At 36 hours after transfection with miRNA mimics or siRNAs, the Prime-Script RT reagent Kit (TaKaRa, Dalian, Liaoning Province, China) was used to synthesize reverse-transcribed complementary DNA (cDNA). PCR was conducted using the SYBR Premix Ex Taq kit (TaKaRa). RT-PCR was performed on an ABI7900 thermocycler (Applied Biosystems, Foster, CA, USA). qPCR primers were designed using NCBI Primer Blast and were manufactured by Sangon Biotech (Shanghai, China). Cand 1 primer sequence: forward: 5′-CCA GTC ACA GAT CAG CTC CA-3′; reverse: 5′-CCT CAT GTG GAA CAC ACG TC-3′; product size: 119 bp. The reaction system was as follows: 10 μL SYBR® Premix Ex TaqTM (2×), 2 μL PCR primer, 0.4 μL ROX Reference Dye, 1 μL product from RT reaction, and ddH2O to 20 μL. The PCR reaction was as follows: initial denaturation at 95°C for 2 minutes; 45 cycles of denaturation at 95°C for 15 seconds, annealing elongation at 60°C for 1 minute; final elongation at 95°C for 15 seconds, 60°C for 1 minute. Each sample was run in triplicate in each assay. β-Actin was used as the endogenous control. The relative expression level was calculated using the comparative 2−ΔΔCt methods (Livak and Schmittgen, 2001).
Cell migration assay
SC migration was investigated using Transwell chambers with 8-mm-deep pores (Costar, Cambridge, MA, USA). The bottom surface of each membrane was coated with 10 mg/mL fibronectin (Sigma). At 36 hours after transfection with miR-148b-3p mimics (20 mM), miR-148b-3p inhibitors (100 mM), or siRNAs (100 mM), 100 μL SCs (1 × 106 cells/mL) were re-suspended in DMEM and transferred to the top chambers of each Transwell (Mantuano et al., 2008).
The lower chambers were loaded with 500 μL complete medium. After 24 hours, a cotton swab was used to clean the upper surface of each membrane. Migrated cells on the bottom surface of the Transwell membrane were stained with 0.1% crystal violet and quantified using a DMR inverted microscope (Leica Microsystems Bensheim, Germany). Assays were performed three times in triplicate wells. A total of 10 fields were randomly sampled per well. The average number of crystal violet-stained cells per field was determined.
Luciferase reporter assay
Potential mRNA targets of miR-148b-3p were predicted by Target Scan and microarray. Cand1 was finally chosen from the intersection of the prediction and microarray. We obtained the 3′-UTR sequence of Cand1 from the genomic DNA and sub-cloned the region directly downstream of the luciferase gene stop codon in the luciferase reporter vector. Different p-Luc-UTR luciferase reporter vectors were obtained from PCR amplification of the 3′-UTR sequence of Cand1 using appropriate primers. Cand1-3′ UTR primer sequence: forward: 5′-CCG GAA TTC ACG TGT GTT CCA CAT GAG-3′; reverse: 5′-CCG CTC GAG AAA GTT TTA ACA TTT TAA TCC-3′; product size: 336 bp. The 3′-UTR sequences were confirmed by sequencing.
HEK293T cells were transfected with p-Luc-UTR (30 ng), miRNA mimics (5 pmol), and Renilla (5 ng) in each well of 96-well plates using the Lipofectamine 2000 transfection system (Invitrogen). At 48 hours after incubation, activities of firefly and Renilla luciferases were measured in the cell lysates using the dual-luciferase reporter assay system (Promega, Madison, WI, USA).
Statistical analysis
All data are expressed as the mean ± SD. Statistical analyses were performed by SPSS 18.0 software (SPSS, Chicago, IL, USA). The Student's t-test was used to compare the difference of intergroup data. A value of P < 0.05 was considered statistically significant.
