Abstract
Pseudomonas aeruginosa employs its type VI secretion system (T6SS) as a highly effective and tightly regulated weapon to deliver toxic molecules to target cells. T6SS-secreted proteins of P. aeruginosa can be detected in the sputum of cystic fibrosis (CF) patients, who typically present a chronic and polymicrobial lung infection. However, the mechanism of T6SS activation in the CF lung is not fully understood. Here we demonstrate that extracellular DNA (eDNA), abundant within the CF airways, stimulates the dynamics of the H1-T6SS cluster apparatus in Pseudomonas aeruginosa PAO1. Addition of Mg2+ or DNase with eDNA abolished such activation, while treatment with EDTA mimicked the eDNA effect, suggesting that the eDNA-mediated effect is due to chelation of outer membrane-bound cations. DNA-activated H1-T6SS enables P. aeruginosa to nonselectively attack neighboring species regardless of whether or not it was provoked. Because of the importance of the T6SS in interspecies interactions and the prevalence of eDNA in the environments that P. aeruginosa inhabits, our report reveals an important adaptation strategy that likely contributes to the competitive fitness of P. aeruginosa in polymicrobial communities.
INTRODUCTION
Cystic fibrosis (CF) is a common lethal inherited disease in Caucasians (1). Disease symptoms are caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene that result in pleiotropic effects, including the accumulation of a dehydrated, viscous mucus layer in the patient's respiratory tract (2–4). This thickened mucus impairs the normal bronchopulmonary clearance and permits chronic microbial colonization of the respiratory tract (5, 6). The microbial clearance defect leads to an intense, sustained recruitment of the neutrophilic inflammatory response, resulting in tissue destruction, promotion of the decline of lung function, and patient mortality (7–9).
Although a large diversity of species comprise a dynamic CF microbiome, which varies through disease progression and between patients (10–18), Pseudomonas aeruginosa is one of the predominant pathogens present in CF patients according to the 2013 CF Foundation patient registry report (https://www.cff.org/). Chronic infection with P. aeruginosa and its conversion to an alginate-hyperproducing, mucoid phenotype is associated with progressive decline in lung function, increased risk of hospitalization, and reduced survival (19–22). P. aeruginosa can also cause serious infections to patients suffering from burns and immunocompromised diseases (23).
P. aeruginosa employs the type 6 secretion system (T6SS) to deliver toxic antimicrobial and antieukaryote effectors to target cells (24–28). Commonly found in Gram-negative bacteria, the T6SS is an organelle consisting of a cytosolic contractile outer sheath, a membrane complex, and an inner tube (27, 29). Sheath contraction ejects the inner tube and its associated effector proteins to the extracellular environment (30–32). The inner tube is composed of stacks of hemolysin corregulated proteins (Hcp), which have been found in the sputum of CF patients infected with P. aeruginosa (27). After contraction, the sheath is recognized and disassembled by an ATPase ClpV (31, 33). The dynamic activities of T6SS assembly, contraction, and disassembly can be visualized using fluorescence tags to ClpV or sheath proteins (31). P. aeruginosa model strain PAO1 possesses three T6SS clusters that are tightly regulated at transcriptional and posttranslational levels (24, 27, 34–36). For example, RetS (regulator of exopolysaccharide and type III secretion) negatively regulates the transcription of the H1-T6SS cluster (27, 37); thus, the retS mutant displays enhanced H1-T6SS activities (31). Although the H1-T6SS is dormant in the wild-type P. aeruginosa strain, its dynamic activity is rapidly induced when cells sense extracellular assaults, including the T6SS activity (31, 34), conjugation (38), and polymyxin B activity (38). A multicomponent signaling cascade, TagQRST-PpkA-PppA-Fha1, is required for the precise retaliatory response that activates the firing of H1-T6SS at the exact site of external assault (34, 38). Although the H2-T6SS and the H3-T6SS clusters are implicated in both antimicrobial and antieukaryote activities (26, 39, 40), their contribution to bacterial killing is substantially less effective than that of the H1-T6SS, as exhibited by prey cell tolerance of the P. aeruginosa mutant lacking the H1-T6SS (25, 38).
Extracellular DNA (eDNA) is present in many different environments, including soil, water, sediment, and gastrointestinal and respiratory tracts (41–44). The physiological functions of eDNA include horizontal gene transfer, nutrient source, and biofilm formation (43, 45, 46). As a negatively charged macromolecule, eDNA in excess amounts also displays antimicrobial effects by chelating membrane-bound cations (47, 48). In the viscous CF lung mucus, eDNA is accumulated to a considerable level (up to 20 mg/ml) (1, 41, 42, 44). The DNA in CF sputum is largely derived from neutrophils, including the presence of neutrophil extracellular traps (NETs) that are deployed to combat infection of the lung (47, 49). However, eDNA in the matrix of laboratory-grown biofilms is microbial in origin and results from cell lysis or excretion of membrane vesicles (46, 50).
