Abstract
Glutamate (Glu) and glutamine (Gln) play an important role in neuronal regulation and are of value as MRS-observable diagnostic biomarkers. In this study the relative concentrations of these metabolites have been measured in multiple regions in the normal brain using a short-TE whole-brain MRSI measurement at 3T combined with a modified data analysis approach that used spatial averaging to obtain high-SNR spectra from atlas-registered anatomic regions or interest. By spectral fitting of high-SNR spectra this approach yielded reliable measurements across a wide volume of the brain. Spectral averaging also demonstrated increased SNR and improved fitting accuracy for the sum of Glu and Gln (Glx) compared to individual voxel fitting. Results in 26 healthy controls showed relatively constant Glu/Cr and Gln/Cr throughout the cerebrum, although with increased values in the anterior cingulum and paracentral lobule, and increased Gln/Cr in the superior motor area. The deep gray-matter regions of thalamus, putamen, and pallidum show lower Glu/Cr compared to cortical white-matter regions. Lobar measurements demonstrated reduced Glu/Cr and Gln/Cr in the cerebellum as compared to the cerebrum, where white-matter regions show significantly lower Glu/Cr and Gln/Cr as compared to gray-matter regions across multiple brain lobes. Regression analysis showed no significant effect of gender on Glu/Cr and Gln/Cr measurement, however, Glx/Cr ratio was found to be significantly negatively correlated to age in some lobar brain regions. In summary, this methodology provides the spectral quality necessary for reliable separation of Glu, and Gln at 3T from a single MRSI acquisition enabling generation of regional distributions of metabolites over a large volume of the brain, including cortical regions.
Keywords: Glutamate, Glutamine, MRSI, Spectroscopy, Aging
Graphical Abstract
This study reports relative concentrations of Glu, and Gln over multiple regions in the normal brain using a single short-TE whole-brain MRSI measurement combined with a modified data analysis approach that uses atlas-based spatial averaging to obtain high-SNR spectra. The primary advantage of this methodology is that it provides the spectral quality necessary for reliable separation of Glu, and Gln at 3T from multiple brain regions using a single MRSI acquisition.
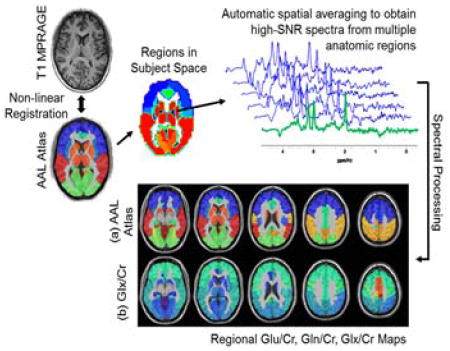
1. Introduction
Glutamate and glutamine are relatively abundant amino acids in the brain that are critical for neuronal function (1,2). The glutamate-glutamine cycle controls the supply of the neurotransmitter glutamate by synaptic release of glutamate or γ-aminobutyric acid (GABA), which is taken up by astrocytes to release glutamine that is used in the re-synthesis of glutamate or GABA. Alteration of this cyclic nature of glutamate (Glu) and glutamine (Gln) has been shown to play an important role in regulation of various neurological disorders, including epilepsy, multiple sclerosis, traumatic brain injury, schizophrenia, and brain tumors.
Both Glu and Gln can be detected in vivo using 1H Magnetic Resonance Spectroscopy (MRS); however, at the commonly available field strengths (≤3T) care must be taken to account for the strongly overlapping multiplet resonances of these two compounds, which also overlap with signals from N-Acetylaspartate (NAA) and GABA. Several techniques have been used to improve discrimination of these overlapping signal contributions, primarily based on single-voxel spectroscopy (SVS) PRESS or STEAM acquisitions (3–5), including TE-averaging (6), 2D correlation (7), and J-resolved spectroscopy (8); however, the increase in acquisition time and complexity of these methods has limited their application in clinical studies. Quantitation of Glu and Gln concentrations is also possible using conventional short TE SVS (5) provided that data are acquired with sufficient signal-to-noise and spectral quality. However, these requirements can be difficult to achieve for routine clinical measurements and therefore the analysis is frequently limited to measurement of the combined Glu+Gln (Glx) signal. Since spectroscopic imaging studies are widely implemented using higher spatial resolution (i.e. smaller voxel volumes) than SVS, studies at 3T have been limited to mapping Glx distributions (9), although individual Glu and Gln maps have been demonstrated at 7T (10). MRI based techniques using amine group-water Chemical Exchange Saturation Transfer (GluCEST) have also been reported (11).
Glu and Gln concentrations have been shown to be altered in a number of diseases and psychiatric disorders. For example, Glu/NAA is elevated in gray-matter with Rett’s syndrome (12); Glx is altered with multiple sclerosis (MS) (13); Gln is altered in normal-appearing brain tissue in subjects with hepatic encephalopathy and other liver diseases (14); glutamatergic dysregulation in schizophrenia (15) and depression and mania (16). Glu also has a role in initiation, spread, and maintenance of epileptic seizures (17) and Glx may serve as a biomarker for diagnosis of brain lesions (18,19).
For Glu and Gln to be of value as a diagnostic biomarker the normal concentration levels must be known; however, previous studies have shown that Glu and Gln levels change with age (20,21), are different in gray- and white-matter, and in brain regions such as the thalamus and cerebellum (22–24). Therefore, to optimize the sensitivity as a biomarker the subject age and brain location must also be taken into account.
Although MRS is an effective technique for non-invasive quantification of Glu and Gln the limitations of SVS methods for sampling multiple brain regions, or the lower signal-to-noise ratio (SNR) of MRSI methods, limits the potential clinical applications. The aim of this study is to measure relative concentrations of Glu, and Gln over multiple regions in the normal brain using a single whole-brain MRSI measurement at 3T combined with a modified data analysis approach. In addition, the relative sensitivity for measurement of the combined Glx value is also assessed. The method combines a short-TE MRSI acquisition with spatial averaging to obtain high-SNR spectra from multiple anatomic regions. In addition, by accounting for the tissue content in multiple voxels the results are presented for lobar white- and gray-matter measures.
2. Methods
2.1 Participants
Twenty-six healthy normal control subjects, 16 female and 10 male, from age 26 to 60 years (median 36 years) were recruited from the local community. Subjects completed a self-reporting questionnaire to indicate absence of neurological or psychological disease or injury and all MRIs were confirmed to be without any structural abnormalities via visual inspection. Informed consent was acquired from each subject and the protocol was approved by the human subjects’ research review boards at the University of Miami.
