Abstract
Phyllodes tumours (PTs) are breast fibroepithelial lesions that are graded based on histological criteria as benign, borderline or malignant. PTs may recur locally. Borderline PTs and malignant PTs may metastasize to distant sites. Breast fibroepithelial lesions, including PTs and fibroadenomas, are characterized by recurrent MED12 exon 2 somatic mutations. We sought to define the repertoire of somatic genetic alterations in PTs and whether these may assist in the differential diagnosis of these lesions. We collected 100 fibroadenomas, 40 benign PTs, 14 borderline PTs and 22 malignant PTs. Six, 6 and 13 benign, borderline and malignant PTs respectively and their matched normal tissue were subjected to targeted massively parallel sequencing (MPS) using the MSK-IMPACT sequencing assay. Recurrent MED12 mutations were found in 56% of PTs; in addition, mutations affecting cancer genes (e.g. TP53, RB1, SETD2 and EGFR) were exclusively detected in borderline and malignant PTs. We found a novel recurrent clonal hotspot mutation in the TERT promoter (−124 C>T) in 52% and TERT gene amplification in 4% of PTs. Laser capture microdissection revealed that these mutations were restricted to the mesenchymal component of PTs. Sequencing analysis of the entire cohort revealed that the frequency of TERT alterations increased from benign (18%), to borderline (57%) and to malignant PTs (68%; P<0.01), and TERT alterations were associated with increased levels of TERT mRNA (P<0.001). No TERT alterations were observed in fibroadenomas. An analysis of TERT promoter sequencing and gene amplification distinguished PTs from fibroadenomas with a sensitivity and a positive predictive value of 100% (CI 95.38%–100%) and 100% (CI 85.86%–100%), respectively, and a sensitivity and a negative predictive value of 39% (CI 28.65%–51.36%) and 68% (CI 60.21%–75.78%), respectively. Our results suggest that TERT alterations may drive the progression of PTs, and may assist in the differential diagnosis between PTs and fibroadenomas.
Keywords: mutation, gene amplification, promoter, phyllodes tumour, massively parallel sequencing, telomerase
INTRODUCTION
Phyllodes tumours (PTs) are rare fibroepithelial lesions (accounting for 0.3% to 0.5% of breast neoplasms) which are characterized by a proliferation of stromal cells with varying cellularity and atypia, resulting in the formation of leaf-like projections protruding into cleft-like or cystically-dilated spaces lined by epithelium [1]. PTs are graded as benign, borderline or malignant based on the levels of stromal cellularity, nuclear atypia, tumour borders, proliferation rate, and the presence of stromal overgrowth [1]. Accurate grading of PTs is clinically relevant because local recurrence rates, although largely related to surgical margins, increase with grade, being documented in 5%–21%, 14%–46% and 18%–65% of benign PTs, borderline PTs and malignant PTs, respectively [2–4]. Although approximately 29% of malignant PTs display metastatic behaviour, distant metastases have also been documented in 2%–3% and up to 11% of benign PT and borderline PTs, respectively [2,4–6]. The treatment of PTs consists of surgical excision with clear margins. Patients with malignant PTs may receive adjuvant chemotherapy, however the response rates to current systemic therapies are only modest [7].
Despite their rarity, PTs pose diagnostic challenges[8,9]. Grading of these lesions is not uncommonly challenging, as is the distinction between benign PTs and cellular fibroadenomas, a variant of fibroadenoma that mimics PTs[1,9]. This distinction has important clinical implications, because fibroadenomas are considered to have limited potential for growth and progression to a more aggressive phenotype. Thus lumpectomy alone or radiologic follow-up are adequate treatment options for fibroadenomas. In the latest World Health Organization (WHO) classification [1], it has been acknowledged that in some circumstances a generic diagnosis of cellular fibroepithelial neoplasm may be appropriate, recognizing our inability to classify these lesions correctly and predict their chance of recurrence and/or progression.