Interestingly, P. aeruginosa can sense some unknown signals released by killed P. aeruginosa cells during interspecies competition and can respond by increasing the expression of the H1-T6SS, the response termed P. aeruginosa response to antagonism (PARA) (51). Similarly, the H1-T6SS is transcriptionally induced when P. aeruginosa is cultivated in CF mucus (52) and secretion of Hcp is readily detectable in clinical CF sputum samples (27). However, the signals that trigger T6SS activation in CF are not well understood. Here we provide evidence that sputum isolated from adult CF patients enhances the dynamic activities of the H1-T6SS of P. aeruginosa within seconds of exposure, suggesting a posttranslational response. This response is mechanistically and temporarily distinct from the PARA (51), which also stimulates the expression of the H1-T6SS. We determined that H1-T6SS activation results from perturbation of the bacterial outer membrane caused by extracellular DNA (eDNA), a common macromolecule enriched in the CF sputum. We demonstrated that eDNA-mediated chelation of membrane-bound cations was sufficient to trigger H1-T6SS assembly. The addition of excess magnesium cations suppresses eDNA-mediated T6SS activation. Importantly, we demonstrate that deletion of the TagQRST signal-sensing pathway abolishes activation of the H1-T6SS. We further show that the eDNA-induced T6SS activities result in nonselective killing of neighboring bacterial species. Given the prevalence of eDNA in different environments, our results suggest that eDNA may function as a signaling molecule that contributes to the competitive fitness of P. aeruginosa.
MATERIALS AND METHODS
Strains and growth conditions.
P. aeruginosa PAO1 and its derivative ClpV1-green fluorescent protein (ClpV1-GFP), tagT, retS, and T6SS mutants, and the vasK mutant of Vibrio cholerae V52 were described previously (27, 34, 53). Burkholderia cepacia ATCC 17759 and ATCC 25416 were from laboratory stock. The V52 vasK mutant was transformed with an mCherry2-expressing pBAD24 plasmid to differentiate V52 vasK genes from P. aeruginosa GFP-labeled cells. Escherichia coli PIR1 (Invitrogen) and Sm10 λpir were used for cloning and conjugation, respectively. Chromosomal fusions of TssB1, TssB2, and TssB3 to superfolder GFP (sfGFP) were constructed using crossover PCR and homologous recombination, as previously described (31). The stop codon of tssB1, tssB2, or tssB3 was replaced by sfgfp in frame. Allelic replacement in PAO1 was performed using the pEXG2 suicide plasmid (54). All constructs were verified by sequencing.
Bacteria were routinely cultured aerobically at 37°C in LB medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, and 0.5% [wt/vol] NaCl). The following antibiotics and chemicals were used when appropriate: carbenicillin (100 μg/ml), streptomycin (100 μg/ml), irgasan (25 μg/ml), gentamicin (30 μg/ml), polymyxin B (50 μg/ml), and arabinose (0.1% [wt/vol]).
Extracellular DNA isolation.
Upon enrollment in the Calgary Adult Cystic Fibrosis Clinic (CACFC), all patients consent to the regular submission and banking of sputum and bacterial isolates from each and every encounter. Ethical approval for this study was granted (Conjoint Health Research Ethics Board [CHREB] REB15-0854 and REB15-2744).
Fresh sputum samples from 5 unrelated CF adults who visited the CACFC were isolated, resuspended separately, and thoroughly mixed in 0.5× phosphate-buffered saline (PBS) buffer (Invitrogen) (pH 7.4) to obtain a final extracellular DNA concentration of approximately 300 to 500 μg/ml per sputum. Each resuspended sputum sample was heat treated at 100°C for 20 min to remove any heat-labile components. DNA was extracted from each of the sputum samples, P. aeruginosa, or neutrophils using a Wizard Genomic DNA purification kit (Promega) following the kit instructions. DNA was eluted to a final concentration of 500 μg/ml. Salmon sperm DNA (USB, MJS BioLynx) was used at a concentration of 0.4% (wt/vol) in 0.5× PBS.
Extracellular DNA and bacterial competition assay.
P. aeruginosa (killer) and B. cepacia (prey) were grown aerobically overnight in LB. Killer cells were subcultured in fresh LB at a 1:50 dilution and grown to an optical density at 600 nm (OD600) of 0.8. Cells were centrifuged and resuspended in LB or LB with 0.4% DNA medium. Killer cells and prey cells were then mixed at a 10:1 ratio and incubated on LB or LB with 0.4% DNA plates at 37°C for 3 h and 6 h. At each time point, cells were recovered in 0.5 ml LB solution, serially diluted, and plated on LB medium containing polymyxin B (50 μg/ml) to select against P. aeruginosa and for B. cepacia survival.
Fluorescence microscopy and data analysis.
Overnight cultures were subcultured into fresh LB and cultivated aerobically at 37°C to an OD600 of 0.6. Cells were collected and concentrated 10 times by centrifugation. Cell pellets were resuspended in 50 μl of 0.5× PBS containing 0.4% (wt/vol) salmon sperm DNA or 10 ng/μl bacterial/neutrophil genomic DNA. Resuspended bacteria were spotted on a thin pad of 1% agarose–0.5× PBS and were covered with a glass coverslip. Cells were imaged at room temperature immediately to assess fluorescence foci. For the interaction between the V. cholerae vasK mutant and PAO1, round V. cholerae cells were quantified after 45 min of coincubation. A total of 36 30-by-30-μm fields of view of P. aeruginosa and V. cholerae were captured for the production of round cells and at least 24 fields of view for the quantification of ClpV-GFP foci. Data were generated through the analysis of the fields of view from three separate biological replicates.
