Abstract
Regeneration or replacement of lost cardiomyocytes within the heart has the potential to revolutionize cardiovascular medicine. Numerous methodologies have been used to achieve this aim, including the engraftment of bone marrow- and heart-derived cells as well as the identification of modulators of adult cardiomyocyte proliferation. Recently, the conversion of human somatic cells into induced pluripotent stem cells and induced cardiomyocyte-like cells has transformed potential approaches toward this goal, and the engraftment of cardiac progenitors derived from human embryonic stem cells into patients is now feasible. Here we review recent advances in our understanding of the genetic and epigenetic control of human cardiogenesis, cardiac differentiation, and the induced reprogramming of somatic cells to cardiomyocytes. We also cover genetic programs for inducing the proliferation of endogenous cardiomyocytes and discuss the genetic state of cells used in cardiac regenerative medicine.
Keywords: induced pluripotent stem cells, embryonic stem cells, cardiomyocyte, heart, epigenetics, developmental biology
INTRODUCTION
Heart disease is the leading cause of death in the developed world, accounting for one in four deaths in the United States (http://www.cdc.gov/heartdisease/facts.htm). A vast number of cardiac diseases are characterized by the acute or progressive loss of the cardiomyocytes that form the heart muscle and for which the adult heart has little or no regenerative capacity (5). The generation of new cardiomyocytes to replace these lost cells has focused on two paradigms: (a) cell replacement therapy, in which new cardiomyocytes are engrafted into the heart, and (b) the proliferation of endogenous cells in the heart, using factors or cells that stimulate endogenous cell turnover. These two methodologies are not mutually exclusive, and it is possible that the therapeutic effect of cell replacement therapies depends on a combination of proliferation, prevention of cell death, and improved function of endogenous cardiomyocytes. Advances in human cardiac cell isolation, differentiation, reprogramming, and engraftment to achieve these aims have progressed at a rapid pace. In approximately 15 years, we have proceeded from the first attempts at engrafting human bone marrow–derived cells into the heart, to trials using adult heart-derived cardiac progenitor cells (10, 79), and to the possibility of in vivo direct reprogramming of cells to cardiomyocytes within the heart (98). Recently, the first clinical trial using human embryonic stem cell (hESC)-derived cardiac progenitor cells (CPCs) was initiated (82). In addition, advances in bioengineering have vastly improved strategies for producing multilayered patches consisting of mixtures of heart cell types to enhance engraftment (142), and continuing clinical trials in cardiovascular medicine have provided a mature pipeline for the progression from cells generated in vitro to therapies (59).
Many of the advances in this field have resulted from our growing understanding of heart development. Human heart development is challenging to study, as primary adult cardiomyo-cytes are difficult to isolate and can only be maintained in active cell culture for a few weeks. This has forced the cardiovascular research community to look for other mammalian models to understand cardiomyocyte biology, focusing largely on murine and avian cardiogenesis. More recently, researchers have begun to study cardiomyocytes derived from human pluripotent stem cells (hPSCs), which include both hESCs and human induced pluripotent stem cells (hiPSCs). Despite this progress, we must greatly improve our understanding of the genetics and epigenetics of human heart development and repair before safe and effective therapies for human cardiac regenerative medicine can be developed on a large scale.
GENETIC CONTROL OF CARDIAC DEVELOPMENT
Over the past two decades, studies have elucidated the transcription factor networks and signaling pathways that control the four specific processes that result in heart formation: specification of the cardiac mesoderm, determination of the bilateral symmetry of the heart fields, patterning of the heart fields, and terminal cardiac differentiation (Figure 1). Understanding these pathways is crucial for generating therapies for the failing heart, allowing more extensive strategies for cardiac differentiation, cardiomyocyte subtype specification, and cell maturation, as well as identifying true cardiac stem cell populations. Much of our knowledge is based on the mouse, but despite the homology between human and mouse models, many important differences exist. For examples, the human heart is more than 1,000 times larger than that of the mouse (132); the human heart begins to contract at approximately day 22 after fertilization versus approximately day 9 in the mouse heart; and murine heart septation is complete in 2 weeks, whereas this process takes 2 months in humans (132). The difficulty of studying human embryology meant that relatively little was known about the genetic control of human cardiogenesis until mouse findings could be validated with studies of hPSC cardiac differentiation.
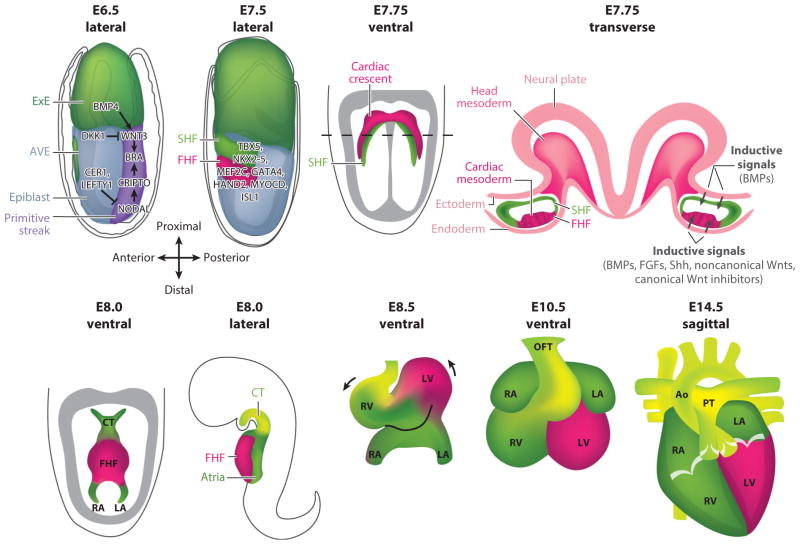
Figure 1.
Development of the murine heart as a model for human cardiogenesis. On embryonic day (E) 6.5, BMP4 signaling from the extraembryonic ectoderm (ExE) upregulates T (brachyury) expression via WNT3 signaling. NODAL signaling from the epiblast also upregulates T expression via CRIPTO. Inhibitory DKK1 and CER1/LEFTY1 from the anterior visceral endoderm (AVE) restrict WNT3 and NODAL, respectively, confining the primitive streak to the posterior embryo. On E7.5 (lateral view) a portion of the mid-streak mesoderm expressing T-induced Mesp1 undergoes epithelial-to-mesenchymal transition and moves bilaterally around the embryo from the primitive streak to the anterior side of the embryo to form the first and secondary heart fields (FHF and SHF, respectively). On E7.75 (ventral view), the FHF forms the cardiac crescent that is ventral to the SHF progenitor to maintain a precursor state. As shown in the transverse view on E7.75, signaling from the endoderm induces FHF differentiation and signaling from the neural plate/ectoderm to maintain SHF progenitors. On E8.0, the FHF forms the heart tube and begins to contract; the SHF is specific to the conotruncus (CT) and left and right atria (LA and RA). On E8.5, heart looping begins, and the left ventricle (LV) and right ventricle (RV) are spatially localized. On E10.5, the LA and RA are localized, and the outflow tract (OFT) is formed. On E14.5, the four-chambered heart and valves are completed, and the pulmonary trunk (PT) and aorta (Ao) are formed. Abbreviation: BMPs, bone morphogenetic proteins.
Early cardiac progenitors are assigned to a specific developmental path prior to or shortly after the initiation of gastrulation (34), the process by which the three germ layers (ectoderm, mesoderm, and endoderm) are formed (reviewed in 117; Figure 1). The induction of mesoderm, the germ layer from which the heart forms, begins with higher concentrations of NODAL in the proximal epiblast on mouse embryonic day (E)5.0. NODAL diffuses to the neighboring extraembryonic ectoderm (ExE) to maintain high levels of BMP4 expression, which is a secondary signal for mesoderm formation (1). Simultaneously, NODAL signals maintain the expression of embryonic visceral endoderm–related genes such as Lhx1, Fgf5, Fgf8, Bmp2, Otx2, and Foxa2. BMP4 from the ExE diffuses and induces WNT3 expression in the proximal epiblast. At E5.5, a signaling center on the tip of the distal visceral endoderm migrates anteriorly and expresses NODAL antagonists (LEFTY1 and CER1) and a WNT antagonist (DKK1), restricting NODAL and WNT signaling to the posterior epiblast (103). At E5.75, WNT3 induces the expression of mesoendodermal transcription factors such as T (brachyury), MIXL1, and EOMES, which, along with NODAL and the growth factors FGF4 and FGF8, help form the primitive streak.
At E6.25, T and EOMES induce the expression of MESP1, which is thought to be a master regulator of cardiac progenitor specification (11). It was once thought to be an exclusive marker of mesoderm committed to the cardiac fate, although more recently it has been demonstrated to be present in mesoderm-committed hematopoietic and skeletal muscle lineages as well (22). MESP1 regulates many of the key transcription factors implicated in epithelial-to-mesenchymal transition, such as SNAI1, which downregulates CDH1 (E-cadherin) to allow for cell movement (76). Cardiac progenitors expressing MESP1 and the surface markers KDR, PDGFRA, and CXCR4 (12) transition out of the primitive streak and migrate bilaterally, looping around either side of the embryo in the lateral plate mesoderm to converge at the midline, the site of the future heart. Two distinct progenitor cell populations that arise from the cardiac mesoderm are called first and second heart fields (FHFs and SHFs, respectively), and they provide different regional contributions to the developing heart. The FHF (from the lateral anterior splanchnic mesoderm) predominantly forms the left ventricle, with a minor contribution to the atria and conduction system, whereas the SHF (from the splanchnic pharyngeal mesoderm) gives rise to the right ventricle, outflow tract, and atria (14; Figure 1). Importantly, recent experiments using temporal clonal labeling have demonstrated that, prior to initiation of the respective gene regulatory networks, MESP1+ cardiac mesoderm progenitors have already divided into those cells that will generate the FHF and those that will generate the SHF (70).