2.2 Imaging Protocol
Subjects underwent a MR study at 3T (Siemens Medical Solutions, Erlangen, Germany) that included a T1-weighted MRI and volumetric MRSI. T1-weighted imaging was carried out using a 3D Magnetization Prepared Rapid Acquisition Gradient Echo (MPRAGE) sequence with 1.0 mm isotropic or 0.9 x 0.9 x 0.7 mm3 resolution; TR/TE/TI=2300/2.41/930 ms; FA, 9°; NEX, 1; image matrix, 256 x 256 or 320 x 216. Whole-brain MRSI was acquired using echo-planar acquisition with spin-echo excitation; TR/TE = 1551/17.6 ms; non-selective lipid inversion-nulling with TI = 198 ms; a FOV of 280 x 280 x 180 mm3; matrix size of 50x50 with 18 slices; echo train length of 1000 points; bandwidth of 2500 Hz; reduced k-space sampling (acceleration factor = 0.7), and a nominal voxel volume of 0.313 cc and acquisition time of 17 min (25,26). Acquisition included a water reference measurement that was interleaved with the metabolite signal acquisition.
2.3 Spectral Processing
MRSI reconstruction and analysis was carried out using the MIDAS package (26,27). Processing included corrections for B0 shifts, generation of white-matter, gray-matter, and CSF tissue segmentation maps using FSL FAST (28), lipid k-space extrapolation (29), and linear registration between T1-weighted MR and whole-brain MRSI. Metabolite maps were interpolated to 64x64x32 points, for an interpolated voxel volume of 0.107 cc. Following spatial smoothing the effective voxel volume was 1.55 cc. Correction for frequency shifts due to B0 inhomogeneity was done using two steps. First, the spectra were frequency and phase corrected using a time-domain phase correction function obtained from the water reference MRSI. Second, the spectra were analyzed using a parametric spectral modeling procedure (30) and the final data were corrected for the phase and frequency offset values obtained from the fitting.
To improve detection of Glu and Gln spectra were averaged by summing voxels within a region of interest (ROI). These regions were defined in subject space using inverse spatial transformation of a spatial reference brain defined in MNI space (31) that was associated with a brain atlas. Two atlases were used, a lobar atlas defined for frontal, temporal, parietal, and occipital lobes and the cerebellum (9 regions) (26) and a version of the AAL atlas (32) that was modified to combine small brain regions to provide volumes suitable for the lower resolution of the MRSI measurement and which resulted in 47 regions covering cortical and subcortical gray-matter structures. Inverse spatial transformations were obtained via a two-step process. Initially, the interleaved water reference image was registered to the pre-contrast T1-weighted image using rigid registration (26,33). Secondly, the subject’s T1-weighted image was registered to the atlas spatial reference using a non-rigid registration. The combined transformation was then computed and the corresponding inverse transformation applied to obtain the atlas regions in the subject space (34) at MRSI resolution. In light of the fact that the final MRSI resolution is larger than the final interpolated voxel size, the partial volume contributions that may occur with the spatial transformation of individual atlas regions were removed by thresholding at a level of 50% volume, and the results converted to a binary mask. Prior to averaging, poor quality spectra were excluded using a linewidth of greater than 10 Hz and a voxel CSF fraction of greater than 20% as criteria to reject a voxel. Additionally, for the lobar measurements spectra were generated for gray- and white-matter using a threshold of >80% content from each respective tissue.
Following inverse spatial transformation of the atlas into subject space the integrated spectra from multiple ROIs were automatically created using functionality provided in the MIDAS package and spectral analysis carried out using the FITT program (30) for NAA, total creatine (creatine and phosphocreatine, denoted Cr), total choline (choline, glycerophosphocholine, and phosphocholine denoted Cho), myo-inositol, lactate and Glu and Gln. A separate analysis was carried out using Glx, assuming a fixed ratio of Glu:Gln of 4:1 (30). The baseline was characterized using wavelet smoothing without inclusion of macromolecular resonances, which are considered to be negligible due to the use of the lipid-inversion nulling used in the acquisition. The results of the ROI-averaged spectra were excluded from further analysis if the Glx or Glu spectral fitting showed an uncertainty as measured by Cramer-Rao lower bounds (CRLB) exceeding 20%. To account for the difficulty in spectral fitting of the lower concentration of Gln a rejection criterion for CRLB >40% was used. As a final quality control step the ROI-integrated spectra were visually inspected and any exhibiting excessive linewidth, or large lipid or water signals were removed.
Normalization of metabolite signal across subjects remains a challenging task due to variations in tissue T1, water content (M0) and bias fields, therefore, in order to account for these variations between subjects mean Glu, Gln and Glx values normalized to Cr are reported.
2.4 Data Analysis
The mean regional Glx/Cr, Glu/Cr and Gln/Cr along with the mean CRLB for 47 regions of the AAL atlas and white- and gray-matter of lobar brain regions are reported. Student’s t-test analysis was carried out to compare the mean Glx/Cr, Glu/Cr and Gln/Cr in the gray- and white-matter regions of the lobar atlas. The average Glu/Gln ratio was calculated for each brain lobe as the ratio of the mean values across all subjects and the confidence interval estimated using the method of Fieller (35).
To evaluate improvements in spectral quality from signal averaging the SNR was estimated as the ratio of the height of the NAA peak to the standard deviation of the noise signal estimated between 0 to 1.2 ppm. It is possible that this spectral region may contain signal contributions from subcutaneous lipid contamination, which would increase the measured noise value and result in reduced SNR values.
Cramer-Rao maps were also generated for Glx quantitation for each AAL region and compared with Glx mapping by spectral fitting of all individual spectra. The ratio of the mean CRLB of fitting Glx in individual spectra of a region to the CRLB of fitting Glx in the corresponding summed regional spectra is termed as the regional improvement factor (IF):
where, n are the number of regions in the AAL atlas (n =47) and CRLBGlx denotes the CRLB of fitting of Glx.
Additionally, general linear models were used to find correlations between Glx/Cr, Glu/Cr, and Gln/Cr with age, controlling for the effect of gender. If a first pass analysis revealed that gender was not a significant factor in a linear model correlation between metabolite ratios and age then a simple Pearson correlation was used. False Discovery Rate (FDR) based correction was used for accounting multiple comparisons (36). An FDR corrected p-value (q) less than 0.05 was considered significant for the correlations.
