Abstract
Purpose
In patients with neuroblastoma (NB), treatment with anti-GD2 monoclonal antibody (mAb) directs natural killer (NK) cell–mediated antibody-dependent cellular cytotoxicity (ADCC) against tumor cells. However, tumor cytotoxicity is attenuated by ligation of inhibitory killer immunoglobulin-like receptors (KIRs) by HLA class I molecules. KIR3DL1 polymorphism influences its ability to engage HLA-Bw4 ligands. We tested the hypothesis that poorly interacting combinations of KIR3DL1 and HLA ligands are more permissive of mAb-mediated antitumor effect.
Methods
KIR3DL1 and HLA-B subtyping were performed with a multiplex intermediate-resolution polymerase chain reaction assay for a cohort of 245 patients who were treated with antibody 3F8 for high-risk NB. Patient outcomes were analyzed according to expected degree of interaction between KIR3DL1 and HLA-B subtypes and grouped as strong, weak, or noninteractors. A comparison of NK response to 3F8 mAb opsonized NB cells between strong- and noninteracting donors was performed by flow cytometry.
Results
KIR3DL1 and HLA-B subtype combinations associated with noninteraction as a result of lack of receptor expression [KIR3DL1(−)], failure of interaction with inhibitory ligands [KIR3DS1(+)], or absence of KIR ligands resulted in significantly improved overall and progression-free survival. Patients with KIR3DL1 and HLA-B subtype combinations that were predictive of weak interaction had superior outcomes compared with those that were predictive of strong interaction; however, both groups were inferior to those with noninteracting subtype combinations. In vitro analysis of 3F8-mediated ADCC showed that KIR3DL1(−) and 3DS1(+) NK cells were insensitive to inhibition by HLA-Bw4–expressing NB targets.
Conclusion
We conclude that KIR3LD1 and HLA-B allele combinations can have a prognostic impact on patient survival after treatment with anti-GD2 mAb that relies on NK-ADCC. The survival advantage seen in noninteracting combinations supports the therapeutic disinhibition of individuals with strongly interacting KIR and ligand pairs.
INTRODUCTION
Of patients with neuroblastoma (NB), > 50% have high-risk disease at diagnosis, and despite intense multimodal therapy, long-term survival is poor.1-6 Monoclonal antibody (mAb) therapies directed at disialoganglioside GD2 have been a major advancement in the treatment of NB.7-10 Natural killer cells (NK cells), which mediate ADCC (antibody-dependent cellular cytotoxicity), are vital to the efficacy of anti-GD2 antibody therapy.11-15
NK cells are lymphocytes with innate antiviral and antitumor capacity whose potency is determined by the balance of various activating and inhibitory signals delivered via cell-surface receptors. The most well-studied receptors that are known to modulate NK activity are the killer immunoglobulin-like receptors (KIRs).16 KIRs direct a process of education, whereby NK cells that express inhibitory KIR for self-HLA are licensed and become armed with effector functions against target cells that lack self-HLA.17 NK cells that fail to encounter cognate HLA as self are unlicensed; however, with a sufficient activating signal, these populations have the potential to mount an effector response.14,18-20
Among patients with NB who are treated with the anti-GD2 mAb 3F8, patients who lack HLA class I ligands for their inhibitory KIR (missing ligand) exhibit significantly improved outcomes compared with patients who possess all KIR ligands. Of note, the absence of the HLA-Bw4 ligand for KIR3DL1 is associated with the greatest survival benefit.14,15 Considering the allelic diversity that exists for both KIR3DL1 and HLA-Bw4, we hypothesized that the varying strengths of interaction between their protein products might influence the potency of NK response to 3F8.21-24 The KIR3DL1 locus encodes both activating (KIR3DS1) and inhibitory alleles.16 On the basis of patterns of expression and homology, inhibitory KIR3DL1 alleles can be categorized as high (KIR3DL1-h), low (KIR3DL1-l), and null (KIR3DL1-n) surface expression subtypes.24-26 A dimorphism at position 80 of HLA-Bw4 between isoleucine (80I) or threonine (80T) results in variable binding specificity and the inhibition of different KIR3DL1 subtypes.22,23,27
We investigated whether subtype combinations of KIR3DL1 and HLA-B with no, weak, and strong interaction could influence patient outcome after treatment with 3F8. Lack of interaction as a result of a lack of ligand or of cell-surface receptor was associated with greatest survival. Weak interaction was associated with intermediate survival, and strong interaction was associated with the poorest survival. These data may aid in the identification of patients with NB who are likely to benefit from treatment with 3F8 as well as those for whom alternative therapies should be explored.
METHODS
Patients
We performed a retrospective analysis of 245 patients with NB who were treated with 3F8 at Memorial Sloan Kettering Cancer Center (MSKCC) between 1994 and 2007. Antibody therapy was initiated after multimodal therapy that included chemotherapy (Appendix Table A1, online only), surgery, and radiation therapy,28 with or without autologous stem cell transplantation. Patients treated in clinical trials NCT00072358, NCT00002560, and NCT00040872 received intravenous 3F8 plus granulocyte-macrophage colony-stimulating factor (GM-CSF; Appendix Table A2, online only).9 Investigators obtained consent for therapy, specimen collection, and analysis as approved by the MSKCC Institutional Review Board.
Peripheral Blood Mononuclear Cells
Peripheral blood mononuclear cells (PBMCs) collected from patients or anonymous healthy volunteer donors from the New York Blood Center were isolated by Ficoll (New York, NY) density gradient centrifugation. Additional research consent for New York Blood Center samples was waived. PBMCs were cryopreserved in liquid nitrogen.
HLA Typing
Genomic DNA was extracted from PBMCs or marrow mononuclear cells by using the QIAmp DNA Blood Mini Kit according to the manufacturer instructions (Qiagen, Germantown, MD). Patient HLA typing was performed by a combination of HLA serology, sequence-specific polymerase chain reaction (PCR), and PCR-specific oligonucleotide probe. HLA genotyping for healthy donors was performed by HistoGenetics (Ossining, NY).
KIR3DL1 and HLA-B Subtyping and Combined Group Assignments
KIR3DL1 subtyping was performed by using an intermediate-resolution, multiplex PCR-based typing method, which categorizes alleles by surface density without assigning specific alleles.26 KIR3DL1 alleles were classified as KIR3DL1-high (KIR3DL1-h), KIR3DL1-low (KIR3DL1-l), KIR3DL1-null (KIR3DL1-n), and activating KIR3DS1 (KIR3DL1-s) subtypes.26 Compound KIR3DL1 subtypes, whose known surface expression profiles and binding affinities provide predicted inhibitory strengths, were based on previously described classifications22,23,25,29: KIR3DL1-N (KIR3DL1-n/n, or -n/s); KIR3DL1-L (KIR3DL1-l/l, -l/h, -n/l, and -l/s); KIR3DL1-H (KIR3DL1-h/h, -n/h, and -h/s); and 3DS1 (-s/s).29
HLA-B alleles were assigned as HLA-Bw6, Bw4-80I, or Bw4-80T epitopes, per the Immuno Polymorphism database.30 Compound epitopes were categorized as: group Bw4-80I (Bw4-80I/Bw4-80I, Bw4-80I/Bw4-80T, or Bw4-80I/Bw6); group Bw4-80T (Bw4-80T/Bw4-80T or Bw4-80T/Bw6); and group Bw6 (Bw6/Bw6).