Static images were captured by a Leica DMI 4000B inverted microscope equipped with an Orca R2 digital camera and Metamorph software for image acquisition using the 100× objective. The following excitation and emission filters were used for green fluorescence (excitation [Ex], 490-nm wavelength/20; emission [Em], 525/36). Images were formatted and analyzed using Imaris 7.0.0 imaging software. Time course image series were captured by a Nikon Ti-E inverted motorized microscope with a Perfect Focus system and Plan Apo 100× oil Ph3 DM (numerical aperture [NA], 1.4) objective lens. Spectra X light engine (Lumencore), ET-GFP (Chroma 49002), and ET-mCherry (Chroma 49008) filter sets were used to excite and filter fluorescence. A Photometrics CoolSNAP HQ2 camera (pixel size, 60 nm) and NIS Elements 4.0 were used to record images. For time course experiments, images were captured every 10 s for 5 min. The Fiji platform was used for all image analysis and manipulations (55). The image intensity and contrast were normalized across the time course by the Fiji autocorrectional macros. Image movement over the time course was corrected using StackReg transformation when necessary. The Fiji macro “Spectrum” temporal-color code was used to visualize localization of fluorescent foci over a 5-min time course. Merged phase-contrast and fluorescence images are presented.
RESULTS
Extracellular DNA in CF sputum elicits T6SS activation.
Although substrates of the normally dormant H1-T6SS can be detected and T6SS genes are upregulated in the CF sputum (27, 52), the regulatory signals are unknown. To visually confirm T6SS activation in the sputum and differentiate transcriptional and posttranscriptional effects, we used a previously constructed ClpV1-GFP strain to monitor the dynamics of H1-T6SS (27, 31). When cells were exposed to sputum samples from five adult CF patients (Fig. 1A), the activity of the H1-T6SS was induced, as measured by the formation of ClpV1-GFP foci (Fig. 1). Sputum derived from the CF lung is a complex mixture of proteoglycans, lipids, extracellular DNA (eDNA), microbial and host cells, and debris (41, 44, 49). Therefore, to determine the activating signal of T6SS, CF sputum was boiled to denature any protein components and kill any microbial or eukaryotic cells persisting in the milieu. Boiled sputum maintained its capacity to induce the formation of ClpV1-GFP foci, indicating that at least a portion of the contributing molecules responsible for T6SS activation were not heat labile (Fig. 1A). Given that total eDNA is present in considerable proportions (up to 20 mg/ml) in the CF lung sputum (42) and is heat stable, we extracted sputum-derived eDNA and challenged P. aeruginosa by the use of this purified component. Sputum-derived eDNA was capable of inducing ClpV1-GFP foci to a level comparable to that seen with the untreated and heat-treated CF sputum, suggesting that eDNA is a major component of sputum that elicits T6SS activation in P. aeruginosa (Fig. 1).
FIG 1.
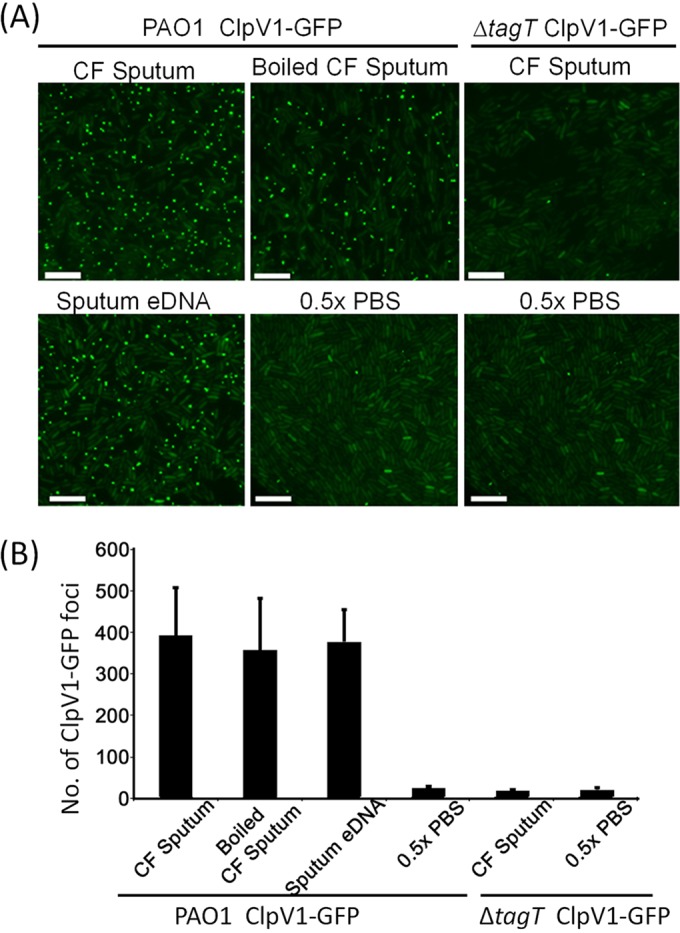
Cystic fibrosis sputum induces T6SS dynamics in P. aeruginosa. (A) T6SS activation of the P. aeruginosa wild-type strain or a ΔtagT mutant expressing ClpV1-GFP exposed to crude or heat-treated cystic fibrosis sputum or to sputum-derived eDNA. Results are representative of sputum samples derived from five adult cystic fibrosis patients. Imaging experiments were repeated independently three times per CF sputum sample, and representative images are presented (bar, 10 μm). (B) Quantification of active P. aeruginosa T6SS events as measured by the number of ClpV1-GFP foci present in the field of view (FOV). Each bar represents the mean of results from 15 fields of view with 350 to 600 P. aeruginosa cells in each field ± standard deviation (SD). Data are pooled from five independent cystic fibrosis sputum samples.
Because the H1-T6SS is known to be activated when cells sense external attacks and induction is dependent on the cell envelope-associated TagQRST signal transducing cascade (34, 38), we tested whether CF sputum-induced activation of the H1-T6SS is controlled similarly. Using the tagT mutant that cannot sense attacks (24, 34), we found that the ClpV1-GFP foci were occasionally formed in the CF sputum but that the number of foci was substantially reduced in the tagT mutant (Fig. 1).