3. Results
Spectral Quality Assessment
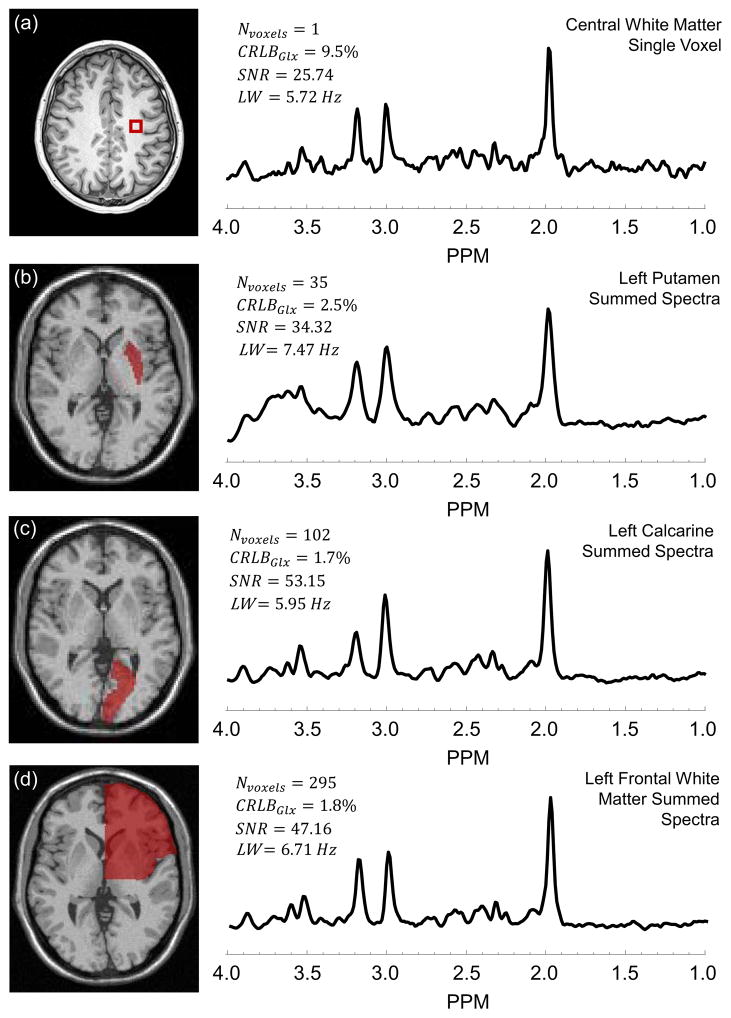
In Figure 1 are shown example spectra and their corresponding analysis results, including a single voxel spectrum in the central white-matter (a) and summed spectra for the left putamen (b), left calcarine cortex (c), and left frontal white-matter (d). It can be seen from Figure 1 that the summed spectra have a higher SNR as compared to single-voxel spectra and are fit with improved accuracy (lower CRLB) for Glx. An improvement of 53% and 77% in SNR was obtained for the integrated spectrum over the mean SNR of the individual voxels in the left putamen and left calcarine cortex regions, respectively. It can also be seen that excellent linewidth is maintained even from large tissue volumes, which is a consequence of spectral averaging being carried out following B0 inhomogeneity correction and the small nominal spatial resolution (0.313 cc), for which the linewidth in individual voxels is typically very good. Detailed fitting results for the summed spectra of the left calcarine region are shown in supplementary Figure 1.
Figure 1.
Example spectrum for a single voxel in the central white-matter (a) and summed spectra for the left putamen (b), left calcarine (c), and left frontal white-matter (d). With each spectrum is given the corresponding number of voxels summed (NVoxels), Cramer-Rao bounds of fitting of Glx (CRLBGlx), linewidth (LW), and signal-to-noise ratio (SNR). The corresponding regions are indicated on the MRI shown to the left of each spectrum.
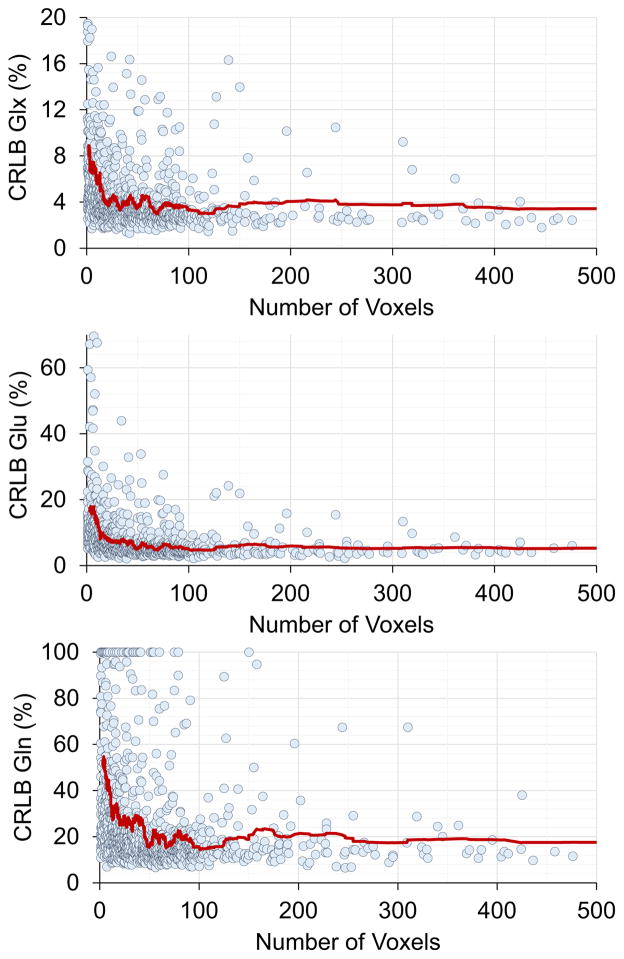
Generally, summed spectra from larger regions show improved SNR and smaller CRLB values than for smaller regions; although this will also be impacted by the number of spectra that pass the quality evaluation with each ROI. Figure 2 shows the relationship between CRLB of the spectral fitting for Glu, Gln and Glx as a function of the number of spectra summed, for all regions of the AAL atlas for all subjects. These plots indicate that a large fraction of the ROIs (96%) result in CRLBs for fitting of Glu and Glx under the acceptable threshold level of 20%, whereas for Gln 14.4% of the ROIs did not meet the CRLB threshold of 40%. While the quality of fitting is clearly improved with increasing number voxel count outliers occur and visual examination of these ROIs indicate that most of the voxels showing poor Gln CRLBs were found in the frontal regions with increased linewidths as compared to the whole brain average but otherwise with no visually-evident loss in quality.
Figure 2.
Relationship between CRLBs for spectral fitting of Glx, Glu, and Gln and number of voxels in a region. Trend lines are generated using a 50-point moving average filter to illustrate the approximate relationship between CRLB and number of voxels. Results indicate that the average CRLBs produced by the spectral averaging technique reduce as the size of the region is increased.