Target Cells and Culture Conditions
The NB cell lines SKNBE(1)n and SKNMM were derived from patient tumors, and they express the KIR ligands HLA-C1, HLA-C2, and HLA-Bw4. NB cells were cultured in RPMI 1640 medium that was supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin and they were incubated at 37°C with 5% CO2. To induce class I HLA expression in NB cell lines, recombinant human interferon-gamma (IFN-γ; PeproTech, Rocky Hill, NJ) was added at 1,000 U/mL per day for 72 hours. Frozen PBMCs from healthy donors were thawed, cultured in medium that was supplemented with human IL-2 (proleukin; Prometheus Laboratories, San Diego, CA) at 200 IU/mL, and incubated at 37°C with 5% CO2 for 12 to 16 hours before experiments.
NK Activation and Flow Cytometry
NK cell activation was measured by including anti-LAMP1 antibody (CD107a, BD Biosciences, San Jose, CA) during NK:target cell cocultures. PBMCs (5 × 105 cells/well) were cultured with targets at a 5:1 ratio, with or without 1.0 μg/mL 3F8, in a 96-well V-bottom plate in complete RPMI 1640. After a 4-hour incubation, PBMCs were stained with live/dead fixable stain (Life Technologies, Grand Island, NY) and the following mAbs: anti-CD3 (OKT-3; BioLegend, San Diego, CA); anti-CD56 (N901; Beckman Coulter, Brea, CA); anti-KIR3DL1 (DX9; BioLegend); anti-3DL1/S1 (Z27; Beckman Coulter); anti-NKG2A (Z199; Beckman Coulter); anti-KIR2DL1/S1 (EB6B; Beckman Coulter); and anti-KIR2DL2/L3/S2 (GL183; Beckman Coulter). To identify NK that expresses intracellular KIR3DL1-n, PBMCs were fixed and permeabilized for 10 minutes (Fix & Perm; Life Technologies), then stained for 1 hour at room temperature with anti-KIR3DL1 clone 177407 (Beckman Coulter). HLA class I expression in NB cell lines was evaluated with anti–HLA-Bw4 (REA274; Miltenyi Biotec, San Diego, CA) and anti–HLA-C (DT-9; NCI, Frederick, MD).24 The effect of disrupting KIR3DL1/HLA-B ligation was evaluated by using anti-3DL1 (DX9) and anti–HLA-B,C (clone 4E; MSKCC mAb core facility) antibodies. Multicolor flow cytometry was performed by using an LSR Fortessa (BD Biosciences). Data analysis was performed by using FlowJo 9.7 software (Treestar, Ashland, OR).
Statistical Methods
Overall survival (OS) was defined as the time interval between the start of 3F8 therapy and the date of last follow-up or death. Progression-free survival (PFS) was defined as the time interval between the start of 3F8 therapy and the earliest date of disease progression, death, or last follow-up when no event was observed in the patient. The Kaplan-Meier method and the log-rank test were used to estimate and compare OS and PFS for KIR3DL1 and HLA-B subtype interaction and clinical factors. The permutation log-rank test was used when the number of events was less than five.31 Univariable estimates of hazard ratio (HR) and 95% CIs were computed by using the partial likelihood from the Cox proportional hazards regression model.
A weighted partial likelihood estimate of the average HR was computed to determine OS and PFS risks associated with KIR3DL1/HLA-B subtypes after adjustment for statistically significant clinical factors (P ≤ .05) of age, LDH, disease status, and GM-CSF route (GM-CSF administration route for OS only). Variables that were found not to be statistically significant included the presence of bone marrow disease, metastatic bone disease, and MYCN amplification. The weighted partial likelihood was applied to account for the lack of proportional hazards in risk models.32
P values for paired comparisons of activation in NK that express specific KIRs were generated from a permutation distribution that was derived from the two-sample mean difference. An unpaired Student t test was used to compare NK inhibition. Statistical analyses were performed in software packages SAS (SAS/STAT User’s Guide, Version 9.2; SAS Institute, Cary, NC), library clinfun, coxphw in R version 2.13 (The R Foundation for Statistical Computing), and Prism 6 (GraphPad Software, La Jolla, CA) for Mac.
RESULTS
Patient and Disease Characteristics
To investigate the impact of KIR3DL1 and HLA-B interactions in response to 3F8 in patients with NB, we analyzed KIR3DL1 and HLA-B subtype combinations in a cohort of 245 patients with high-risk NB who were treated with 3F8 at MSKCC from October 1994 to December 2007. Patient and disease characteristics are listed in Table 1. Patients treated with and without autologous stem cell transplantation were analyzed as a single group as it was previously determined that KIR and HLA effects were similar in both treatment groups.14 One hundred and seventeen patients were alive at the last follow-up, with a median follow-up of 102.9 months (95%CI, 96.6 to 111.8 months; range, 27.7 to 232.7 months).
Table 1.
Patient and Disease Characteristics (N = 245)
| Patient Characteristic | No. of Patients |
| INSS neuroblastoma disease stage | |
| 4 | 243 |
| 3 | 2 |
| Age, years | |
| < 1.5 | 21 |
| > 1.5 | 224 |
| MYCN status | |
| Nonamplified | 142 |
| Amplified | 77 |
| Unknown | 26 |
| LDH, U/mol | |
| < 1,500 | 128 |
| > 1,500 | 66 |
| Unknown | 51 |
| Bone marrow metastases | |
| No | 24 |
| Yes | 221 |
| Bone metastases | |
| No | 56 |
| Yes | 189 |
| Disease status at 3F8 therapy | |
| First CR/VGPR | 108 |
| Second CR/VGPR | 19 |
| Primary refractory | 86 |
| Secondary refractory | 4 |
| Other* | 26 |
| KIR3DL1 status | |
| Positive | 230 |
| Negative† | 15 |
| Treatment regimen | |
| ASCT + 3F8 | 168 |
| 3F8 alone | 77 |
Abbreviations: ASCT, autologous stem cell transplantation; CR, complete response; INSS, International Staging System for Neuroblastoma; LDH, lactate dehydrogenase; VGPR, very good partial response.
Patients who received 3F8 in combination with induction chemotherapy.
KIR3DL1-negative patients were considered homozygous for KIR3DS1.