Results
Effects of miR-148b-3p on Schwann cell migration in vitro
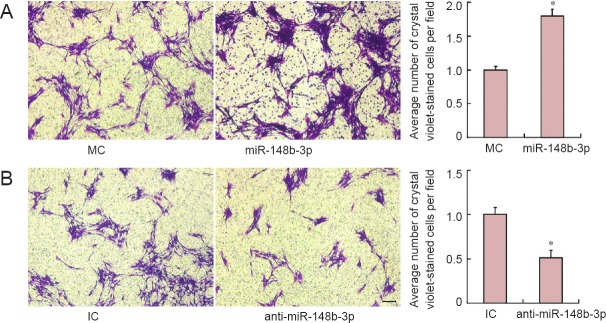
We investigated whether miR-148b-3p played a part in the regeneration of peripheral nerves. Primary SCs were transfected with mimic control and miR-148b-3p mimic, and then added to Transwell inserts 36 hours later. At 24 hours after cell culture, crystal violet staining showed that miR-148b-3p mimic significantly promoted SC migration compared with the control (P < 0.05; Figure 1A). Silencing miR-148b-3p decreased SC migration when transfected with the inhibitor control and miR-148b-3p inhibitor (P < 0.05; Figure 1B). These results indicated that miR-148b-3p increased SC migration in vitro.
Figure 1.
Effects of miR-148b-3p on Schwann cell migration in vitro (crystal violet staining).
(A) Transwell assays show improved migration of Schwann cells transfected with miR-148b-3p mimic (miR-148b-3p) compared with cells transfected with MC. (B) Migration of Schwann cells transfected with miR-148b-3p inhibitor (anti-miR-148b-3p) significantly decreases compared with cells transfected with IC. Scale bar: 50 μm. Schwann cells stained with crystal violet. *P < 0.05, vs. MC or IC (mean ± SD, Student's t-test). MC: Mimic control; IC: inhibitor control.
miR-148b-3p induced inhibition of Cand1 expression by targeting the 3′-UTR region
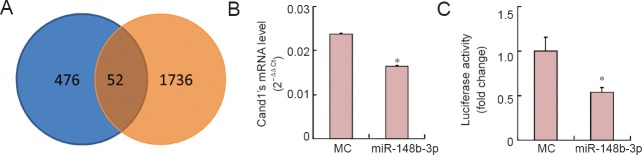
To investigate the underlying molecular mechanisms of miR-148b-3p initiating SC migration, potential mRNA targets of miR-148b-3p were selected by cross-referencing programs (Target Scan) and microarray results. A total of 476 potential target genes were predicted by software, and 1,736 down-regulated genes after transfection with miR-148b-3p mimics of SC were obtained (P < 0.05; Figure 2A). Among the 52 genes in the intersection of the two predictions, Cand1 was finally selected as a potential target of miR-148b-3p. Furthermore, a luciferase reporter construct was made by inserting the Cand1 3′-UTR containing the predicted target site of miR-148b-3p into the luciferase reporter gene. The relative luciferase activity was repressed by nearly 50% by miR-148b-3p (Figure 2B). These results demonstrated that miR-148b-3p specifically repressed Cand1 expression through the predicted target site in the Cand1 3′-UTR. qRT-PCR analysis further demonstrated that miR-148b-3p dramatically suppressed endogenous mRNA expression of Cand1 when the SCs were transfected with miR-148b-3p mimics (Figure 2C). These results suggested that miR-148b-3p reduced Cand1 expression by targeting the 3′-UTR region.
Figure 2.
miR-148b-3p-induced inhibition of Cand1 expression by targeting the 3′-untranslated region.
(A) A total of 476 potential target genes of miR-148b-3p were predicted by software in the blue area (left), and 1,736 down-regulated genes were obtained from microarray after transfection with miR-148b-3p mimics of SCs in the orange area (right), resulting in a total of 52 genes in the intersection of the two predictions. (B) Cand1 mRNA expression is down-regulated by miR-148b-3p mimics (miR-148b-3p) compared with MC (luciferase reporter assay). (C) Measurement of the relative activity of luciferase after co-transfecting the p-Luc-UTR vectors with miR-148b-3p mimics (miR-148b-3p) or MC into 293T cells (luciferase reporter assay). *P < 0.05, vs. MC (mean ± SD, Student's t-test). Cand1: Cullin-associated and neddylation-dissociated 1; MC: mimic control.