PTs have recently been shown to harbour somatic mutations affecting MED12, RARA, FLNA, SETD2 and KMT2D, followed by the acquisition of additional mutations in cancer-associated genes in borderline and malignant PTs, including TP53, RB1, EGFR and NF1 [10,11]. Most recently, TERT promoter mutations have been found to be associated with MED12 mutations in PTs, suggesting that these mutations may act in cooperation [12]. Here, we sought to characterize the repertoire of somatic genetic alterations in PTs and to define whether these genetic alterations may be employed in the differential diagnosis between PTs and fibroadenomas.
MATERIAL AND METHODS
Cases
The archives of the Department of Pathology of Memorial Sloan Kettering Cancer Center (MSKCC) were searched for PTs diagnosed and surgically removed at our institution between January 1996 and July 2015. The diagnostic slides and formalin-fixed paraffin-embedded (FFPE) tissue blocks of 40 benign PTs, 14 borderline PTs and 22 malignant PTs were retrieved. In addition a series of 100 consecutive fibroadenomas was retrieved from the pathology archives of MSKCC. Samples were anonymized prior to analysis, and the study was approved by the MSKCC Institutional Review Board. Informed consent was obtained following the protocol approved by the Institutional Review Board. All cases including all tumour sections were independently reviewed by four pathologists with expertise in breast pathology (MM, ME, FCG and JSR-F), and classified according to the latest WHO criteria [1]. For discordant cases, a consensus diagnosis was achieved on a multi-head microscope.
Power Calculation
If we assume that PTs are driven by a recurrent genetic alteration in a way akin to MED12 exon 2 mutations in fibroadenomas, and this driver event would be present in at least ≥50% of cases, with 6 benign PTs, 6 borderline PTs and 13 malignant PTs, we would have >89%, >89% and >99% power, respectively, to identify a recurrent event (i.e. in two or more cases). Based on the report of Cani, et al. [11], we anticipated that a higher number of non-synonymous somatic mutations would be identified in malignant PTs. With 13 samples, we would be able to detect genes recurrently mutated in >25% of samples at >80% power.
Microdissection and nucleic acid extraction
For all cases except MaPT19, MaPT20 and MaPT22, 8 μm-thick sections representative of the tumour and normal tissue were stained with nuclear fast red and microdissected using a sterile needle under a stereomicroscope (Olympus SZ61), to ensure a tumour cell content >80% and that the normal tissue was devoid of any neoplastic cells as previously described[13]. DNA extraction from microdissected tumour samples and normal adjacent tissues was performed separately using the DNeasy Blood and Tissue Kit (Qiagen) and total RNA extraction from microdissected tumour samples was performed using the RNeasy FFPE Kit (Qiagen), according to the manufacturers’ guidelines. DNA and RNA quantification was performed using the Qubit Fluorometer (Invitrogen).
Targeted capture massively parallel sequencing
Tumour and normal DNA samples from six benign, six borderline and 10 malignant PTs were subjected to targeted capture massively parallel sequencing at the MSKCC Integrated Genomics Operation (IGO), using the Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) assay [14] targeting all exons of 410 cancer genes harbouring actionable mutations and non-coding regions of selected genes. Extracted DNA (250ng) was used to prepare barcoded sequence libraries (New England Biolabs, KapaBiosystems) as previously described [14] (Supplementary Methods online). Details of the MSK-IMPACT analysis for the detection of somatic mutations are provided in the Supplementary Methods online [15–23]. Allele-specific copy number alterations (CNAs) were identified using FACETS [24] (Supplementary Methods online). Sequencing data for these PTs have been deposited to the NCBI Sequence Read Archive under the accession SRP062618.