DNA-mediated chelation of outer membrane-bound cations activates the T6SS.
We next sought to characterize the mechanism by which eDNA elicits the T6SS activation. Previous research has shown that eDNA exhibits dose-dependent killing of P. aeruginosa by acting as a cation chelator that removes membrane-stabilizing divalent cations from the bacterial cell surface (42, 47, 48). Addition of excess divalent cations, including Mg2+ and Ca2+, protects cells from eDNA-mediated lethality (48). Therefore, we postulated that removal of outer membrane-bound cations by eDNA triggers the T6SS activation. To test this, we treated cells with eDNA and excess Mg2+ divalent cations (Fig. 2). Consistent with the cation-chelating hypothesis, treatment with excess cations abolished the eDNA-induced T6SS activation in P. aeruginosa. Importantly, divalent cations did not suppress T6SS activities in the H1-T6SS hyperactive P. aeruginosa ΔretS ClpV1-GFP strain (31) (Fig. 2B). In addition, DNA that was degraded due to DNase treatment also failed to activate the H1-T6SS (Fig. 2).
FIG 2.
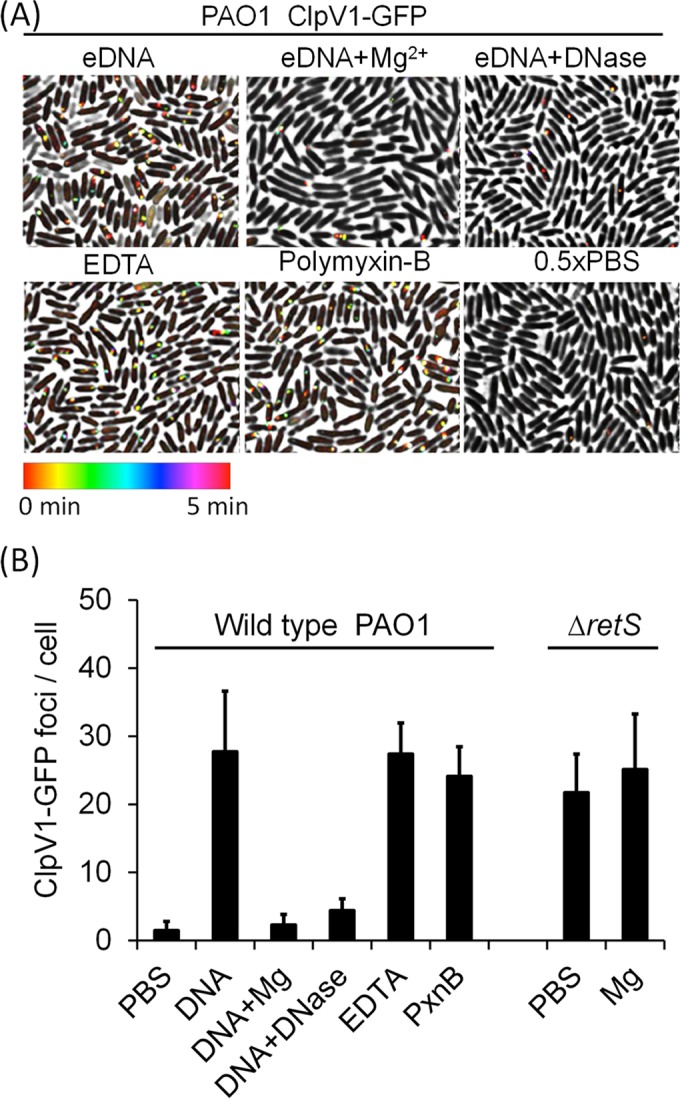
T6SS activation occurs through extracellular DNA-induced cation chelation. (A) Activation of formation of T6SS ClpV1-GFP foci in response to eDNA or to eDNA treated with excess divalent Mg2+ or DNase, EDTA, or polymyxin B (PxnB). Wild-type (WT) P. aeruginosa cells were imaged every 10 s starting immediately after they were spotted onto 0.5× PBS agarose pads. Bacteria were resuspended in 0.5× PBS containing 0.4% (wt/vol) eDNA or were resuspended in 0.5× PBS and subsequently spotted onto pads containing 0.2 mM EDTA or 2 μg/ml polymyxin B. ClpV1-GFP localization was followed every 10 s for 5 min, and the results were temporally color coded. (B) The total number of ClpV1-GFP foci was divided by the number of cells for each field of cells to determine the average number of foci per cell in a 5-min period. Each bar represents the mean of results from 10 fields with 400 to 600 cells in each field ± SD. To test whether the repression of T6SS activity by Mg2+ is independent of membrane damage, the PAO1 retS mutant, known to exhibit activated T6SS, was tested in the presence or absence of excess Mg2+. The T6SS activity in the retS mutant was not affected by Mg2+ treatment.
Because eDNA is thought to perturb the bacterial cell surfaces in a fashion analogous to that seen with the cation chelator EDTA (47, 48, 56), we then tested whether EDTA could induce the T6SS activity in P. aeruginosa (Fig. 2). Indeed, EDTA treatment resulted in activation of the H1-T6SS, within seconds, to a level comparable to that seen with eDNA (Fig. 2). Exposure to polymyxin B, a cation-independent pore-forming antibiotic, was equally capable of inducing formation of P. aeruginosa ClpV1-GFP foci, consistent with previous results (38). The rapid response upon exposure to eDNA, EDTA, and polymyxin B highlights the importance of membrane integrity in regulating the P. aeruginosa T6SS activation through posttranslational control (Fig. 2).