The relationship between different cardiac progenitor cell populations is becoming better understood (Figure 2). When FHF progenitors reach the future site of the heart, they coalesce to form the cardiac crescent. Recently, HCN4 has been proposed as a marker for the FHF (112), and SHF progenitors are identified by Isl1 expression (85). ISL1+ cells lie medial and posterior to the crescent and remain in an undifferentiated progenitor state until their incorporation into the heart, as a result of inhibitory WNT signals from the midline (68). Along with expression in the SHF, Isl1 is detected within the FHF at the crescent stage (97). Isl1−/− mutants still form a primitive cardiac tube; the mutant phenotype primarily affects the SHF, confirming ISL1’s major role in SHF development. ISL1+ progenitors are multipotent and capable of giving rise to cardiomyocytes, smooth muscle cells, and endothelial cells (85). By E8.0, the FHF cells form a primitive heart tube, consisting of an interior layer of endocardial cells and an exterior layer of myocardial cells, and begin to contract. The heart tube contains an anterior arterial pole and a posterior venous pole. Heart tube extension is driven by the addition of SHF cells to these poles, resulting in rapid elongation and a rightward looping at E8.5 as a consequence of uneven growth and remodeling, which leads to the formation of primitive ventricles and atria (Figure 1). In total, ISL1+ SHF cells are responsible for generating more than two-thirds of the heart.
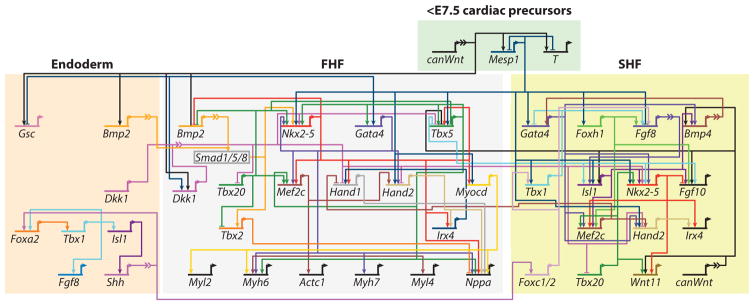
Figure 2.
Gene regulatory network for heart field specification. A current model of the relationship between genes downstream of MESP1 that control first heart field (FHF) and second heart field (SHF) specification, derived from published gene regulation relationships. At the top is shown signaling from the mesodermal cardiac progenitors. On the left is shown the contribution of the overlying endoderm. Genes are represented by their regulatory region and their transcriptional start site. Information from other genes is processed within the regulatory region. The transcriptional start site of a gene indicates expression and influences transcription of other genes. Arrowheads indicate activation, and bar heads indicate inhibition of gene transcription. Broken lines represent intercellular signaling with an integrated signal transduction cascade. Figure adapted with permission from Reference 50. Abbreviation: E, embryonic day.
GENE REGULATORY NETWORKS IN THE FIRST AND SECOND HEART FIELDS
Complex temporal and spatially controlled gene regulatory events govern the signaling networks downstream of MESP1 that regulate cardiogenesis. Mature models of the gene regulatory networks involved in heart field specification have now been developed (50; Figure 3). These events are controlled by intrinsic and extrinsic signals and include crosstalk between the FHF and SHF. Extrinsic signaling includes BMP (bone morphogenetic protein) signaling from the underlying anterior ectoderm as well as BMP, FGF, SHH, and noncanonical WNT signaling, along with inhibition of canonical and WNT signaling, from the overlying anterior endoderm (110; Figure 1). Downstream of MESP1, the FHF and SHF are governed by both shared and distinct genetic programs. Myocardial regulatory genes that are activated in both the FHF and SHF include those encoding the transcription factors Gata4 and Nkx2-5, considered secondary master regulators (Figure 3).
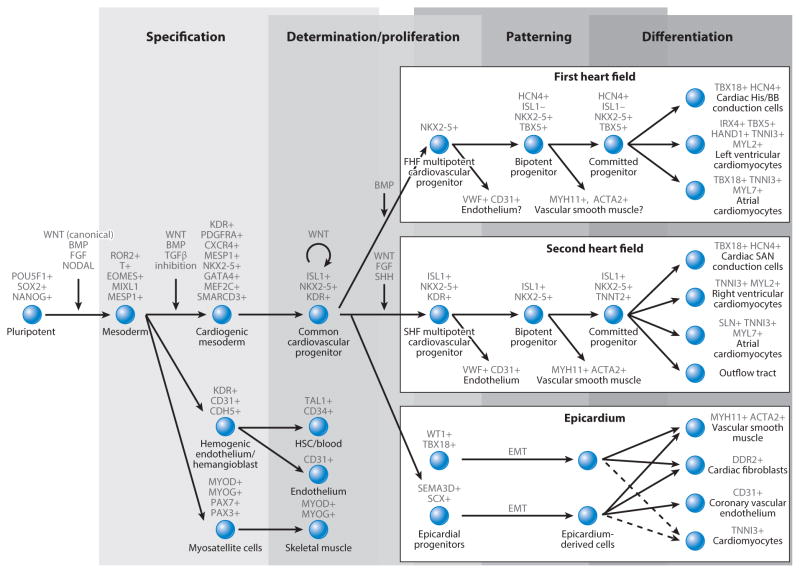
Figure 3.
A schematic demonstrating the relationship of the three major cardiac lineages, subsequent sublineages, and marker expression for each cell type. Progression from pluripotent cells to the separation of the cardiac, hematopoietic, and skeletal muscle lineages is shown. The cardiogenic mesoderm progresses to a tripotent common cardiovascular progenitor capable of forming the first heart field (FHF), second heart field (SHF), and epicardium. The FHF box (top right) shows the HCN4+ FHF progenitor, the unknown contribution to the endothelial and vascular smooth muscle lineages, the major contribution to the left ventricle, and minor contributions to the cardiac bundle of His/bundle branches (His/BB) and atria. The SHF box (middle right) shows the confirmed contribution of ISL1+ progenitors to the endothelium and vascular smooth muscle and the major contribution to the atria, right ventricle, sinoatrial node (SAN) conduction cells, and the outflow tract. Finally, the lineages of the epicardium include two demonstrated epicardial progenitor populations (bottom right), which can progress through epithelial-to-mesenchymal transition (EMT) to form vascular smooth muscle, cardiac fibroblasts, and coronary vascular endothelium. The suggested potential to generate cardiomyocytes is denoted by a dotted line. Abbreviation: BMP, bone morphogenetic protein; HSC, hematopoietic stem cell.
In the FHF, MESP1-induced expression of DKK1 and other downstream inhibitors of WNT/β-catenin signaling in the anterior endoderm induces expression of HEX (30). This in turn induces expression of NKX2-5 and the T-box transcription factor TBX5 (43), expressed only in the FHF. Both NKX2-5 and TBX5 synergistically induce MYOCD to (a) drive FHF cardiac differentiation and (b) activate connexin 40 (GJA5) and NPPA to induce the differentiation of the cardiac chambers and cardiac trabeculation in the ventricles (51; Figure 3).
In the SHF, the MESP1 target Foxh1 (11) is directly upstream of Isl1 and Mef2c. Mef2c is also a direct target of NKX2-5, ISL1, and GATA4 (37). FOXH1 and MEF2C appear to initiate a cascade of cardiac transcription factors in the SHF with proteins such as SMYD1, which regulates Hand2 expression. HAND2, in turn, upregulates Irx4 and Fgf10 (131). In the anterior SHF, a second cascade is controlled by TBX1, which activates FGF8 (93; Figure 3).
In addition to the two heart fields, the proepicardial organ, a transient extracardiac cluster of cells that arises as an outgrowth of the coelomic mesothelium at the ventrocaudal base of the developing heart, gives rise to the epicardium. A subset of these epicardial cells invades the underlying heart tube and contributes to various lineages within the developing heart itself (147). Adult epi-cardial cells can undergo epithelial-to-mesenchymal transition to generate cardiac and adventitial fibroblasts as well as coronary smooth muscle cells. It has also been proposed that proepicardial cells, marked by expression of Wt1/Tbx18 or Sema3d/Scx, are able to generate cardiomyocytes (62; Figure 2).
DNA METHYLATION AND HISTONE MODIFICATIONS
Along with the gene regulatory networks described above, several epigenetic factors play a critical role in modulating cardiac lineage differentiation and specification, providing an additional level of control to the complex temporally and spatially controlled gene regulatory events that govern cardiac differentiation. For example, genome-wide analysis of the developing mouse heart at E11.5 and E14.5 has identified >1,500 differentially methylated genes. Of these, 79 display correlated expression changes, including genes such as Has2, whose product regulates Myocd, Tbx20, Hand1, and Mef2c (20). This suggests an important role for DNA methylation in cardiac development.
Histone modifications are also closely linked to cardiac specification. For example, the his-tone acetyltransferase (HAT) p300 is essential to cardiac development, as it contributes to Gata4, Mef2c, and Srf expression (114). Knockouts of p300 are embryonically lethal at E9.0–11.0, owing to reduced trabeculation and expression of cardiac structural proteins (141). In later cardiac development, the HAT KAT6A (previously known as MOZ) is linked to the activation of Tbx1 expression (128). Histone deacetylases (HDACs) work in an opposite manner to HATs, removing lysine acetylation and resulting in more condensed chromatin and lower gene expression. Mouse double mutants lacking both Hdac1 and Hdac2 demonstrate neonatal lethality as a result of ar-rhythmias and dilated cardiomyopathy (84). Similarly, knockout of the class III HDAC-encoding gene Sirt1 leads to perinatal or postnatal lethality, as a result of septal defects (26).
Chromatin condensation can also be produced by histone methylation, which is controlled by histone methyltransferases (HMTs) and histone demethylases (HDMs). Loss of the HMT Smyd1 is embryonic lethal, owing to disrupted cardiomyocyte maturation and right ventricular hypoplasia (46). Similarly, the HMT WHSC1 is involved in repressing Nkx2-5 via H3K36me3 (88). Conversely, RAE28, a member of polycomb repressive complex 1 (PRC1), helps maintain the level of Nkx2-5 (107). The HDM JARID2 is a key regulator of late cardiac development, as shown by the death of endothelial-specific Jarid2−/− mice in the uterus or immediately after birth. Deaths occur from deficiencies such as ventricular septal defects, double outlet right ventricles, and hypertrabeculation associated with noncompaction of the ventricular wall. The underlying mechanism is the repression of endocardially expressed Notch and Nrg1 through modification of H3K9 (86).