Lobar Measurements
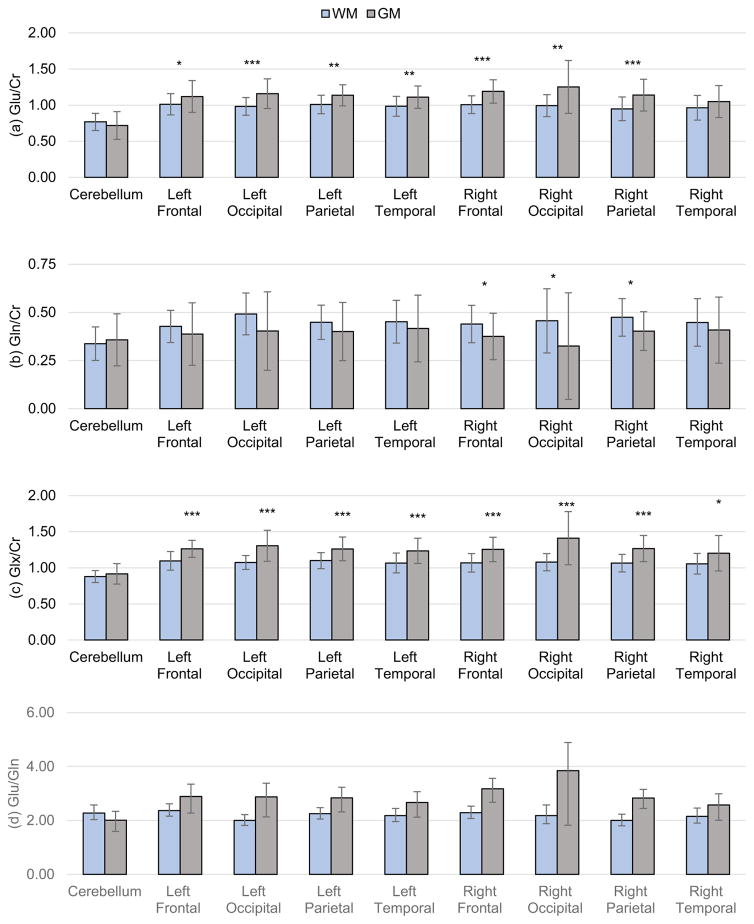
Mean Glx/Cr, Glu/Cr, Gln/Cr, and Glu/Gln ratios for white- and gray-matter over lobar volumes are shown in Figure 3 and supplementary Tables 1 and 2. Gray-matter regions show a significantly higher (p<0.05) Glx/Cr as compared to white-matter regions for all lobes of the brain except in the cerebellum. Glu/Cr in gray-matter was also significantly higher than that of white-matter in the left frontal (p = 0.044), occipital (p < 0.001), parietal (p = 0.016) and temporal (p = 0.003) lobes and in the right frontal (p < 0.001), occipital (p = 0.002) and parietal (p < 0.001) lobes. Gray-matter Gln/Cr was significantly increased relative to white-matter in the right frontal (p = 0.038), occipital (p = 0.044) and temporal (p = 0.013) lobes. There were no significant differences between left and right for any measured value.
Figure 3.
Mean lobar Glu/Cr (a), Gln/Cr (b), and Glx/Cr (c) measurements for gray- and white-matter regions with error bars denoting standard deviation in measurements. Significance of difference between gray- and white-matter ratios are shown by asterisks (* p<0.05, ** p < 0.01, *** p<0.001). (d) Glu/Gln ratio for gray- and white-matter lobar brain regions with error bars denoting confidence interval (CI) calculated using the Fieller method.
The Glu/Gln ratio in each lobar region is shown in Figure 3d. These again reflect the differences of the Glu/Cr and Gln/Cr ratios in gray- and white-matter but without significant differences between regions, for mean values of 2.1 and 3.0 in cerebral white- and gray-matter respectively.
AAL Atlas Measurements
Manual quality assessment of the ROI-averaged spectra using the modified AAL atlas resulted in exclusion of 3.1% of the datasets, which were primarily located in the superior motor area, anterior cingulum and paracentral lobule. Of these, exclusion was based on excessive lipid contamination. The number of voxels selected for each region after applying the voxel exclusion criteria, averaged across all subject, ranged from 3.3 to 387.6, with the least number of voxels in the pallidum and the maximum in the cerebellum. Out of the total of 1123 regional average spectra across the 26 subjects, 95 (8.45%) had less than 5 voxels averaged. These regions were primarily located in caudate, pallidum, paracentral, and superior motor area. This number is governed not only by the size of the region but also the exclusion criteria mentioned in section 2.3 for maintaining the quality of the averaged spectra.
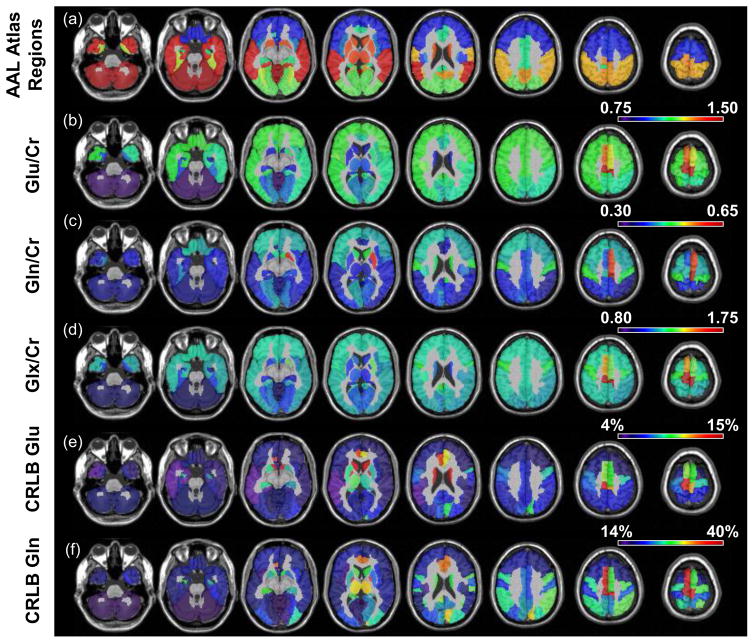
In Figure 4 and supplemental Table 3 are shown results for the regional analysis using the AAL atlas, with Figure 4a showing the brain parcellation regions. For Figures 4b–4d the brain regions have been color coded according to the mean regional values determined for that region, for Glu/Cr, Gln/Cr and Glx/Cr, respectively. These results indicate that Glu/Cr and Gln/Cr are fairly constant throughout the cerebrum, although with increased values in the anterior cingulum and paracentral lobule, and increased Gln in the superior motor area. The deep gray-matter regions of the thalamus, putamen, and pallidum show lower Glu/Cr compared to cortical white-matter regions. The lowest values are observed in the cerebellum, which is due to the increased Cr relative to the cerebrum (27).
Figure 4.
Maps showing the AAL anatomic regions (a), and mean regional values for Glu/Cr (b), Gln/Cr (c), Glx/Cr (d), Glu Cramer-Rao (e) and Gln Cramer-Rao (f), for each of the atlas regions.