Recapitulation of miR-148b-3p effects by Cand1 knockdown in Schwann cells
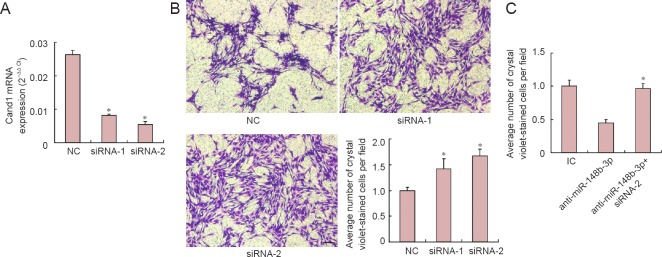
Two specific small interfering RNAs (siRNAs) against Cand1 were synthesized. The results showed that siRNA-1 and siRNA-2 both inhibited Cand1 expression compared with the negative control (P < 0.05; Figure 3A). The Transwell assay showed that siRNA-1 and siRNA-2 both promoted SC migration, although the effect of siRNA-2 was more obvious (P < 0.05; Figure 3B). To further determine whether down-regulation of Cand1 directly mediated miR-148b-3p-induced SC migration, SCs were transfected with miR-148b-3p inhibitor with or without siRNA-2 against Cand1 (P < 0.05). As shown in Figure 3C, anti-miR-148b-3p significantly decreased SC migration. Conversely, a significant increase in cell migration was detected in groups co-transfected with miR-148b-3p inhibitor and siRNA-2 (P < 0.05). These results suggested that inhibition of Cand1 expression rescued the migration suppression induced by the miR-148b-3p inhibitor.
Figure 3.
Recapitulation of miR-148b-3p effects by Cand1 knockdown in Schwann cells.
(A) Cand1 mRNA expression is down-regulated after transfection with Cand1 siRNA-1 (siRNA-1) or Cand1 siRNA-2 (siRNA-2) compared with siRNA control (NC) in Schwann cells. (B) The Transwell assay suggests that Cand1 siRNA-1 and siRNA-2 both significantly increase Schwann cell migration compared with NC. Schwann cells stained with crystal violet. (C) Transwell assay for Schwann cell migration following transfection with miR-148b-3p (anti-miR-148b-3p) with or without Cand1 siRNA-2. Cand1 silencing reverses suppression of miR-148b-3p inhibitor on Schwann cell migration. *P < 0.05, vs. NC or IC (mean ± SD, Student's t-test). Scale bar: 50 μm. Cand1: Cullin-associated and neddylation-dissociated 1; NC: negative control; IC: inhibitor control.
Discussion
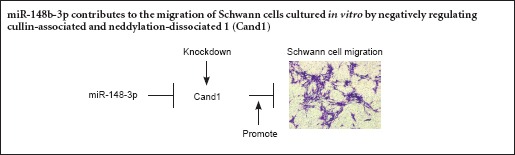
The in vitro role of miR-148b-3p in SCs was explored in this study. Transfection with miR-148b-3p mimics or inhibitors revealed that miR-148b-3p improved SC migration. Cand1, a negative regulator of SKP1-Cullin1-F-box ubiquitin ligases, has the direct target region of miR-148b-3p. Decreased Cand1 expression can promote SC migration. These data showed that increased expression of miR-148b-3p promotes SC migration by reducing Cand1 expression.
During nerve regeneration after peripheral nerve injury, miRNAs provide a powerful mechanism for post-transcriptional control of gene expression. Microarray analysis revealed miRNAs with significant expression changes, such as miR-9, miR-132, miR-182, Let-7, miR-221, and miR-222. Our previous studies showed that miR-9 inhibits SC migration by targeting Cthrc1 (Zhou et al., 2014); miR-221 and miR-222 promote SC proliferation and migration by targeting LASS2 (Yu et al., 2012b); miR-182 inhibits SC proliferation and migration by targeting FGF9 and NTM following sciatic nerve injury (Yu et al., 2012a); and Let-7 reduces SC proliferation and migration by targeting NGF (Li et al., 2015). Liu et al. (2015) showed that inhibition of miR-148b stimulates cell proliferation, enhances chemosensitivity, and increases cell metastasis and angiogenesis in vitro. Another study confirmed that miR-148b suppresses hepatocellular carcinoma cell proliferation and invasion by targeting the WNT1/β-catenin pathway (Zhang et al., 2015). However, the mechanisms of miR-148b-3p are different from miR-148b, and miR-148b-3p has been shown to increase proliferation of breast cancer cell lines (Aure et al., 2013). Proliferation of breast cancer cell lines can also be increased by miR-148b-3p (Jiang et al., 2015). The results from the present study showed another function of miR-148b-3p increased SC migration in vitro by targeting Cand1.