ATP-dependent chromatin remodeling complexes alter the nucleosome packaging by moving nucleosomes along the DNA, expelling or exchanging histones to activate or repress genes. The chromatin remodelers BRG1/BRM-associated factor (BAF) complex and polybromo BRG1-associated factor (PBAF) have been most extensively studied in cardiac development. BAF complexes consist of 11 subunits plus one of two possible ATPases, BRG1 (SMARCA4) or BRM (SMARCA2), which have different promoter associations. BRG1 interacts with Nkx2-5, Gata4, Tbx5, and Tbx20 (116). During development, BRG1 activates Myh7 (βMHC) while repressing Myh6 (αMHC). BRG1 is reactivated upon cardiac stress in adult cardiomyocytes and induces a MYH6-to-MYH7 shift. Brg1+/− mice have a range of mild cardiac defects (48). A subunit of the BAF complex, BAF60C (SMARCD3), is restricted to the developing heart from E7.5. Baf60c knockout is embryonic lethal in mice at E10.0, resulting from outflow tract defects and hypoplastic right ventricles (73). Polybromo 1 (PBRM1/BAF180), a prominent subunit of the PBAF complex, is also involved in late cardiogenesis by potentiating nuclear receptors such as RXRA, VDR, and PPARG, resulting in transcriptional activation related to chamber specification (130).
microRNAs IN CARDIAC DEVELOPMENT
MicroRNAs (miRNAs) are a class of small, noncoding RNAs of ~22 nucleotides in length. They primarily function post-transcriptionally by interacting with the 3′ untranslated region (UTR) of specific target mRNAs. Nearly 1,881 precursors and 2,588 mature human miRNAs have been described to date (http://www.miRBase.org). Disrupting all miRNA expression in the early embryonic heart using Nkx2-5-Cre-mediated mutation of the miRNA-maturing endoribonuclease DICER1 leads to improperly compacted ventricular myocardium (146). Similarly, Myh6-Cre-mediated conditional deletion of DICER1 causes postnatal lethality, as a result of dilated cardiomyopathy and heart failure (24). These studies suggest that many miRNAs play crucial roles in cardiac development.
miR-1 is the most abundant miRNA in the mammalian heart; it accounts for 40% of all miRNA in the murine heart (102). Genes encoding miR-1 (miR1-1, miR1-2, and miR-206) and miR-133 (miR-133a-1, miR-133a-2, and miR-133b) family members are generated as pairs from a common bicistronic precursor transcript. The miR-1-1/miR-133a-2 cluster is located in an intergenic region on human chromosome 20, whereas the miR-1-2/miR-133a-1 cluster is positioned in the antisense orientation within an intron of the Mib1 gene on human chromosome 18. The third cluster, miR-206/miR-133, is expressed primarily in somites during skeletal muscle development. The miR-1/miR-133a clusters are regulated by several important myogenic transcription factors, including SRF, MEF2C, and NKX2-5 (99). Deletion of miR-1-2 causes lethality between E15.5 and birth, as a result of ventricular septal defects (146). Mice lacking either miR-133a-1 or miR-133a-2 do not display any overt developmental phenotype, but deletion of both miRNAs causes a fetal heart phenotype of variable penetrance with ventricular septal defects (75). The phenotype of miR-1-2 mutants has primarily been ascribed to upregulation of Hand2, Irx5, and Kcnd2 (146). The phenotype of miR-133a double mutants has been primarily ascribed to the loss of miR-133a-mediated repression of Ccnd2 and Srf (75). Overexpression of miR-1-2 under the control of the Myh7 promoter is associated with thin-walled ventricles, heart failure, and embryonic lethality at E13.5. This is in part owing to defective ventricular cardiomyocyte proliferation from the inhibition of HAND2 and also increased arrhythmias caused by the repression of KCNJ2 and GJA1 (136). More recent studies have shown that knockout of miR-1-1/miR-133a-2 and miR-1-2/miR-133a-1 results in upregulation of genes characteristic of immature cardiomyocytes (135), indicating that miR-1 and miR-133a are responsible for the differentiation of early cardiomyocyte progenitors into fetal cardiomyocytes via a negative feedback loop with MYOCD.
Embedded within their respective introns, three muscle-specific myosin heavy chain genes (Myh6, Myh7, and Myh7b) encode a family of miR-208a, miR-208b, and miR-499 miRNAs called myomiRs. The expression of myomiRs parallels the expression of their respective host genes during development: miR-208a/Myh7 expression levels decline rapidly after birth, whereas miR-499/Myh6 and miR-208b/Myh7b expression levels are relatively low during embryogenesis and increase postnatally (125). miR-208a, miR-208b, and miR-499 have identical or nearly identical seed sequences, suggesting that the myomiRs probably share common target genes and functions. The presence of these miRNAs allows the myosin genes to act more broadly to control muscle gene expression and performance. Genetic loss-of-function studies in mice have revealed that myomiRs are not essential for cardiac development but are indispensable for mediating some aspects of the cardiac stress response (125). The role of miRNAs in cardiomyocyte proliferation is discussed further below.
LONG NONCODING RNAs IN CARDIAC DEVELOPMENT
Like most mRNAs, long noncoding RNAs (lncRNAs) are longer than 200 nucleotides, 5′ capped, and polyadenylated, although they have limited coding potential. Recently, 321 lncRNAs were identified in the heart, 157 of which were differentially expressed during cardiac development (80). Although this field is relatively new, several lncRNAs have been validated as having roles in cardiac differentiation. Braveheart (Bvht) is abundant in both mouse ESCs and in mouse ESCs differentiating to the cardiac lineage. It is essential for murine cardiac differentiation and acts upstream of Mesp1 (65) by interacting with SUZ12. SUZ12 is a part of the polycomb repressive complex 2 (PRC2), which catalyzes H3K27me3. However, it remains to be seen if Bvht is required for in vivo cardiac development. Bvht appears to be a mouse-specific lncRNA, as sequence alignment does not identify human Bvht homologs. Another novel lncRNA, Fendrr, is expressed in the mouse lateral plate mesoderm; loss of Fendrr is embryonic lethal at E13.75 in mice (47), resulting in defects in cardiac morphogenesis. Like Bvht, Fendrr interacts with PRC2, which promotes H3K4me3.
THE DYNAMIC PLURIPOTENT STATE AND EFFECTS ON CARDIAC DIFFERENTIATION
In 1981, Evans & Kaufman (41) demonstrated that pluripotent mouse embryonic stem cells (mESCs) can be derived from mouse blastocysts. In 1998, Thomson et al. (118) demonstrated that hESCs can be derived from human blastocyst embryos. In 2006, Takahashi & Yamanaka (113) demonstrated that mouse induced pluripotent stem cells can be generated from mouse fibroblasts by overexpressing Pou5f1 (Oct4), Sox2, Klf4, and Myc. This was quickly followed by the generation of hiPSCs in 2007 (143). hESCs and hiPSCs, collectively referred to as hPSCs, can theoretically be cultured indefinitely in vitro with the potential to differentiate into all ~220 somatic cell types in the human body. In the past decade, significant progress has been made in understanding the intricacies of the highly complex gene regulatory networks that govern human pluripotency and differentiation potential (reviewed in 29).
hESCs were first derived on a feeder layer of mouse embryonic fibroblasts in media containing fetal bovine serum (FBS) (118). Because of substantial progress that has been made in developing feeder-free culture systems and chemically defined cell culture media eliminating variables such as serum and other animal-derived products, the regulatory networks that control in vitro human pluripotency are now well understood. High levels of FGF2 are required to stimulate the MAPK/ERK and PI3K/AKT pathways and prevent differentiation (2); TGFβ1/activin A/NODAL (54) is required to stimulate the SMAD2/3 pathway and maintain high levels of NANOG expression, and insulin or insulin-like growth factor (IGF) signaling thorough PI3K/AKT is required for cell survival and proliferation (4). The role of WNT signaling in hPSCs is more enigmatic, owing to contradictory results among published studies. Only recently has it been demonstrated that WNT signaling is not required for the maintenance of pluripo-tency and that inhibition of WNT in hPSCs actually biases differentiation toward the ectodermal lineages (31).
Human pluripotency has been characterized by the expression of three fundamental regulators of pluripotency: OCT4, SOX2, and NANOG. All three proteins participate in a reciprocal regulatory feed-forward loop with each other and co-occupy a substantial portion of their target genes (13). As part of this core gene regulatory network, the OCT4/SOX2 dimer acts as a common transcription factor for all three genes (OCT4, SOX2, and NANOG). NANOG is thought to have a more complex role, with recent data suggesting that it is autorepressive, its influence on OCT4 and SOX2 expression is minimal (87), and high levels of OCT4 may repress NANOG. NANOG is not essential for the indefinite self-renewal of mESCs (21), but forced expression of NANOG can maintain mESC pluripotency in the absence of LIF, which neither Oct4 nor Sox2 expression can do (21). In individual mESCs, NANOG expression levels transiently and reversibly fluctuate (58). This heterogeneity is controlled by dynamic reversion from NANOG-low to NANOG-high states, with differing differentiation capacities (58). NANOG heterogeneity in mESCs cultured in serum and LIF without feeders is substantially reduced when using 2i culture conditions with a GSK3B and MEK inhibitor (42). This suggests that more tightly controlled culture conditions may remove these heterogeneity-induced artifacts, and that more suitable conditions for culturing hPSCs may yet be discovered. In hESCs, OCT4, SOX2, and NANOG have different roles in preventing differentiation, with SOX2 repressing the primitive streak, NANOG repressing neuronal ectoderm, and OCT4 required for BMP4-induced mesendoderm induction (129). NANOG is essential for cardiac mesoderm induction by both BMP4/activin A and GSK3B inhibitors (83). The endogenous expression of POU5F1 (OCT4) and NANOG has a well-established role in mesoderm induction (144), and endogenous levels of mesoderm inducers, such as NODAL, BMP4, and WNT3A (61, 92), affect cardiomyocyte differentiation.
