Abstract
The pathogenicity of Clostridium difficile is linked to its ability to produce two toxins: TcdA and TcdB. The level of toxin synthesis is influenced by environmental signals, such as phosphotransferase system (PTS) sugars, biotin, and amino acids, especially cysteine. To understand the molecular mechanisms of cysteine-dependent repression of toxin production, we reconstructed the sulfur metabolism pathways of C. difficile strain 630 in silico and validated some of them by testing C. difficile growth in the presence of various sulfur sources. High levels of sulfide and pyruvate were produced in the presence of 10 mM cysteine, indicating that cysteine is actively catabolized by cysteine desulfhydrases. Using a transcriptomic approach, we analyzed cysteine-dependent control of gene expression and showed that cysteine modulates the expression of genes involved in cysteine metabolism, amino acid biosynthesis, fermentation, energy metabolism, iron acquisition, and the stress response. Additionally, a sigma factor (SigL) and global regulators (CcpA, CodY, and Fur) were tested to elucidate their roles in the cysteine-dependent regulation of toxin production. Among these regulators, only sigL inactivation resulted in the derepression of toxin gene expression in the presence of cysteine. Interestingly, the sigL mutant produced less pyruvate and H2S than the wild-type strain. Unlike cysteine, the addition of 10 mM pyruvate to the medium for a short time during the growth of the wild-type and sigL mutant strains reduced expression of the toxin genes, indicating that cysteine-dependent repression of toxin production is mainly due to the accumulation of cysteine by-products during growth. Finally, we showed that the effect of pyruvate on toxin gene expression is mediated at least in part by the two-component system CD2602-CD2601.
INTRODUCTION
Clostridium difficile is a Gram-positive spore-forming obligate anaerobe and the major cause of nosocomial diarrhea associated with antibiotic therapy. The symptoms of C. difficile infection (CDI) vary from mild diarrhea to life-threatening pseudomembranous colitis, a severe form of CDI (1). Virulent C. difficile strains produce two large toxins: an enterotoxin (TcdA) and a cytotoxin (TcdB). The tcdA and tcdB genes are clustered within a single chromosomal region, called the pathogenicity locus (PaLoc), with three accessory genes: tcdR, tcdE, and tcdC. The expression of the toxin genes is controlled through the coordinated action of the alternative sigma factor TcdR and its antagonist factor, TcdC (2–4). The tcdE gene encodes a holin-like protein that is required for toxin release (5).
The spectrum of diseases caused by C. difficile depends on host factors and, for the severe forms, on the level of toxins produced, suggesting that the regulation of toxin synthesis is a critical determinant of C. difficile pathogenicity (6). Toxin production starts when C. difficile cultures enter the stationary growth phase (7) and is modulated in response to various environmental signals. Exposure to subinhibitory concentrations of antibiotics, a temperature of 37°C, biotin limitation, or the presence of butyric acid stimulates toxin production (8, 9). In contrast, the presence of rapidly metabolized carbon sources, such as glucose and butanol, or amino acids, such as cysteine and proline, inhibits toxin synthesis (7, 10–12). Some of the molecular mechanisms regulating C. difficile toxin synthesis in response to environmental signals have been elucidated (13–16). It has been shown that CodY, the global regulator involved in the adaptive response to nutrient limitation, represses toxin gene expression by binding to the tcdR promoter region (14, 17) and that glucose-dependent repression of toxin production is mediated by CcpA, the global regulator of carbon catabolite repression (CCR) (13). This repression is the result of the direct binding of CcpA to a cis-acting catabolite response element (cre site) that is present in the regulatory regions of the tcdA, tcdB, tcdR, and tcdC genes, with the strongest affinity observed for the tcdR promoter (18). Toxin gene expression also depends on transcriptional factors, such as SigH and Spo0A, which control the transition to the postexponential growth phase and the initiation of sporulation (15, 16).
Changes in colonic flora after antibiotic treatment lead to the modification of metabolic pools, which affects the spore germination and cell growth of C. difficile (19). Specifically, the levels of several phosphotransferase system (PTS) sugars, such as mannitol and sorbitol, and amino acids, such as proline, cysteine, and cystine, the cysteine dimer, increase during gut dysbiosis. These compounds are metabolized by C. difficile and may serve as metabolic signals that are detected by regulators to coordinate adaptation, growth, and virulence factor production during gut colonization.
Among the amino acids that downregulate toxin production in C. difficile strains, cysteine is the most potent (11, 12). Links between bacterial virulence and cysteine metabolism have been described in several pathogenic bacteria. In Clostridium perfringens and Bordetella pertussis, toxin synthesis is regulated in response to cysteine availability (20, 21). Additionally, genes involved in sulfur metabolism are induced when Mycobacterium tuberculosis, Yersinia ruckeri, Staphylococcus aureus, and Nesseiria meningitidis interact with human cells (22–24). In addition, loss-of-function mutations in genes involved in cysteine biosynthesis or degradation affect the virulence for some of these pathogens (23–26). Finally, the master regulator of cysteine metabolism in S. aureus, CymR, plays an important role in both the stress response and the control of bacterial virulence (27).
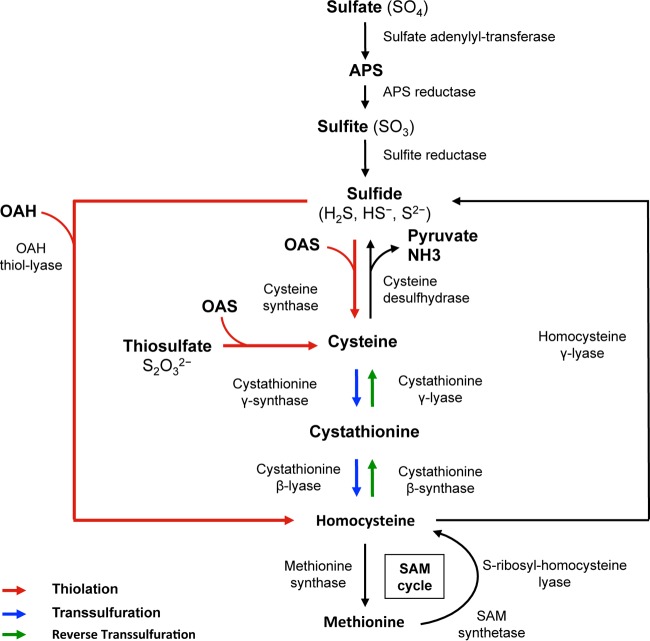
The sulfur-containing amino acid cysteine is central to bacterial physiology. This amino acid is a precursor of methionine and of several coenzymes (biotin, thiamine, coenzyme A [CoA], and coenzyme M). Cysteine is also the sulfur donor for the biogenesis of the iron-sulfur (Fe-S) clusters that are found in the catalytic site of several enzymes and assists in protein folding and assembly by forming disulfide bonds. Moreover, cysteine-containing proteins (thioredoxin and glutaredoxin) and molecules (glutathione, bacillithiol, and mycothiol) are important in protecting cells against oxidative stress (28, 29). Two major cysteine biosynthetic pathways are present in microorganisms: (i) the thiolation pathway, which directly incorporates sulfide or thiosulfate into O-acetyl l-serine (OAS), and (ii) the reverse transsulfuration pathway, which converts homocysteine into cysteine via a cystathionine intermediate (Fig. 1) (30, 31). Homocysteine is synthesized from methionine using the S-adenosyl-methionine (SAM) recycling pathway, while sulfide arises mostly from the reduction of sulfate.
FIG 1.
Schematic overview of sulfur metabolism in bacteria. APS, adenylyl sulfate; OAS, O-acetylserine; OAH, O-acetylhomoserine; SAM, S-adenosyl-methionine.
Due to the reactivity of its thiol group, the intracellular concentration of cysteine must be tightly controlled. The pathways responsible for depleting free cysteine include those that incorporate cysteine into molecules (proteins, methionine, Fe-S clusters, and vitamins) and those that degrade or export it (30). Cysteine can also be catabolized by cysteine desulfhydrases or cysteine desulfidase, producing pyruvate and hydrogen sulfide (H2S) (Fig. 1) (24, 32). Finally, a large variety of molecular mechanisms participate in fine-tuning cysteine metabolism in response to environmental changes. These systems include regulation by the premature termination of transcription at T-box systems in response to the level of charge of tRNACys (33) or by several transcriptional regulators, including activators of the LysR family (30) and CymR, a repressor of the Rrf2 family (34, 35).
To understand the molecular mechanisms involved in the cysteine response, we performed a reconstruction of C. difficile sulfur metabolism and analyzed the global effect of cysteine on gene expression. Then, we showed that cysteine-dependent repression of toxin production requires SigL. Moreover, we observed that the production of pyruvate and H2S decreased in the sigL mutant compared to that in the wild-type strain. Interestingly, addition of pyruvate to the growth medium of the wild-type and the sigL mutant strains repressed toxin gene transcription, suggesting that the effect of cysteine on toxin production is due, at least in part, to the accumulation of cysteine by-products resulting from cysteine degradation. Finally, we showed that the regulation of toxins by exogenous pyruvate is mediated by a two-component system (TCS) through a still uncharacterized mechanism.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The C. difficile strains used in this study are described in Table 1. Escherichia coli strain NEB 10-beta (BioLabs) and E. coli strain HB101(RP4) were used, respectively, for cloning and as a donor strain for C. difficile conjugation experiments. C. difficile strains were grown anaerobically (5% H2, 5% CO2, 90% N2) in PY medium (20 g/liter Bacto peptone, 10 g/liter yeast extract, 0.4% [2 ml/liter] CaCl2, 0.025% [4 ml/liter] resazurin, 0.05% [10 ml/liter] hemin, 0.05% [1 ml/liter] vitamin K, and 40 ml/liter of salts solution containing 1 g/liter K2HPO4, 1 g/liter KH2PO4, 10 g/liter NaHCO2, 2 g/liter NaCl, and 0.2 g/liter MgSO4 · 7H2O), PYC (PY with 10 mM cysteine), or PYHC medium (PY with 10 mM homocysteine) (12). After 9 h of cell growth, 15 mM acetate or 10 mM pyruvate, Na2S, or formate was added to the PY medium. When necessary, cefoxitin (25 μg/ml), thiamphenicol (15 μg/ml), or erythromycin (2.5 μg/ml) was added to C. difficile cultures. E. coli strains were grown in Luria-Bertani (LB) broth. When indicated, ampicillin (100 μg/ml) or chloramphenicol (15 μg/ml) was added to the culture medium. Additionally, 200 ng/ml of anhydrotetracycline (Atc) was used to induce the Ptet promoter of the pRPF185 vector derivatives in C. difficile (36). The sulfur-free minimal medium was as previously described (20), with the addition of 0.3 g/liter of proline. The concentrations of the sulfur sources added are indicated in Table 2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic | Gene type | Plasmid or resistance typea | Origin or reference |
|---|---|---|---|---|
| Strains | Background | Knockout or overexpressed gene | Plasmid | |
| 630Δerm | 87 | |||
| M7404 | BI/NAP1/027 | 2 | ||
| M7404 (tcdC+) | BI/NAP1/027 | pDLL17 (tcdC+) | 2 | |
| VPI 10463 | Virginia Polytechnic Institute | |||
| CDIP001 | 630Δerm | CD1287 (fur)::erm | This study | |
| CDIP106 | 630Δerm | CD0278::erm | This study | |
| CDIP107 | 630Δerm | CD2023::erm | This study | |
| CDIP110 | 630Δerm | CD2065::erm | This study | |
| CDIP217 | 630Δerm | CD3176 (sigL)::erm | This study | |
| CDIP342 | 630Δerm | CD3176 (sigL)::erm | pDIA6309 | This study |
| CDIP540 | 630Δerm | CD1594 (cysK)::erm | This study | |
| CDIP656 | 630Δerm | pDIA6456 | This study | |
| CDIP657 | 630Δerm | CD2602::erm | This study | |
| JIR8094 | 88 | |||
| CDIP100 | JIR8094 | CD1064 (ccpA)::erm | 13 | |
| LB-CD15 | JIR8094 | CD1275 (codY)::erm | pBL92 | L. Bouillaut |
| Plasmids | Vector | Cloned gene | Resistance | Origin |
| pDIA5906 | pMTL007 | Intron CD1287 (fur) | Cmr Tmr | This study |
| pDIA6309 | pMTL84121 | CD3176 (sigL) | Cmr Tmr | This study |
| pDIA6450 | pMTL007 | Intron CD0278 | Cmr Tmr | This study |
| pDIA6451 | pMTL007 | Intron CD2065 | Cmr Tmr | This study |
| pDIA6452 | pMTL007 | Intron CD2023 | Cmr Tmr | This study |
| pDIA6453 | pMTL007 | Intron CD3176 | Cmr Tmr | This study |
| pDIA6454 | pMTL007 | Intron CD2602 | Cmr Tmr | This study |
| pDIA6455 | pMTL007 | Intron CD1594 (cysK) | Cmr Tmr | This study |
| pDIA6456 | pRPF185 | Antisense CD3029 (malY) | Cmr Tmr | This study |
Cm, chloramphenicol; Tm, thiamphenicol; Erm, erythromycin.