HLA-B Subtypes Impact Survival After Treatment With 3F8
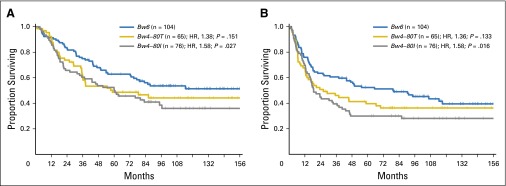
HLA typing revealed that 58% of patients (n = 141) possessed at least one allele that exhibited the Bw4 epitope (HLA-Bw4), whereas 42% of patients (n = 104) were homozygous for alleles that exhibited the Bw6 epitope (Bw6), which was consistent with published population frequencies.33 Patients who were homozygous for Bw6 alleles and who exhibited KIR3DL1 are missing ligand for KIR3DL1, which is associated with superior outcomes.14,15,34 Patients with HLA-Bw4 were further segregated by amino acid at Bw4 position 80. In univariable analysis, patients with Bw4-80T had an observed reduction in OS and PFS compared with Bw6 patients (OS: HR, 1.38; 95% CI, 0.89 to 2.14; P = .151; and PFS: HR, 1.36; 95% CI, 0.91 to 2.03; P = .133); and OS and PFS among patients with Bw4-80I were demonstrably worse compared with Bw6 patients (OS: HR, 1.58; 95% CI, 1.05 to 2.38; P = .027; and PFS: HR, 1.58; 95% CI, 1.09 to 2.30; P = .016; Fig 1). On multivariable analysis, inferior outcomes observed for Bw4-80T (OS: HR, 1.63; 95% CI, 0.93 to 2.84; P = .085; and PFS: HR, 1.94; 95% CI, 1.14 to 3.30; P = .014) and Bw4-80I (OS: HR, 2.23; 95% CI, 1.34 to 3.71; P = .002; and PFS: HR, 2.43; 95% CI, 1.53 to 3.87; P ≤ .001) were maintained.
Fig 1.
HLA-B subtypes influence outcomes in patients with neuroblastoma (NB) who are treated with 3F8. Among patients with NB, 245 who received treatment with 3F8 were segregated on the basis of HLA-B epitope groups. Kaplan-Meier curves for (A) overall survival and (B) progression-free survival among patients encoding Bw6 (blue lines), Bw4-80T (gold lines), and Bw4-80I (gray lines) are shown. HR, hazard ratio.
KIR3DL1 Subtypes Do Not Impact Survival Independent of HLA-B
Among patients, 94% (n = 230) carried at least one allele of KIR3DL1 and 6% (n = 15) were homozygous for KIR3DS1. KIR3DL1 subtyping was completed on genomic DNA from all patients and revealed KIR3DL1 subtype frequencies of 48.4% (n = 237), 15.9% (n = 78), 16.3% (n = 80), and 19.4% (n = 95) for KIR3DL1-h, KIR3DL1-l, KIR3DL1-n, and KIR3DL1-s, respectively, which was consistent with published frequencies.26,35 Patients were then categorized on the basis of composite subtype, with KIR3DL1-H, -L, -N, and KIR3DS1 representing 57.6% (n = 141), 26.9% (n = 66), 9.4% (n = 23), and 6.1% (n = 15), respectively. No one KIR3DL1 subgroup was associated with either OS or PFS in patients who were treated with 3F8 (data not shown).
Patients With KIR3DL1-N, KIR3DS1, or HLA-Bw6 Experience Similar Outcomes
Activated by CD16 engagement but refractory to inhibition by induced HLA on NB cells, unlicensed NK cells, such as KIR3DL1+ cells in Bw6 patients, seem to be the primary effectors for the antitumor benefit of anti-GD2 mAb.14 We hypothesized that KIR3DL1 subtypes that are unable to engage HLA-Bw4 on tumor cells might be also associated with favorable outcomes. The KIR3DL1-n protein is improperly folded and retained intracellularly,36,37 and KIR3DS1 does not interact with HLA-Bw4.27,38 A univariable comparison of HLA-Bw4+ and KIR3DL1-N or KIR3DS1 patients (n = 23) with Bw6 patients (n = 104) revealed similar outcomes (OS: HR, 1.14; 95% CI, 0.56 to 2.33; P = .721; and PFS: HR, 1.12; 95% CI, 0.59 to 2.14; P = .724), which highlighted the potential benefit of the noninteracting receptor populations.
Strength of KIR3DL1 and HLA-B Interactions Predict 3F8 Therapeutic Efficacy
The high degree of polymorphism among KIR3DL1 and HLA-B subtypes, when combined, produce a spectrum of NK response capacity.22,39 Most notably, strongly interacting combinations are associated with superior protection against HIV progression29 but greater susceptibility to leukemic relapse after hematopoietic stem cell transplantation (manuscript in submission). We examined whether subtype combinations might also titrate the sensitivity of NK cells to inhibition by HLA-Bw4 presented in NB cells and influence the efficacy of 3F8 therapy in patients.
Patients were segregated on the basis of the relative strengths with which their KIR3DL1 and HLA-B subtypes were predicted to interact: strong, weak, or noninteracting.29 Strong interaction pairings were composed of KIR3DL1-H + Bw4-80I or KIR3DL1-L + Bw4-80T, and weak interacting pairings were composed of KIR3DL1-H + Bw4-80T or KIR3DL1-L + Bw4-80I. Because patients with Bw6, Bw4 + KIR3DL1-N, and Bw4 + KIR3DS1 exhibited similar outcomes after 3F8 therapy, we grouped together these subtype configurations as noninteracting pairs (Appendix Table A3, online only).
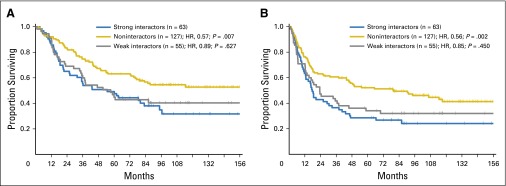
Univariable analysis demonstrated that, compared with strongly interacting combinations (n = 63), the noninteracting combinations (n = 127) had the most favorable OS outcomes (HR, 0.57; 95% CI, 0.38 to 0.85; P = .007) and PFS outcomes (HR, 0.56; 95% CI, 0.39 to 0.81; P = .002; Fig 2). Even when the Bw6 patients were excluded from the group of noninteractors, which left only patients with KIR3DL1-N and KIR3DS1 + HLA-Bw4 (n = 23), similar trends toward benefit were observed (OS: HR, 0.51; 95% CI, 0.25 to 1.06; P = .070; and PFS: HR, 0.52 95% CI, 0.27 to 1.00; P = .052). Patients with weakly interacting pairs (n = 55) demonstrated a trend toward intermediate protection, with lower mortality and relapse compared with the strongly interacting group (OS: HR, 0.89; 95% CI, 0.56 to 1.42; P = .627; and PFS: HR, 0.85; 95% CI, 0.55 to 1.30; P = .450).
Fig 2.
KIR3DL1 and HLA-B subtype combinations predict outcomes in patients with neuroblastoma (NB) who are treated with 3F8 on the basis of strength of interaction. Among patients with NB, 245 who received treatment with 3F8 were grouped according to expected strength of interaction of KIR3DL1 and HLA-B into strongly interacting pairs (blue lines), weakly interacting pairs (gray lines), and noninteracting pairs (gold lines). Kaplan-Meier curves for (A) overall survival and (B) progression-free survival are shown. HR, hazard ratio.