Activation by eDNA is specific to H1-T6SS.
P. aeruginosa possesses three T6SS clusters (H1, H2, and H3) that have been shown to be involved in targeting prokaryotic and eukaryotic organisms and to be triggered in response to various environmental cues (26, 34, 40). Therefore, we sought to determine whether the other two T6SS clusters (H2 and H3) of P. aeruginosa are activated by eDNA treatment. We individually labeled the TssB1, TssB2, and TssB3 sheath proteins with sfGFP to monitor sheath dynamics. We found that T6SS foci were occasionally formed but that exposure to eDNA failed to activate formation of TssB2-sfGFP or TssB3-sfGFP foci (Fig. 3A).
FIG 3.
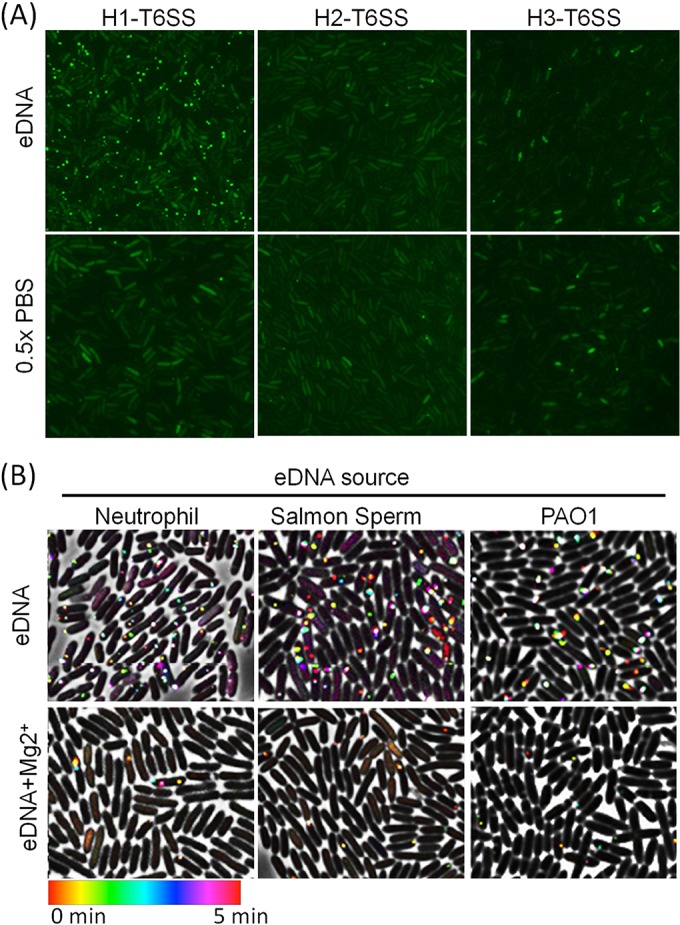
Prokaryotic and eukaryotic extracellular DNA activates H1-T6SS. (A) eDNA promotes activation of H1-T6SS but not H2 or H3 in P. aeruginosa. Mid-log-phase P. aeruginosa cells were imaged immediately following the addition of 0.4% (wt/vol) salmon sperm DNA or 0.5× PBS. For each sample or condition, 15 images were taken. Representative images are presented. (B) eDNA derived from human neutrophils, salmon sperm, or P. aeruginosa elicits activation of H1-T6SS. Mid-log-phase P. aeruginosa ClpV1-GFP was imaged every 10 s for 5 min immediately following the addition of 10 μg/ml of eDNA or eDNA supplemented with 10 mM Mg2+.
Activation of H1-T6SS by prokaryotic and eukaryotic DNA.
The CF lung is highly enriched in DNA derived primarily from neutrophils, including the presence of NETs that trap and kill bacteria (47, 49, 57). It was recently shown that P. aeruginosa lysates induce the H1-T6SS of kin cells but that the signal is unknown (51). Therefore, we sought to decipher whether DNA isolated from human neutrophil cells or P. aeruginosa is capable of inducing P. aeruginosa H1-T6SS dynamics. We also included DNA from salmon sperm to test if the activation is specific to CF-related DNA or is due to the intrinsic cation-chelating nature of DNA. Indeed, we found that eDNA, regardless of the source, activated the H1-T6SS in P. aeruginosa (Fig. 3B).
Extracellular DNA induces nonselective attack in P. aeruginosa.
The H1-T6SS is tightly regulated to stay dormant and is able to carry out precise retaliatory attacks only when P. aeruginosa is provoked by a neighboring T6SS active species (31, 34). Considering that eDNA is also present in many diverse environments, including soil and water (43), eDNA-mediated activation of H1-T6SS would allow P. aeruginosa to nonselectively attack neighboring species regardless of being or not being provoked.
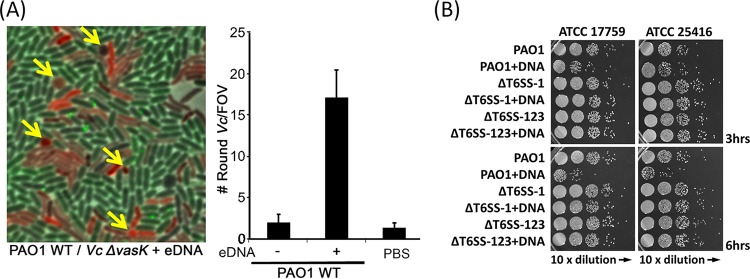
To test this prediction, we used the vasK mutant of V. cholerae, a T6SS null mutant previously demonstrated to be incapable of activating the H1-T6SS response and thus to be able to coexist with P. aeruginosa (34). We found that coincubation of P. aeruginosa, V. cholerae vasK, and eDNA led to a significant increase in V. cholerae cell rounding, suggesting that eDNA was a sufficient trigger to promote P. aeruginosa T6SS-H1 attack (Fig. 4A). Importantly, coincubation of the two bacterial species in the absence of eDNA or incubation of V. cholerae vasK with eDNA alone resulted in few round V. cholerae cells (Fig. 4A).