In Figures 4e and 4f are shown mean regional CRLB values for Glu and Gln, averaged over all subjects, indicating the quality of the spectral fitting. It is seen that Glu fitting is well below 20% but in some locations the Gln result showed CRLB of higher than 40%. In contrast, the CRLB values for fitting of Glu and Gln in individual voxel spectra were both considerably greater than the thresholds of 20% and 40%, respectively. Performance for fitting of individual voxel spectra was improved when using Glx, for which the mean CRLB, averaged over the whole brain, was found to be 16.5 ± 3%, with the worst performance seen in the caudate (mean CRLB of 22.5% for left and 21.1% for right) and superior motor areas (mean CRLB of 26.5% for left and 23.0% for right). When using the ROI-averaged spectra for fitting of Glx the mean CRLB, averaged over all regions, was 4.5 ± 1.3%, which corresponded to a mean improvement factor of 3.7 ± 0.7 relative to quantification of Glx in individual voxels. Results for regional improvement factors are presented in supplementary Table 4. Supplementary Table 4 also shows the mean Glx/Cr values obtained by fitting the ROI-averaged spectra as compared to the mean of fitting individual spectra in a given ROI. On average, fitting individual spectra resulted in a 23.0 ± 5.0% lower estimation of Glx/Cr.
Age Correlations
Regression analysis revealed that gender was not a significant factor in any linear model. Simple correlation analysis showed statistically significant correlations between Glx/Cr ratios and age in the left parietal (r = −0.564, q = 0.021) and right temporal (r = −0.494, q = 0.010) white-matter regions, and gray-matter regions in the left (r = −0.523, q = 0.030) and right (r = −0.502, q = 0.039) parietal lobes, and in the right occipital lobe including the lingual gyrus (r = −0.554, q = 0.026). Glu/Cr was found to be inversely correlated to age only in the right posterior cingulum cortex (r = −0.544, q = 0.024), whereas Gln/Cr was found to be negatively associated with age for the left temporal lobe (r = −0.541, q = 0.023). These findings likely reflect the moderate increase of Cr with age (37).
4. Discussion
This report has presented a technique for estimation of Glu and Gln in multiple anatomically-defined brain regions from a volumetric MRSI measurement at 3T. The strength of the technique lies in the use of anatomically-driven spatial averaging of multivoxel spectra to enable regional metabolite measures to be obtained with increased accuracy. When combined with a short-TE spin-echo acquisition and signal averaging over the 17-min MRSI acquisition, this methodology provides the spectral quality necessary for reliable separation of Glu, and Gln at 3T, while also providing information from multiple brain regions, including cortical regions.
Previous studies that have reported regional measures for Glu and Gln have relied on single voxel methods and few studies have used MRSI based methods. A summary of studies that have reported Glu/Cr and Gln/Cr in different brain regions is presented in Table 1. Mean Glu/Cr ratios have been reported in the range of 0.96 – 1.52 and 0.72 – 1.16 for pure gray-matter and white-matter respectively (3,6,38). The present study showed a mean cerebral Glu/Cr of 0.99±0.02 and 1.14±0.06 in the white-matter and gray-matter regions, which are consistent with other studies. There is good agreement with this study in the measurement of Glu/Cr in the precentral gyrus; however, this study reports lower Glu/Cr ratio in the occipital region (1.09 ± 0.12) compared to the studies by Frias-Martinez et al. (39,40). However, a large variation in Glu/Cr is seen across studies in the parietal and occipital regions with studies reporting ratios as low as 0.9 (8) to 1.48 (39). Glu/Cr measures in the anterior cingulate showed good concordance with reported values but slightly lower values in the posterior cingulate. However, it should be noted that measurements of Glu/Cr in the anterior cingulate region show a large range from 1.12 – 1.31 (5,41). The mean Gln/Cr in the parieto-occipital cortex region was 0.38, with CRLB under 20%, which was slightly higher than those found by Jensen et al. (8). Studies by Frias-Martinez et al. have shown similar ratio for Gln/Cr in the occipital region using JPRESS and L-COSY techniques (39,40).
Table 1.
Comparison of Regional Glu/Cr and Gln/Cr in Different Studies.
| Study | Method | Region | Glu/Cra | Gln/Cra |
|---|---|---|---|---|
| Gonenc et al. (56) | Multi-TE J-resolved | Precentral Gyrus | 1.11 ± 0.11 | 0.43 ± 0.08 |
| Jensen et al. (8) | JPRESS | Parieto-Occipital | 0.9 ± 6% | 0.25 ± 14% |
| PRESS | 1.24 ± 8% | 0.17 ± 30% | ||
| Frias-Martinez et al. (40) | JPRESS ProFit | Occipital Region | 1.24 ± 0.12 | 0.42 ± 0.06 |
| Frias-Martinez et al. (39) | L-COSY Profit | Occipital Region | 1.48 ± 0.31 | 0.43 ± 0.01 |
| Schulte et al.* (57) | J-resolved Profit | Parietal Lobe | 1.28 | 0.21 |
| Waddell et al. (58) | Difference Spectroscopy | Anterior Cingulate | 1.16 ± 0.10 | NA |
| Wijtenburg et al. (5) | Phase Rotation STEAM | Anterior Cingulate | 1.12 ± 0.08 | 0.33 ± 0.08 |
| Wijtenburg et al.* (41) | Phase Rotation STEAM | Anterior Cingulate | 1.31 | 0.30 |
| Posterior Cingulate | 1.32 | 0.31 | ||
| Present Study$ | Short-TE MRSI | Precentral Gyrus | 1.14 ± 0.27 | 0.43 ± 0.11 |
| Parietal Cortex | 1.10 ± 0.17 | 0.37 ± 0.12 | ||
| Occipital Cortex | 1.09 ± 0.12 | 0.38 ± 0.12 | ||
| Anterior Cingulate | 1.06 ± 0.30 | 0.36 ± 0.19 | ||
| Posterior Cingulate | 1.14 ± 0.18 | 0.42 ± 0.16 |
Ratios are reported as mean ± standard deviation.
Standard deviation of measure not available.
Values are averaged across the hemispheres.