On multivariable analysis, the benefit of noninteracting KIR3DL1/HLA-Bw4 pairs versus strongly interacting pairs was maintained for both OS (HR, 0.41; 95% CI, 0.25 to 0.65; P ≤ .001) and PFS (HR, 0.43; 95% CI, 0.28 to 0.66; P ≤ .001), and the weakly interacting pairs were again associated with a trend toward intermediate outcomes (OS: HR, 0.55; 95% CI, 0.32 to 0.96; P = .036; and PFS: HR, 0.68; 95% CI, 0.40 to 1.15; P = .153; Table 2). Because of the heterogeneity of this cohort, we performed a similar analysis on a more uniform subset of International Neuroblastoma Staging System stage 4 patients, age > 18 months, and in first complete response or very good partial response (n = 66). The findings observed within this subset recapitulated the above findings, as noninteractors demonstrated superior OS (HR, 0.40; 95% CI, 0.18 to 0.85; P = .018) and PFS (HR, 0.46; 95% CI, 0.22 to 0.97; P = .040), and weak interactors displayed a trend toward intermediate outcomes (OS: HR, 0.53; 95% CI, 0.19 to 1.43; P = .209; and PFS: HR, 0.61; 95% CI, 0.25 to 1.48; P = .275) compared with strong interactors (Appendix Table A4, online only).
Table 2.
OS and PFS Associated With KIR3DL1 and HLA-B Subtype Pairs Among Patients Who Received 3F8 for High-Risk Neuroblastoma
| KIR3DL1/HLA-B Pairs | No. | Univariable Analysis | Multivariable Analysis* | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OS | PFS | OS | PFS | ||||||
| OS/PFS | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | |
| Strong interacting | 63/63 | 1 | 1 | 1 | 1 | ||||
| Weak interacting | 55/55 | 0.89 (0.56 to 1.42) | .627 | 0.85 (0.55 to 1.30) | .450 | 0.55 (0.32 to 0.96) | .036 | 0.68 (0.40 to 1.15) | .153 |
| Noninteracting | 127/127 | 0.57 (0.38 to 0.85) | .007 | 0.56 (0.39 to 0.81) | .002 | 0.41 (0.25 to 0.65) | < .001 | 0.43 (0.28 to 0.66) | < .001 |
NOTE. Strong interacting: KIR3DL1-H + Bw4-80I or KIR3DL1-L + Bw4-80T; weak interacting: KIR3DL1-H + Bw4-80T or KIR3DL1-L + Bw4-80I; and noninteracting: KIR3DL1-N + any HLA-B, 3DS1 + any HLA-B, and Bw6 + any KIR3DL1.
Abbreviations: HR, hazard ratio; OS, overall survival; PFS, progression-free survival.
Adjusted for age, lactate dehydrogenase, granulocyte-macrophage colony-stimulating factor route (in OS only) and disease status at 3F8.
KIR3DL1-n and KIR3DS1(+) NK Cells Are Activated by 3F8 and Resist Inhibition by HLA-Bw4
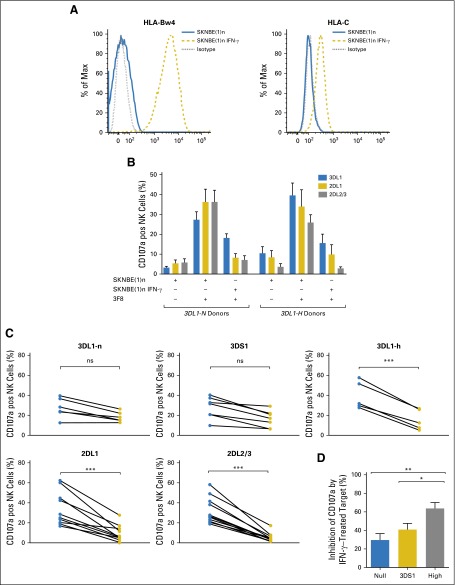
NB primary tumor cells and cell lines typically express low surface HLA at rest40,41; however, treatment-induced inflammation results in upregulation of HLA class I molecules, which provides ligand for the inhibition of licensed NK cells.14,42 We hypothesized that this would negatively impact the activity of cells that bear KIR3DL1 on the NK surface but would minimally impact the activity of noninteracting populations. To test this hypothesis, we compared the responsiveness of NK cells that express KIR3DL1-n, KIR3DS1, or KIR3DL1-h toward the Bw4-80I+ NB cell line SKNBE(1)n. To mimic an autologous setting, Bw4-80I+ donors were selected for study. To induce expression of HLA class I, we treated the cell line with IFN-γ (Fig 3A).14
Fig 3.
KIR3DL1-n+ and KIR3DS1+ natural killer cells (NK cells) are resistant to inhibition by neuroblastoma (NB) cells that express HLA-Bw4-80I. (A) Incubation of SKNBE(1)n with interferon-gamma (IFN-γ) results in increased expression of HLA-Bw4 and HLA-C. (B) Peripheral blood mononuclear cells from healthy Bw4-80I+ donors were stimulated with 3F8 and the KIR3DL1 ligand-matched NB cell line SKNBE(1)n with or without IFN-γ pretreatment. CD107a degranulation of NK cells that exclusively express KIR2DL1 (gold bars), KIR2DL2/3 (gray bars), or KIR3DL1 (blue bars) was compared between KIR3DL1-N (n = 6) and 3DL1-H (n = 5) donors. (C) Paired comparison of CD107a degranulation against IFN-γ–untreated (blue circles) and IFN-γ–treated (gold circles) SKNBE(1)n among NK populations that express specific killer immunoglobulin-like receptors with P value generated from a permutation distribution. (D) Percent inhibition among NK cells that exclusively express KIR3DL1-n (blue bars), KIR3DS1 (gold bars), or KIR3DL1-h (gray bars) was calculated by determining the change in CD107a degranulation in response to 3F8 against IFN-γ–untreated and –treated SKNBE(1)n targets and compared by unpaired Student t test. Data represent three independent trials with four to six donors per group, per trial. ns, nonsignificant. *P < .05; **P ≤ .01; ***P ≤ .001.