TABLE 2.
Growth of C. difficile strain 630Δerm in minimal medium containing different sulfur sources
| Sulfur source (concn) | Growth ata: |
|
|---|---|---|
| 16 h | 48 h | |
| Sulfate (4 mM) | − | − |
| Sulfite (4 mM) | − | − |
| Sulfide (4 mM) | + | + |
| Thiosulfate (4 mM) | − | + |
| Cysteine (4 mM) | + | + |
| Cystine (2 mM) | − | + |
| Glutathione (2 mM) | + | + |
| Cystathionine (2 mM) | − | + |
| Homocysteine (2 mM) | + | + |
| Methionine (1.5 mM) | − | − |
+, growth; −, no growth.
Dot blot analysis.
Crude extracts were obtained using the FastPrep (MP Biomedicals) cell lysis system (speed, 6; time, 40 s; performed twice), followed by centrifugation (10 min at 4°C) to remove cell debris. For dot blot experiments, 20 ng (VPI 10463) or 200 ng (630Δerm, M7404, and M7404 tcdC+ strains) of proteins from the crude extracts was directly spotted onto a nitrocellulose membrane (Hybond-C Extra; Amersham Biosciences). The membranes were blocked with 5% (wt/vol) nonfat dried milk in Tris-buffered saline (TBS) supplemented with 0.2% (vol/vol) Tween 20 (TBST) for 1 h at room temperature. The membranes were then incubated for 90 min at 37°C with the TcdA antibody (PCG-4; Santa Cruz Biotechnology) and visualized as described by Antunes et al. (13).
Cell cultures and cytotoxicity assays.
Vero cells were cultured in Dulbecco's modified Eagle's medium (DMEM [Gibco]) supplemented with 5% (vol/vol) fetal calf serum and a 1% ready-to-use solution (vol/vol) of penicillin (10,000 U ml−1) and streptomycin (10 mg ml−1) (Sigma) at 37°C in a 5% CO2 atmosphere. For cytotoxicity assays, cells were grown until confluence in 96-well plates and incubated with 2-fold serially diluted C. difficile crude extracts in DMEM. After 24 h at 37°C, the cytopathic effect was evaluated using an optical microscope. Positive toxin reactions were indicated by the characteristic rounding of Vero cells. The titer of each sample corresponds to the well containing 50% round Vero cells.
Detection and quantification of hydrogen sulfide and pyruvate production.
H2S production was detected using lead-acetate paper (Macherey-Nagel), which turns black in the presence of this compound. Cells were grown in PY, PYC, or PYHC to an optical density at 600 nm (OD600) of 0.7. Then, the lead-acetate paper was placed at the bottom of the flask for 1 min to 1 h at 37°C, depending on the experiment. H2S production was further quantified as previously described (31, 37). Briefly, 5 ml of the 630Δerm strain culture was introduced into a flask with an alkaline agar layer enriched with zinc acetate and incubated for 1 h at 37°C. The OD670 was measured against an H2O blank. The amount of H2S was calculated using a standard curve of Na2S. For pyruvate quantification, cells were grown in PY or PYC for 10 h at 37°C, and pyruvate was quantified in the supernatant using a Sigma pyruvate assay kit. The final pyruvate concentration was standardized using the OD600 of the bacterial cultures.
Estimation of the intracellular amino acid content.
The intracellular concentrations of amino acids were estimated using high-pressure liquid chromatography (HPLC) (31, 38). Briefly, cells were suspended in a sulfosalicylic acid buffer (3% final concentration) and disrupted using a Fastprep apparatus (MP Biomedicals). After centrifugation, supernatant samples were analyzed by cation-exchange chromatography, followed by ninhydrin postcolumn derivatization as previously described (31).
Zymogram.
Zymograms were used to detect the cysteine desulfhydrase and homocysteine γ-lyase activities. Native protein crude extracts (40 and 100 μg, respectively) were run on a nondenaturing protein gel (12% polyacrylamide in Tris-glycine buffer). After electrophoresis, the gel was incubated at 37°C for 1 to 4 h in a Tris solution [50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 0.5 mM Pb(NO3)2, and 5 mM dithiothreitol (DTT)] with 0.4 mM pyridoxal-5-phosphate (PLP) containing either 10 mM l-cysteine or 10 mM homocysteine as previously described (32). H2S formed by the cysteine desulfhydrase or homocysteine γ-lyase activity precipitates as insoluble PbS.
Construction of C. difficile mutants.
The ClosTron gene knockout system (39) was used to inactivate genes encoding Fur (CD1287), SigL (CD3176), CysK (CD1594), and a TCS sensor histidine kinase (CD2602), as well as several regulators of unknown function (CD2065, CD0278, and CD2023 [Table 1]). As described in Fig. S1 in the supplemental material, primers were designed to retarget the group II intron of pMTL007 to these genes (see Table S1 in the supplemental material) and were used to generate a 353-bp DNA fragment by overlap PCR according to the manufacturer's instructions. These PCR products were cloned into the HindIII and BsrGI restriction sites of pMTL007 and were verified by DNA sequencing using the pMTL007-F and pMTL007-R primers (see Table S1). The derived pMTL007 plasmids were transformed into E. coli strain HB101(RP4) and transferred by conjugation into the C. difficile strain 630Δerm. C. difficile transconjugants were selected by subculture on brain heart infusion (BHI) agar containing thiamphenicol (15 μg/ml), and the integration of the group II intron RNA into genes was induced and selected by plating onto BHI agar containing erythromycin (2.5 μg/ml). The chromosomal DNA of the transconjugants was extracted using the InstaGene kit (Bio-Rad), and PCR with the primers ErmRAM-F and ErmRAM-R (see Table S1) was used to confirm the erythromycin-resistant phenotype due to the splicing of the group I intron from the group II intron following integration (see Fig. S1A). The insertion of the group II intron into target genes was verified by Southern blotting (see Fig. S1C) and by PCRs (see Fig. S1B) with primers flanking the 5′ ends of genes (see Table S1) and EBSu primer. To knock down malY (CD3029) expression, a DNA fragment comprising the 5′ untranslated region (UTR) and the beginning of the CD3029 open reading frame (−38 to +154 from the ATG start codon) was amplified by PCR and cloned between the XhoI and BamHI sites of the pRPF185 vector (36) to generate pDIA6456 expressing the 5′ end of malY in the antisense orientation under the control of the ATc-inducible Ptet promoter. This plasmid was transferred by conjugation into C. difficile strain 630Δerm. To complement the sigL mutant, the sigL gene and its promoter (−193 to +1380 from the ATG start codon) were amplified by PCR using the appropriate primers (see Table S1). The PCR fragment was cloned into the XhoI and BamHI sites of pMTL84121 (40) to generate plasmid pDIA6309. This plasmid was transferred by conjugation into the C. difficile sigL mutant (CDIP217), yielding strain CDIP342.
All experiments conducted with the mutants were standardized versus the wild type for the culture growth (OD600) and the protein concentration of the samples or by using a reference gene for the quantitative reverse transcriptase PCR (qRT-PCR) assays.
RNA isolation and quantitative real-time PCR.
C. difficile strains were grown in PY or PYC for 10 h. Total RNA extraction was performed using the FastRNA Pro Blue kit and a FastPrep apparatus according to the manufacturer's instructions (MP Biomedicals) as previously described (13). To synthesize cDNA, 1 μg of total RNA was heated at 70°C for 10 min in the presence of 1 μg of hexamer oligonucleotide primers (pdN6; Roche). RNAs were then reverse transcribed for 2 h at 37°C using an avian myeloblastosis virus (AMV) reverse transcriptase (RT) (Promega), 20 mM deoxynucleoside triphosphates (dNTPs), and 40 U of RNasin (Promega). Reverse transcriptase was inactivated by heating at 85°C for 5 min. Real-time quantitative RT-PCR was performed in a 20-μl reaction volume containing 20 ng of cDNAs, FastStart SYBR green master mix (ROX; Roche), and 200 nM gene-specific primers (see Table S1 in the supplemental material). Amplification and detection were performed as previously described (13). The quantity of each cDNA was normalized to the quantity of the cDNA of the DNA polymerase III (Pol III) gene (CD1305). The relative change in gene expression was recorded as the ratio to normalized target concentrations (threshold cycle [ΔΔCT]) (41). The Shapiro-Wilk test was performed to test the normality of the replicates for each condition (see Table S3 in the supplemental material). When a population of the two conditions was normally distributed, a t test was used: otherwise, we used a Mann-Whitney test as indicated in the figure legends. A P value of ≤0.05 was considered significant.
Microarray design for the C. difficile genome, DNA array hybridization, and data analysis.
The microarray of the C. difficile strain 630 genome was designed as previously described (15) (GEO database accession no. GPL10556). The transcriptomic analysis was performed with four different RNA preparations and a dye swap method. First, 10 μg of total RNA was reverse transcribed in cDNA using the SuperScript Indirect cDNA labeling system kit (Invitrogen) and Cy3 or Cy5 fluorescent dye (GE Healthcare) according to the manufacturer's recommendations. Labeled DNA hybridization to microarrays and array scanning were performed as previously described (15). All slides were analyzed using the R and limma software (Linear Model for Microarray Data) from the Bioconductor project (www.bioconductor.org). For each slide, we corrected for background with the “normexp” method (42), which resulted in strictly positive values and reduced variability in the log ratios for genes with low hybridization signal levels. Then, we normalized each slide by the “loess” method (43). To test for differential expression, we used Bayesian adjusted t statistics and performed the multiple testing correction of Benjamini and Hochberg based on the false discovery rate (FDR) (44). A gene was considered to be differentially expressed when the P value was <0.05.
Raw sequence analysis.
The presence of the TCS locus (CD2602 to CD26021) was inferred from raw sequences of 2,424 published strains (Sequence Read Archive [SRA] accession no. PRJEB2039, PRJEB4556, PRJEB3010, PRJEB190 to PRJEB216, PRJEB6600 to PRJEB6602, and PRJEB6575). For that purpose, we mapped the sequencing reads of each strain onto the nucleotide sequence of the TCS locus using Bowtie (1). A strain was considered to contain the TCS locus when the coverage was above 80%.
Microarray data accession number.
The complete experimental data set has been deposited in the GEO database under accession no. GSE22423.
RESULTS AND DISCUSSION
Cysteine-dependent repression of PaLoc genes.
It has been shown that toxin synthesis is repressed by cysteine in the high-toxin-level-producing strain VPI 10463 (12). To determine whether the effect of cysteine on toxin synthesis is strain dependent, we measured the effect of cysteine on toxin production in several C. difficile backgrounds (Table 1), such as strains 630Δerm and M7404 (a NAP1/027 epidemic strain), as well as an M7404 derivative strain carrying a wild-type copy of the tcdC gene on the pDLL17 plasmid (2); VPI 10463 was used as a control. All of the strains grew similarly in PY with or without cysteine. Cell crude extracts were obtained from these four strains after 10 h of growth in PY or PYC, and toxin production was assayed by Vero cell cytotoxicity assays, which predominantly assess TcdB, and protein dot blot analysis using a specific antibody raised against TcdA. Cytotoxic activity was lower in cells grown in the presence of cysteine (Fig. 2A) than in cells grown without cysteine, with 25- to 125-fold decreased cytotoxicity for strains 630Δerm, M7404, and M7404(pDLL17-tcdC) and 16,000-fold decreased cytotoxicity for strain VPI 10463. Moreover, TcdA accumulation was strongly reduced in the presence of cysteine in all of the strains tested (Fig. 2B). These results suggest that cysteine-dependent repression of toxin production is conserved among the C. difficile strains. Cysteine repressed toxin synthesis in both the epidemic 027 strain M7404, which does not express functional TcdC, and in its derivative strain that contains a wild-type tcdC gene (2). Thus, the effect of cysteine on toxin production is not mediated by TcdC.
FIG 2.