Taken together, these findings suggest that careful control over the pluripotent state, specifically with regard to NANOG expression, and further improvements in pluripotent culture methodologies, such as inhibition of heterogeneity-inducing pathways, are still required. By contrast, methodologies for reprogramming have not been demonstrated to have long-term influences on subsequent differentiation, although our knowledge of the changes in the transcriptome and epigenome over extended periods of culture is still relatively primitive.
REGULATION OF CARDIAC DIFFERENTIATION OF HUMAN PLURIPOTENT STEM CELLS
Protocols for the cardiac differentiation of hPSCs have evolved continuously over the past 15 years. Early protocols involved forming spheroid embryoid bodies from clusters of pluripotent stem cells in the presence of FBS and had differentiation efficiencies of ~5–10% (63). In 2006, a growth factor-directed human cardiac differentiation protocol without coculture, which used activin A and FGF2 for mesoderm induction, raised the efficacy to ~25% (15). Over time, these methodologies matured, using additional mesoderm induction growth factors from developmental biology, such as BMP4 and WNT3A (120, 137). This resulted in 10-fold higher levels of T (brachyury), with shortened differentiation timelines (18). Finally, it was demonstrated that mesoderm inductive growth factors could be replaced by the small molecule, GSK3B inhibitor CHIR99021 (45).
Subsequent to mesoderm induction, a major breakthrough was made in cardiac specification. This breakthrough hinged on the addition of the WNT inhibitor DDK1 (137), which mimics the biphasic WNT signaling seen in the embryo. DDK1 was quickly replaced by small molecule WNT signaling inhibitors (133). Inhibition of other pathways, such as the BMP, TGFβ, FGF, p38 MAPK, and NOTCH pathways, and activation of the SHH pathway enhances the efficiency of in vitro cardiac differentiation (reviewed in 16). Typically, however, differentiation of embryoid bodies in this manner produces fewer than 70% contracting cardiomyocytes.
A second system for in vitro hPSC cardiomyocyte differentiation is the adherent monolayer-based method (69), which uses activin A treatment followed by BMP4. Importantly, the medium is not changed for 4 days to allow buildup of potential secreted factors (e.g., DKK1). This novel protocol results in a differentiation efficiency of ~30% cardiomyocytes, although initially it was only successful in a single hESC line. Improvements were made to this system, such as the addition of WNT3A and DKK1, similar to the development of the embryoid body method (92). The monolayer strategy, along with biphasic WNT induction with a small molecule GSK3B inhibitor, followed by a WNT inhibitor, leads to a 95% yield of spontaneously contracting cardiomyocytes (72). Recently, the monolayer system has evolved into a chemically defined methodology (17). Despite being induced by a GSK3B inhibitor, cardiac differentiation depends on the induction of endogenous BMP4 and activin/NODAL/TGFβ signaling. This suggests that in vitro differentiation protocols are highly dependent on the same signaling and gene expression pathways required in vivo. Indeed, during differentiation, hPSCs progress through the same T (brachyury)+, MESP1+, NKX2-5+, MEF2C+, TBX5+, and finally MYH6+and MYH7+ populations seen in development (17), and no evidence exists that the gene regulatory networks that control hPSC differentiation are different from those that control human development.
HUMAN PLURIPOTENT STEM CELL–DERIVED CARDIOVASCULAR PROGENITORS
An alternative approach to hPSC-derived cardiomyocyte differentiation is the generation of mul-tipotent cardiovascular progenitor cells (CVPCs) from hPSCs (19). Cells can be captured in the CVPC state by simultaneously inhibiting GSK3B, BMP, and NODAL/activin/TGFβ signaling and can proliferate for extended periods while maintaining the potential to generate cardiomyocytes, smooth muscle cells, and endothelial cells. Although this work has yet to be replicated, this technique may offer a simplified route for generating large numbers of cardiovascular cell types by starting at an intermediate step (Figure 3).
CARDIOMYOCYTE SUBTYPE SPECIFICATION
After cardiomyocyte generation, the next major step is determining how to produce the specific cardiomyocyte subtypes found in the heart, such as ventricular, atrial, and nodal cells. Subtype isolation could prove useful for transplantation purposes, as hPSC-derived ventricular cardiomyocytes could be used to repair ischemic damage to the ventricular myocardium following myocardial infarction. Likewise, hPSC-derived nodal cells could be used to restore proper cardiac rhythm and electrical conduction, and hPSC-derived atrial cells could be used to screen for atrial specific drugs (33). As assessed by patch clamp electrophysiology, common cardiac differentiation protocols produce predominantly ventricular cells, with ~15–20% atrial cells and ~5% nodal cells. The goal of producing specific subtypes has been hampered by the different atrial and ventricular markers in mouse and human models. For example, in mice, MYL7 (MLC2A) is expressed in the atria during development and MYL2 (MLC2V) is expressed in the ventricles; in humans, MYL7 is expressed throughout the atria, ventricles, and outflow tract and only MYL2 is specific for the ventricles (28). Reporter gene constructs such as MLC2V-GFP have been used to isolate ventricular cells from hPSCs (8). Similarly, sarcolipin (SLN)-tdTomato has been used to isolate atrial cells (56). Other genes that are subtype specific include IRX4 (ventricular), HCN4 and TBX18 (nodal), and NPPA (ANF) and NPPB (BNF) (ventricular and atrial). Modification of existing differentiation protocols to skew cardiomyocyte subpopulations has also been successful. For example, using factors such as the small molecule AG1478 to inhibit neuregulin signaling can increase the nodal population to ~50% (148). Similarly, inhibiting retinoic acid signaling using the small molecule BMS-189453 increases the ventricular population to 83%, and additional retinoic acid increases the atrial population to 94% (145).
CARDIOMYOCYTE MATURATION
It is now well established that cardiomyocytes generated from hPSCs by any of the current differentiation protocols are of a fetal phenotype, in terms of automaticity, proliferation, metabolism, structure, calcium handling, and electrophysiological properties (139). These hPSC cardiomyocytes (hPSC-CMs) can mature over time in culture (>80 days) (78). Methodologies for improving cardiomyocyte maturation include the addition of triiodothyronine (T3) (140), which binds the THRA and THRB receptors and upregulates MYH7, SERCA2a (ATP2A2), and phospholamban (PLN), along with numerous ion channels. The lack of Kir2.1, an inward-rectifier potassium ion channel thought to be a main contributor to the observed immature electrophysiological properties of hPSC-CMs, and the forced expression of KCNJ2, which encodes Kir2.1, are effective at inducing maturation (74). Other techniques, such as adrenergic stimulation, electrical stimulation, stretch stimulation, and micropatterning, have also been employed to improve cell maturity. Another approach being actively studied is to force hPSC-CMs to a mature metabolic profile by switching metabolism from glycolysis to oxidative phosphorylation. This is done by eliminating L-glucose from the culture media and replacing it with D-galactose and oleic and palmitic fatty acids (101).
CARDIOMYOCYTE PROLIFERATION
Adult human cardiomyocytes are terminally differentiated, meaning that they exhibit little potential to naturally reenter the cell cycle. Developing cardiomyocytes can undergo bi- and multinucleation without completing cytokinesis. One study, which utilized carbon-14 incorporated into adult human myocardium during the nuclear testing era that ended in 1963, revealed less than 5% turnover of adult cardiomyocytes (5). Lower vertebrates such as newts and zebrafish have the capacity to fully regenerate the heart after amputation of up to 25% of the ventricle (96), likely via dedifferentiation and subsequent proliferation of pre-existing cardiomyocytes. Likewise, neonatal mice up to one week old retain the ability to regenerate their hearts through cardiomyocyte proliferation following induced injury; this ability is absent in adult mice (94). Thus, elucidating the process by which adult cardiomyocytes can be induced to proliferate, potentially by reinitializing this program, may offer an additional route to regenerating lost myocardium.
Several in vivo and in vitro studies have demonstrated modest artificial stimulation of cardiomyocyte cell cycle reentry through injury models, genetic modification, and mitogen stimulation (Figure 4). Overexpression of critical cell cycle regulating cyclin proteins, such as CCNB1, induces cell cycle reentry, DNA synthesis, and the multinucleation of cardiomyocytes (7). For potential translational purposes, growth factors or small molecules that induce proliferation are of significant interest. GSK3B inhibitors that promote Wnt signaling increase cardiomyocyte cell cycle reentry (121). A small molecule inhibitor of p38 MAPK, SB203580, promotes cytokinesis in adult mouse myocytes through the dedifferentiation of sarcomeric structures (40). High-throughput, small molecule screens using large chemical libraries have also identified compounds that increase proliferation in terminally differentiated mESC-CMs by more than twofold; these compounds include GSK3B and CAMK2D inhibitors and ERK and RAF activators (123). In vitro and in vivo treatment of cardiomyocytes with growth factors and peptides can activate cardiomyocyte proliferation to a limited extent as well. Periostin treatment activates adult rat ventricular cardiomyocyte proliferation through downstream PI3K signaling (67). This compound improves ventricular remodeling, enhances myocardial repair, and stimulates angiogenesis following induced injury (67). Likewise, the growth factor neuregulin induces cell cycle reentry in cardiomyocytes by activating tyrosine kinase signaling downstream of the ERBB4 receptor (6). Neuregulin also enhances post-myocardial infarction regeneration in a mouse model, through the proliferation of surviving myocytes (6). Insulin-like growth factor 1 (IGF1) increases proliferation of terminally differentiated hESC-CMs and mESC-CMs in vitro by triggering PI3K/Akt signaling (81). Similarly, intrauterine injection of granulocyte colony stimulating factor (GCSF) promotes cardiomyocyte proliferation, as well as in vitro proliferation of mPSC-CMs, by activating the JAK/STAT signaling pathway (106). Recently, the Hippo-YAP signaling pathway has garnered attention for its role in regulating cardiac growth and cardiomyocyte proliferation by inhibiting Wnt pathway signaling (49). Hippo-YAP signaling is linked to the PI3K/Akt signaling pathway through interaction with the YAP effector Pik3cb (75). α-Catenins directly regulate YAP signaling, and the inhibition of α-catenins enhances myocardial regeneration post-injury (71).