Cand1 has been shown to remold the SKP1-Cullin1-F-box repertoire in response to changing growth conditions (Zemla et al., 2013), and Cand1 has also been shown to be a negative regulator in the proliferation of lymph node carcinoma of prostate cells (Murata et al., 2010). The present study explored whether miR-148b-3p and Cand1 affected SC proliferation, and the results showed no change in SC proliferation, regardless of whether expression of miR-148b-3p or Cand1 was altered.
In summary, Cand1 suppressed migration of SCs, and the results showed a direct interaction between Cand1 and miR-148b-3p. SC proliferation and migration can affect myelination, suggesting that further studies are needed to determine the effects of Cand1 on the myelin of axons. The results from the present study offer a novel target to study SC migration, and provide evidence for a role for Cand1 in peripheral nerve regeneration, as well as cancer diagnosis and treatment.
Footnotes
Funding: This study was supported by the National Key Basic Research Program of China, No. 2014CB542202; the National High-Tech R&D Program of China (863 Program), No. 2012AA020502; the National Natural Science Foundation of China, No. 81130080, 81371389 and 81571198; the Natural Science Foundation of Nantong University of China, No. 13040397; the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflicts of interest: None declared.
Plagiarism check: This paper was screened twice using Cross-Check to verify originality before publication.
Peer review: This paper was double-blinded and stringently reviewed by international expert reviewers.
Copyedited by Cooper C, Hindle A, Wang J, Qiu Y, Li CH, Song LP, Zhao M
References
- Aure MR, Leivonen SK, Fleischer T, Zhu Q, Overgaard J, Alsner J, Tramm T, Louhimo R, Alnaes GI, Perala M, Busato F, Touleimat N, Tost J, Borresen-Dale AL, Hautaniemi S, Troyanskaya OG, Lingjaerde OC, Sahlberg KK, Kristensen VN. Individual and combined effects of DNA methylation and copy number alterations on miRNA expression in breast tumors. Genome Biol. 2013;14:R126. doi: 10.1186/gb-2013-14-11-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer J, O’Connor T, Tiberi C, Rehrauer H, Weis J, Aguzzi A. Ablation of Dicer from murine Schwann cells increases their proliferation while blocking myelination. PLoS One. 2010;5:e12450. doi: 10.1371/journal.pone.0012450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, 3rd, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Jiang X, Du L, Wang L, Li J, Liu Y, Zheng G, Qu A, Zhang X, Pan H, Yang Y, Wang C. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int J Cancer. 2015;136:854–862. doi: 10.1002/ijc.29041. [DOI] [PubMed] [Google Scholar]
- Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009;106:13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, van den Oord EJ, Riley BP, Kendler KS, Vladimirov VI. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124:183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kury P, Stoll G, Muller HW. Molecular mechanisms of cellular interactions in peripheral nerve regeneration. Curr Opin Neurol. 2001;14:635–639. doi: 10.1097/00019052-200110000-00013. [DOI] [PubMed] [Google Scholar]
- Li S, Wang X, Gu Y, Chen C, Wang Y, Liu J, Hu W, Yu B, Wang Y, Ding F, Liu Y, Gu X. Let-7 microRNAs regenerate peripheral nerve regeneration by targeting nerve growth factor. Mol Ther. 2015;23:423–433. doi: 10.1038/mt.