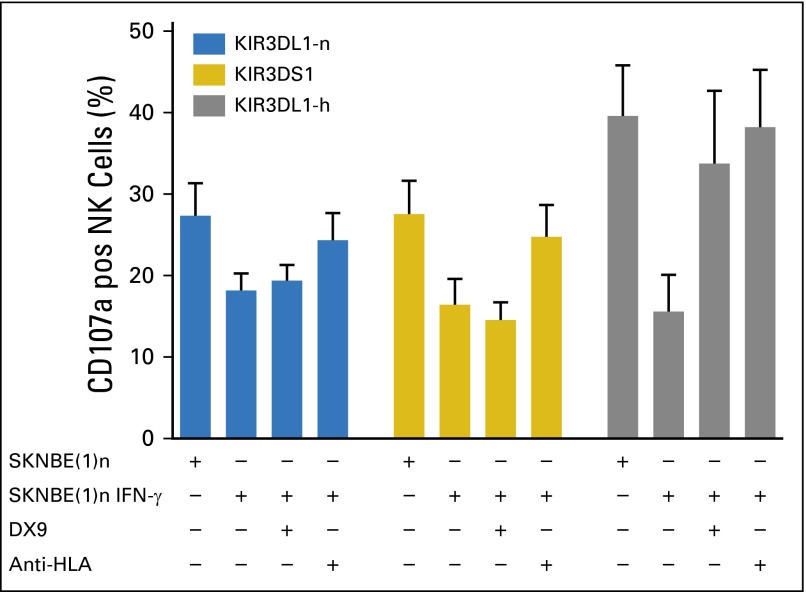
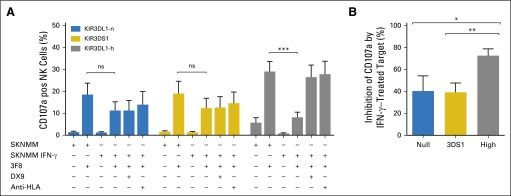
Multiparametric flow cytometry enabled the evaluation of NK cells that express KIR3DL1-h, 3DS1, or intracellular KIR3DL1-n, exclusive of all other inhibitory KIRs and NKG2A. CD107a was used to quantitate degranulation in response to 3F8-opsonized NB cells. In the presence of 3F8, KIR3DL1-h(+) NK cells were highly activated against resting NB cells but were inhibited by IFN-γ–treated, HLA-Bw4–expressing NB cells (Figs 3B-3D). Similarly, upregulation of HLA-C dramatically interfered with the reactivity of NK cells that express KIR2DL1, KIR2DL2, or KIR2DL3, the inhibitory KIR specific for the HLA-C ligand (Figs 3B and 3C). Of note, KIR3DL1-n(+) and 3DS1(+) NK populations reacted to NB cells in the presence of 3F8 and were only mildly inhibited by IFN-γ–treated target cells (Figs 3C and 3D). Inhibition was released by the addition of anti-HLA mAb but not anti-KIR3DL1 (DX9), which suggested that their inhibition may be mediated by other HLA-specific receptors, such as the leukocyte immunoglobulin-like receptor family of receptors (Fig 4).43 In contrast, KIR3DL1-h(+) cells were more dramatically inhibited, and the addition of either DX9 or anti-HLA antibodies rescued the functional response of KIR3DL1-h(+) NK, which indicated that KIR3DL1/HLA-Bw4 interactions were the primary source of inhibition of KIR3DL1-h(+) NK cells (Fig 4). Similar results were seen with the Bw4-80I+ target NB cell line SKNMM (Appendix Fig A1, online only).
Fig 4.
Blocking KIR3DL1-h interaction with Bw4-80I rescues natural killer cells (NK cells) from inhibition. Peripheral blood mononuclear cells from healthy donors were stimulated with 3F8 and KIR3DL1 ligand-matched cell line SKNBE(1)n with or without interferon-gamma (IFN-γ) pretreatment. Effect of addition of KIR3DL1 monoclonal antibody (DX9) and anti–HLA-B-C antibody on CD107a degranulation was evaluated in NK cells that exclusively express KIR3DL1-n (blue bars), KIR3DS1 (gold bars), and KIR3DL1-h (gray bars). Data represent two independent trials with four to six donors per group, per trial.
DISCUSSION
Treatment of NB with anti-GD2 mAb is most successful in patients who lack HLA ligands for at least one inhibitory KIR,14,15 with the greatest benefit observed among those who lack HLA-Bw4 ligands for KIR3DL1.14 KIR3DL1 and HLA-B ligands mediate NK cell inhibition with varying efficiency,22,23,27 which supports the hypothesis that the strength with which KIR3DL1 and HLA-B subtypes interact determines the extent of NK inhibition and, consequently, the antitumor benefit of 3F8 therapy. We demonstrate that individuals with KIR3DL1 and HLA-B subtypes that do not interact at the cell surface exhibit the greatest OS and PFS and that those with weakly and strongly interacting combinations were associated with intermediate and poor 3F8 antitumor effect, respectively. Thus, although maximizing the NK cell activation for ADCC should enhance NB tumor clearance,44-46 considerations should also be given to specific KIR and HLA interactions, which could influence the efficacy of mAb therapy. In this study, we demonstrate that NK effectiveness against cancer may be tuned at the allotype level and not necessarily abolished by tumor expression of HLA-Bw4. Comprehensive in vitro studies are still needed, however, to model inhibitory strengths of receptor–ligand allotypes.
KIR3DL1-n(+) NK cells have been associated with NK cell education36,37 and delayed HIV progression,29 which suggests that intracellular sequestration of the receptor does not impede the ability of these NK cells to recognize and respond to target cells. Similarly, despite its sequence homology to KIR3DL1 and expression on the NK cell surface, KIR3DS1 does not interact with HLA-Bw4 molecules.27,38 Nevertheless, KIR3DS1(+) cells respond to stimulation with target cells27,47,48 and have been associated with protection from graft-versus-host disease,49 HIV29,50 and hepatitis C virus,51 which suggests that either the presence of KIR3DS1 or the absence of KIR3DL1 is relevant to disease control. Here, we extend these findings to the setting of mAb treatment of NB, for which the ability of these NK cells to respond to activating signals, but not inhibitory ligands, results in increased disease control and survival among patients with NB who receive 3F8. The effect of KIR3DL1 and HLA-B subtypes and anti-GD2 therapy should be confirmed in a prospective multicenter trial of a homogeneous cohort, and future studies should examine these interactions in patients who are treated without mAb.
Because other therapeutic antibodies engage NK cells for ADCC,52 including rituximab and trastuzumab, KIR3DL1 and HLA-B subtyping may have a broader use. Indeed, the lack of NK inhibition in patients with lymphoma who are treated with rituximab is associated with higher survival, making KIR3DL1 and HLA-B subtyping potentially relevant.53,54 For patients with an unfavorable genomic profile, alternative treatment options could be considered to improve outcomes. Strategies to augment NK-ADCC can also be considered via adoptive cell therapies or allogeneic hematopoietic cell transplantation with donors selected for NK cells not easily inhibited by recipient HLA. Therapeutic antibodies designed to disrupt KIR and HLA interactions may enhance NK-mediated ADCC in patients with strongly interacting KIR3DL1 and HLA-B pairs, as evidenced in vitro by the restoration of the effector function in the KIR3DL1-h expressing NK cells after antibody blockade during the KIR3DL1 and Bw4 interaction. These antibodies are an area of ongoing clinical interest.
Acknowledgment
We thank the neuroblastoma team at Memorial Sloan Kettering Cancer Center, Chi Hang Cheung for technical assistance, and Mary Carrington, MD, for the DT9 hybridoma used to produce the anti–HLA-C monoclonal antibody.
GLOSSARY TERMS
- ADCC (antibody-dependent cell-mediated cytotoxicity):
a mechanism of cell-mediated immunity whereby an effector cell of the immune system actively lyses a target cell that has been bound by specific antibodies.
- CD16:
a component of the low-affinity Fc receptor involved in phagocytosis and antibody-dependent cellular cytotoxicity. CD16 is found on neutrophils, natural killer cells, and macrophages.
- natural killer cells (NK cells):
cells that belong to the innate immune system and are specialized to kill target cells that are either infected with viruses or host cells that have become cancerous. CD56 is a surface marker specific to NK cells.
Appendix
Fig A1.