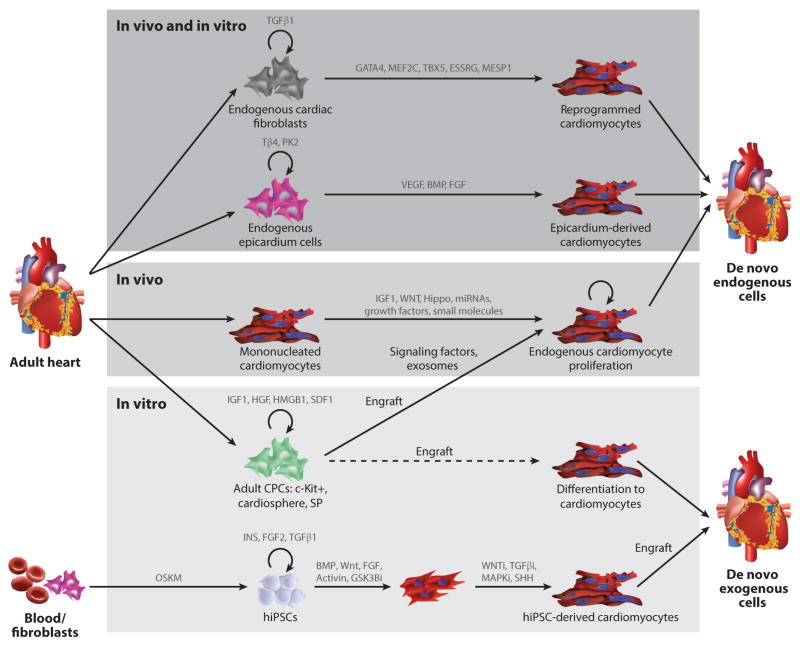
Figure 4.
Methodologies for cardiac regeneration. Shown first (top) are methods that can function both in vivo and in vitro, including directed reprogramming of fibroblasts to cardiomyocytes using exogenously expressed transcription factors and the induction of epicardium cell differentiation to cardiomyocytes using growth factors. These two cell types exist in the heart and can be isolated and induced to proliferate in vitro. A second category of methodologies functions only in vivo (middle). This includes inducing the proliferation of endogenous cardiomyocytes. A third category consists of methodologies that function only in vitro (bottom). This includes various types of adult cardiac progenitor cells (CPCs) such as c-Kit+, cardiosphere, and side population (SP) cells, which show the potential to engraft and produce signaling factors and/or exosomes that induce endogenous cardiomyocyte survival or proliferation and promote engraftment and in vivo differentiation to cardiomyocytes. Also shown is the generation of human induced pluripotent stem cells (hiPSCs) from patient peripheral blood mononuclear cells or fibroblasts by using OCT4, SOX2, KLF4, and MYC (OSKM) reprogramming factors. This is followed by subsequent differentiation to cardiomyocytes and engraftment into the heart. Abbreviations: BMP, bone morphogenetic growth factor; FGF, fibroblast growth factor; hiPSCs, human induced pluripotent stem cells, VEGF, vascular endothelial growth factor.
miRNAs also contribute to mammalian heart regeneration by activating regeneration programs conserved in zebrafish and by directly regulating cardiomyocyte proliferation (77). Notably, miR-590, miR-199a, and the miR-17-92 cluster enhance mouse cardiomyocyte proliferation when introduced in vitro or in vivo (23). Similarly, miR-499 enhances differentiation of cardiomyocytes from human cardiomyocyte progenitor cells (108, 134). Members of the miR-15 cluster have contrasting effects on cardiac function: They promote regeneration after myocardial infarction, function as biomarkers for cardiac ischemia, and regulate cardiomyocyte mitotic arrest after birth in mice (95). The miR-133a family promotes cardiomyocyte proliferation while simultaneously suppressing smooth muscle gene expression in the murine heart (77). Inhibition of miR-378 has a cardioprotective effect, shielding cardiomyocytes against hypoxia-induced apoptosis by upregulat-ing insulin-like growth factor 1 receptor (IGF1R) expression and promoting the AKT pro-survival signaling cascade (66). Other miRNAs, such as miR-29, slow the progression of cardiac fibrosis post-myocardial infarction by inducing apoptosis and reducing secretion of extracellular matrix in proliferating cardiac fibroblasts, a major source of fibrotic tissue that can interfere with cardiac rhythm propagation and function (126). Finally, antisense oligonucleotides called antagomiRs have been utilized to silence the function of miRNAs such as miR-21 to protect mice against cardiac fibrosis (119). Thus, cardiomyocyte and cardiac fibroblast proliferation and function can be regulated by miRNAs, small molecules, and growth factors alike. In vitro and in vivo regulation of cell cycle reentry, cardiomyocyte proliferation, and mammalian cardiac regeneration are all burgeoning fields at the frontier of cardiovascular biology.
DIRECT REPROGRAMMING
Although the induction of pluripotency in somatic cells is a recent discovery, direct transformation of one somatic cell type into another is not a new concept. For example, overexpression of the skeletal muscle gene Myod1 is sufficient to induce transformation of fibroblasts and non-fibroblasts into skeletal muscle cells (32). Additionally, expression of the gene encoding the chromatin remodeling complex subunit Smarcd3 (Baf60c) in conjunction with the known cardiogenic transcription factor genes Tbx5 and Gata4 can transform murine non-cardiac mesoderm into functional car-diomyocytes (115). However, direct reprogramming of murine cardiac fibroblasts and other adult cell types into cardiomyocytes using cardiac-specific transcription factors (e.g., Gata4, Mef2c, and Tbx5) represents another source of de novo cardiomyocytes for disease modeling and therapeutic purposes (52) (Figure 4). The authors showed that cardiac fibroblasts from Myh6-GFP transgenic mice expressed GFP 7 days after transduction with these factors, suggesting that reprogramming to a cardiomyocyte fate had occurred. Transduced cells expressed cardiac-specific markers, although fewer than 10% of the induced cardiomyocytes were able to spontaneously contract. Recently, human fibroblast-to-cardiomyocyte transdifferentiation was shown using a cocktail of GATA4, MEF2C, and TBX5, supplemented with ESRRG and MESP1 (44). Human induced cardiomyocytes express cardiac-specific genes, and approximately 20% of the reprogrammed cells produce calcium transients. The addition of MYOCD and ZFPM2 improves transdifferentiation efficiency, although the overall fibroblast-to-cardiomyocyte yield remains less than 20%. This suggests that, following an adverse event such as myocardial infarction, human cardiac fibroblasts may be coaxed to become functional cardiomyocytes in vivo by direct reprogramming, although significant advancement and refinement of current technology will be needed to demonstrate this.
An alternative partial reprogramming method of generating cardiomyocytes from murine fibro-blasts has been demonstrated using exogenous expression of Oct4, Sox2, and Klf4, commonly used for reprogramming to pluripotency (38). The authors of this study employed a JAK/STAT pathway inhibitor to prevent partially reprogrammed cells from achieving true pluripotency, maintaining a plastic intermediate state that could then be directed to cardiomyocyte differentiation using BMP4. Cardiomyocytes generated using this method express typical cardiac-specific markers, generate action potentials, and contract spontaneously. If these partially reprogrammed cells can be maintained in culture and expanded in a similar fashion to true pluripotent intermediates, such as hiPSCs, then it may be possible to create large numbers of cells primed for cardiac differentiation without the time and cost of hiPSC generation. It remains to be seen whether this methodology translates to human cells; because the JAK/STAT pathway does not have the same role in human pluripotency, an alternative method of preventing full reprograming will likely be necessary.
Transdifferentiated cardiomyocytes raise the possibility of reprogramming cardiac fibroblasts in vivo for heart regeneration using direct delivery of these transcription factors. By converting cardiac fibroblasts into functional cardiomyocytes, we may be able to restore contractile function following an adverse cardiac event. Qian et al. (98) have demonstrated that up to 15% of cells infected with a retrovirus expressing Gata4, Mef2C, and Tbx5 are converted into cardiomyocyte-like cells in the mouse heart after myocardial infarction, with notable improvements in cardiac ejection fraction and a decrease in scar size evident following myocardial infarction. Similarly, Inagawa and colleagues (53) have shown that following induced myocardial infarction in a mouse model, injection of a retrovirus expressing Gata4, Mef2c, and Tbx5 is able to convert cardiac fibroblasts from the infarct region into cardiomyocyte-like cells that exhibit sarcomeres. Song et al. (111) have shown that supplementing Gata4, Mef2c, and Tbx5 expression with Hand2 leads to more efficient in vivo generation of cardiomyocyte-like cells from cardiac fibroblasts. Treating ischemic mouse myocardium with a cocktail of select miRNAs also promotes the direct conversion of cardiac fibroblasts into cardiomyocytes in vivo (55). Altogether, these studies demonstrate that it is possible to generate cardiomyocytes via the in vivo reprogramming of cardiac fibroblasts and that such treatment could improve cardiac function post-myocardial infarction. However, in vivo conversion rates of cardiac fibroblasts into functional cardiomyocytes remain low, suggesting that the optimal stoichiometric combination of cardiac direct reprogramming factors remains to be determined.
Currently, the efficiency of direct and partial reprogramming of somatic cells into cardiomyo-cytes is poor in comparison with what is possible with hiPSC differentiation. Transdifferentiation is limited by the amount of cellular starting material that can be subjected to viral transduction, resulting in a substantially smaller number of cardiomyocytes produced in comparison to hiPSC methods. One advantage of direct reprogramming is that it does not require a true pluripotent intermediate, avoiding the risk of residual pluripotent cells and teratoma formation. However, these studies (53, 98) utilized integrating retro/lentiviruses, reducing their utility for in vivo translational purposes.