For MaPT19, MaPT20 and MaPT22, targeted capture massively parallel sequencing was performed using the MSK-IMPACT v.3 (MaPT19–20) and MSK-IMPACT v.5 (MaPT22) clinical sequencing assays, in a Clinical Laboratory Improvement Amendments (CLIA)-certified environment, targeting all coding exons of 275 and 300 genes, respectively, as well as non-coding regions of selected genes as described by Cheng et al [14]. Mutation and CNA detection in the clinical setting was performed as described in Cheng et al [14] (Supplementary Methods online). The list of 227 genes concurrently present in MSK-IMPACT (410 genes), MSK-IMPACTv3 and MSK-IMPACTv5 is presented in Supplementary Table S1. The agreement between the different MSK-IMPACT assays employed for the detection of mutations in the 227 genes targeted by all targeted capture panels is >98% (data not shown).
The likelihood of a mutation to be considered pathogenic was assessed on the basis of a combination of mutation function predictors [25–28] that has been shown to have a high negative predictive value [29], coupled with the presence of the mutated gene in the cancer gene lists described by Kandoth, et al. (127 significantly mutated genes) [30], the Cancer Gene Census [31] or Lawrence, et al. (Cancer5000-S gene set) [32] (Supplementary Methods online).
The cancer cell fraction (CCF) of each mutation was inferred using ABSOLUTE (v1.0.6)[33,34] (Supplementary Methods online)[35].
PCR amplification and Sanger sequencing
Hotspot somatic mutations in the MED12 (exon 2) gene and in the TERT promoter were investigated by Sanger sequencing in the entire cohort of fibroadenomas and PTs as previously described [13] (Supplementary Methods online).
Validation of mutations identified by MSK-IMPACT using amplicon resequencing
Somatic mutations in the MED12 (exon 2) gene and in the TERT promoter were further validated using amplicon resequencing in 72 of the 76 PTs (95%) and in a subset of fibroadenomas (23/100) included in this study. For amplicon resequencing, 10 ng of genomic DNA was amplified with primers described in Supplementary Table S2 using the AmpliTaq Gold 360 Master Mix Kit as described in the Supplementary Methods online.
Quantitative reverse transcription PCR (qRT-PCR)
100 ng DNase-treated RNA was reverse transcribed using the SuperScript VILO kit (Invitrogen, Life Technologies). qRT-PCR was performed in triplicate for each sample using the TERT TaqMan Assay-on-Demand (IDs: Hs00972656_m1; Applied Biosystems, Life Technologies) on the StepOnePlus Real-Time PCR System (Applied Biosystems) as previously described [36,37] (Supplementary Methods online).
TERT copy number assay
Copy number variations affecting the TERT gene were further validated using the TaqMan copy number assay (IDs: Hs06005815_cn; Applied Biosystems) on the StepOnePlus Real-Time PCR System (Applied Biosystems) as per the manufacturer’s guidelines. RNase P (ID: 4403326; Applied Biosystems) was employed for assay normalization. All reactions were performed in quadruplicate. Assessment of copy number status was performed using CopyCaller software (Applied Biosystems) as recommended.
Quantitative PCR method for measuring telomere length
Telomere length was quantified in triplicate by quantitative PCR as previously described by Callaghan, et al.[38] using 40 ng of genomic DNA per sample per reaction (Supplementary Methods online).
TeloFISH
Telomere fluorescence in situ hybridization analysis (TeloFISH) using representative FFPE sections from 14 PTs (list of cases in the Supplementary Methods) was performed as previously described [39] (Supplementary Methods online). For each case, signals were counted in 100 morphologically unequivocal neoplastic cells, and the results were compared using the Mann-Whitney U test.
Southern blot for telomere length determination
For Southern blot analysis of telomere length, representative 8 μm-thick sections were cut from frozen samples of four PTs (two with TERT promoter mutations (MaPT03 and BoPT02), one with TERT amplification (MaPT06), one with wild-type TERT (BoPT11)), and subjected to microdissection under a stereomicroscope as described above. DNA extraction from microdissected samples and DNA quantification were performed as described above. Length of terminal restriction fragment (TRF) was assessed using the TeloTAGGG telomere length assay kit (Roche Molecular Biochemical), following the manufacturer’s instructions and as previously described [40] (Supplementary Methods online).