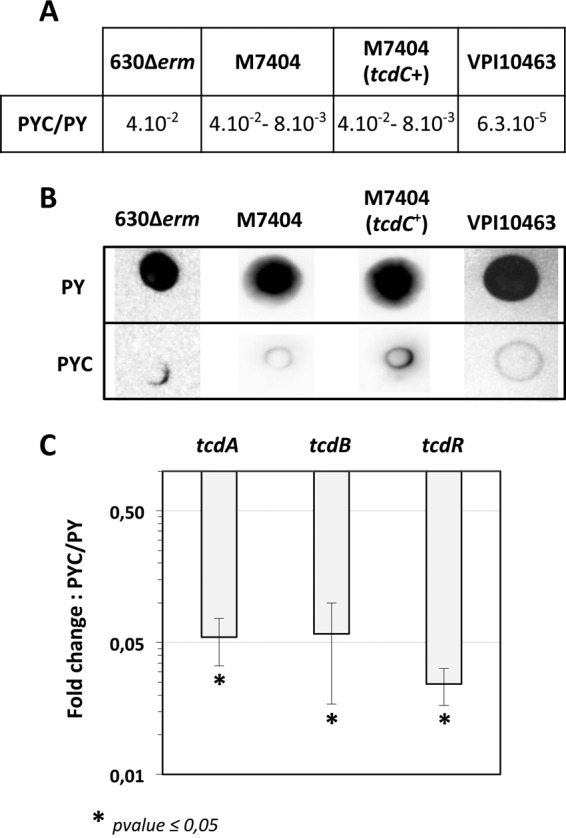
Effect of cysteine on toxin production in different C. difficile strains. (A) Cytotoxicity assays on Vero cells. Twofold serial dilutions of intracellular bacterial crude extracts were performed, and the dilutions were added to a 96-well plate of confluent Vero cells. The toxin titer corresponds to the lowest dilution of C. difficile crude extracts required for >50% cell rounding. Cytotoxicity results are presented as the ratio of the toxin titers of bacterial cells grown in the presence of cysteine (PYC) to those of bacterial cells grown in the absence of cysteine (PY). (B) TcdA dot blot analysis. The crude extracts of C. difficile strains (200 ng for strains 630Δerm, M7404, and M7404 complemented with pDLL17-tcdC and 20 ng for strain VPI 10463) were probed with anti-TcdA antibodies as described in Materials and Methods. The results presented are representative of crude extracts tested from at least three independent experiments. (C) Transcript levels of the tcdR, tcdA, and tcdB genes in strain 630Δerm grown in the presence or absence of cysteine. Results are presented as the ratio of the mRNA level (arbitrary units) of each gene in bacterial cells grown in the presence of cysteine (PYC) to that of each gene in cells grown in the absence (PY) of cysteine. The results are the averages from at least three independent experiments, and error bars are the standard deviations from the mean values. The statistical analysis was performed by using a t test (tcdA and tcdR) or a Mann-Whitney test (tcdB).
To determine whether the effect of cysteine on toxin production occurred at the transcriptional level, we performed qRT-PCR experiments for the tcdA, tcdB, and tcdR genes using strain 630Δerm (Fig. 2C). After 10 h of growth, the transcript levels of tcdA and tcdB decreased 18- and 17-fold, respectively, in the presence of cysteine. We also observed that the expression of the tcdR gene encoding the alternative sigma factor required for toxin gene transcription decreased 40-fold when cysteine was added (Fig. 2C). These data are in agreement with the results obtained by Karlsson et al. (11), suggesting that toxin gene transcription is repressed by cysteine through negative regulation of tcdR.
Reconstruction of sulfur metabolism in C. difficile.
An understanding of sulfur metabolism was a prerequisite to elucidating how cysteine negatively regulates toxin production. To reconstitute the sulfur metabolism pathways, we first searched for all of the gene homologs to the genes involved in sulfur assimilation pathways in other firmicutes (30) in the complete genome sequence of the reference C. difficile strain 630 (45) (Fig. 3). All genes identified are conserved in the VPI 10463 and NAP1/027 epidemic strains (see Table S4 in the supplemenal material). Then, to support the metabolic reconstruction and to obtain new insights into the physiology of C. difficile, we tested the ability of strain 630Δerm to grow in minimal media with different sulfur sources (Table 2).
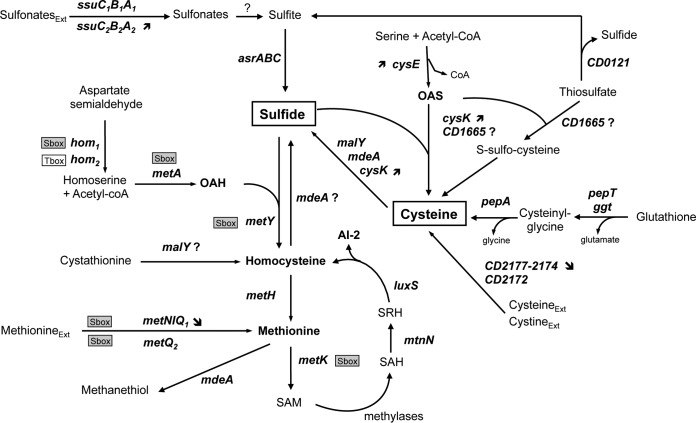
FIG 3.
Reconstruction of sulfur metabolism in C. difficile. Genes of strain 630Δerm are renamed on the basis of B. subtilis orthologs. The genes and their products are as follows: cysE, serine O-acetyltransferase (CD1595); cysK, OAS-thiol-lyase (CD1594); asrABC, anaerobic sulfite reductase (CD2231 to -2233); ssuCBA1, ABC-transport system sulfonates (CD1482 to -1484); ssuCBA2, ABC-transport system sulfonates (CD2989 to -2991); metA, homoserine acetyl-transferase (CD1826); metY, OAH thiol-lyase (CD1825); malY, cystathionine β-lyase (CD3029); metH, cobalamin-dependent methionine synthase (CD3596); metK, SAM synthetase (CD0130); mtnN, adenosylhomocysteine nucleosidase (CD2611); luxS, S-ribosylhomocysteine lyase (CD3598); mdeA, methionine γ-lyase (CD3577), metNIQ1, ABC transport system methionine (CD1489 to -1491); pepT, peptidase T (CD1046); pepA, leucine aminopeptidase (CD1300). Other abbreviations: AI-2, autoinducer 2; OAS; O-acetylserine; OAH, O-acetylhomoserine; SAM, S-adenosyl-methionine; SAH, S-adenosyl-homocysteine; SRH, S-ribosyl-homocysteine; Ext, external. As indicated, an S-box motif is located upstream of the metY-metA and metNIQ1 operons and of the metQ2 and metK genes, suggesting that they are controlled by a SAM-dependent riboswitch (55). A T box is present upstream of hom-CD1580.
Strain 630Δerm cannot grow when sulfate is the only sulfur source (Table 2). This finding is consistent with the absence of genes involved in the first steps of the sulfate assimilation pathway leading to sulfite (Fig. 1). In contrast, strain 630Δerm was able to grow in the presence of sulfide or thiosulfate (Table 2), indicating that C. difficile can synthesize cysteine from these compounds, probably through the CysE/CysK thiolation pathway (Fig. 3). Cysteine can also be produced from glutathione, a sulfur source utilized by strain 630Δerm (Table 2). PepT and PepA are probably involved in the degradation of glutathione to form cysteine (Fig. 3). However, the pathway of glutathione synthesis from cysteine that is found in C. perfringens (20) is absent in C. difficile. Strain 630Δerm can also grow with cysteine as the sole sulfur source, indicating that methionine is efficiently produced from this compound. As shown in Fig. 3, methionine is synthesized from homocysteine, likely through the cobalamine-dependent methionine synthase MetH. The two main pathways of homocysteine production in bacteria are transsulfuration and thiolation (Fig. 1) (30). Both pathways involve PLP-dependent enzymes: transsulfuration requires a cystathionine γ-synthase and a cystathionine β-lyase, while thiolation requires an O-acetyl-homoserine (OAH)-thiol-lyase (Fig. 1). In the genome of strain 630, three PLP-dependent enzymes were identified by their similarities: MetY, MalY, and MdeA (46). MetY contains an amino acid insertion specific to OAH-thiol-lyases, MalY is a cystathionine β-lyase of the PatB/MalY family (32), and MdeA is a probable methionine γ-lyase. However, no cystathionine γ-synthase is present in C. difficile, suggesting that a functional transsulfuration pathway is absent and that C. difficile synthesizes both methionine and cysteine by thiolation pathways.
Homocysteine and cysteine degradation.
Strain 630Δerm can grow in the presence of homocysteine and, to a lesser extent, in the presence of cystathionine but cannot use methionine as the sole sulfur source (Table 2). The ability to use homocysteine is surprising because the reverse transsulfuration pathway, which involves a cystathionine β-synthase and a cystathionine γ-lyase (Fig. 1), is absent in C. difficile (31, 47). Growth in the presence of homocysteine could be explained by the existence of a homocysteine γ-lyase, allowing the production of H2S from homocysteine and its possible conversion into cysteine (Fig. 3). Using lead-acetate paper, we detected the production of H2S during the growth of strain 630Δerm in PY plus homocysteine (PYHC), but not in PY alone (Fig. 4A). When we performed zymography using homocysteine as a substrate, we detected a single band in crude extracts of strain 630Δerm grown in PY, PYC, and PYHC (Fig. 4B), suggesting that homocysteine γ-lyase activity is induced under all of the growth conditions used. Among the PLP-dependent enzymes encoded by the C. difficile genome, MdeA shares significant similarities with the methionine γ-lyases of Citrobacter freundii (48) and of Brevibacterium linens. Interestingly, the methionine γ-lyase of B. linens also has homocysteine γ-lyase activity (49), making MdeA a probable candidate for the production of H2S from homocysteine and the degradation of methionine to form methanethiol in C. difficile (Fig. 3), as previously proposed (50).
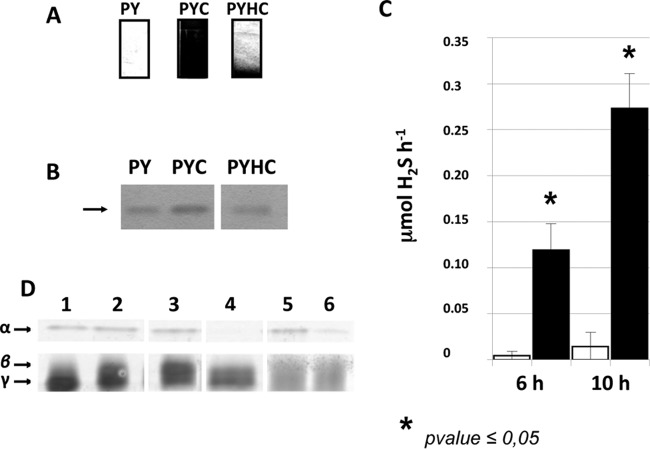
FIG 4.
Hydrogen sulfide production in strain 630Δerm. (A) Detection of H2S production using lead-acetate paper. H2S production was evaluated in the media PY, PY plus cysteine (PYC), and PY plus homocysteine (PYHC). The production of H2S yielded a black color due to the formation of PbS. (B) Detection of the homocysteine γ-lyase activity on a zymogram. Crude extracts of strain 630Δerm grown in PY, PYC, or PYHC were loaded on a native polyacrylamide gel (12%) and incubated with 10 mM homocysteine. Homocysteine γ-lyase was detected by the formation of insoluble PbS via the release of H2S. Lanes of the zymogram have been reorganized from the same image to present data chronologically. (C) Quantitative detection of H2S after 6 or 10 h of growth of strain 630Δerm in PY (white boxes) or PYC (black boxes). H2S production was measured using the quantitative methylene blue method, as described in Materials and Methods. The statistical analysis was performed by using the Mann-Whitney test for all genes. (D) Detection of cysteine desulfhydrase activities on a zymogram. Crude extracts of strain 630Δerm (lanes 1 and 2), 630Δerm::cysK (lane 3), 630Δerm::sigL (lane 4), 630Δerm(pRPF185) (lane 5), and 630Δerm(pDIA6456-ASmalY) (lane 6). The strains were grown in PY (lane 1) or PYC (lanes 2 to 6). Samples were charged on a native polyacrylamide gel (12%) and incubated with 10 mM cysteine. The cysteine desulfhydrases were detected by the formation of insoluble PbS formed by the release of H2S. The results presented are representative of at least three independent experiments. Lanes of the zymogram have been reorganized from the same image to present data chronologically.
In bacteria, cysteine is usually catabolized by cysteine desulfhydrases (32), producing H2S, pyruvate, and ammonia (Fig. 1). We detected high production of H2S during the growth of strain 630Δerm in PYC (Fig. 4A). Indeed, the quantification of H2S showed a 20- to 30-fold increase of H2S production when cysteine was added to the medium (Fig. 4C). This result clearly indicated that cysteine is efficiently degraded in C. difficile. To detect the cysteine desulfhydrase activities, we performed zymography using l-cysteine as the substrate. We detected two bands (α and γ) in the crude extract of strain 630Δerm grown in PY (Fig. 4D, lane 1). Interestingly, when strain 630Δerm was grown in PYC, we detected an additional band (β), indicating that synthesis of this desulfhydrase enzyme was induced by cysteine (Fig. 4D, lane 2). In B. subtilis, PatB/MalY- and CysK-type enzymes have cysteine desulfhydrase activities (32). To determine whether CysK of C. difficile is a cysteine desulfhydrase, we inactivated the cysK gene in strain 630Δerm using the ClosTron system (see Fig. S1 in the supplemental material). The zymogram profile obtained with the cysK mutant strain grown in PYC is similar to that obtained with the 630Δerm strain (Fig. 4D, lane 3). This finding suggests that CysK is not a major cysteine desulfhydrase in C. difficile under the conditions tested, although we cannot exclude a role for CysK in cysteine degradation. We failed to inactivate the gene encoding the PatB/MalY enzyme, probably because of its essentiality for C. difficile (51). Thus, to evaluate whether PatB/MalY is a cysteine desulfhydrase, we constructed a PatB/MalY-depleted strain using an antisense strategy (36, 52). Compared to the strain carrying the control plasmid (Fig. 4D, lane 5), the PatB/MalY-depleted strain (Fig. 4D, lane 6) displayed a decreased intensity of the α band, suggesting that MalY has cysteine desulfhydrase activity. However, the enzymes with cysteine desulfhydrase activity corresponding to the γ and β bands on the zymogram (Fig. 4D) remain to be identified.