Work with adeno-associated virus, specifically AAV-9 using a cardiac-specific promoter, has shown promise for non-integrating, cardiac-specific gene expression (138). The experimental complexity and the potential for depletion of endogenous cardiac fibroblasts, smooth muscle cells, and endothelial cells during in vivo transdifferentiation and partial reprogramming must be addressed; however, as non-myocytes are also critical to proper cardiac function. In addition, artificial overexpression or potential secretion of the transgenic GMT factors (Gata4, Mef2c, and Tbx5) could elicit an immune response that recognizes these reprogrammed cells as foreign, thereby leading to rejection. Finally, although AAV is currently the viral vector of choice for introducing factors such as GMT into human tissue, gene therapy clinical trials have reported adverse inflammatory responses associated with its use (57). Thus, it may be advantageous to consider non-viral vectors for introducing GMT factors as well.
HUMAN PLURIPOTENT STEM CELL-DERIVED CARDIOMYOCYTES IN REGENERATIVE MEDICINE
Ever since the initial generation of cardiomyocytes from hPSCs, there has been hope that these cells would be suitable for cell replacement therapies. Some of the earliest experiments demonstrated that hPSC-CMs could be injected into the nude rat heart along with pro-survival factors to improve cardiac function (69). In a guinea pig model, these cells electrically couple and, surprisingly, even suppress cardiac arrhythmias (105). In nonhuman primates, these cells can extensively remuscularize the ischemia-reperfusion model infarcted heart (27), although this causes significant cardiac arrhythmias. Overall, the survival of injected cells is poor; to overcome this issue, researchers have incorporated biomaterials and tissue engineering approaches (60). Recently, a fibrin patch containing hPSC-derived cardiomyocytes, endothelial cells, smooth muscle cells, and the growth factor IGF1 was shown to improve cardiac function in a porcine ischemia-reperfusion model (142). In addition, Menasché et al. (82) have begun the first clinical trial of hESC-CMs in patients with severe left ventricular systolic dysfunction and an ejection fraction of ≤35%. Cells will be engrafted as a fibrin patch sutured to the epicardium of the infarcted area and covered with a pericardial flap. In this protocol, which was optimized in nonhuman primates, cells are generated using BMP2 followed by an FGF inhibitor, and they are purified using SSEA1 (9). During the preclinical development phase of this protocol, efficacy was assessed using nude rats, and surgical applicability was assessed using sheep. The efficacy and safety of this methodology in humans, particularly with regard to the arrhythmias seen in nonhuman primate models, have yet to be confirmed.
CARDIAC PROGENITOR CELLS IN REGENERATIVE MEDICINE
Although the adult human heart lacks the capacity to regenerate in any meaningful manner, it may harbor a resident population of CPCs that can be isolated and maintained long term in culture. CPCs have the ability to generate cardiomyocytes, smooth muscle cells, and endothelial cells. Different pools of CPCs have been identified, largely in mice, including c-Kit+cells, cardiosphere-derived cells, Sca-1 (LY6A)+cells, and side population (SP) cells (Figure 4).
CPCs were first identified in rat heart using the surface marker c-Kit (KIT or CD117) (3). Human c-Kit+cells are reported to be self-renewing, clonogenic, multipotent, and capable of generating cardiomyocytes, endothelial cells, and smooth muscle cells, although this finding has not been universal; the ability of c-Kit+cells to differentiate to cardiomyocytes may be a rare event (124). Engraftment of c-Kit+cells into the heart induces regeneration in the infarcted rodent ventricle (39). A major trial involving these cells called Cardiac Stem Cell Infusion in Patients with Ischemic Cardiomyopathy (SCIPIO) is ongoing (10).
Cardiospheres, derived from human cardiac biopsies cultured in FGF2, EGF, cardiotrophin-1, and thrombin (109), are highly clonogenic, multicellular microtissues. Cardiospheres contain a core of primitive, proliferating cells and an outer layer of mesenchymal/stromal cells and differentiating cells that express cardiomyocyte proteins and c-Kit, Sca-1, CD31, CD34, and CX43 (109). A clinical trial using these cells is underway called Cardiosphere-Derived Autologous Stem Cells to Reverse Right Ventricular Dysfunction (CADUCEUS) (79).
Sca-1 has also been used as a human CPC marker. Sca-1 cells isolated from the adult human heart are cardiac multipotent when exposed to the demethylating agent 5-azacytidine (127). The expression of Sca-1 in heart sections is restricted to the stromal compartment of the myocardium, potentially indicating endothelial progenitors (122), which are thought to possess cardiac differentiation capacity. Although Sca-1+cells are abundant in the heart, only a small subset can differentiate to cardiomyocytes (122). Intriguingly, no human homolog of Sca-1 exists, although it is likely that functional orthologs do. Therefore, the relationship between murine and human Sca-1+cells is unknown at this point.
Finally, SP cells are identified by their expression of the transporters ABCG2 and/or ABCB1, which export the DNA dye Hoechst 33342 to allow selection of these cells by flow cytometry. SP cells reside in the human left atrium (104). Rodent SP cells are Sca-1+, CD34+, CD31−, and CD45−, and they differentiate to cardiomyocytes, endothelial cells, and smooth muscle cells (91). When delivered intravenously, SP cells also migrate to areas of myocardial injury in a rat model of cryoinjury (91).
A comparison of gene expression in mouse c-Kit+, Sca-1+, and SP cells shows different levels of cardiac commitment in CPCs, with c-Kit+cells being the most primitive, SP cells intermediate, and Sca-1+cells displaying the transcriptional profile closest to cardiomyocytes (35). Because SP and Sca-1+cells share the Sca-1 antigen but not the c-Kit surface marker, they may represent different states of the same progenitor cell. Whether any of these adult heart-derived cells can differentiate into true beating cardiomyocytes in the heart is a much-contested issue (89). It is possible that these cells release paracrine molecules, such as exosomes, that induce the growth of endothelial cells and result in blood vessel formation and reduced ischemia (90) Additionally, CPCs release exosomes to act on cardiomyocytes by reducing oxidative stress and consequently apoptosis (25).
FUTURE AIMS OF THE FIELD
Our knowledge of genetic and epigenetic events during cell reprogramming, cardiac development, and cardiomyocyte differentiation is advancing rapidly. The combination of rapid genome editing in hiPSCs using clustered regularly interspaced short palindromic repeats (CRISPRs) (100) and transcription activator-like effector nucleases (TALENs) (36); highly efficient, chemically defined cardiac differentiation techniques (17); and established murine models of cardiac development allows interrogation of the effectors of the various stages of cardiac differentiation and cardiomyocyte proliferation. Simultaneously, our ability to generate billions of hiPSC-CMs cheaply and efficiently using suspension-based protocols (64) contributes to resolving outstanding issues regarding engraftment of hiPSC-CMs into the heart. Major questions concerning direct reprograming in vivo include how to improve efficiency and whether this technique may be applied to human cardiac regeneration. Finally, more progress must be made in understanding how adult CPCs affect regenerative function.
Acknowledgments
We thank B. Wu, A. Sturzu, and E. Matsa for their comments on this manuscript. We gratefully acknowledge funding support from NIH Pathway to Independence Award K99HL121177 and AHA Beginning Grant-in-Aid 14BGIA20480329 to P.W.B.; AHA Predoctoral Fellowship 13PRE15770000 and NSF Graduate Research Fellowship Program DGE-114747 to A.S.; and AHA Established Investigator Award 14420025, and NIH grants U01 HL099776, U01 HL107393, R01 HL113006, R01 HL123968, and R01 HL126527 to J.C.W. Owing to space limitations, we are unable to include all of the important citations relevant to this subject; we apologize to those investigators whose work was omitted here.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Paul W. Burridge, Email: paul.burridge@northwestern.edu.
Joseph C. Wu, Email: joewu@stanford.edu.