FIG 4.
Extracellular DNA triggers P. aeruginosa T6SS attack. (A) Coincubation of 0.4% (wt/vol) salmon sperm DNA and P. aeruginosa ClpV1-GFP leads to rounding of the V. cholerae V52 ΔvasK strain (T6SS null mutant). Arrows point to examples of round V. cholerae cells. The average number of round V. cholerae cells per field of view (FOV; error bars represent ± standard deviations) is shown for each mixture from 35 images. (B) Competition between P. aeruginosa and B. cepacia strains (ATCC 17759 and ATCC 25416). Cultures of killer and prey cells were mixed at a ratio of 10:1 on LB medium with or without 0.4% (wt/vol) DNA. Survival of B. cepacia was examined by serial plating on LB polymyxin B plates. ΔT6SS-1, the tssB1 deletion mutant; ΔT6SS-123, the tssB1 tssB2 tssB3 triple deletion mutant.
We then examined the effect of eDNA on the interaction between P. aeruginosa and B. cepacia, another frequent opportunistic pathogen that can coinhabit the CF lung (58). We tested the survival of two Burkholderia cepacia strains after coincubation on solid LB medium in the presence or absence of eDNA. Results showed that the eDNA treatment substantially reduced the survival of B. cepacia when it was mixed with wild-type PAO1 but not when it was mixed with the T6SS mutants (Fig. 4B). These data suggest that P. aeruginosa could nonselectively attack other species regardless of being or not being provoked in eDNA-abundant environments.
DISCUSSION
P. aeruginosa has been shown to adapt to the complex polymicrobial environment in the CF lung through genetic mutations and regulatory changes, but characterization of the molecular mechanism for P. aeruginosa persistence in the context of CF multispecies interaction remains elusive. Here we demonstrated that eDNA, abundant in the CF lung, promotes P. aeruginosa H1-T6SS firing through chelating surface-bound cations and causing outer membrane instability. This activation enables P. aeruginosa to nonselectively attack neighboring microbial species, thereby expanding its target range. Because eDNA is ubiquitously present in diverse environments, including biofilms and bodily fluids, eDNA-mediated H1-T6SS activation likely occurs under many other DNA-enriched conditions.
Gram-negative bacteria possessing lipopolysaccharide (LPS) on the cell outer surface are negatively charged, and the stability of the outer membrane requires the presence of divalent cations to neutralize the highly charged environment (42, 56). We found that treatment with eDNA and EDTA activates the H1-T6SS dynamics and that addition of excess magnesium cations abolishes such activation. We also confirmed previous findings indicating that treatment with the membrane-targeting antibiotic polymyxin B induces the H1-T6SS dynamics (38). Collectively, these results support the contention of the current model that outer membrane instability is a major signal perceived by P. aeruginosa, leading to orchestration of an immediate retaliatory H1-T6SS attack (24, 34).
A recent report indicated that P. aeruginosa lysates, prepared by microbial interaction or sonication, can trigger a defensive response in the remaining P. aeruginosa kin population, termed the P. aeruginosa response to antagonism (PARA), but that the signaling molecule(s) involved remains unknown (51). However, it is important to delineate that eDNA exposure results in rapid (occurring on the order of seconds to minutes) firing of the H1-T6SS whereas PARA occurs over hours (51) independently of the TagQRST pathway. While a diffusible unknown signal of kin lysis leads to a longer-term, sustained PARA response, our results obtained using eDNA and EDTA suggest that external insults promoting outer membrane instability result in an immediate retaliatory response. This is consistent with previous observations from studies using the T6SS attack and polymyxin B (34, 38).
The DNA present in the CF lung contributes to the high viscosity of CF sputum by forming a gel-like matrix with other macromolecules that affects lung function and antibiotic efficacy (48, 59). Recombinant human DNase 1 (rhDNase) has been used to treat CF patients to reduce the viscosity and increase mucus clearance (60). This treatment results in moderate to significant improvements in lung function and reductions in exacerbation frequency (61–63). Interestingly, rhDNase treatment does not affect the overall bacterial load but does causes a mild decrease in the amount of P. aeruginosa present in the CF lung (61, 64). Our findings with respect to DNA-mediated H1-T6SS activation suggest that rhDNase treatment may reduce the fitness of P. aeruginosa in competing with other microbes, thereby shifting the microbial community. A systematic evaluation of microbiota changes during rhDNase treatment may help clinicians to fully understand the variation in rhDNase treatment efficacies.
In conclusion, our study demonstrated that the H1-T6SS of P. aeruginosa can be activated by native and heat-treated CF sputum, eDNA, and EDTA. In comparison with the previously described precise retaliatory response to aggressive competing species (34), eDNA-mediated activation may represent an important nonselective strategy that promotes the competitive fitness of P. aeruginosa against passive neighboring species in environments enriched with membrane-targeting agents, including divalent-cation chelators and polymyxin B.