Laser capture microdissection
Representative 8 μm-thick sections from five cases (BePT10, BoPT02, BoPT03, BoPT07, MaPT01) were cut and mounted on Arcturus PEN membrane glass slides (Applied Biosystems) and microdissected using the Leica LMD 6500 System (Supplementary Methods online).
Comparative analysis of the frequencies of the TERT −124 C>T promoter hotspot mutation and TERT gene amplification in fibroadenomas and PTs
To assess the ability to distinguish PTs from fibroadenomas and to distinguish benign PTs from fibroadenomas based on TERT promoter mutational status and/or TERT gene amplification status, we calculated the sensitivity, specificity, positive predictive value and negative predictive value based on the presence or absence of the TERT −124 C>T promoter mutation and/or TERT gene amplification. 95% confidence intervals were calculated according to the efficient-score method (corrected for continuity) as previously described [41] based on the method outlined by Wilson [42] (http://vassarstats.net/clin1.html).
Statistical analysis
The Chi-square test (χ2 test) and the Fisher’s exact test were employed for the comparison of categorical variables. Kruskal-Wallis test was employed to calculate the differences between the groups (i.e. benign PTs, borderline PTs and malignant PTs). All statistical analyses were carried out using IBM SPSS Statistics v.20 (IBM) or R v3.1.2. Two-tailed p-values <0.05 were considered statistically significant.
RESULTS
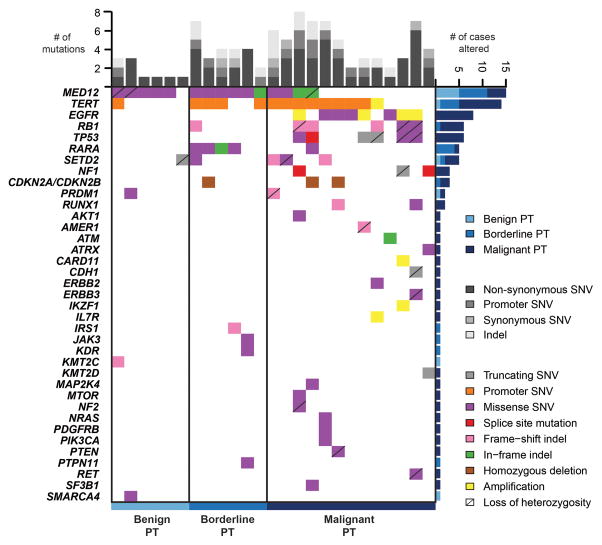
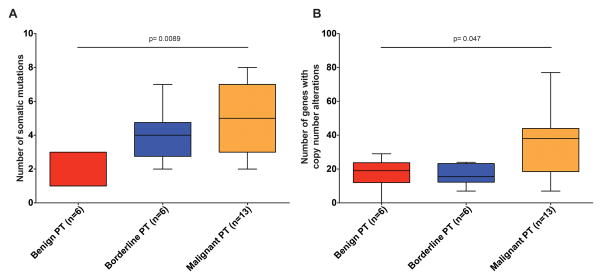
Here we subjected six benign PTs, six borderline PTs and 13 malignant PTs (diagnoses based on concordant reviews by four pathologists with expertise in breast pathology (ME, MM, FCG and JSR-F)) to high-depth targeted capture massively parallel sequencing using MSK-IMPACT [14], a sequencing assay interrogating the coding regions of up to 410 genes, and intronic regions and promoters of selected genes. With a median depth of coverage of 560x (range 308–1114x; Supplementary Table S3), MSK-IMPACT analysis revealed that the number of somatic mutations (mean 2 (range 1–3), mean 4 (range 2–7) and mean 5 (range 2–8) in benign PT, borderline PT and malignant PT, respectively; Figure 1) and gene CNAs (Supplementary Figure S1) significantly increased with the grade of the lesions (Figure 2, Supplementary Table S4; p=0.0089 and p=0.047, respectively, Kruskal-Wallis tests). Consistent with previous observations [43], MED12 exon 2 somatic mutations were present in 56% of PTs, but were significantly less frequent in malignant PTs (31%) than in borderline PTs (83%) and benign PTs (83%) (p=0.01, Fisher’s exact test; Figure 1, Supplementary Table S5). This is in contrast with the results of Tan, et al. [10], where a similar frequency of MED12 mutations was found in PTs regardless of their histologic grades. MED12 exon 2 mutations were found to be clonal (i.e. inferred to be present in virtually all cancer cells analyzed in a given sample, see Supplementary Methods online) in all but one PT harbouring these somatic mutations (Supplementary Figure