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Xu Y, Wei S, Gao W, Chen L, Zhou T, Wang Z, Ying M, Zheng Q. microRNA-148b suppresses hepatic cancer stem cell by targeting neuropilin-1. Biosci Rep. 2015;35:e00229. doi: 10.1042/BSR20150084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu A, Huang Z, Zhang C, Zhang X, Zhao J, Zhang H, Zhang Q, Wu S, Yi X. Differential expression of microRNAs in dorsal root ganglia after sciatic nerve injury. Neural Regen Res. 2014;9:1031–1040. doi: 10.4103/1673-5374.133164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantuano E, Inoue G, Li X, Takahashi K, Gaultier A, Gonias SL, Campana WM. The hemopexin domain of matrix metalloproteinase-9 activates cell signaling and promotes migration of schwann cells by binding to low-density lipoprotein receptor-related protein. J Neurosci. 2008;28:11571–11582. doi: 10.1523/JNEUROSCI.3053-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Takayama K, Katayama S, Urano T, Horie-Inoue K, Ikeda K, Takahashi S, Kawazu C, Hasegawa A, Ouchi Y, Homma Y, Hayashizaki Y, Inoue S. miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis. 2010;13:356–361. doi: 10.1038/pcan.2010.32. [DOI] [PubMed] [Google Scholar]
- Navarro X, Vivo M, Valero-Cabre A. Neural plasticity after peripheral nerve injury and regeneration. Prog Neurobiol. 2007;82:163–201. doi: 10.1016/j.pneurobio.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Ngeow WC. Scar less: a review of methods of scar reduction at sites of peripheral nerve repair. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2010;109:357–366. doi: 10.1016/j.tripleo.2009.06.030. [DOI] [PubMed] [Google Scholar]
- Pereira JA, Baumann R, Norrmen C, Somandin C, Miehe M, Jacob C, Luhmann T, Hall-Bozic H, Mantei N, Meijer D, Suter U. Dicer in Schwann cells is required for myelination and axonal integrity. J Neurosci. 2010;30:6763–6775. doi: 10.1523/JNEUROSCI.0801-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol Psychiatry. 2011;69:180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- Verrier JD, Semple-Rowland S, Madorsky I, Papin JE, Notterpek L. Reduction of Dicer impairs Schwann cell differentiation and myelination. J Neurosci Res. 2010;88:2558–2568. doi: 10.1002/jnr.22418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Murashov AK. Molecular mechanisms of peripheral nerve regeneration: emerging roles of microRNAs. Front Physiol. 2013;4:55. doi: 10.3389/fphys.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Zhou S, Wang Y, Ding G, Ding F, Gu X. Profile of microRNAs following rat sciatic nerve injury by deep sequencing: implication for mechanisms of nerve regeneration. PLoS One. 2011;6:e24612. doi: 10.1371/journal.pone.0024612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding F, Gu X. miR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. Nucleic Acids Res. 2012a;40:10356–10365. doi: 10.1093/nar/gks750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Zhou S, Wang Y, Qian T, Ding G, Ding F, Gu X. miR-221 and miR-222 promote Schwann cell proliferation and migration by targeting LASS2 after sciatic nerve injury. J Cell Sci. 2012b;125:2675–2683. doi: 10.1242/jcs.098996. [DOI] [PubMed] [Google Scholar]
- Zemla A, Thomas Y, Kedziora S, Knebel A, Wood NT, Rabut G, Kurz T. CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat Commun. 2013;4:1641. doi: 10.1038/ncomms2628. [DOI] [PubMed] [Google Scholar]
- Zhang JG, Shi Y, Hong DF, Song M, Huang D, Wang CY, Zhao G. MiR-148b suppresses cell proliferation and invasion in hepatocellular carcinoma by targeting WNT1/beta-catenin pathway. Sci Rep. 2015;5:8087. doi: 10.1038/srep08087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Gao R, Hu W, Qian T, Wang N, Ding G, Ding F, Yu B, Gu X. MiR-9 inhibits Schwann cell migration by targeting Cthrc1 following sciatic nerve injury. J Cell Sci. 2014;127:967–976. doi: 10.1242/jcs.131672. [DOI] [PubMed] [Google Scholar]
- Zou H, Ho C, Wong K, Tessier-Lavigne M. Axotomy-induced Smad1 activation promotes axonal growth in adult sensory neurons. J Neurosci. 2009;29:7116–7123. doi: 10.1523/JNEUROSCI.5397-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]