ACKNOWLEDGMENTS
We thank John Mekalanos for providing bacterial strains. We are grateful to the Advancing Canadian Wastewater Assets (ACWA) initiative for providing infrastructure and equipment support. We thank Alex Le and Linh Lam for providing reagents and general support.
We declare that we have no conflicts of interest.
This work was supported by a Canadian Institutes of Health Research (CIHR) operating grant and an Alberta Livestock and Meat Agency (ALMA) grant to T.G.D. and by a Cystic Fibrosis Canada grant to S.L. T.G.D. is also supported by a Government of Canada Research Chair award, a Canadian Foundation for Innovation grant (CFI-JELF), and an Alberta Innovation and Advanced Education grant. S.L. held the Westaim-Alberta Science and Research Authority (ASRA) Chair in Biofilm Research. M.W. was supported by a Cystic Fibrosis Canada Postdoctoral Fellowship. M.J.Q.W. was supported by an Ecosystem and Public Health scholarship. L.T. was supported by an Alberta Innovates Health Solutions (AIHS) postdoctoral fellowship.
REFERENCES
- 1.White R, Woodward S, Leppert M, O'Connell P, Hoff M, Herbst J, Lalouel JM, Dean M, Vande Woude G. 1985. A closely linked genetic marker for cystic fibrosis. Nature 318:382–384. doi: 10.1038/318382a0. [DOI] [PubMed] [Google Scholar]
- 2.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. 1989. Identification of the cystic fibrosis gene: genetic analysis. Science 245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 3.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL. 1989. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 4.Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N. 1989. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science 245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 5.Matsui H, Grubb BR, Tarran R, Randell SH, Gatzy JT, Davis CW, Boucher RC. 1998. Evidence for periciliary liquid layer depletion, not abnormal ion composition, in the pathogenesis of cystic fibrosis airways disease. Cell 95:1005–1015. doi: 10.1016/S0092-8674(00)81724-9. [DOI] [PubMed] [Google Scholar]
- 6.Ratjen F, Döring G. 2003. Cystic fibrosis. Lancet 361:681–689. doi: 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- 7.Chmiel JF, Davis PB. 2003. State of the art: why do the lungs of patients with cystic fibrosis become infected and why can't they clear the infection? Respir Res 4:8. doi: 10.1186/1465-9921-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramsey BW. 1996. What is the role of upper airway bacterial cultures in patients with cystic fibrosis? Pediatr Pulmonol 21:265–266. doi:. [DOI] [PubMed] [Google Scholar]
- 9.Sanders DB, Bittner RCL, Rosenfeld M, Hoffman LR, Redding GJ, Goss CH. 2010. Failure to recover to baseline pulmonary function after cystic fibrosis pulmonary exacerbation. Am J Respir Crit Care Med 182:627–632. doi: 10.1164/rccm.200909-1421OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bittar F, Richet H, Dubus J-C, Reynaud-Gaubert M, Stremler N, Sarles J, Raoult D, Rolain J-M. 2008. Molecular detection of multiple emerging pathogens in sputa from cystic fibrosis patients. PLoS One 3:e2908. doi: 10.1371/journal.pone.0002908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coenye T, Goris J, Spilker T, Vandamme P, LiPuma JJ. 2002. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J Clin Microbiol 40:2062–2069. doi: 10.1128/JCM.40.6.2062-2069.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guss AM, Roeselers G, Newton ILG, Young CR, Klepac-Ceraj V, Lory S, Cavanaugh CM. 2011. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J 5:20–29. doi: 10.1038/ismej.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, Kaess H, Deterding RR, Accurso FJ, Pace NR. 2007. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci U S A 104:20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison F. 2007. Microbial ecology of the cystic fibrosis lung. Microbiology 153:917–923. doi: 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- 15.Parkins MD, Sibley CD, Surette MG, Rabin HR. 2008. The Streptococcus milleri group—an unrecognized cause of disease in cystic fibrosis: a case series and literature review. Pediatr Pulmonol 43:490–497. doi: 10.1002/ppul.20809. [DOI] [PubMed] [Google Scholar]
- 16.Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD. 2004. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol 42:5176–5183. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. 2008. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci U S A 105:15070–15075. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahab AA, Janahi IA, Marafia MM, El-Shafie S. 2004. Microbiological identification in cystic fibrosis patients with CFTR I1234V mutation. J Trop Pediatr 50:229–233. doi: 10.1093/tropej/50.4.229. [DOI] [PubMed] [Google Scholar]
- 19.Emerson J, Rosenfeld M, McNamara S, Ramsey B, Gibson RL. 2002. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr Pulmonol 34:91–100. doi: 10.1002/ppul.10127. [DOI] [PubMed] [Google Scholar]
- 20.Henry RL, Mellis CM, Petrovic L. 1992. Mucoid Pseudomonas aeruginosa is a marker of poor survival in cystic fibrosis. Pediatr Pulmonol 12:158–161. doi: 10.1002/ppul.1950120306. [DOI] [PubMed] [Google Scholar]
- 21.Kosorok MR, Zeng L, West SE, Rock MJ, Splaingard ML, Laxova A, Green CG, Collins J, Farrell PM. 2001. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr Pulmonol 32:277–287. doi: 10.1002/ppul.2009.abs. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Kosorok MR, Farrell PM, Laxova A, West SEH, Green CG, Collins J, Rock MJ, Splaingard ML. 2005. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA 293:581–588. doi: 10.1001/jama.293.5.581. [DOI] [PubMed] [Google Scholar]