KIR3DL1-n+ and KIR3DS1+ natural killer cells (NK cells) are resistant to inhibition by the HLA-Bw4-80I–expressing neuroblastoma (NB) cell line, SKNMM. (A) CD107a degranulation to the NB cell line, SKNMM, among NK cells that exclusively express KIR3DL1-n (blue bars), KIR3DS1 (gold bars), and KIR3DL1-h (gray bars). Conditions included presence and/or absence of 3F8, pretreatment of tumor with interferon-gamma (IFN-γ), and presence and/or absence of anti-HLA or anti-KIR3DL1 (DX9) mAb. Comparison of NK degranulation against IFN-γ–untreated and –treated SKNMM among NK populations that express specific KIR with P value generated from a permutation distribution. (B) Percent inhibition among NK cells that exclusively express KIR3DL1-n (blue bars), KIR3DS1 (gold bars), or KIR3DL1-h (gray bars) was calculated by determining the change in CD107a degranulation in response to 3F8 against IFN-γ–untreated and –treated SKNMM targets and compared by unpaired Student t test. Data represent two independent trials with four to six donors per group, per trial. ns, nonsignificant. *P < .05; **P ≤ .01; ***P ≤ .001.
Table A1.
Induction Chemotherapy Before 3F8 Therapy
| Regimen | No. of Patients |
|---|---|
| High-risk induction: COGA3973-based therapy10 | 186 |
| High-risk induction: POG9640-based therapy* | 40 |
| High-risk induction: per European Group Protocols (United Kingdom, Germany)†1 | 7 |
| Other | 12 |
NOTE. In addition to induction chemotherapy, all patients received maximal surgical resection and radiotherapy to primary sites and bulky sites of disease.
Granger M, et al: Pediatr Blood Cancer 59:902-907, 2012.
Pearson AD, et al: Lancet Oncol 9:247-256, 2008.
Table A2.
Comparison of 3F8 Dosing and Supportive Treatment as Determined by Protocol
| 3F8 Protocol | No. | 3F8 Dose/Cycle, mg/m2 | GM-CSF | Isotretinoin 160 mg/m2/d* | Other |
|---|---|---|---|---|---|
| NCT00002560† | 104 | 100 | IV‡ | 14 d × 6 cycles | N/A |
| NCT000723587 | 115 | 100 | SQ | 14 d × 6 cycles | N/A |
| NCT0004087228 | 26 | 50 | SQ | 14 d × 6 cycles | Oral etoposide§ |
Abbreviations: GM-CSF, granulocyte-macrophage colony-stimulating factor; IV, intravenous; N/A, not applicable; SQ, subcutaneous.
Matthay KK, et al: N Engl J Med 341:1165-1173, 1999.
Kushner BH, et al: J Clin Oncol 19:4189-4194, 2001.
Patients received IV GM-CSF during weeks of 3F8 infusion.
Patients treated in NCT00040872 received adjuvant oral etoposide 50 mg/m2/d for 21 days alternating with cycles of 3F8 for four cycles.
Table A3.
Summary of Composite KIR3DL1 and HLA-B Subtype Pairs
| KIR3DL1 + HLA-B Subtype Pair | No. |
|---|---|
| Strong interaction | 63 |
| h/h + Bw4-80I | 24 |
| h/s + Bw4-80I | 12 |
| n/h + Bw4-80I | 11 |
| l/l + Bw4-80T | 4 |
| l/s + Bw4-80T | 4 |
| l/h + Bw4-80T | 8 |
| Weak interaction | 55 |
| h/h + Bw4-80T | 15 |
| h/s + Bw4-80T | 6 |
| l/l + Bw4-80I | 3 |
| l/s + Bw4-80I | 6 |
| l/h + Bw4-80I | 9 |
| n/h + Bw4-80T | 13 |
| n/l + Bw4-80I | 3 |
| No interaction | 127 |
| n/n + Bw4-80I | 1 |
| n/s + Bw4-80I | 3 |
| n/n + Bw4-80T | 7 |
| n/s + Bw4-80T | 4 |
| n/n + Bw6 | 2 |
| n/s + Bw6 | 6 |
| h/h + Bw6 | 25 |
| h/s + Bw6 | 18 |
| n/h + Bw6 | 17 |
| l/l + Bw6 | 5 |
| l/s + Bw6 | 6 |
| l/h + Bw6 | 3 |
| n/l + Bw6 | 15 |
| 3DS1+ Bw6 | 7 |
| 3DS1 + Bw4-80I | 4 |
| 3DS1 + Bw4-80T | 4 |
Table A4.
OS and PFS Associated With KIR3DL1 and HLA-B Subtype Pairs Among Patients International Staging System for Neuroblastoma Stage 4, > 18 Months, and in First Complete Response/Very Good Partial Response When Receiving 3F8 for High-Risk Neuroblastoma
| KIR3DL1/HLA-B Pairs | No. | OS Multivariable Analysis | PFS Multivariable Analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Strong interacting | 24 | 1 | 1 | ||
| Weak interacting | 16 | 0.53 (0.19 to 1.43) | .209 | 0.61 (0.25 to 1.48) | .275 |
| Noninteracting | 36 | 0.40 (0.18 to 0.85) | .018 | 0.46 (0.22 to 0.97) | .040 |
NOTE. Multivariable analyses adjusted for age, lactate dehydrogenase only, granulocyte-macrophage colony-stimulating factor route of administration not significant on univariable analysis. Strong interacting: KIR3DL1-H + Bw4-80I or KIR3DL1-L + Bw4-80T; weak interacting: KIR3DL1-H + Bw4-80T or KIR3DL1-L + Bw4-80I; and noninteracting: KIR3DL1-N + any HLA-B, 3DS1 + any HLA-B, and Bw6 + any KIR3DL1.
Abbreviations: HR, hazard ratio; OS, overall survival; PFS, progression-free survival.
Footnotes
Processed as a Rapid Communication manuscript.
Supported by the National Institutes of Health Grant No. CA164365 and by Alex’s Lemonade Stand (K.C.H.), National Institutes of Health Grant No. CA008748 (J.Z. and G.H.), National Institutes of Health Grant No. CA008748 to Memorial Sloan Kettering Cancer Center for core facility support, and St Baldrick’s Foundation (C.J.F.).