Finally, we demonstrated that both homocysteine and cysteine are actively catabolized by C. difficile. Interestingly, it has recently been shown that when C. difficile grows in minimal medium with Casmino Acids, cysteine is consumed immediately and sulfide is produced (53). This finding is in complete agreement with our results. Thus, we propose that sulfide is a central compound of sulfur metabolism in C. difficile, as it is the direct precursor of both methionine and cysteine, as well as the major degradation product of the sulfur-containing amino acids homocysteine and cysteine (Fig. 3).
Global analysis of gene expression in response to cysteine.
To determine the global impact of cysteine on gene expression and to elucidate the mechanism of cysteine-dependent repression of toxin production, we performed a comparative transcriptional analysis of strain 630Δerm grown in PY or PYC at the onset of the stationary phase (10 h). In the presence of 10 mM cysteine, 6% of the genome (201 genes) was differentially expressed with a fold change of ≥2 (see Table S2 in the supplemental material). Among these genes, 120 and 81 were upregulated and downregulated, respectively. The major expression changes were seen in genes encoding cell-surface-associated proteins and proteins involved in sulfur, amino acid, carbon, and energy metabolism as well as in iron uptake (see Table S2). The transcriptomic analysis confirmed that toxin gene expression decreased in the presence of cysteine. In addition, exposure of C. difficile to high cysteine concentrations strongly induced the expression of genes encoding heat shock proteins belonging to both class I (HrcA dependent), such as the groESL and hrcA operons, and class III (CtsR dependent), such as the ctsR and the clpB operons. We validated the transcriptomic analysis by performing qRT-PCR with a selection of 12 representative genes. The results confirmed the microarray data (see Table S2).
Regulation of genes involved in sulfur metabolism and in thiol protection by cysteine.
As expected, the expression of genes related to sulfur metabolism, including transporters of amino acids, was controlled by cysteine. The metQ1 gene encoding the methionine-binding protein of an ABC transporter (54, 55) (Fig. 3) was less strongly expressed in PYC. This gene is probably regulated by an S-box riboswitch in the promoter region of the metNIQ1 operon, like most of the genes required for methionine uptake (Fig. 3) (55, 56). The ABC transporter system composed of CD2177, CD2176, CD2175, CD2174, and CD2172 is likely involved in the uptake of cystine and/or cysteine in C. difficile (Fig. 3). CD2177 and CD2174 share similarities with the cystine-binding proteins of E. coli and Bacillus subtilis (57), while CD2176 and CD2175 are similar to the l-cystine permeases of E. coli and B. subtilis. The expression of all of the genes encoding this ABC transporter decreased 2.5- to 4-fold in PYC, as is usually observed for cysteine/cystine transporters. In contrast, the expression of the ssuA2 and ssuC2 genes, which encode proteins sharing similarities with sulfonate ABC transporters, increased in the presence of cysteine (Fig. 3).
The expression of cysK, encoding the OAS-thiol-lyase, and cysE, encoding the serine acetyl-transferase was induced 40- to 50-fold in the presence of cysteine (Fig. 3; see Table S2 in the supplemental material). The upregulation of cysKE expression in PYC is surprising because CysK and CysE, which are required for cysteine biosynthesis, are usually induced during cysteine limitation, as previously observed in C. perfringens, B. subtilis, E. coli, and Salmonella (20, 30, 34). However, in the presence of high cysteine concentrations, CysK contributes to cysteine degradation rather than cysteine synthesis (58). Under these conditions, CysE activity is inhibited by feedback, as established in several bacteria and plants (38, 59). Nonetheless, the role of CysK in cysteine metabolism remains to be clarified.
Finally, we observed that genes involved in thiol protection were induced in the presence of cysteine (see Table S2 in the supplemental material). They encode two thioredoxins (CD1690 and CD2355), a thioredoxin reductase (CD1691), and a thiol peroxidase (CD1822). The induction of genes involved in thiol protection and in the stress response suggests that cysteine or its derivative products (e.g., H2S) stress C. difficile. However, the addition of 10 mM cysteine to the PY medium did not affect C. difficile growth and cell viability (data not shown), while this amino acid is toxic in other bacteria, such as E. coli and B. subtilis (60; I. Martin-Verstraete, unpublished results). The expression of the stress-responsive genes in relation to the absence of cysteine toxicity in C. difficile may be the result of an adaptation to an anaerobic lifestyle.
Induction of fur and Fur-regulated genes in the presence of cysteine.
The ferric uptake regulator (Fur) protein is an iron response repressor that controls the expression of genes involved in iron transport in bacteria (61, 62). The CD1287 protein shares 48% identity with the Fur protein of B. subtilis. To demonstrate that CD1287 corresponds to Fur, we constructed a CD1287 mutant strain using the ClosTron system (see Fig. S1 in the supplemental material). Then, we tested the effect of CD1287 disruption on the level of transcription of the feoB1 and fhuD genes by qRT-PCR. In B. subtilis, FeoB1 and FhuD participate in ferrous iron and ferrichrome uptake, respectively (61, 63). We showed that the addition of 200 μM dipyridyl, a ferrous iron chelator, to the growth medium increased the transcript level of the CD1287, fhuD, and feoB1 genes and that transcription of feoB1 and fhuD increased 3,500- and 45-fold, respectively, in the CD1287 mutant compared to the wild-type strain (data not shown). These results strongly indicate that CD1287 is the Fur repressor in C. difficile, as recently demonstrated (64).
From our global transcriptomic analysis, we found that the presence of cysteine in the medium induced the Fur regulon, including fur and genes encoding transporters of ferrous iron and ferrichrome (Table 3). Using the Fur-binding site of B. subtilis (61), we detected a potential Fur box upstream of approximately 20 genes that are differentially expressed in PYC, including fur, feoB1, cysK, and fhuD, as well as genes encoding proteins of unknown function, such as CD2992, CD1485, CD2499, and CD2881 (Table 3). The consensus Fur box for C. difficile (Fig. 5A), deduced from the putative Fur-binding motifs present in the regulatory region of these genes, is highly similar to that defined by Ho et al. (64). We then tested the effect of cysteine on the transcription of some of these Fur targets by qRT-PCR in both 630Δerm and a fur mutant strain. In the presence of cysteine, the transcript levels of the fur, feoB1, cysK, fhuD, and CD2992 genes increased 3.2-, 750-, 56-, 12-, and 10-fold, respectively, in strain 630Δerm (Fig. 5B), a result consistent with the transcriptome data (Table 3). The cysteine-dependent upregulation of feoB1, fhuD, and CD2992 was abolished in the fur mutant, indicating that the effect of cysteine is mediated by the Fur repressor. Interestingly, the induction of cysK transcription by cysteine was not completely abolished in the fur mutant and was only 5-fold lower than it was in strain 630Δerm (Fig. 5B). In addition, in the absence of cysteine, the transcript level of cysK was 4.5-fold higher in the fur mutant than in strain 630Δerm (data not shown). As a Fur box is located in the promoter region of the cysK gene, the regulation of cysK by cysteine is complex, involving both direct regulation by the Fur repressor and control by a still-uncharacterized regulator. While very few data concerning the control of cysK expression by Fur are available (65), the cysteine-dependent regulation of CysK synthesis in C. difficile seems to be atypical.
TABLE 3.
List of the Fur regulon genes differentially expressed in PY and PYC with a putative Fur box in their promoter region
| Gene | Protein function | PYC/PY ratio | Fur box positiona |
|---|---|---|---|
| CD1287 (fur) | Ferric uptake regulation protein | 3.3 | −38 |
| CD1477 (feoA) | Ferrous iron transport protein A | 64.1 | |
| CD1478 (feoA1) | Ferrous iron transport protein A1 | 107.0 | |
| CD1479 (feoB1) | Ferrous iron transport protein B1 | 127.1 | −60 |
| CD1480 | Putative exported protein | 162.6 | |
| CD1745A (feoA) | Ferrous iron transport protein A | 16.7 | −30 |
| CD3273 (feoA3) | Ferrous iron transport protein A | 13.4 | −30 |
| CD3274 (feoB3) | Ferrous iron transport protein B | 12.1 | |
| CD2878 (fhuD) | ABC transporter, ferrichrome substrate-binding protein | NAb | −47 |
| CD2875 (fhuC) | Ferrichrome ABC transporter | 3.0 | |
| CD1594 (cysK) | O-Acetyl-serine sulfhydrylase | 43.0 | −162 |
| CD1595 (cysE) | Serine acetyltransferase | 44.0 | |
| CD1999 (fldX) | Flavodoxin | 28.5 | −158 |
| CD1777 | Putative arsenate reductase | 3.3 | −80 |
| CD1485 | Conserved hypothetical protein | 6.9 | −34 |
| CD2499 | Conserved hypothetical protein | 15.4 | −34 |
| CD2881 | Conserved hypothetical protein | 2.6 | −58 |
| CD2992 | Conserved hypothetical protein | 2.5 | −36 |
| CD2991 | ABC transporter, sulfonate-permease | 3.5 | |
| CD2989 | ABC transporter, sulfonate-extracellular solute-binding protein | 4.8 |
The position of the Fur box is indicated according to the translational start site of the corresponding gene.
NA, not detected in transcriptome.
FIG 5.
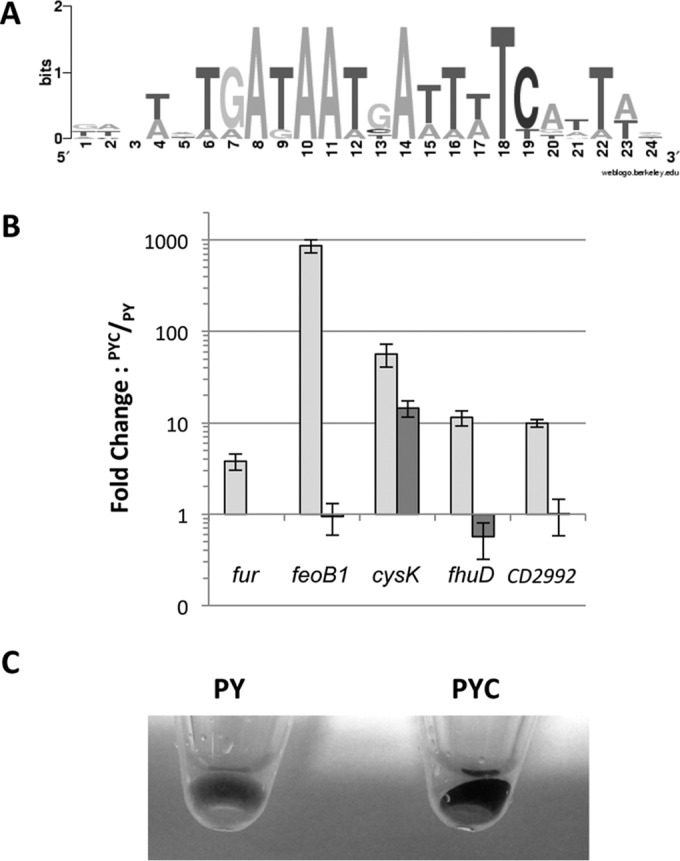
Analysis of Fur-regulated genes induced in the presence of cysteine. (A) Consensus sequence of the Fur box motif of C. difficile. The sequence logo was created by the alignment of putative Fur-regulated genes induced in the presence of cysteine on the WebLogo website (http://weblogo.berkeley.edu). The height of the nucleotides is proportional to their frequency. (B) Effect of cysteine on the transcript level of fur (CD1287), feoB1 (CD1479), cysK (CD1594), fhuD (CD2878), and CD2992 in 630Δerm and the 630Δerm::fur. The 630Δerm (white boxes) and 630Δerm::fur (black boxes) strains were grown for 10 h in PY or PYC. qRT-PCR results are presented as the ratio of the amount of mRNA (arbitrary units) of each gene in bacterial cells grown in PYC to that of each mRNA in the bacterial cells grown in PY. Data are the averages from at least three independent experiments, and error bars are the standard deviations from the mean values. (C) Aspect of the bacterial pellet of strain 630Δerm grown for 10 h in PY or in PYC. The black precipitate is due to FeS precipitation.