S2). Malignant PTs were found to harbour likely pathogenic somatic genetic alterations affecting known cancer genes, including TP53, RB1, PTEN, NF1, CDKN2A, NRAS, MTOR, EGFR and SF3B1 (Figure 1, Supplementary Table S5), some of which are potentially targetable, including homozygous deletions of CDKN2A, amplifications of EGFR (Supplementary Figure S3), mutations affecting the ligand binding and tyrosine kinase domains of EGFR, and hotspot mutations affecting PIK3CA (His1047Arg), NRAS (Gln61Lys), ERBB2 (Val777Leu) and ERBB3 (Val104Leu; Supplementary Table S5). A comparison between the results of the massively parallel sequencing performed in this study and in Tan, et al.[10] revealed differences in the prevalence of somatic mutations affecting TP53, RB1 and EGFR (Supplementary Table S6); no differences, however, were observed when mutation frequencies were adjusted according to histologic grade (Supplementary Table S6).
Figure 1. Somatic mutations, gene amplifications and homozygous deletions in phyllodes tumours of the breast.
Non-synonymous somatic mutations, gene amplifications and homozygous deletions identified in the 25 phyllodes tumours of the breast subjected to targeted capture massively parallel sequencing. Each column represents one sample; genes are reported in rows. Only the 227 genes consistently present in the three targeted capture panels are included. Alteration types are color-coded according to the legend. The presence of loss of heterozygosity of the wild-type allele of a mutated gene is represented by a diagonal bar. Bar charts (top) indicate the number of non-synonymous, promoter and synonymous somatic single nucleotide variants (SNVs), as well as the number of somatic insertions and deletions (indels) for each sample. Bar charts (right) show the number of cases harbouring non-synonymous or promoter somatic SNVs, indels, gene amplifications or homozygous deletions in a given gene, color-coded based on the grade of the lesions.
Figure 2. Comparison of the frequency of somatic mutations and of genes with copy number alterations in phyllodes tumours stratified according to histological grade.
A, number of somatic mutations identified in phyllodes tumours (PTs) stratified histologically as benign, borderline and malignant. Error bars, S. D. of the mean. B, number of genes with copy number alterations identified in phyllodes tumours stratified histologically as benign, borderline and malignant. Error bars, S. D. of the mean. Statistical comparisons were performed using Kruskal-Wallis test.
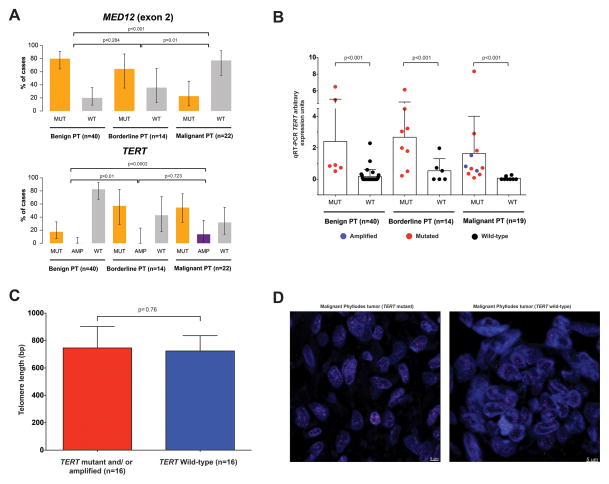
Our analysis revealed the presence of recurrent hotspot somatic mutations affecting the promoter region of TERT (−124 C>T; Supplementary Figure S4) in 52% and high-level amplification of the TERT gene in 4% of all PTs (Figure 1, Supplementary Figure S3), which were confirmed by high-depth targeted amplicon sequencing, Sanger sequencing and/or quantitative PCR (Supplementary Figure S5, Supplementary Table S7). The TERT −124 C>T promoter hotspot mutation was found to be clonal in all but one case (Supplementary Figure S2).