For the regional spectra that were included in the final analysis of this study the CRLB values were in the range of 4% to 15% for Glu, and 14% to 40% (mean 23±11%) for Gln, which are comparable to those provided by short-TE SVS measurements at 3T (3). Although the use of integrated volumes from a MRSI measurement entails decreased detection sensitivity relative to a SVS measurement for the equivalent tissue volume (42), the primary advantage of the MRSI approach is to sample multiple regions in a single measurement. The ROI-averaged MRSI approach additionally enables sampling of complex tissue volumes, automated ROI selection using image registration methods, separation of mean white- and gray-matter values, and removes the requirement of a predefined placement of sampled volumes. The variant of the MRSI measurement implemented for this study adds additional sensitivity losses from the use of spatial oversampling (43) and lipid inversion-nulling (44); however, these methods represent trade-offs that benefit spectral quality by decreasing linewidth, nulling macromolecular resonances, and enable sampling of the cortical surface regions. The advantage of signal integration after individual-voxel spectra have been corrected for B0 inhomogeneity has been shown to result in decreased linewidth relative to a SVS measurement (45). While the acquisition time for the MRSI measurement is longer than that needed for a SVS measurement it becomes comparable to that required for two SVS measurements that include a water reference measurement and shimming at each location. Additional considerations include the increased susceptibility to subject motion and the processing requirements of the volumetric MRSI measurement. In summary, this study indicates that despite the sensitivity losses associated with the MRSI measurement the proposed method can provide comparable reliability to SVS measurements while offering a much larger brain coverage. This approach therefore enables widespread metabolic changes to be studied, where SVS measurements would be limited due to its localized sampling, with improved quality relative to that obtained from individual MRSI voxels.
Many clinical MRS studies report the value of Glx in light of the known difficulty for separating the individual Glu and Gln contributions, and the increased reliability of the Glx estimation is demonstrated in this study by the lower CRLB of spectral fitting. These results support the use of Glx for studies of brain disorders where separate Gln estimation is not required; however, although this result will be dominated by the larger proportion of Glu it will nevertheless be subject to quantitation errors in light of the varying ratio of Glu/Gln, which ranged from 2.0 in left-occipital white-matter to 3.9 in right occipital gray-matter.
Previous studies have demonstrated the use of spectral averaging of in vivo MRSI data prior to spectral fitting for improving the quality of analysis. Zhu et al. first demonstrated the increase in spectral SNR by coherently summing spectra in a ROI following phase and frequency correction (46). Corrigan et al. showed that averaging data within a ROI before spectral fitting significantly improved the measurement of lactate and that this provided an advantage over analysis of low SNR single-voxel spectra, which can lead to incorrect estimations of signal amplitude and CRLB values (47). Mandl et al. used a similar approach to create a single spectra for the entire cingulum bundle by averaging MRSI voxels within a volume that was identified using fiber tracking (48). Ratiney et al. used spatial averaging over small regions to improve SNR for metabolite T1 measurement (49). These previous studies were implemented for 2D MRSI data only, and the current report describes the first implementation using volumetric MRSI data that has been combined with a brain atlas.
A limitation of this study is the absence of absolute concentration estimates for Glu and Gln (or their combination). This requires measurement of a known reference signal, which was not available for this study, and ideally, knowledge of relaxation rates and receive and transmit bias fields, which can be problematic to obtain for MRSI studies. However, previous MRSI studies have reported absolute concentrations of Glu by using assumed values for tissue water content and relaxation rates and taking a ratio to a separate MRSI acquisition of the full tissue water signal, and results have been presented for a single axial slice (9,38,50). However, limiting the analysis to report metabolite ratios has the advantage of simpler acquisition and processing requirements, which can be more conveniently implemented for clinical studies. Additionally, since the reference signal is acquired simultaneously with the metabolite signal, systemic errors from sources such as partial volume from cerebrospinal fluid and variable flip angles are removed (51). Moreover, it has been shown that metabolite ratios across subjects are more robust than other spectral quantitation techniques with less than 10% variation with a single-site and 15% across sites (52). On the other hand, metabolite ratios are ambiguous in nature, making it impossible to determine if an abnormality is due to changes in the numerator or denominator metabolite. Although Cr has been generally adopted as the reference metabolite there are regional variations (e.g. reduced Cr in cerebellum) (27) and it is not uncommon to find altered concentrations in pathological conditions (53–55).
An additional limitation of the proposed atlas-based spectral averaging technique is that metabolite concentrations across the selected region, and optionally for each tissue type within the volume, are assumed to be similar; however, this assumption may not hold in the presence of pathologic changes such as tumor infiltration and necrosis. Another potential limitation of this study is the absence of GABA and glutathione from the spectral fitting model, which also have overlapping peaks with Glu and Gln. However, the concentrations of these compounds have been reported to be less than one-fifth that of Glu and less than two-thirds that of Gln (5) and therefore the potential impact is expected to be small.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by National Institute of Health (NIH) grants R01CA172210, R01EB016064 and R21CA170284.
Non-standard Abbreviations List
- SVS
Single Voxel Spectroscopy
- GABA
γ-aminobutyric acid
- AAL
Automated Anatomical Labeling
- CRLB
Cramer-Rao lower bounds
References
- 1.Erecinska M, Silver IA. Metabolism and role of glutamate in mammalian brain. Prog Neurobiol. 1990;35(4):245–296. doi: 10.1016/0301-0082(90)90013-7. [DOI] [PubMed] [Google Scholar]