The induction of the Fur regulon by cysteine suggests that the presence of cysteine in the growth medium mimics the conditions of iron depletion. A black precipitate appears when strain 630Δerm is grown in PYC (Fig. 5C). This finding is consistent with the production of high levels of H2S via cysteine degradation by cysteine desulfhydrases (Fig. 4C), which probably leads to the formation of this black deposit from iron-sulfide precipitation. This phenomenon is often described in anaerobic waste collection systems (66). Therefore, iron depletion due to the precipitation of iron in the presence of excess sulfide can explain the induction of the Fur-regulated genes.
Regulation by cysteine of carbon and energy metabolism.
The ability of C. difficile to use a wide range of carbohydrates might be important during infection. Accordingly, Antunes et al. (13) demonstrated the existence of links between carbon metabolism and toxin production. The addition of cysteine to the medium increased the expression of several genes of carbon metabolism, including genes encoding phosphotransferase systems (PTS) and genes encoding enzymes involved in the second part of glycolysis (Fig. 6A; see Table S2 in the supplemental material).
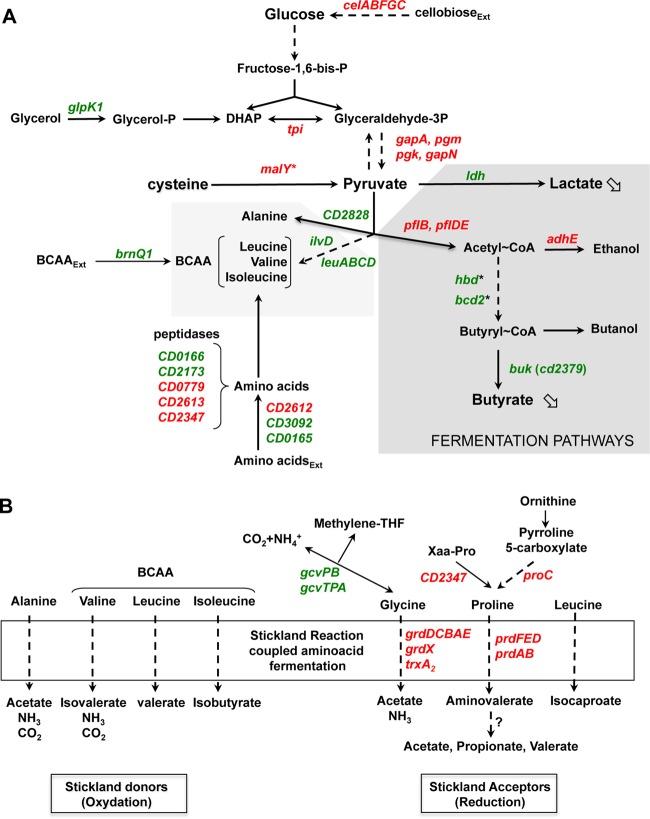
FIG 6.
Overview of C. difficile genes involved in carbon and amino acid metabolism that are differentially expressed in the presence of cysteine. Genes that are up- and downregulated in the presence of cysteine in the transcriptome analysis are indicated in red and green, respectively. An asterisk indicates that the differential transcript level was detected by qRT-PCR. (A) Carbon metabolism and fermentation pathways. Assignments of genes (listed with their products) regulated by cysteine availability are as follows: tpi, triosephosphate isomerase; gapA/gapN, glyceraldehyde-3-phosphate dehydrogenase; pgk, phosphoglycerate kinase; pgm, 2,3-bisphosphoglycerate-mutase; celABC, PTS cellobiose; celF, cellobiose-6-P hydrolase; ldh, lactate dehydrogenase; adhE, aldehyde-alcohol dehydrogenase; pflB and pflD, pyruvate formate lyase; pflE and pflA, pyruvate formate lyase activating enzyme; thiA1, acetyl-CoA acetyltransferase; bcd2, butyryl-CoA dehydrogenase; hbD2, 3-hydroxybutyryl-CoA dehydrogenase; crt2, 3-hydroxybutyryl-CoA dehydratase; buk, butyrate kinase; malY, cysteine desulfhydrase; ilvD, dihydroxy-acid dehydratase; leuA, 2-isopropylmalate synthase; leuB, 3-isopropylmalate dehydrogenase; leuC, 3-isopropylmalate dehydratase large subunit; leuD, 3-isopropylmalate dehydratase small subunit; brnQ1, BCAA transporter. (B) Stickland reactions and associated metabolism. Assignments of genes regulated in response to cysteine availability are as follows: grdDCBAEX, glycine reductase complex; prdEDBA, proline reductase; prdF, proline racemase; CD2347, putative Xaa-Pro dipeptidase; proC, pyrroline-5-carboxylate reductase, gcvPB, glycine decarboxylase; gcvTPA, bifunctional glycine dehydrogenase/aminomethyl transferase protein.
The expression of genes involved in the fermentation pathways of C. difficile was also modulated by the presence of cysteine (Fig. 6A). Thus, the expression of ldh and buk, which encode lactate dehydrogenase and one butyrate kinase, respectively, decreased, while the expression of genes encoding pyruvate formate lyases and an alcohol dehydrogenase increased in the presence of cysteine. Surprisingly, the bcd2 operon, which is involved in the production of butyryl coenzyme A (butyryl-CoA) from acetyl-CoA (Fig. 6A), was not differentially expressed in the transcriptome analysis. However, when we tested the effect of cysteine on the expression of the bcd2 and hbd2 genes by qRT-PCR, we showed that their transcript levels decreased 5.5- and 6-fold in PYC compared to PY, respectively. This result is in agreement with the results of a proteome analysis performed in strain VPI 10463 (12), showing that the production of enzymes involved in the conversion of acetyl-CoA to butyryl-CoA (Bcd2, Crt2, and Hbd) decreases when cells are grown in the presence of cysteine. To evaluate the impact of cysteine on fermentation pathways, we quantified the end products of fermentation in strain 630Δerm grown over 48 h in PY or PYC by gas-liquid chromatography. The amounts of lactate and butyrate were reduced 4- and 6-fold, respectively, in the presence of cysteine (see Fig. S2 in the supplemental material), as observed in strain VPI 10463 (12). This result is consistent with the downregulation of ldh, buk, and bcd2 operon expression. After 48 h of growth, butyric acid production was high in PY, leading to a final concentration of 5 mM compared to less than 1 mM in PYC (see Fig. S2). Interestingly, the addition of butyric acid to the growth medium enhances toxin production in strain VPI 10463 (12). Thus, the addition of cysteine to the medium may indirectly control toxin production at least partly via its influence on butyric acid production. However, the molecular mechanisms of the regulation of toxin synthesis in response to butyric acid availability remain to be determined.
Control of amino acid metabolism by cysteine.
To analyze the impact of cysteine on amino acid metabolism, we compared the transcriptome and the pools of amino acids obtained from strain 630Δerm grown in PY or PYC. A total of 32 genes involved in peptide or amino acid metabolism were differentially expressed under these two conditions (see Table S2 in the supplemental material), while the intracellular concentrations of leucine, tyrosine, alanine, valine, phenylalanine, and glutamic acid were increased in the presence of cysteine (Table 4). The expression of several genes involved in peptide degradation (CD0779, CD2613, CD2347, CD2173, and CD0166) and amino acid uptake (CD2612, CD3092, and CD0165) was differentially regulated when cysteine was added to the medium (Fig. 6A). In C. difficile, amino acid catabolism by the Stickland reactions can be a primary source of energy when bacteria are grown with amino acids as the sole carbon and nitrogen sources (67). Stickland reactions couple the metabolism of a pair of amino acids, of which one serves as the Stickland donor (alanine, valine, leucine, or isoleucine) and is oxidatively deaminated or decarboxylated to generate ATP and reducing power (NADH), and the second serves as the Stickland acceptor (glycine, proline, hydroxyproline, or leucine) and is reduced or reductively deaminated, regenerating NAD+ from NADH (Fig. 6B). The genes encoding the glycine reductase (grd) and d-proline reductase (prd) operons, which are involved in the reduction of the Stickland acceptors glycine and proline, were induced up to 10-fold in the presence of cysteine (Fig. 6B; see Table S2). Interestingly, the expression of proC (CD3281), which is involved in the conversion of ornithine into proline, and of CD2347, which encodes a peptidase sharing similarities with Xaa-Pro dipeptidases and potentially generates free proline for use in the Stickland reactions (67), was also increased in the presence of cysteine.
TABLE 4.
Effect of cysteine addition on the intracellular concentration of amino acids in strain 630Δerm
| Amino acid | Amino acid concn (μmol/liter) |
PYC/PY ratiob | |
|---|---|---|---|
| PY | PYC | ||
| Upregulated in presence of cysteine | |||
| Leucine | NDa | 16.25 ± 1.6 | + |
| Tyrosine | ND | 20 ± 1.2 | + |
| Alanine | 13.5 ± 0.7 | 748 ± 40 | 55 |
| Valine | 5.3 ± 0.5 | 56.8 ± 1.3 | 10 |
| Phenylalanine | 2.6 ± 0.5 | 25.75 ± 5 | 10 |
| Glutamic acid | 19.3 ± 2 | 121.9 ± 9.5 | 6.5 |
| Aminobutyric acid | 8.5 ± 0.5 | 46.35 ± 0.8 | 5.5 |
| Threonine | 1.1 ± 0.1 | 2.4 ± 0.4 | 2.2 |
| Serine | 4 ± 0 | 9.85 ± 0.15 | 2.5 |
| Asparagine | 29.7 ± 2.6 | 59.7 ± 1.6 | 2 |
| Methionine | 4.85 ± 0.35 | 11.65 ± 1.65 | 2.5 |
| Downregulated in presence of cysteine | |||
| Cystathionine | 1.2 ± 0.2 | ND | − |
| Glutamine | 11.8 ± 0.4 | 6 ± 0.1 | 0.5 |
| Hydroxyproline | 23.3 ± 1.5 | 13.9 ± 0.5 | 0.6 |
ND, not detectable.
+, detected only in PYC medium; −, detected only in PY medium.
In strain 630Δerm grown in the presence of cysteine, we observed a substantial accumulation of alanine (Table 4), a by-product of cysteine catabolism. Indeed, cysteine is first converted into pyruvate through cysteine desulfhydrases; pyruvate is then converted into alanine by alanine aminotransferases (Fig. 6A). CD2828, which shares similarities with an alanine aminotransferase characterized in E. coli (68), is a good candidate for this activity. However, CD2828 was repressed in PYC (see Table S2 in the supplemental material). The negative control of CD2828 expression in the presence of a high intracellular concentration of alanine might explain this downregulation.
Several genes involved in the biosynthesis of branched-chain amino acids (BCAAs) were also repressed by cysteine (Fig. 6A; see Table S2 in the supplemental material). The expression of ilvD involved in the BCAA synthesis from pyruvate and of the leuABCD operon, which is involved in synthesis of leucine, was downregulated 5- to 10-fold in the presence of cysteine. Interestingly, the transcription of brnQ1, which encodes a BCAA transporter (Fig. 6A), was also decreased in PYC. A T box specific to leucine (T boxLeu) is present in the promoter region of the leuABCD operon and of the leuS gene, indicating that these genes are probably induced during leucine starvation via premature termination of transcription (56, 69). We note that ilvD, leuABCD, and brnQ1 belong to the CodY regulon, which is involved in the adaptive response to nutrient limitation (17). Thus, the increase in the concentration of valine and leucine when cysteine is added (Table 4) might lead to the repression of genes involved in BCAA biosynthesis and uptake through their control by a T boxLeu or by CodY. In B. subtilis, changes in the rate of endogenous isoleucine, leucine, and valine synthesis modulate the expression of CodY-regulated genes (70). In addition, for Clostridum sticklandii, using amino acids as the carbon and energy sources, cysteine is one of the six amino acids that is preferentially degraded, while valine, leucine, and isoleucine are used later, suggesting that certain amino acids regulate the metabolism of others (71). Cysteine is also one of the three amino acids that are preferentially used by C. difficile (53). Our results suggest that the presence of cysteine may delay the use of other amino acids, such as BCAAs, which are known to act as corepressors of CodY (17). Accordingly, we observed that 31 CodY-regulated genes were repressed in strain 630Δerm when cysteine was added (see Table S2), suggesting that cysteine has an impact on CodY activity.
Involvement of regulators in the cysteine-dependent repression of toxin production.