- 23.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 24.Ho BT, Dong TG, Mekalanos JJ. 2014. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RRS, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang F, Waterfield NR, Yang J, Yang G, Jin Q. 2014. A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 15:600–610. doi: 10.1016/j.chom.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 27.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basler M. 5 October 2015. Type VI secretion system: secretion by a contractile nanomachine. Philos Trans R Soc Lond B Biol Sci doi: 10.1098/rstb.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basler M, Mekalanos JJ. 2012. Type 6 secretion dynamics within and between bacterial cells. Science 337:815. doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. 2013. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. 2009. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J 28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol 9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 36.Silverman JM, Austin LS, Hsu F, Hicks KG, Hood RD, Mougous JD. 2011. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol 82:1277–1290. doi: 10.1111/j.1365-2958.2011.07889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 38.Ho BT, Basler M, Mekalanos JJ. 2013. Type 6 secretion system-mediated immunity to type 4 secretion system-mediated gene transfer. Science 342:250–253. doi: 10.1126/science.1243745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. 2013. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496:508–512. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sana TG, Hachani A, Bucior I, Soscia C, Garvis S, Termine E, Engel J, Filloux A, Bleves S. 2012. The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J Biol Chem 287:27095–27105. doi: 10.1074/jbc.M112.376368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chernick WS, Barbero GJ. 1959. Composition of tracheobronchial secretions in cystic fibrosis of the pancreas and bronchiectasis. Pediatrics 24:739–745. [PubMed] [Google Scholar]
- 42.Lewenza S. 2013. Extracellular DNA-induced antimicrobial peptide resistance mechanisms in Pseudomonas aeruginosa. Front Microbiol 4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorenz MG, Wackernagel W. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev 58:563–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews LW, Spector S, Lemm J, Potter JL. 1963. Studies on pulmonary secretions. I. The over-all chemical composition of pulmonary secretions from patients with cystic fibrosis, bronchiectasis, and laryngectomy. Am Rev Respir Dis 88:199–204. [DOI] [PubMed] [Google Scholar]
- 45.Finkel SE, Kolter R. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J Bacteriol 183:6288–6293. doi: 10.1128/JB.183.21.6288-6293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 47.Halverson TWR, Wilton M, Poon KKH, Petri B, Lewenza S. 2015. DNA is an antimicrobial component of neutrophil extracellular traps. PLoS Pathog 11:e1004593. doi: 10.1371/journal.ppat.1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mulcahy H, Charron-Mazenod L, Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lethem MI, James SL, Marriott C, Burke JF. 1990. The origin of DNA associated with mucus glycoproteins in cystic fibrosis sputum. Eur Respir J 3:19–23. [PubMed] [Google Scholar]
- 50.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol 59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 51.LeRoux M, Kirkpatrick RL, Montauti EI, Tran BQ, Peterson SB, Harding BN, Whitney JC, Russell AB, Traxler B, Goo YA, Goodlett DR, Wiggins PA, Mougous JD. 2 February 2015. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. Elife doi: 10.7554/eLife.05701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cattoir V, Narasimhan G, Skurnik D, Aschard H, Roux D, Ramphal R, Jyot J, Lory S. 2013. Transcriptional response of mucoid Pseudomonas aeruginosa to human respiratory mucus. mBio 3:e00410-12. doi: 10.1128/mBio.00410-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang L, Liang X, Moore R, Dong TG. 18 November 2015. Commentary: the icmF3 locus is involved in multiple adaptation- and virulence-related characteristics in Pseudomonas aeruginosa PAO1. Front Cell Infect Microbiol doi: 10.3389/fcimb.2015.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicas TI, Hancock RE. 1983. Alteration of susceptibility to EDTA, polymyxin B and gentamicin in Pseudomonas aeruginosa by divalent cation regulation of outer membrane protein H1. J Gen Microbiol 129:509–517. [DOI] [PubMed] [Google Scholar]
- 57.Dwyer M, Shan Q, D'Ortona S, Maurer R, Mitchell R, Olesen H, Thiel S, Huebner J, Gadjeva M. 2014. Cystic fibrosis sputum DNA has NETosis characteristics and neutrophil extracellular trap release is regulated by macrophage migration-inhibitory factor. J Innate Immun 6:765–779. doi: 10.1159/000363242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60:539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Potter JL, Matthews LW, Spector S, Lemm J. 1965. Complex formation between basic antibiotics and deoxyribonucleic acid in human pulmonary secretions. Pediatrics 36:714–720. [PubMed] [Google Scholar]
- 60.Shak S, Capon DJ, Hellmiss R, Marsters SA, Baker CL. 1990. Recombinant human DNase I reduces the viscosity of cystic fibrosis sputum. Proc Natl Acad Sci U S A 87:9188–9192. doi: 10.1073/pnas.87.23.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomson AH. 1995. Human recombinant DNase in cystic fibrosis. J R Soc Med 88(Suppl 25):24–29. [PMC free article] [PubMed] [Google Scholar]
- 62.Tyrrell JC, Lewis PA, Sheldon CD, Connett G. 1999. rhDNase in cystic fibrosis. Thorax 54:752. doi: 10.1136/thx.54.8.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zach MS. 1996. The role of recombinant human DNase in the treatment of patients with cystic fibrosis: many promises, more problems. Thorax 51:750–755. doi: 10.1136/thx.51.7.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rochat T, Pastore FD, Schlegel-Haueter SE, Filthuth I, Auckenthaler R, Belli D, Suter S. 1996. Aerosolized rhDNase in cystic fibrosis: effect on leucocyte proteases in sputum. Eur Respir J 9:2200–2206. doi: 10.1183/09031936.96.09112200. [DOI] [PubMed] [Google Scholar]