- 2.Miladinovic T, Nashed MG, Singh G. Overview of glutamatergic dysregulation in central pathologies. Biomolecules. 2015;5(4):3112–3141. doi: 10.3390/biom5043112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramadan S, Lin A, Stanwell P. Glutamate and glutamine: A review of in vivo MRS in the human brain. NMR Biomed. 2013;26(12):1630–1646. doi: 10.1002/nbm.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snyder J, Wilman A. Field strength dependence of PRESS timings for simultaneous detection of glutamate and glutamine from 1.5 to 7T. J Magn Reson. 2010;203(1):66–72. doi: 10.1016/j.jmr.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Wijtenburg SA, Knight-Scott J. Very Short Echo Time Improves the Precision of Glutamate Detection at 3T in H-1 Magnetic Resonance Spectroscopy. J Magn Reson Imaging. 2011;34(3):645–652. doi: 10.1002/jmri.22638. [DOI] [PubMed] [Google Scholar]
- 6.Hurd R, Sailasuta N, Srinivasan R, Vigneron DB, Pelletier D, Nelson SJ. Measurement of brain glutamate using TE-averaged PRESS at 3T. Magn Reson Med. 2004;51(3):435–440. doi: 10.1002/mrm.20007. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe H, Takaya N, Mitsumori F. Simultaneous observation of glutamate, gamma-aminobutyric acid, and glutamine in human brain at 4.7 T using localized two-dimensional constant-time correlation spectroscopy. NMR Biomed. 2008;21(5):518–526. doi: 10.1002/nbm.1223. [DOI] [PubMed] [Google Scholar]
- 8.Jensen JE, Licata SC, Ongur D, Friedman SD, Prescot AP, Henry ME, Renshaw PF. Quantification of J-resolved proton spectra in two-dimensions with LCModel using GAMMA-simulated basis sets at 4 Tesla. NMR Biomed. 2009;22(7):762–769. doi: 10.1002/nbm.1390. [DOI] [PubMed] [Google Scholar]
- 9.Posse S, Otazo R, Caprihan A, Bustillo J, Chen H, Henry PG, Marjanska M, Gasparovic C, Zuo C, Magnotta V, Mueller B, Mullins P, Renshaw P, Ugurbil K, Lim KO, Alger JR. Proton echo-planar spectroscopic imaging of J-coupled resonances in human brain at 3 and 4 Tesla. Magn Reson Med. 2007;58(2):236–244. doi: 10.1002/mrm.21287. [DOI] [PubMed] [Google Scholar]
- 10.Henning A, Fuchs A, Murdoch JB, Boesiger P. Slice-selective FID acquisition, localized by outer volume suppression (FIDLOVS) for (1)H-MRSI of the human brain at 7 T with minimal signal loss. NMR Biomed. 2009;22(7):683–696. doi: 10.1002/nbm.1366. [DOI] [PubMed] [Google Scholar]
- 11.Cai K, Haris M, Singh A, Kogan F, Greenberg JH, Hariharan H, Detre JA, Reddy R. Magnetic resonance imaging of glutamate. Nat Med. 2012;18(2):302–306. doi: 10.1038/nm.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan JW, Lane JB, Hetherington H, Percy AK. Rett syndrome: 1H spectroscopic imaging at 4.1 Tesla. J Child Neurol. 1999;14(8):524–528. doi: 10.1177/088307389901400808. [DOI] [PubMed] [Google Scholar]
- 13.Cianfoni A, Niku S, Imbesi SG. Metabolite findings in tumefactive demyelinating lesions utilizing short echo time proton magnetic resonance spectroscopy. AJNR Am J Neuroradiol. 2007;28(2):272–277. [PMC free article] [PubMed] [Google Scholar]
- 14.Kreis R, Ross BD, Farrow NA, Ackerman Z. Metabolic disorders of the brain in chronic hepatic encephalopathy detected with H-1 MR spectroscopy. Radiology. 1992;182:19–27. doi: 10.1148/radiology.182.1.1345760. [DOI] [PubMed] [Google Scholar]
- 15.Theberge J, Al-Semaan Y, Williamson PC, Menon RS, Neufeld RWJ, Rajakumar N, Schaefer B, Densmore M, Drost DJ. Glutamate and glutamine in the anterior cingulate and thalamus of medicated patients with chronic schizophrenia and healthy comparison subjects measured with 4.0-T proton MRS. Am J Psychiat. 2003;160(12):2231–2233. doi: 10.1176/appi.ajp.160.12.2231. [DOI] [PubMed] [Google Scholar]
- 16.Yuksel C, Ongur D. Magnetic resonance spectroscopy studies of glutamate-related abnormalities in mood disorders. Biol Psychiatry. 2010;68(9):785–794. doi: 10.1016/j.biopsych.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petroff OA, Errante LD, Rothman DL, Kim JH, Spencer DD. Glutamate-glutamine cycling in the epileptic human hippocampus. Epilepsia. 2002;43(7):703–710. doi: 10.1046/j.1528-1157.2002.38901.x. [DOI] [PubMed] [Google Scholar]
- 18.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123(9):3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollingworth W, Medina LS, Lenkinski RE, Shibata DK, Bernal B, Zurakowski D, Comstock B, Jarvik JG. A systematic literature review of magnetic resonance spectroscopy for the characterization of brain tumors. AJNR Am J Neuroradiol. 2006;27(7):1404–1411. [PMC free article] [PubMed] [Google Scholar]
- 20.Pouwels PJW, Brockmann K, Kruse B, Wilken B, Wick M, Hanefeld F, Frahm J. Regional age dependence of human brain metabolites from infancy to adulthood as detected by quantitative localized proton MRS. Pediatr Res. 1999;46(4):474–485. doi: 10.1203/00006450-199910000-00019. [DOI] [PubMed] [Google Scholar]
- 21.Raininko R, Mattsson P. Metabolite concentrations in supraventricular white matter from teenage to early old age: A short echo time 1H magnetic resonance spectroscopy (MRS) study. Acta Radiol. 2010;51(3):309–315. doi: 10.3109/02841850903476564. [DOI] [PubMed] [Google Scholar]
- 22.Haga KK, Khor YP, Farrall A, Wardlaw JM. A systematic review of brain metabolite changes, measured with 1H magnetic resonance spectroscopy, in healthy aging. Neurobiol Aging. 2009;30(3):353–363. doi: 10.1016/j.neurobiolaging.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Kaiser LG, Schuff N, Cashdollar N, Weiner MW. Age-related glutamate and glutamine concentration changes in normal human brain: 1H MR spectroscopy study at 4 T. Neurobiol Aging. 2005;26(5):665–672. doi: 10.1016/j.neurobiolaging.2004.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segovia G, Porras A, Del Arco A, Mora F. Glutamatergic neurotransmission in aging: A critical perspective. Mech Ageing Dev. 2001;122(1):1–29. doi: 10.1016/s0047-6374(00)00225-6. [DOI] [PubMed] [Google Scholar]
- 25.Sabati M, Sheriff S, Gu M, Wei J, Zhu H, Barker PB, Spielman DM, Alger JR, Maudsley AA. Multivendor implementation and comparison of volumetric whole-brain echo-planar MR spectroscopic imaging. Magn Reson Med. 2015;74(5):1209–1220. doi: 10.1002/mrm.25510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maudsley AA, Darkazanli A, Alger JR, Hall LO, Schuff N, Studholme C, Yu Y, Ebel A, Frew A, Goldgof D, Gu Y, Pagare R, Rousseau F, Sivasankaran K, Soher BJ, Weber P, Young K, Zhu X. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19(4):492–503. doi: 10.1002/nbm.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maudsley AA, Domenig C, Govind V, Darkazanli A, Studholme C, Arheart K, Bloomer C. Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI) Magn Reson Med. 2009;61(3):548–559. doi: 10.1002/mrm.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imag. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 29.Haupt CI, Schuff N, Weiner MW, Maudsley AA. Removal of lipid artifacts in 1H spectroscopic imaging by data extrapolation. Magn Reson Med. 1996;35(5):678–687. doi: 10.1002/mrm.1910350509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soher BJ, Young K, Govindaraju V, Maudsley AA. Automated spectral analysis III: Application to in vivo proton MR spectroscopy and spectroscopic imaging. Magn Reson Med. 1998;40(6):822–831. doi: 10.1002/mrm.1910400607. [DOI] [PubMed] [Google Scholar]