LITERATURE CITED
- 1.Ang SL, Constam DB. A gene network establishing polarity in the early mouse embryo. Semin Cell Dev Biol. 2004;15:555–61. doi: 10.1016/j.semcdb.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong L, Hughes O, Yung S, Hyslop L, Stewart R, et al. The role of PI3K/AKT, MAPK/ERK and NFβ signalling in the maintenance of human embryonic stem cell pluripotency and viability highlighted by transcriptional profiling and functional analysis. Hum Mol Genet. 2006;15:1894–913. doi: 10.1093/hmg/ddl112. [DOI] [PubMed] [Google Scholar]
- 3.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–76. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 4.Bendall SC, Stewart MH, Menendez P, George D, Vijayaragavan K, et al. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature. 2007;448:1015–21. doi: 10.1038/nature06027. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, et al. Evidence for car-diomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–70. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 7.Bicknell KA, Coxon CH, Brooks G. Forced expression of the cyclin B1-CDC2 complex induces proliferation in adult rat cardiomyocytes. Biochem J. 2004;382:411–16. doi: 10.1042/BJ20031481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bizy A, Guerrero-Serna G, Hu B, Ponce-Balbuena D, Willis BC, et al. Myosin light chain 2–based selection of human iPSC-derived early ventricular cardiac myocytes. Stem Cell Res. 2013;11:1335–47. doi: 10.1016/j.scr.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blin G, Nury D, Stefanovic S, Neri T, Guillevic O, et al. A purified population of multipotent cardiovascular progenitors derived from primate pluripotent stem cells engrafts in postmyocardial infarcted nonhuman primates. J Clin Investig. 2010;120:1125–39. doi: 10.1172/JCI40120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolli R, Chugh AR, D’Amario D, Loughran JH, Stoddard MF, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–57. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Bondue A, Tannler S, Chiapparo G, Chabab S, Ramialison M, et al. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J Cell Biol. 2011;192:751–65. doi: 10.1083/jcb.201007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myo-cardial cells. Nat Rev Genet. 2005;6:826–35. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 15.Burridge PW, Anderson D, Priddle H, Barbadillo Munoz MD, Chamberlain S, et al. Improved human embryonic stem cell embryoid body homogeneity and cardiomyocyte differentiation from a novel V-96 plate aggregation system highlights interline variability. Stem Cells. 2007;25:929–38. doi: 10.1634/stemcells.2006-0598. [DOI] [PubMed] [Google Scholar]
- 16.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripo-tent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–60. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burridge PW, Thompson S, Millrod MA, Weinberg S, Yuan X, et al. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLOS ONE. 2011;6:e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao N, Liang H, Huang J, Wang J, Chen Y, et al. Highly efficient induction and long-term maintenance of multipotent cardiovascular progenitors from human pluripotent stem cells under defined conditions. Cell Res. 2013;23:1119–32. doi: 10.1038/cr.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chamberlain AA, Lin M, Lister RL, Maslov AA, Wang Y, et al. DNA methylation is developmentally regulated for genes essential for cardiogenesis. J Am Heart Assoc. 2014;3:e000976. doi: 10.1161/JAHA.114.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–34. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 22.Chan SS, Shi X, Toyama A, Arpke RW, Dandapat A, et al. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell. 2013;12:587–601. doi: 10.1016/j.stem.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, et al. mir-17–92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. 2013;112:1557–66. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. PNAS. 2008;105:2111–16. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Wang Y, Pan Y, Zhang L, Shen C, et al. Cardiac progenitor–derived exosomes protect ischemic myocardium from acute ischemia/reperfusion injury. Biochem Biophys Res Commun. 2013;431:566–71. doi: 10.1016/j.bbrc.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. PNAS. 2003;100:10794–99. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong JJ, Yang X, Don CW, Minami E, Liu YW, et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–77. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuva de Sousa Lopes SM, Hassink RJ, Feijen A, van Rooijen MA, Doevendans PA, et al. Patterning the heart, a template for human cardiomyocyte development. Dev Dyn. 2006;235:1994–2002. doi: 10.1002/dvdy.20830. [DOI] [PubMed] [Google Scholar]
- 29.Dalton S. Signaling networks in human pluripotent stem cells. Curr Opin Cell Biol. 2013;25:241–46. doi: 10.1016/j.ceb.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.David R, Brenner C, Stieber J, Schwarz F, Brunner S, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10:338–45. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- 31.Davidson KC, Adams AM, Goodson JM, McDonald CE, Potter JC, et al. Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. PNAS. 2012;109:4485–90. doi: 10.1073/pnas.1118777109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 33.Devalla HD, Schwach V, Ford JW, Milnes JT, El-Haou S, et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol Med. 2015;7:394–410. doi: 10.15252/emmm.201404757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devine WP, Wythe JD, George M, Koshiba-Takeuchi K, Bruneau BG. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife. 2014;3:e03848. doi: 10.7554/eLife.03848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dey D, Han L, Bauer M, Sanada F, Oikonomopoulos A, et al. Dissecting the molecular relationship among various cardiogenic progenitor cells. Circ Res. 2013;112:1253–62. doi: 10.1161/CIRCRESAHA.112.300779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, et al. A TALEN genome-editing system for generating human stem cell–based disease models. Cell Stem Cell. 2013;12:238–51. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–42. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 38.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, et al. Conversion of mouse fibroblasts into cardio-myocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–22. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 39.Ellison GM, Vicinanza C, Smith AJ, Aquila I, Leone A, et al. Adult c-kitpos cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell. 2013;154:827–42. doi: 10.1016/j.cell.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 40.Engel FB, Schebesta M, Duong MT, Lu G, Ren S, et al. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19:1175–87. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–56. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 42.Filipczyk A, Gkatzis K, Fu J, Hoppe PS, Lickert H, et al. Biallelic expression of Nanog protein in mouse embryonic stem cells. Cell Stem Cell. 2013;13:12–13. doi: 10.1016/j.stem.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 43.Foley AC, Mercola M. Heart induction by Wnt antagonists depends on the homeodomain transcription factor Hex. Genes Dev. 2005;19:387–96. doi: 10.1101/gad.1279405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu JD, Stone NR, Liu L, Spencer CI, Qian L, et al. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013;1:235–47. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez R, Lee JW, Schultz PG. Stepwise chemically induced cardiomyocyte specification of human embryonic stem cells. Angew Chem Int Ed. 2011;50:11181–85. doi: 10.1002/anie.201103909. [DOI] [PubMed] [Google Scholar]
- 46.Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 47.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–14. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hang CT, Yang J, Han P, Cheng HL, Shang C, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–61. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herrmann F, Gross A, Zhou D, Kestler HA, Kuhl M. A Boolean model of the cardiac gene regulatory network determining first and second heart field identity. PLOS ONE. 2012;7:e46798. doi: 10.1371/journal.pone.0046798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, et al. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28:276–80. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- 52.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inagawa K, Miyamoto K, Yamakawa H, Muraoka N, Sadahiro T, et al. Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circ Res. 2012;111:1147–56. doi: 10.1161/CIRCRESAHA.112.271148. [DOI] [PubMed] [Google Scholar]
- 54.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFβ/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–82. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 55.Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–73. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Josowitz R, Lu J, Falce C, D’Souza SL, Wu M, et al. Identification and purification of human induced pluripotent stem cell–derived atrial-like cardiomyocytes based on sarcolipin expression. PLOS ONE. 2014;9:e101316. doi: 10.1371/journal.pone.0101316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaiser J. Gene therapy. Side effects sideline hemophilia trial. Science. 2004;304:1423–25. doi: 10.1126/science.304.5676.1423b. [DOI] [PubMed] [Google Scholar]
- 58.Kalmar T, Lim C, Hayward P, Munoz-Descalzo S, Nichols J, et al. Regulated fluctuations in Nanog expression mediate cell fate decisions in embryonic stem cells. PLOS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karantalis V, DiFede DL, Gerstenblith G, Pham S, Symes J, et al. Autologous mesenchymal stem cells produce concordant improvements in regional function, tissue perfusion, and fibrotic burden when administered to patients undergoing coronary artery bypass grafting: the Prospective Randomized Study of Mesenchymal Stem Cell Therapy in Patients Undergoing Cardiac Surgery (PROMETHEUS) trial. Circ Res. 2014;114:1302–10. doi: 10.1161/CIRCRESAHA.114.303180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karikkineth BC, Zimmermann WH. Myocardial tissue engineering and heart muscle repair. Curr Pharm Biotechnol. 2013;14:4–11. doi: 10.2174/138920113804805322. [DOI] [PubMed] [Google Scholar]
- 61.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, et al. Stage-specific optimization of Activin/Nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–40. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 62.Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, et al. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev Cell. 2012;22:639–50. doi: 10.1016/j.devcel.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Investig. 2001;108:407–14. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kempf H, Olmer R, Kropp C, Ruckert M, Jara-Avaca M, et al. Controlling expansion and car-diomyogenic differentiation of human pluripotent stem cells in scalable suspension culture. Stem Cell Rep. 2014;3:1132–46. doi: 10.1016/j.stemcr.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knezevic I, Patel A, Sundaresan NR, Gupta MP, Solaro RJ, et al. A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor: implications in postnatal cardiac remodeling and cell survival. J Biol Chem. 2012;287:12913–26. doi: 10.1074/jbc.M111.331751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuhn B, del Monte F, Hajjar RJ, Chang YS, Lebeche D, et al. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. 2007;13:962–69. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 68.Kwon C, Arnold J, Hsiao EC, Taketo MM, Conklin BR, Srivastava D. Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. PNAS. 2007;104:10894–99. doi: 10.1073/pnas.0704044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–24. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 70.Lescroart F, Chabab S, Lin X, Rulands S, Paulissen C, et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat Cell Biol. 2014;16:829–40. doi: 10.1038/ncb3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J, Gao E, Vite A, Yi R, Gomez L, et al. α-Catenins control cardiomyocyte proliferation by regulating Yap activity. Circ Res. 2015;116:70–79. doi: 10.1161/CIRCRESAHA.116.304472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. PNAS. 2012;109:E1848–57. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–12. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 74.Lieu DK, Fu JD, Chiamvimonvat N, Tung KC, McNerney GP, et al. Mechanism-based facilitated maturation of human pluripotent stem cell–derived cardiomyocytes. Circ Arrhythm Electrophysiol. 2013;6:191–201. doi: 10.1161/CIRCEP.111.973420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin Z, Zhou P, von Gise A, Gu F, Ma Q, et al. Pi3kcb links Hippo-YAP and PI3K-AKT signaling pathways to promote cardiomyocyte proliferation and survival. Circ Res. 2015;116:35–45. doi: 10.1161/CIRCRESAHA.115.304457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lindsley RC, Gill JG, Murphy TL, Langer EM, Cai M, et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3:55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, et al. MicroRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–54. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lundy SD, Zhu WZ, Regnier M, Laflamme MA. Structural and functional maturation of cardio-myocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LE, et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379:895–904. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matkovich SJ, Edwards JR, Grossenheider TC, de Guzman Strong C, Dorn GW., II Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. PNAS. 2014;111:12264–69. doi: 10.1073/pnas.1410622111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McDevitt TC, Laflamme MA, Murry CE. Proliferation of cardiomyocytes derived from human embryonic stem cells is mediated via the IGF/PI 3-kinase/Akt signaling pathway. J Mol Cell Cardiol. 2005;39:865–73. doi: 10.1016/j.yjmcc.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Menasché P, Vanneaux V, Fabreguettes JR, Bel A, Tosca L, et al. Towards a clinical use of human embryonic stem cell–derived cardiac progenitors: a translational experience. Eur Heart J. 2015;36:743–50. doi: 10.1093/eurheartj/ehu192. [DOI] [PubMed] [Google Scholar]