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Authors' disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
See accompanying editorial on page 2437
AUTHOR CONTRIBUTIONS
Conception and design: Christopher J. Forlenza, Jeanette E. Boudreau, Jean-Benoît Le Luduec, Nai-Kong V. Cheung, Katharine C. Hsu
Financial support: Katharine C. Hsu
Administrative support: Nai-Kong V. Cheung, Katharine C. Hsu
Provision of study materials or patients: Nai-Kong V. Cheung, Katharine C. Hsu
Collection and assembly of data: Christopher J. Forlenza, Elizabeth Chamberlain, Nai-Kong V. Cheung
Data analysis and interpretation: Christopher J. Forlenza, Jeanette E. Boudreau, Junting Zheng, Jean-Benoît Le Luduec, Glenn Heller, Nai-Kong V. Cheung, Katharine C. Hsu
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
KIR3DL1 Allelic Polymorphism and HLA-B Epitopes Modulate Response to Anti-GD2 Monoclonal Antibody in Patients With Neuroblastoma
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Christopher J. Forlenza
No relationship to disclose
Jeanette E. Boudreau
Patents, Royalties, Other Intellectual Property: Patent pending on the KIR3DL1 multiplex PCR assay that is used for subtyping in this manuscript
Junting Zheng
No relationship to disclose
Jean-Benoît Le Luduec
No relationship to disclose
Elizabeth Chamberlain
No relationship to disclose
Glenn Heller
Consulting or Advisory Role: Millennium Pharmaceuticals
Nai-Kong V. Cheung
Consulting or Advisory Role: AstraZeneca, MedImmune
Patents, Royalties, Other Intellectual Property: ScFv constructs of anti-GD2 antibodies (Inst), therapy-enhancing glucan (Inst), use of mAb 8H9 (Inst), methods for preparing and using scFv (Inst), GD2 peptide mimics (Inst), methods for detecting MRD (Inst), anti-GD2 antibodies (Inst), generation and use of HLA-A2–restricted peptide-specific mAbs and CARs (Inst), high-affinity anti-GD2 antibodies (Inst), multimerization technologies (Inst), bispecific HER2 and CD3 binding molecules (Inst), affinity matured hu8H9 (Inst), anti-chondroitin sulfate proteoglycan 4 antibodies and uses thereof (Inst), ROR2 antibodies (Inst), T-cell receptor–like antibody agents specific for EBV latent membrane protein 2A peptide presented by human HLA (Inst)
Katharine C. Hsu
Patents, Royalties, Other Intellectual Property: Patent application on the KIR3DL1 multiplex PCR assay that is used for subtyping in this manuscript
REFERENCES
- 1.Berthold F, Boos J, Burdach S, et al. Myeloablative megatherapy with autologous stem-cell rescue versus oral maintenance chemotherapy as consolidation treatment in patients with high-risk neuroblastoma: A randomised controlled trial. Lancet Oncol. 2005;6:649–658. doi: 10.1016/S1470-2045(05)70291-6. [DOI] [PubMed] [Google Scholar]
- 2.Matthay KK, Yanik G, Messina J, et al. Phase II study on the effect of disease sites, age, and prior therapy on response to iodine-131-metaiodobenzylguanidine therapy in refractory neuroblastoma. J Clin Oncol. 2007;25:1054–1060. doi: 10.1200/JCO.2006.09.3484. [DOI] [PubMed] [Google Scholar]
- 3.Ladenstein R, Pötschger U, Hartman O, et al. 28 years of high-dose therapy and SCT for neuroblastoma in Europe: Lessons from more than 4000 procedures. Bone Marrow Transplant. 2008;41(suppl 2):S118–S127. doi: 10.1038/bmt.2008.69. [DOI] [PubMed] [Google Scholar]
- 4.Pasqualini C, Dufour C, Goma G, et al. Tandem high-dose chemotherapy with thiotepa and busulfan-melphalan and autologous stem cell transplantation in very high-risk neuroblastoma patients. Bone Marrow Transplant. 2016;51:227–231. doi: 10.1038/bmt.2015.264. [DOI] [PubMed] [Google Scholar]
- 5. doi: 10.1200/JCO.2007.13.8925. Matthay KK, Reynolds CP, Seeger RC, et al: Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: A Children’s Oncology Group study. J Clin Oncol 27:1007-1013, 2009 [Erratum: J Clin Oncol 32:1862-1863, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ladenstein RL, Poetschger U, Luksch R, et al. Busulphan-melphalan as a myeloablative therapy (MAT) for high-risk neuroblastoma: Results from the HR-NBL1/SIOPEN trial. J Clin Oncol. 2011;29(abstr 2) [Google Scholar]
- 7.Cheung IY, Hsu K, Cheung NK. Activation of peripheral-blood granulocytes is strongly correlated with patient outcome after immunotherapy with anti-GD2 monoclonal antibody and granulocyte-macrophage colony-stimulating factor. J Clin Oncol. 2012;30:426–432. doi: 10.1200/JCO.2011.37.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu AL, Gilman AL, Ozkaynak MF, et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med. 2010;363:1324–1334. doi: 10.1056/NEJMoa0911123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung NK, Cheung IY, Kushner BH, et al. Murine anti-GD2 monoclonal antibody 3F8 combined with granulocyte-macrophage colony-stimulating factor and 13-cis-retinoic acid in high-risk patients with stage 4 neuroblastoma in first remission. J Clin Oncol. 2012;30:3264–3270. doi: 10.1200/JCO.2011.41.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreissman SG, Seeger RC, Matthay KK, et al. Purged versus non-purged peripheral blood stem-cell transplantation for high-risk neuroblastoma (COG A3973): A randomised phase 3 trial. Lancet Oncol. 2013;14:999–1008. doi: 10.1016/S1470-2045(13)70309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barker E, Mueller BM, Handgretinger R, et al. Effect of a chimeric anti-ganglioside GD2 antibody on cell-mediated lysis of human neuroblastoma cells. Cancer Res. 1991;51:144–149. [PubMed] [Google Scholar]
- 12.Zeng Y, Fest S, Kunert R, et al. Anti-neuroblastoma effect of ch14.18 antibody produced in CHO cells is mediated by NK-cells in mice. Mol Immunol. 2005;42:1311–1319. doi: 10.1016/j.molimm.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 13.Honsik CJ, Jung G, Reisfeld RA. Lymphokine-activated killer cells targeted by monoclonal antibodies to the disialogangliosides GD2 and GD3 specifically lyse human tumor cells of neuroectodermal origin. Proc Natl Acad Sci USA. 1986;83:7893–7897. doi: 10.1073/pnas.83.20.7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarek N, Le Luduec JB, Gallagher MM, et al. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Invest. 2012;122:3260–3270. doi: 10.1172/JCI62749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venstrom JM, Zheng J, Noor N, et al. KIR and HLA genotypes are associated with disease progression and survival following autologous hematopoietic stem cell transplantation for high-risk neuroblastoma. Clin Cancer Res. 2009;15:7330–7334. doi: 10.1158/1078-0432.CCR-09-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Poursine-Laurent J, Truscott SM, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 18.Anfossi N, André P, Guia S, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Yu J, Heller G, Chewning J, et al. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J Immunol. 2007;179:5977–5989. doi: 10.4049/jimmunol.179.9.5977. [DOI] [PubMed] [Google Scholar]