- 31.Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM) Neuroimage. 1995;2(2):89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- 32.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 33.Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recogn. 1999;32(1):71–86. [Google Scholar]
- 34.Studholme C, Hill DL, Hawkes DJ. Automated three-dimensional registration of magnetic resonance and positron emission tomography brain images by multiresolution optimization of voxel similarity measures. Med Phys. 1997;24(1):25–35. doi: 10.1118/1.598130. [DOI] [PubMed] [Google Scholar]
- 35.Fieller EC. Some problems in interval estimation. J Roy Stat Soc B. 1954;16(2):175–185. [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - A practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
- 37.Maudsley AA, Govind V, Arheart KL. Associations of age, gender and body mass with 1H MR-observed brain metabolites and tissue distributions. NMR Biomed. 2012;25(4):580–593. doi: 10.1002/nbm.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai SY, Otazo R, Posse S, Lin YR, Chung HW, Wald LL, Wiggins GC, Lin FH. Accelerated proton echo planar spectroscopic imaging (PEPSI) using GRAPPA with a 32-channel phased-array coil. Magn Reson Med. 2008;59(5):989–998. doi: 10.1002/mrm.21545. [DOI] [PubMed] [Google Scholar]
- 39.Frias-Martinez E, Rajakumar N, Liu X, Singhal A, Banakar S, Lipnick S, Verma G, Ramadan S, Kumar A, Thomas MA. ProFit-based quantitation of cerebral metabolites using 2D L-COSY at 3T. proceedings of the International Society of Magnetic Resonance in Medicine; Toronto, Canada. 2008. p. 691. [Google Scholar]
- 40.Frias-Martinez E, Rajakumar N, Ramadan S, Banakar S, Liu X, Singhal A, Thomas MA. A pilot comparison of 2D and 1D MR spectroscopic quantitation of metabolites in healthy human brain at 3T. proceedings of the International Society of Magnetic Resonance in Medicine; Toronto, Canada. 2008. p. 1603. [Google Scholar]
- 41.Wijtenburg SA, Gaston FE, Spieker EA, Korenic SA, Kochunov P, Hong LE, Rowland LM. Reproducibility of phase rotation STEAM at 3T: focus on glutathione. Magn Reson Med. 2014;72(3):603–609. doi: 10.1002/mrm.24959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edelstein WA, Glover GH, Hardy CJ, Redington RW. The intrinsic signal-to-noise ratio in NMR imaging. Magn Reson Med. 1986;3:604–618. doi: 10.1002/mrm.1910030413. [DOI] [PubMed] [Google Scholar]
- 43.Ebel A, Maudsley AA. Improved spectral quality for 3D MR spectroscopic imaging using a high spatial resolution acquisition strategy. Magn Reson Imaging. 2003;21(2):113–120. doi: 10.1016/s0730-725x(02)00645-8. [DOI] [PubMed] [Google Scholar]
- 44.Ebel A, Govindaraju V, Maudsley AA. Comparison of inversion recovery preparation schemes for lipid suppression in 1H MRSI of human brain. Magn Reson Med. 2003;49(5):903–908. doi: 10.1002/mrm.10444. [DOI] [PubMed] [Google Scholar]
- 45.Ding XQ, Maudsley A, Sabati M, Sheriff S, Lanfermann H. Comparison of a short-TE whole brain MR spectroscopic imaging to single voxel spectroscopy for measurement of metabolite concentrations in human brain. proceedings of the International Society of Magnetic Resonance in Medicine; Milan, Italy. 2014. p. 2901. [Google Scholar]
- 46.Zhu XP, Young K, Ebel A, Soher BJ, Kaiser L, Matson G, Weiner WM, Schuff N. Robust analysis of short echo time (1)H MRSI of human brain. Magn Reson Med. 2006;55(3):706–711. doi: 10.1002/mrm.20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corrigan NM, Richards TL, Friedman SD, Petropoulos H, Dager SR. Improving 1H MRSI measurement of cerebral lactate for clinical applications. Psychiat Res. 2010;182(1):40–47. doi: 10.1016/j.pscychresns.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandl RC, van den Heuvel MP, Klomp DW, Boer VO, Siero JC, Luijten PR, Hulshoff Pol HE. Tract-based magnetic resonance spectroscopy of the cingulum bundles at 7 T. Hum Brain Mapp. 2012;33(7):1503–1511. doi: 10.1002/hbm.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ratiney H, Noworolski SM, Sdika M, Srinivasan R, Henry RG, Nelson SJ, Pelletier D. Estimation of metabolite T1 relaxation times using tissue specific analysis, signal averaging and bootstrapping from magnetic resonance spectroscopic imaging data. Magn Reson Mater Phy. 2007;20(3):143–155. doi: 10.1007/s10334-007-0076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srinivasan R, Cunningham C, Chen A, Vigneron D, Hurd R, Nelson S, Pelletier D. TE-averaged two-dimensional proton spectroscopic imaging of glutamate at 3 T. Neuroimage. 2006;30(4):1171–1178. doi: 10.1016/j.neuroimage.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 51.Barker PB, Bizzi A, De Stefano N, Gullapalli R, Lin DDM. Clinical MR spectroscopy: Techniques and applications. Cambridge: Cambridge University Press; 2009. [Google Scholar]
- 52.Webb PG, Sailasuta N, Kohler SJ, Raidy T, Moats RA, Hurd RE. Automated single-voxel proton MRS: Technical development and multisite verification. Magn Reson Med. 1994;31(4):365–373. doi: 10.1002/mrm.1910310404. [DOI] [PubMed] [Google Scholar]
- 53.Inglese M, Li BS, Rusinek H, Babb JS, Grossman RI, Gonen O. Diffusely elevated cerebral choline and creatine in relapsing-remitting multiple sclerosis. Magn Reson Med. 2003;50(1):190–195. doi: 10.1002/mrm.10481. [DOI] [PubMed] [Google Scholar]
- 54.Chang L. In vivo magnetic resonance spectroscopy in HIV and HIV-related brain diseases. Reviews in the Neurosciences. 1995;6:365–378. doi: 10.1515/revneuro.1995.6.4.365. [DOI] [PubMed] [Google Scholar]
- 55.Saunders DE. MR spectroscopy in stroke. Br Med Bull. 2000;56(2):334–345. doi: 10.1258/0007142001903256. [DOI] [PubMed] [Google Scholar]
- 56.Gonenc A, Govind V, Sheriff S, Maudsley AA. Comparison of spectral fitting methods for overlapping J-coupled metabolite resonances. Magn Reson Med. 2010;64(3):623–628. doi: 10.1002/mrm.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schulte RF, Boesiger P. ProFit: two-dimensional prior-knowledge fitting of J-resolved spectra. NMR Biomed. 2006;19(2):255–263. doi: 10.1002/nbm.1026. [DOI] [PubMed] [Google Scholar]
- 58.Waddell KW, Zanjanipour P, Pradhan S, Xu L, Welch EB, Joers JM, Martin PR, Avison MJ, Gore JC. Anterior cingulate and cerebellar GABA and Glu correlations measured by (1)H J-difference spectroscopy. Magn Reson Imaging. 2011;29(1):19–24. doi: 10.1016/j.mri.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.