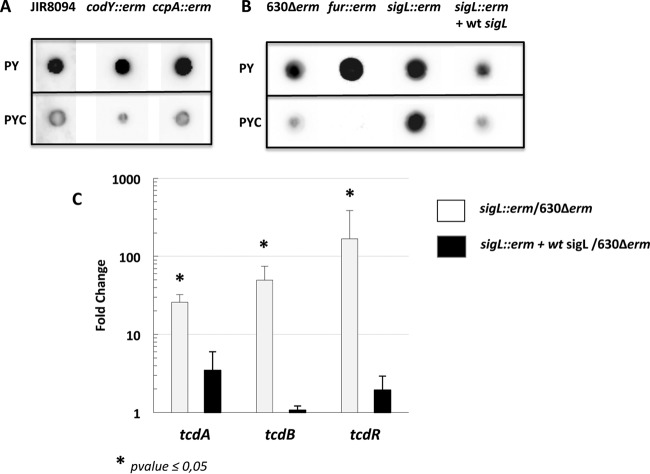
Toxin expression may be under the control of a global regulator that is able to sense cysteine availability. Interestingly, we showed that several genes encoding regulators are regulated in response to cysteine availability. Indeed, CD0278, CD1692, CD2023, and CD2065 were up- or downregulated in the presence of cysteine (see Table S2 in the supplemental material). Using the ClosTron system, we inactivated CD0278, CD2023, and CD2065, but we did not succeed in disrupting CD1692. Compared to the wild-type strain, toxin gene expression was similarly repressed by cysteine in the CD0278, CD2023, and CD2065 mutants (data not shown). However, we cannot exclude the possibility that a still-unidentified regulator intervenes in this control. Alternatively, the effector of cysteine-dependent regulation might be a cysteine by-product that is accumulated during growth in PYC. To discriminate between a direct effect of cysteine and an indirect metabolic effect, we added 10 mM cysteine to the growth medium for 1 h at the onset of the stationary phase. Surprisingly, under this condition limiting the catabolism of cysteine, we observed that the transcription of tcdA, tcdB, and tcdR was increased (see Fig. S3 in the supplememtal material), suggesting that cysteine downregulates toxin production through a product of cysteine degradation (see below). As changes in carbon source and amino acid availability were observed after the growth of C. difficile in the presence of cysteine (Table 4; see Fig. S2 in the supplemental material), we wondered whether toxin synthesis could be controlled by the global regulators CodY and CcpA, which are known to regulate toxin gene expression in response to the levels of the BCAAs and PTS sugars, respectively (13, 14, 18). However, we showed that toxin synthesis is similarly repressed by cysteine in the codY or ccpA mutant and wild-type strains (Fig. 7A), indicating that CodY and CcpA do not mediate the control of toxin synthesis by cysteine.
FIG 7.
Role of Fur, CcpA, CodY, and SigL in the cysteine-dependent repression of toxin production. Strains JIR8094, JIR8094::codY, and JIR8094::ccpA (A) and strains 630Δerm, 630Δerm::fur, 630Δerm::sigL, and 630Δerm::sigL(pDIA6309-sigL) (B) were grown for 10 h in PY or PYC. TcdA production was estimated from crude extracts by dot blot analysis using an anti-TcdA antibody. The results are representative of at least three independent experiments. (C) Effect of cysteine on tcdA, tcdB, and tcdR transcript levels in 630Δerm::sigL (white boxes) or 630Δerm::sigL complemented with pDIA6309-sigL (black boxes) versus the wild-type strain, 630Δerm. All strains were grown for 10 h in PYC. qRT-PCR results are presented as the ratio between the amount of the mRNA (arbitrary units) of each gene normalized by the DNA Pol III gene in both 630Δerm::sigL and 630Δerm::sigL complemented with pDIA6309-sigL compared to the mRNA level in the wild-type strain. Data are the averages from at least three independent experiments, and error bars are the standard deviations from the mean values. The statistical analysis was performed by using a t test for all genes, with the exception of tcdB (Mann-Whitney test).
The Fur regulator might also be responsible for the cysteine-dependent regulation of toxin synthesis. Indeed, the expression of many virulence factors in pathogenic bacteria is negatively regulated by Fur in response to iron availability (72). We showed that TcdA production is repressed by cysteine in a fur mutant in the same manner as the wild-type strain (Fig. 7B), indicating that Fur is not involved in the downregulation of toxin production in PYC. This result was in agreement with the absence of the toxin genes in the Fur transcriptome, as recently defined by Ho et al. (64).
Role of SigL in the cysteine-dependent repression of toxin production.
In C. difficile strain VPI 10463, it has been proposed that several proteins induced under toxin-producing conditions (PY) might be controlled by SigL (11). The sigL gene encodes a sigma factor belonging to the SigL/RpoN/σ54 family, which is known to play an important role in metabolism, adaptation, and virulence (73–77). To evaluate the role of SigL in the cysteine-dependent regulation of toxin synthesis, we inactivated the sigL gene (see Fig. S1 in the supplemental material). When we compared the levels of toxin produced between the sigL mutant and the wild-type strain 630Δerm by dot blot analysis, we first observed that TcdA was produced at higher levels in the sigL mutant than in the wild-type strain when the cells were grown in PY medium (Fig. 7B). This effect might be due to decreased competition between SigL and the toxin-specific sigma factor TcdR for the core enzyme of the RNA polymerase, as already proposed for SigH (15). Surprisingly, we observed similar levels of TcdA production in the sigL mutant grown in PY and PYC (Fig. 7B). To determine whether SigL regulates toxin synthesis at the transcriptional level, we tested the transcription of the tcdA, tcdB, and tcdR genes in 630Δerm and sigL mutant strains grown in PYC by qRT-PCR. As shown in Fig. 7C, the transcript levels of the tcdA, tcdB, and tcdR genes were approximately 25- to 50-fold higher in the sigL mutant than in the wild-type strain in the presence of cysteine. Moreover, complementation of the sigL mutant by a wild-type copy of sigL partially restored the cysteine-dependent repression of TcdA production (Fig. 7B) and of PaLoc gene transcription (Fig. 7C). These results indicate that SigL mediates the cysteine-dependent regulation of toxin gene expression. However, using the well-conserved consensus sequence of SigL-dependent promoters (78), we did not find a SigL-type promoter upstream of tcdA, tcdB, and tcdR, suggesting that SigL indirectly regulates the PaLoc genes, probably in response to an increase in the by-products of cysteine degradation. Indeed, using lead-acetate paper, we showed that the production of H2S via cysteine degradation was strongly reduced in the sigL mutant compared to that in strain 630Δerm (Fig. 8A) and was restored by complementation with pDIA6309. Interestingly, according to the zymogram profile obtained with the sigL mutant, we showed that the cysteine desulfhydrase activity of MalY (α band) significantly decreased (Fig. 4D, lane 4). In addition, the expression of malY was 4-fold lower in a sigL mutant than that in the wild-type strain (data not shown). This finding is in agreement with the role of SigL in the control of cysteine degradation in C. difficile. As pyruvate is the first product of cysteine degradation, we measured the extracellular concentration of pyruvate. We observed that in strain 630Δerm, the pyruvate concentration increased more than 2-fold when we added cysteine to the medium. However, the level of pyruvate decreased 8-fold in the sigL mutant compared to the level in the wild-type strain (Fig. 8B). In addition, the sigL mutant strain complemented with the wild-type copy of sigL had an extracellular pyruvate concentration similar to that in strain 630Δerm (Fig. 8B).
FIG 8.
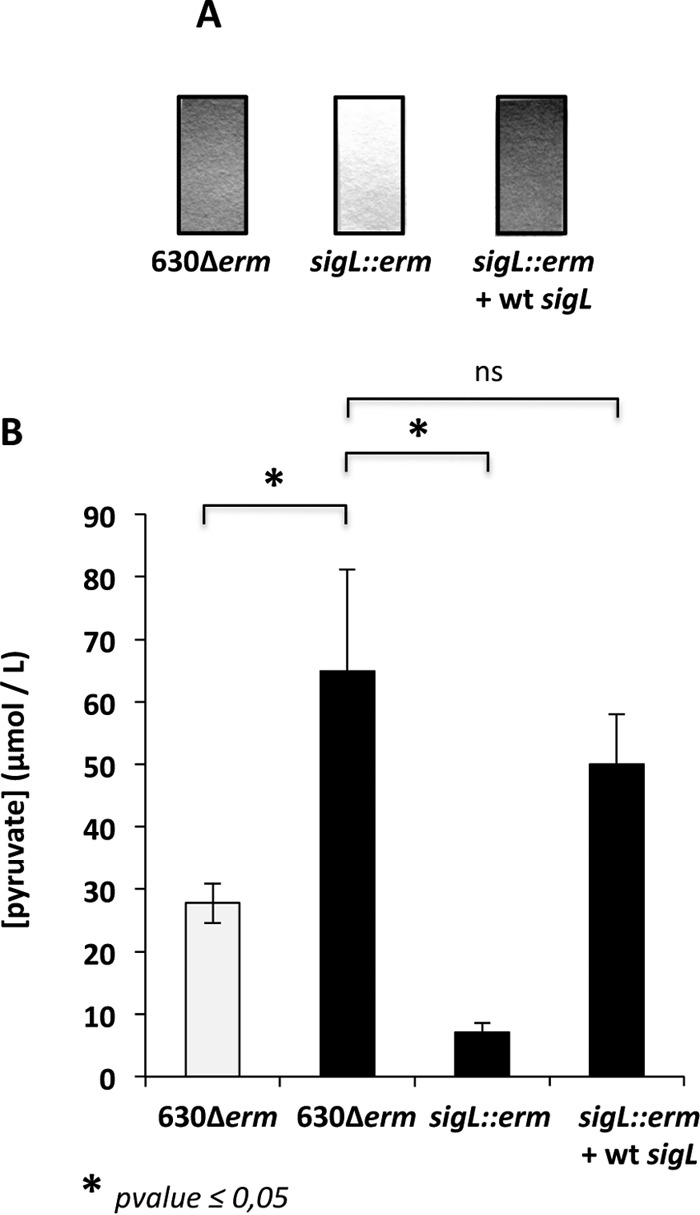
Effect of sigL inactivation on cysteine degradation and pyruvate production. (A) Detection of H2S production in the 630Δerm, 630Δerm::sigL, and 630Δerm::sigL(pDIA6309-sigL) strains grown in PYC by using lead-acetate papers. (B) Quantitative detection of pyruvate in the supernatant of strains 630Δerm, 630Δerm::sigL, and 630Δerm::sigL(pDIA6309-sigL) after 10 h of growth in PY (white boxes) or PYC (black boxes). The statistical analysis was performed by using Mann-Whitney test for all genes. ns, not significant.
Involvement of pyruvate as a signal mediating toxin gene repression in response to cysteine.
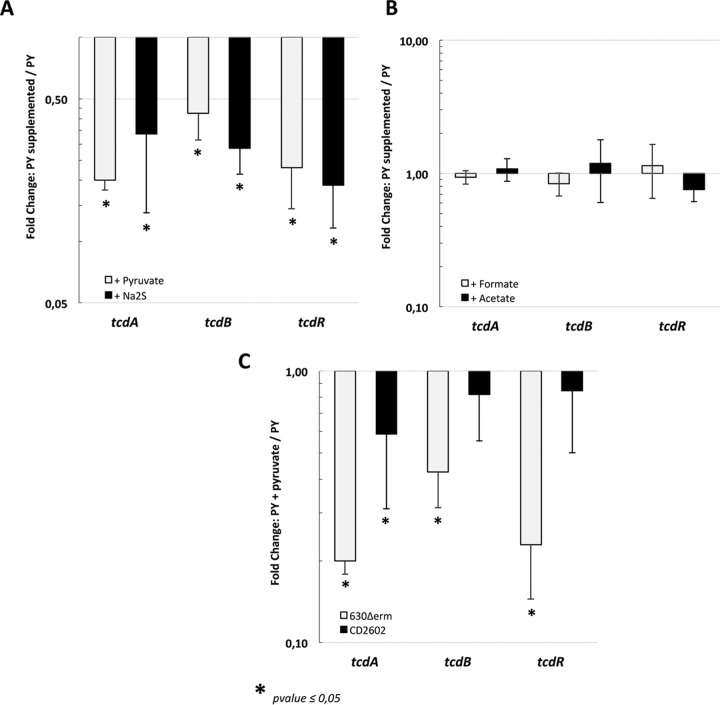
The accumulation of H2S or pyruvate resulting from cysteine degradation during growth may be the signal modulating toxin production. To test this hypothesis, we added 10 mM either Na2S or pyruvate to the PY medium when bacteria reached the stationary growth phase and harvested the cells after 1 h of exposure. The addition of pyruvate or Na2S decreased the transcription of tcdA, tcdB, and tcdR (Fig. 9A). Interestingly, the effect of pyruvate was not abolished when we performed a similar experiment in the sigL mutant (see Fig. S4 in the supplemental material). This finding confirms that cysteine-dependent regulation of toxin production is mainly the consequence of the products of cysteine degradation. We also tested the effect of the pyruvate by-products such as formate and acetate on PaLoc gene transcription. The addition of 10 mM formate or acetate to the growing cells for 1 h did not affect the transcription of tcdA, tcdB, and tcdR (Fig. 9B). Thus, we concluded that pyruvate and probably sulfide are metabolic signals mediating the cysteine-dependent repression of toxin production. As the downregulation of toxin gene expression in the presence of cysteine (Fig. 2C) is more prominent than in the presence of pyruvate or sulfide alone (Fig. 9A), it is possible that the cysteine by-products could have a combined effect on toxin production.