- 83.Mendjan S, Mascetti VL, Ortmann D, Ortiz M, Karjosukarso DW, et al. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell. 2014;15:310–25. doi: 10.1016/j.stem.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 84.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, et al. Multipotent embryonic isl1+progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–65. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 86.Mysliwiec MR, Carlson CD, Tietjen J, Hung H, Ansari AZ, Lee Y. Jarid2 ( Jumonji, AT rich interactive domain 2) regulates NOTCH1 expression via histone modification in the developing heart. J Biol Chem. 2012;287:1235–41. doi: 10.1074/jbc.M111.315945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Navarro P, Festuccia N, Colby D, Gagliardi A, Mullin NP, et al. OCT4/SOX2-independent Nanog autorepression modulates heterogeneous Nanog gene expression in mouse ES cells. EMBO J. 2012;31:4547–62. doi: 10.1038/emboj.2012.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, et al. A histone H3 lysine 36 trimethyltrans-ferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–91. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 89.Nowbar AN, Mielewczik M, Karavassilis M, Dehbi HM, Shun-Shin MJ, et al. Discrepancies in autologous bone marrow stem cell trials and enhancement of ejection fraction (DAMASCENE): weighted regression and meta-analysis. BMJ. 2014;348:g2688. doi: 10.1136/bmj.g2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ong SG, Lee WH, Huang M, Dey D, Kodo K, et al. Cross talk of combined gene and cell therapy in ischemic heart disease: role of exosomal microRNA transfer. Circulation. 2014;130:S60–69. doi: 10.1161/CIRCULATIONAHA.113.007917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oyama T, Nagai T, Wada H, Naito AT, Matsuura K, et al. Cardiac side population cells have a potential to migrate and differentiate into cardiomyocytes in vitro and in vivo. J Cell Biol. 2007;176:329–41. doi: 10.1083/jcb.200603014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Paige SL, Osugi T, Afanasiev OK, Pabon L, Reinecke H, Murry CE. Endogenous Wnt/β-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLOS ONE. 2010;5:e11134. doi: 10.1371/journal.pone.0011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, et al. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–33. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–80. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. PNAS. 2013;110:187–92. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 97.Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–59. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–98. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qian L, Wythe JD, Liu J, Cartry J, Vogler G, et al. Tinman/Nkx2-5 acts via miR-1 and upstream of Cdc42 to regulate heart function across species. J Cell Biol. 2011;193:1181–96. doi: 10.1083/jcb.201006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rana P, Anson B, Engle S, Will Y. Characterization of human induced pluripotent stem cell derived cardiomyocytes: bioenergetics and utilization in safety screening. Toxicol Sci. 2012;130:117–31. doi: 10.1093/toxsci/kfs233. [DOI] [PubMed] [Google Scholar]
- 102.Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res. 2009;105:585–94. doi: 10.1161/CIRCRESAHA.109.200451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Robertson EJ, Norris DP, Brennan J, Bikoff EK. Control of early anterior-posterior patterning in the mouse embryo by TGF-βsignalling. Philos Trans R Soc B. 2003;358:1351–57. doi: 10.1098/rstb.2003.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sandstedt J, Jonsson M, Kajic K, Sandstedt M, Lindahl A, et al. Left atrium of the human adult heart contains a population of side population cells. Basic Res Cardiol. 2012;107:255. doi: 10.1007/s00395-012-0255-7. [DOI] [PubMed] [Google Scholar]
- 105.Shiba Y, Fernandes S, Zhu WZ, Filice D, Muskheli V, et al. Human ES-cell–derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489:322–25. doi: 10.1038/nature11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shimoji K, Yuasa S, Onizuka T, Hattori F, Tanaka T, et al. G-CSF promotes the proliferation of developing cardiomyocytes in vivo and in derivation from ESCs and iPSCs. Cell Stem Cell. 2010;6:227–37. doi: 10.1016/j.stem.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 107.Shirai M, Osugi T, Koga H, Kaji Y, Takimoto E, et al. The Polycomb-group gene Rae28 sustains Nkx2.5/Csx expression and is essential for cardiac morphogenesis. J Clin Investig. 2002;110:177–84. doi: 10.1172/JCI14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sluijter JP, van Mil A, van Vliet P, Metz CH, Liu J, et al. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol. 2010;30:859–68. doi: 10.1161/ATVBAHA.109.197434. [DOI] [PubMed] [Google Scholar]
- 109.Smith RR, Barile L, Cho HC, Leppo MK, Hare JM, et al. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation. 2007;115:896–908. doi: 10.1161/CIRCULATIONAHA.106.655209. [DOI] [PubMed] [Google Scholar]
- 110.Solloway MJ, Harvey RP. Molecular pathways in myocardial development: a stem cell perspective. Cardiovasc Res. 2003;58:264–77. doi: 10.1016/s0008-6363(03)00286-4. [DOI] [PubMed] [Google Scholar]
- 111.Song K, Nam YJ, Luo X, Qi X, Tan W, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Spater D, Abramczuk MK, Buac K, Zangi L, Stachel MW, et al. A HCN4 + cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat Cell Biol. 2013;15:1098–106. doi: 10.1038/ncb2824. [DOI] [PubMed] [Google Scholar]
- 113.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 114.Takaya T, Kawamura T, Morimoto T, Ono K, Kita T, et al. Identification of p300-targeted acetylated residues in GATA4 during hypertrophic responses in cardiac myocytes. J Biol Chem. 2008;283:9828–35. doi: 10.1074/jbc.M707391200. [DOI] [PubMed] [Google Scholar]
- 115.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–11. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguin P, et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tam PP, Loebel DA. Gene function in mouse embryogenesis: get set for gastrulation. Nat Rev Genet. 2007;8:368–81. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- 118.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–47. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 119.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–84. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 120.Tran TH, Wang X, Browne C, Zhang Y, Schinke M, et al. Wnt3a-induced mesoderm formation and cardiomyogenesis in human embryonic stem cells. Stem Cells. 2009;27:1869–78. doi: 10.1002/stem.95. [DOI] [PubMed] [Google Scholar]
- 121.Tseng AS, Engel FB, Keating MT. The GSK-3 inhibitor BIO promotes proliferation in mammalian cardiomyocytes. Chem Biol. 2006;13:957–63. doi: 10.1016/j.chembiol.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 122.Uchida S, De Gaspari P, Kostin S, Jenniches K, Kilic A, et al. Sca1-derived cells are a source of myocardial renewal in the murine adult heart. Stem Cell Rep. 2013;1:397–410. doi: 10.1016/j.stemcr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Uosaki H, Magadum A, Seo K, Fukushima H, Takeuchi A, et al. Identification of chemicals inducing cardiomyocyte proliferation in developmental stage–specific manner with pluripotent stem cells. Circ Cardiovasc Genet. 2013;6:624–33. doi: 10.1161/CIRCGENETICS.113.000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van Berlo JH, Kanisicak O, Maillet M, Vagnozzi RJ, Karch J, et al. c-kit+cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–41. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–73. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. PNAS. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van Vliet P, Roccio M, Smits AM, van Oorschot AA, Metz CH, et al. Progenitor cells isolated from the human heart: a potential cell source for regenerative therapy. Neth Heart J. 2008;16:163–69. doi: 10.1007/BF03086138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Voss AK, Vanyai HK, Collin C, Dixon MP, McLennan TJ, et al. MOZ regulates the Tbx1 locus, and Moz mutation partially phenocopies DiGeorge syndrome. Dev Cell. 2012;23:652–63. doi: 10.1016/j.devcel.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–54. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 130.Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, et al. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18:3106–16. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Watanabe Y, Zaffran S, Kuroiwa A, Higuchi H, Ogura T, et al. Fibroblast growth factor 10 gene regulation in the second heart field by Tbx1, Nkx2-5, and Islet1 reveals a genetic switch for down-regulation in the myocardium. PNAS. 2012;109:18273–80. doi: 10.1073/pnas.1215360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wessels A, Sedmera D. Developmental anatomy of the heart: a tale of mice and man. Physiol Genomics. 2003;15:165–76. doi: 10.1152/physiolgenomics.00033.2003. [DOI] [PubMed] [Google Scholar]
- 133.Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, et al. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell–derived mesoderm. Circ Res. 2011;109:360–64. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wilson KD, Hu S, Venkatasubrahmanyam S, Fu JD, Sun N, et al. Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for miR-499. Circ Cardiovasc Genet. 2010;3:426–35. doi: 10.1161/CIRCGENETICS.109.934281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wystub K, Besser J, Bachmann A, Boettger T, Braun T. miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLOS Genet. 2013;9:e1003793. doi: 10.1371/journal.pgen.1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang B, Lin H, Xiao J, Lu Y, Luo X, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–91. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 137.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–28. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 138.Yang L, Xiao X. Creation of a cardiotropic adeno-associated virus: the story of viral directed evolution. Virol J. 2013;10:50. doi: 10.1186/1743-422X-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yang X, Pabon L, Murry CE. Engineering adolescence: maturation of human pluripotent stem cell–derived cardiomyocytes. Circ Res. 2014;114:511–23. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang X, Rodriguez M, Pabon L, Fischer KA, Reinecke H, et al. Tri-iodo-L-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J Mol Cell Cardiol. 2014;72:296–304. doi: 10.1016/j.yjmcc.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, et al. Gene dosage–dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–72. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 142.Ye L, Chang YH, Xiong Q, Zhang P, Zhang L, et al. Cardiac repair in a porcine model of acute myocardial infarction with human induced pluripotent stem cell–derived cardiovascular cells. Cell Stem Cell. 2014;15:750–61. doi: 10.1016/j.stem.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 144.Zeineddine D, Papadimou E, Chebli K, Gineste M, Liu J, et al. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev Cell. 2006;11:535–46. doi: 10.1016/j.devcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 145.Zhang Q, Jiang J, Han P, Yuan Q, Zhang J, et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–87. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 147.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–13. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhu WZ, Xie Y, Moyes KW, Gold JD, Askari B, Laflamme MA. Neuregulin/ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells. Circ Res. 2010;107:776–86. doi: 10.1161/CIRCRESAHA.110.223917. [DOI] [PMC free article] [PubMed] [Google Scholar]