- 20.Orr MT, Murphy WJ, Lanier LL. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol. 2010;11:321–327. doi: 10.1038/ni.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parham P, Norman PJ, Abi-Rached L, et al. Variable NK cell receptors exemplified by human KIR3DL1/S1. J Immunol. 2011;187:11–19. doi: 10.4049/jimmunol.0902332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. doi: 10.1084/jem.20051884. Yawata M, Yawata N, Draghi M, et al: Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med 203:633-645, 2006 [Erratum: J Exp Med 203:1131, 2006] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175:5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 24.Thomas R, Yamada E, Alter G, et al. Novel KIR3DL1 alleles and their expression levels on NK cells: Convergent evolution of KIR3DL1 phenotype variation? J Immunol. 2008;180:6743–6750. doi: 10.4049/jimmunol.180.10.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardiner CM, Guethlein LA, Shilling HG, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 26.Boudreau JE, Le Luduec JB, Hsu KC. Development of a novel multiplex PCR assay to detect functional subtypes of KIR3DL1 alleles. PLoS One. 2014;9:e99543. doi: 10.1371/journal.pone.0099543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Connor GM, Guinan KJ, Cunningham RT, et al. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 28.Kushner BH, Kramer K, LaQuaglia MP, et al. Reduction from seven to five cycles of intensive induction chemotherapy in children with high-risk neuroblastoma. J Clin Oncol. 2004;22:4888–4892. doi: 10.1200/JCO.2004.02.101. [DOI] [PubMed] [Google Scholar]
- 29.Martin MP, Qi Y, Gao X, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.European Bioinformatics Institute Immuno polymorphism database. https://www.ebi.ac.uk/ipd/
- 31.Heller G, Venkatraman ES. Resampling procedures to compare two survival distributions in the presence of right-censored data. Biometrics. 1996;52:1204–1213. [Google Scholar]
- 32.Xu R, O’Quigley J. Estimating average regression effect under non-proportional hazards. Biostatistics. 2000;1:423–439. doi: 10.1093/biostatistics/1.4.423. [DOI] [PubMed] [Google Scholar]
- 33.Single RM, Martin MP, Gao X, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- 34.Delgado DC, Hank JA, Kolesar J, et al. Genotypes of NK cell KIR receptors, their ligands, and Fcγ receptors in the response of neuroblastoma patients to Hu14.18-IL2 immunotherapy. Cancer Res. 2010;70:9554–9561. doi: 10.1158/0008-5472.CAN-10-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vierra-Green C, Roe D, Hou L, et al. Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR loci in 506 European-American individuals. PLoS One. 2012;7:e47491. doi: 10.1371/journal.pone.0047491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsons MS, Boulet S, Song R, et al. Mind the gap: lack of association between KIR3DL1*004/HLA‐Bw4-induced natural killer cell function and protection from HIV infection. J Infect Dis. 2010;202(suppl 3):S356–S360. doi: 10.1086/655966. [DOI] [PubMed] [Google Scholar]
- 37.Taner SB, Pando MJ, Roberts A, et al. Interactions of NK cell receptor KIR3DL1*004 with chaperones and conformation-specific antibody reveal a functional folded state as well as predominant intracellular retention. J Immunol. 2011;186:62–72. doi: 10.4049/jimmunol.0903657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gillespie GM, Bashirova A, Dong T, et al. Lack of KIR3DS1 binding to MHC class I Bw4 tetramers in complex with CD8+ T cell epitopes. AIDS Res Hum Retroviruses. 2007;23:451–455. doi: 10.1089/aid.2006.0165. [DOI] [PubMed] [Google Scholar]
- 39.Yawata M, Yawata N, Draghi M, et al. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raffaghello L, Prigione I, Airoldi I, et al. Mechanisms of immune evasion of human neuroblastoma. Cancer Lett. 2005;228:155–161. doi: 10.1016/j.canlet.2004.11.064. [DOI] [PubMed] [Google Scholar]
- 41.Wölfl M, Jungbluth AA, Garrido F, et al. Expression of MHC class I, MHC class II, and cancer germline antigens in neuroblastoma. Cancer Immunol Immunother. 2005;54:400–406. doi: 10.1007/s00262-004-0603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neal ZC, Imboden M, Rakhmilevich AL, et al. NXS2 murine neuroblastomas express increased levels of MHC class I antigens upon recurrence following NK-dependent immunotherapy. Cancer Immunol Immunother. 2004;53:41–52. doi: 10.1007/s00262-003-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vitale M, Castriconi R, Parolini S, et al. The leukocyte Ig-like receptor (LIR)-1 for the cytomegalovirus UL18 protein displays a broad specificity for different HLA class I alleles: Analysis of LIR-1 + NK cell clones. Int Immunol. 1999;11:29–35. doi: 10.1093/intimm/11.1.29. [DOI] [PubMed] [Google Scholar]
- 44.Shusterman S, London WB, Gillies SD, et al. Antitumor activity of hu14.18-IL2 in patients with relapsed/refractory neuroblastoma: A Children’s Oncology Group (COG) phase II study. J Clin Oncol. 2010;28:4969–4975. doi: 10.1200/JCO.2009.27.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frost JD, Hank JA, Reaman GH, et al. A phase I/IB trial of murine monoclonal anti-GD2 antibody 14.G2a plus interleukin-2 in children with refractory neuroblastoma: A report of the Children’s Cancer Group. Cancer. 1997;80:317–333. doi: 10.1002/(sici)1097-0142(19970715)80:2<317::aid-cncr21>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 46.Cheung NK, Guo H, Hu J, et al. Humanizing murine IgG3 anti-GD2 antibody m3F8 substantially improves antibody-dependent cell-mediated cytotoxicity while retaining targeting in vivo. OncoImmunology. 2012;1:477–486. doi: 10.4161/onci.19864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carr WH, Rosen DB, Arase H, et al. Cutting edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pascal V, Yamada E, Martin MP, et al. Detection of KIR3DS1 on the cell surface of peripheral blood NK cells facilitates identification of a novel null allele and assessment of KIR3DS1 expression during HIV-1 infection. J Immunol. 2007;179:1625–1633. doi: 10.4049/jimmunol.179.3.1625. [DOI] [PubMed] [Google Scholar]
- 49.Venstrom JM, Gooley TA, Spellman S, et al. Donor activating KIR3DS1 is associated with decreased acute GVHD in unrelated allogeneic hematopoietic stem cell transplantation. Blood. 2010;115:3162–3165. doi: 10.1182/blood-2009-08-236943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Long BR, Ndhlovu LC, Oksenberg JR, et al. Conferral of enhanced natural killer cell function by KIR3DS1 in early human immunodeficiency virus type 1 infection. J Virol. 2008;82:4785–4792. doi: 10.1128/JVI.02449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.López-Vázquez A, Rodrigo L, Martínez-Borra J, et al. Protective effect of the HLA-Bw4I80 epitope and the killer cell immunoglobulin-like receptor 3DS1 gene against the development of hepatocellular carcinoma in patients with hepatitis C virus infection. J Infect Dis. 2005;192:162–165. doi: 10.1086/430351. [DOI] [PubMed] [Google Scholar]
- 52.Seidel UJ, Schlegel P, Lang P. Natural killer cell mediated antibody-dependent cellular cytotoxicity in tumor immunotherapy with therapeutic antibodies. Front Immunol. 2013;4:76. doi: 10.3389/fimmu.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du J, Lopez-Verges S, Pitcher BN, et al. CALGB 150905 (Alliance): Rituximab broadens the antilymphoma response by activating unlicensed NK cells. Cancer Immunol Res. 2014;2:878–889. doi: 10.1158/2326-6066.CIR-13-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terszowski G, Klein C, Stern M. KIR/HLA interactions negatively affect rituximab- but not GA101 (obinutuzumab)-induced antibody-dependent cellular cytotoxicity. J Immunol. 2014;192:5618–5624. doi: 10.4049/jimmunol.1400288. [DOI] [PubMed] [Google Scholar]