FIG 9.
Effect of pyruvate and Na2S on toxin gene expression. (A) Transcript levels of the tcdA, tcdB, and tcdR genes in strain 630Δerm after 1 h of exposure to pyruvate (white boxes) or Na2S (black boxes). The strain was grown in PY for 9 h, and 10 mM pyruvate or 10 mM Na2S was then added to the medium. Cells were centrifuged 1 h later. The statistical analysis was performed by using a t test for all genes, with an exception for tcdB plus Na2S (Mann-Whitney test). (B) Transcript levels of the tcdA, tcdB, and tcdR genes of strain 630Δerm after 1 h of exposure to formate (white boxes) or acetate (black boxes). The strain was grown in PY for 9 h, 10 mM formate or 10 mM acetate was then added to the medium, and cells were centrifuged 1 h later. The statistical analysis was performed by using a t test for all genes, with the exception of tcdB plus acetate (Mann-Whitney test). (C) Transcript levels of the tcdA, tcdB, and tcdR genes of strain 630Δerm (white boxes) and 630Δerm::CD2602 (black boxes) after exposure to pyruvate for 1 h, as described for panel A. qRT-PCR results are presented as the ratio between the amount of mRNA (arbitrary units) of each gene normalized by the DNA Pol III gene from bacterial cells grown in PY supplemented with one of the compounds (pyruvate, Na2S, formate, or acetate) compared to the amount of mRNA in the untreated cells. Data are the averages from at least three independent experiments, and error bars are the standard deviations from the mean values. The statistical analysis was performed by using a t test for all genes, with the exception of tcdR plus pyruvate (Mann-Whitney test).
Identification of a TCS regulating toxin gene expression in response to pyruvate.
Pyruvate is a central metabolite of bacteria, and its cellular concentration is tightly controlled. In a broad range of bacteria, including E. coli and Bacillus licheniformis, pyruvate is excreted into the medium at the end of the exponential growth phase under the conditions of overflow metabolism. This compound is further taken up and metabolized (79, 80). In E. coli, the two-component system (TCS) YpdA-YpdB, which is also present in B. licheniformis (81), reacts predominantly to the presence of exogenous pyruvate and induces the expression of yhjX, which encodes a transporter of the major facilitator superfamily (79). The YpdA/YpdB system probably contributes to nutrient scavenging before entry into the stationary phase. In the genome of all of the C. difficile strains sequenced, we found a TCS (CD2602-CD2601) that is highly similar to YpdA-YpdB. Importantly, the transmembrane receptor domain of CD2602 shares 53% identity and 79% similarity with that of the histidine kinase YpdA, suggesting a common signal for these kinases. To determine whether CD2602-CD2601 is involved in the regulation of toxin gene expression in response to the level of exogenous pyruvate, we inactivated the CD2602 gene in strain 630Δerm (see Fig. S1 in the supplemental material). Then we tested the effect of pyruvate on tcdA, tcdB, and tcdR transcription in the CD2602 mutant. The temporary addition of pyruvate during the growth of the CD2602 mutant had a less pronounced effect on toxin gene transcription than it had in the wild-type strain (Fig. 9C). This result suggests that the transcriptional regulation of tcdA, tcdB, and tcdR in response to pyruvate availability is, at least in part, mediated by the TCS CD2602-CD2601.
Conclusion.
Addition of cysteine to PY medium leads to dramatic changes in the pattern of expression of C. difficile genes involved in several processes, including sulfur and iron metabolism, fermentation, and the stress response. These effects on gene transcription are probably related to modifications of the metabolite pools, as we showed for the repression of toxin production by metabolic changes due to cysteine degradation and transcriptional control by cysteine through a still-uncharacterized regulator. We identified SigL as a major regulator of cysteine-dependent repression of C. difficile toxin production. We found that the levels of H2S and pyruvate resulting from cysteine degradation by cysteine desulfhydrases (32) were decreased in a sigL mutant, which no longer repressed toxin genes in the presence of cysteine. A similar regulation of toxin production through the metabolic conversion of cysteine to sulfate and pyruvate has been observed in B. pertussis (21). SigL also seems to play an important role in the control of pyruvate metabolism in Listeria monocytogenes (82), as observed in C. difficile with a drop in the pyruvate concentration in the sigL mutant. Among the cysteine by-products produced in C. difficile, we demonstrated that the addition of pyruvate or H2S to PY is sufficient to repress toxin gene expression, suggesting that pyruvate and H2S, rather than cysteine, must be metabolic signals regulating toxin production. Interestingly, when strain 630Δerm grows in the presence of cysteine, genes involved in the synthesis of pyruvate from glucose or cysteine are upregulated, while genes required for pyruvate dissimilation leading to butyrate (buk operon) and lactate (ldh) production or involved in the biosynthesis of amino acids or fatty acids from pyruvate and acetyl-CoA, respectively, are downregulated (Fig. 6). This change leads to the accumulation of pyruvate in the extracellular medium (Fig. 7B), where it is probably sensed by the membrane-associated kinase CD2602. Thus, in response to pyruvate, the response regulator CD2601 might negatively control toxin gene expression, either directly or indirectly. Conversely, butyrate, which is known to positively regulate toxin expression (12), is found at a lower concentration in the extracellular medium (see Fig. S2 in the supplemental material), which no longer stimulates toxin synthesis. Recently, it has been shown that C. difficile can grow in all parts of the intestinal tract of a mouse model, while toxins are only produced in the cecum and colon (83). Thus, according to the metabolites present in the small intestine and in the colon, toxin genes might be differentially expressed in the gut. Accordingly, formate and acetate (directly obtained from pyruvate) predominate in the small intestine, while the levels of propionate and butyrate are higher in the colon (84). Such a control has been described in Salmonella enterica serovar Typhimurium: formate acts as a diffusible signal to induce the expression of invasion genes in the small intestine, the site that is preferentially colonized by this enteropathogen, while butyrate is present at higher concentrations in the colon and represses these genes (85, 86). It is tempting to speculate that the high level of pyruvate in the small intestine represses the expression of C. difficile toxin genes, while butyrate mainly present in the colon induces toxin synthesis. Further biochemical studies will be necessary to characterize the signal transduction pathway of the CD2602-CD2601 TCS. Thus, the ability of C. difficile to monitor the pyruvate level to adapt its physiology, metabolism, and virulence might be crucial to the success of a CDI.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from the Institut Pasteur. M. Dancer-Thibonnier and T. Dubois are postdoctoral fellows from the PTR program funded by the Institut Pasteur (PTR256) and the French region Ile-de-France (DIM-Malinf), respectively.
We thank Philippe Bouvet for help with the CPG analysis and Roselyne Garnotel and Sophie Roulin for the intracellular amino acid content assays.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00121-16.
REFERENCES
- 1.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH. 2010. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23:529–549. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter GP, Douce GR, Govind R, Howarth PM, Mackin KE, Spencer J, Buckley AM, Antunes A, Kotsanas D, Jenkin GA, Dupuy B, Rood JI, Lyras D. 2011. The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile. PLoS Pathog 7:e1002317. doi: 10.1371/journal.ppat.1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mani N, Dupuy B. 2001. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc Natl Acad Sci U S A 98:5844–5849. doi: 10.1073/pnas.101126598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matamouros S, England P, Dupuy B. 2007. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol Microbiol 64:1274–1288. doi: 10.1111/j.1365-2958.2007.05739.x. [DOI] [PubMed] [Google Scholar]
- 5.Govind R, Dupuy B. 2012. Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog 8:e1002727. doi: 10.1371/journal.ppat.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akerlund T, Svenungsson B, Lagergren A, Burman LG. 2006. Correlation of disease severity with fecal toxin levels in patients with Clostridium difficile-associated diarrhea and distribution of PCR ribotypes and toxin yields in vitro of corresponding isolates. J Clin Microbiol 44:353–358. doi: 10.1128/JCM.44.2.353-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupuy B, Sonenshein AL. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol 27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 8.Karlsson S, Dupuy B, Mukherjee K, Norin E, Burman LG, Akerlund T. 2003. Expression of Clostridium difficile toxins A and B and their sigma factor TcdD is controlled by temperature. Infect Immun 71:1784–1793. doi: 10.1128/IAI.71.4.1784-1793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deneve C, Delomenie C, Barc MC, Collignon A, Janoir C. 2008. Antibiotics involved in Clostridium difficile-associated disease increase colonization factor gene expression. J Med Microbiol 57:732–738. doi: 10.1099/jmm.0.47676-0. [DOI] [PubMed] [Google Scholar]
- 10.Bouillaut L, Self WT, Sonenshein AL. 2013. Proline-dependent regulation of Clostridium difficile Stickland metabolism. J Bacteriol 195:844–854. doi: 10.1128/JB.01492-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlsson S, Burman LG, Akerlund T. 2008. Induction of toxins in Clostridium difficile is associated with dramatic changes of its metabolism. Microbiology 154:3430–3436. doi: 10.1099/mic.0.2008/019778-0. [DOI] [PubMed] [Google Scholar]
- 12.Karlsson S, Lindberg A, Norin E, Burman LG, Akerlund T. 2000. Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect Immun 68:5881–5888. doi: 10.1128/IAI.68.10.5881-5888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antunes A, Martin-Verstraete I, Dupuy B. 2011. CcpA mediated repression of Clostridium difficile toxin gene expression. Mol Microbiol 79:882–899. doi: 10.1111/j.1365-2958.2010.07495.x. [DOI] [PubMed] [Google Scholar]
- 14.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol Microbiol 66:206–219. doi: 10.1111/j.1365-2958.2007.05906.x. [DOI] [PubMed] [Google Scholar]
- 15.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. 2011. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol 193:3186–3196. doi: 10.1128/JB.00272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Underwood S, Guan S, Vijayasubhash V, Baines SD, Graham L, Lewis RJ, Wilcox MH, Stephenson K. 2009. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J Bacteriol 191:7296–7305. doi: 10.1128/JB.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dineen SS, McBride SM, Sonenshein AL. 2010. Integration of metabolism and virulence by Clostridium difficile CodY. J Bacteriol 192:5350–5362. doi: 10.1128/JB.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antunes A, Camiade E, Monot M, Courtois E, Barbut F, Sernova NV, Rodionov DA, Martin-Verstraete I, Dupuy B. 2012. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res 40:10701–10718. doi: 10.1093/nar/gks864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Theriot CM, Koenigsknecht MJ, Carlson PE Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, Li JZ, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andre G, Haudecoeur E, Monot M, Ohtani K, Shimizu T, Dupuy B, Martin-Verstraete I. 2010. Global regulation of gene expression in response to cysteine availability in Clostridium perfringens. BMC Microbiol 10:234. doi: 10.1186/1471-2180-10-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogdan JA, Nazario-Larrieu J, Sarwar J, Alexander P, Blake MS. 2001. Bordetella pertussis autoregulates pertussis toxin production through the metabolism of cysteine. Infect Immun 69:6823–6830. doi: 10.1128/IAI.69.11.6823-6830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grifantini R, Bartolini E, Muzzi A, Draghi M, Frigimelica E, Berger J, Randazzo F, Grandi G. 2002. Gene expression profile in Neisseria meningitidis and Neisseria lactamica upon host-cell contact: from basic research to vaccine development. Ann N Y Acad Sci 975:202–216. doi: 10.1111/j.1749-6632.2002.tb05953.x. [DOI] [PubMed] [Google Scholar]
- 23.Hatzios SK, Bertozzi CR. 2011. The regulation of sulfur metabolism in Mycobacterium tuberculosis. PLoS Pathog 7:e1002036. doi: 10.1371/journal.ppat.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendez J, Reimundo P, Perez-Pascual D, Navais R, Gomez E, Guijarro JA. 2011. A novel cdsAB operon is involved in the uptake of l-cysteine and participates in the pathogenesis of Yersinia ruckeri. J Bacteriol 193:944–951. doi: 10.1128/JB.01058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shelver D, Rajagopal L, Harris TO, Rubens CE. 2003. MtaR, a regulator of methionine transport, is critical for survival of group B streptococcus in vivo. J Bacteriol 185:6592–6599. doi: 10.1128/JB.185.22.6592-6599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xayarath B, Marquis H, Port GC, Freitag NE. 2009. Listeria monocytogenes CtaP is a multifunctional cysteine transport-associated protein required for bacterial pathogenesis. Mol Microbiol 74:956–973. doi: 10.1111/j.1365-2958.2009.06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soutourina O, Dubrac S, Poupel O, Msadek T, Martin-Verstraete I. 2010. The pleiotropic CymR regulator of Staphylococcus aureus plays an important role in virulence and stress response. PLoS Pathog 6:e1000894. doi: 10.1371/journal.ppat.1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masip L, Veeravalli K, Georgiou G. 2006. The many faces of glutathione in bacteria. Antioxid Redox Signal 8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 29.Zeller T, Klug G. 2006. Thioredoxins in bacteria: functions in oxidative stress response and regulation of thioredoxin genes. Naturwissenschaften 93:259–266. doi: 10.1007/s00114-006-0106-1. [DOI] [PubMed] [Google Scholar]
- 30.Guédon E, Martin-Verstraete I. 2007. Cysteine metabolism and its regulation in bacteria, p 195–218. In Wendisch VF. (ed), Amino acid biosynthesis-pathways, regulation and metabolic engineering. Springer, Berlin, Germany. [Google Scholar]
- 31.Hullo MF, Auger S, Soutourina O, Barzu O, Yvon M, Danchin A, Martin-Verstraete I. 2007. Conversion of methionine to cysteine in Bacillus subtilis and its regulation. J Bacteriol 189:187–197. doi: 10.1128/JB.01273-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auger S, Gomez MP, Danchin A, Martin-Verstraete I. 2005. The PatB protein of Bacillus subtilis is a C-S-lyase. Biochimie 87:231–238. doi: 10.1016/j.biochi.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Gutierrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. 2009. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev 73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Even S, Burguière P, Auger S, Soutourina O, Danchin A, Martin-Verstraete I. 2006. Global control of cysteine metabolism by CymR in Bacillus subtilis. J Bacteriol 188:2184–2197. doi: 10.1128/JB.188.6.2184-2197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soutourina O, Poupel O, Coppee JY, Danchin A, Msadek T, Martin-Verstraete I. 2009. CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulfur source utilization and plays a role in biofilm formation. Mol Microbiol 73:194–211. doi: 10.1111/j.1365-2958.2009.06760.x. [DOI] [PubMed] [Google Scholar]
- 36.Fagan RP, Fairweather NF. 2011. Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem 286:27483–27493. doi: 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez del Castillo Lozano M, Tache R, Bonnarme P, Landaud S. 2007. Evaluation of a quantitative screening method for hydrogen sulfide production by cheese-ripening microorganisms: the first step towards l-cysteine catabolism. J Microbiol Methods 69:70–77. doi: 10.1016/j.mimet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Tanous C, Soutourina O, Raynal B, Hullo MF, Mervelet P, Gilles AM, Noirot P, Danchin A, England P, Martin-Verstraete I. 2008. The CymR regulator in complex with the enzyme CysK controls cysteine metabolism in Bacillus subtilis. J Biol Chem 283:35551–35560. doi: 10.1074/jbc.M805951200. [DOI] [PubMed] [Google Scholar]
- 39.Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Methods 70:452–464. doi: 10.1016/j.mimet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 40.Heap JT, Pennington OJ, Cartman ST, Minton NP. 2009. A modular system for Clostridium shuttle plasmids. J Microbiol Methods 78:79–85. doi: 10.1016/j.mimet.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Breitling R, Armengaud P, Amtmann A, Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 43.Smyth GK, Speed T. 2003. Normalization of cDNA microarray data. Methods 31:265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 44.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc Ser 57:289–300. [Google Scholar]
- 45.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 46.Mehta PK, Christen P. 2000. The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Adv Enzymol Relat Areas Mol Biol 74:129–184. [DOI] [PubMed] [Google Scholar]
- 47.Andre G, Even S, Putzer H, Burguiere P, Croux C, Danchin A, Martin-Verstraete I, Soutourina O. 2008. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res 36:5955–5969. doi: 10.1093/nar/gkn601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manukhov IV, Mamaeva DV, Rastorguev SM, Faleev NG, Morozova EA, Demidkina TV, Zavilgelsky GB. 2005. A gene encoding l-methionine gamma-lyase is present in Enterobacteriaceae family genomes: identification and characterization of Citrobacter freundii l-methionine gamma-lyase. J Bacteriol 187:3889–3893. doi: 10.1128/JB.187.11.3889-3893.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dias B, Weimer B. 1998. Purification and characterization of l-methionine gamma-lyase from Brevibacterium linens BL2. Appl Environ Microbiol 64:3327–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ali V, Nozaki T. 2007. Current therapeutics, their problems, and sulfur-containing-amino-acid metabolism as a novel target against infections by “amitochondriate” protozoan parasites. Clin Microbiol Rev 20:164–187. doi: 10.1128/CMR.00019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dembek M, Barquist L, Boinett CJ, Cain AK, Mayho M, Lawley TD, Fairweather NF, Fagan RP. 2015. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio 6:e02383–14. doi: 10.1128/mBio.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boudry P, Gracia C, Monot M, Caillet J, Saujet L, Hajnsdorf E, Dupuy B, Martin-Verstraete I, Soutourina O. 2014. Pleiotropic role of the RNA chaperone protein Hfq in the human pathogen Clostridium difficile. J Bacteriol 196:3234–3248. doi: 10.1128/JB.01923-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neumann-Schaal M, Hofmann JD, Will SE, Schomburg D. 2015. Time-resolved amino acid uptake of Clostridium difficile 630Deltaerm and concomitant fermentation product and toxin formation. BMC Microbiol 15:281. doi: 10.1186/s12866-015-0614-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hullo MF, Auger S, Dassa E, Danchin A, Martin-Verstraete I. 2004. The metNPQ operon of Bacillus subtilis encodes an ABC permease transporting methionine sulfoxide, d- and l-methionine. Res Microbiol 155:80–86. doi: 10.1016/j.resmic.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 55.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. 2004. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res 32:3340–3353. doi: 10.1093/nar/gkh659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguenec C, Coppee JY, Dupuy B, Martin-Verstraete I. 2013. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet 9:e1003493. doi: 10.1371/journal.pgen.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burguiere P, Auger S, Hullo MF, Danchin A, Martin-Verstraete I. 2004. Three different systems participate in l-cystine uptake in Bacillus subtilis. J Bacteriol 186:4875–4884. doi: 10.1128/JB.186.15.4875-4884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Awano N, Wada M, Mori H, Nakamori S, Takagi H. 2005. Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl Environ Microbiol 71:4149–4152. doi: 10.1128/AEM.71.7.4149-4152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hindson VJ. 2003. Serine acetyltransferase of Escherichia coli: substrate specificity and feedback control by cysteine. Biochem J 375:745–752. doi: 10.1042/bj20030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harris CL. 1981. Cysteine and growth inhibition of Escherichia coli: threonine deaminase as the target enzyme. J Bacteriol 145:1031–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 62.Vasileva D, Janssen H, Honicke D, Ehrenreich A, Bahl H. 2012. Effect of iron limitation and fur gene inactivation on the transcriptional profile of the strict anaerobe Clostridium acetobutylicum. Microbiology 158:1918–1929. doi: 10.1099/mic.0.056978-0. [DOI] [PubMed] [Google Scholar]
- 63.Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. 2006. Role of the Fur regulon in iron transport in Bacillus subtilis. J Bacteriol 188:3664–3673. doi: 10.1128/JB.188.10.3664-3673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho TD, Ellermeier CD. 2015. Ferric uptake regulator Fur control of putative iron acquisition systems in Clostridium difficile. J Bacteriol 197:2930–2940. doi: 10.1128/JB.00098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Torres VJ, Attia AS, Mason WJ, Hood MI, Corbin BD, Beasley FC, Anderson KL, Stauff DL, McDonald WH, Zimmerman LJ, Friedman DB, Heinrichs DE, Dunman PM, Skaar EP. 2010. Staphylococcus aureus Fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect Immun 78:1618–1628. doi: 10.1128/IAI.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nielsen AH, Hvitved-Jacobsen T, Vollertsen J. 2008. Effects of pH and iron concentrations on sulfide precipitation in wastewater collection systems. Water Environ Res 80:380–384. doi: 10.2175/106143007X221328. [DOI] [PubMed] [Google Scholar]
- 67.Jackson S, Calos M, Myers A, Self WT. 2006. Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. J Bacteriol 188:8487–8495. doi: 10.1128/JB.01370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim SH, Schneider BL, Reitzer L. 2010. Genetics and regulation of the major enzymes of alanine synthesis in Escherichia coli. J Bacteriol 192:5304–5311. doi: 10.1128/JB.00738-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. 2008. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA 14:717–735. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brinsmade SR, Kleijn RJ, Sauer U, Sonenshein AL. 2010. Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J Bacteriol 192:6357–6368. doi: 10.1128/JB.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fonknechten N, Chaussonnerie S, Tricot S, Lajus A, Andreesen JR, Perchat N, Pelletier E, Gouyvenoux M, Barbe V, Salanoubat M, Le Paslier D, Weissenbach J, Cohen GN, Kreimeyer A. 2010. Clostridium sticklandii, a specialist in amino acid degradation: revisiting its metabolism through its genome sequence. BMC Genomics 11:555. doi: 10.1186/1471-2164-11-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Troxell B, Hassan HM. 2013. Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol 3:59. doi: 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dalet K, Briand C, Cenatiempo Y, Hechard Y. 2000. The rpoN gene of Enterococcus faecalis directs sensitivity to subclass IIa bacteriocins. Curr Microbiol 41:441–443. doi: 10.1007/s002840010164. [DOI] [PubMed] [Google Scholar]
- 74.Iyer VS, Hancock LE. 2012. Deletion of sigma(54) (rpoN) alters the rate of autolysis and biofilm formation in Enterococcus faecalis. J Bacteriol 194:368–375. doi: 10.1128/JB.06046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mattila M, Somervuo P, Rattei T, Korkeala H, Stephan R, Tasara T. 2012. Phenotypic and transcriptomic analyses of sigma L-dependent characteristics in Listeria monocytogenes EGD-e. Food Microbiol 32:152–164. doi: 10.1016/j.fm.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 76.Okada Y, Okada N, Makino S, Asakura H, Yamamoto S, Igimi S. 2006. The sigma factor RpoN (sigma54) is involved in osmotolerance in Listeria monocytogenes. FEMS Microbiol Lett 263:54–60. doi: 10.1111/j.1574-6968.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- 77.Saldias MS, Lamothe J, Wu R, Valvano MA. 2008. Burkholderia cenocepacia requires the RpoN sigma factor for biofilm formation and intracellular trafficking within macrophages. Infect Immun 76:1059–1067. doi: 10.1128/IAI.01167-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Francke C, Groot Kormelink T, Hagemeijer Y, Overmars L, Sluijter V, Moezelaar R, Siezen RJ. 2011. Comparative analyses imply that the enigmatic sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics 12:385. doi: 10.1186/1471-2164-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fried L, Behr S, Jung K. 2013. Identification of a target gene and activating stimulus for the YpdA/YpdB histidine kinase/response regulator system in Escherichia coli. J Bacteriol 195:807–815. doi: 10.1128/JB.02051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paczia N, Nilgen A, Lehmann T, Gatgens J, Wiechert W, Noack S. 2012. Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb Cell Fact 11:122. doi: 10.1186/1475-2859-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yangtse W, Zhou Y, Lei Y, Qiu Y, Wei X, Ji Z, Qi G, Yong Y, Chen L, Chen S. 2012. Genome sequence of Bacillus licheniformis WX-02. J Bacteriol 194:3561–3562. doi: 10.1128/JB.00572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arous S, Buchrieser C, Folio P, Glaser P, Namane A, Hebraud M, Hechard Y. 2004. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology 150:1581–1590. doi: 10.1099/mic.0.26860-0. [DOI] [PubMed] [Google Scholar]
- 83.Koenigsknecht MJ, Theriot CM, Bergin IL, Schumacher CA, Schloss PD, Young VB. 2015. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun 83:934–941. doi: 10.1128/IAI.02768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Keeney KM, Finlay BB. 2011. Enteric pathogen exploitation of the microbiota-generated nutrient environment of the gut. Curr Opin Microbiol 14:92–98. doi: 10.1016/j.mib.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, Hinton JC, Van Immerseel F. 2006. Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl Environ Microbiol 72:946–949. doi: 10.1128/AEM.72.1.946-949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang Y, Suyemoto M, Garner CD, Cicconi KM, Altier C. 2008. Formate acts as a diffusible signal to induce Salmonella invasion. J Bacteriol 190:4233–4241. doi: 10.1128/JB.00205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hussain HA, Roberts AP, Mullany P. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J Med Microbiol 54:137–141. doi: 10.1099/jmm.0.45790-0. [DOI] [PubMed] [Google Scholar]
- 88.O'Connor JR, Lyras D, Farrow KA, Adams V, Powell DR, Hinds J, Cheung JK, Rood JI. 2006. Construction and analysis of chromosomal Clostridium difficile mutants. Mol Microbiol 61:1335–1351. doi: 10.1111/j.1365-2958.2006.05315.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.