To define the frequency of the TERT −124 C>T promoter hotspot mutation and TERT gene amplification in PTs, we subjected additional 51 PTs to high-depth amplicon sequencing and quantitative PCR, which revealed an increasing frequency of somatic genetic alterations affecting the TERT gene in benign (18%), borderline (57%) and malignant PTs (68%, Figure 3A). By contrast, the rate of MED12 exon 2 somatic mutations in this cohort of PTs, as assessed by both amplicon sequencing and Sanger sequencing, was significantly higher in benign (80%) and borderline (64%) than in malignant PTs (23%; p<0.01, Fisher’s exact test; Figure 3A). Malignant PTs lacking TERT alterations harboured clonal somatic mutations in well-described cancer genes such as EGFR, NF1 and ATRX, and two malignant PTs had concurrent likely pathogenic mutations affecting RB1 and TP53, coupled with loss of heterozygosity of the respective wild-type alleles (Figure 1, Supplementary Figure S2).
Figure 3. MED12 and TERT recurrent somatic genetic alterations in phyllodes tumours of the breast.
A, Frequencies of MED12 exon 2 somatic mutations (upper panel) and TERT −124 C>T promoter hotspot mutations and TERT amplifications (lower panel) in the 76 phyllodes tumours of the breast included in this study. Error bars show 95% confidence intervals. Statistical comparisons were performed using the Fisher’s exact test. AMP, amplified; MUT, mutated; PT, phyllodes tumour; WT, wild-type. B, TERT mRNA levels in phyllodes tumours (PT) harbouring TERT promoter hotspot mutations or gene amplification and in PTs with wild-type TERT as assessed by quantitative real-time PCR (qRT-PCR). Statistical comparisons were performed using the Mann-Whitney U test. Error bars, S. D. of the mean (n = 3 experimental replicates). C, Association of TERT promoter mutations and/or amplifications with telomere length calculated using quantitative PCR [38]. Statistical comparisons performed using the Mann-Whitney U test revealed no differences between the two groups (P=0.76). Error bars, S. D. of the mean (n=3 experimental replicates). D, Representative TeloFISH micrographs of selected phyllodes tumours of the breast with (left panel) and without (right panel) the −124C>T TERT promoter somatic mutation. No differences were detected in the average telomere intensity between phyllodes tumours with and without TERT somatic genetic alterations (p=0.9). Statistical comparisons were performed using the Mann-Whitney U test. Scale bars, 5 μm.
Somatic mutations affecting the promoter of TERT are the most frequent non-coding somatic mutations in cancer [44]. Two TERT promoter hotspot mutations (i.e. −124 C>T and −146 C>T, Supplementary Figure S4)[45] and TERT gene amplification have been reported in various human malignancies [46–51], and shown to result in increased levels of telomerase expression and activity in somatic cells [46,47,52,53]. Consistent with these observations, the levels of TERT mRNA were significantly higher in PTs harbouring the TERT promoter −124 C>T hotspot mutation and/or TERT amplification than in PTs with wild-type TERT (Figure 3B). Importantly, TERT mRNA was undetectable in 80%, 67% and 75% of the benign, borderline and malignant PTs harbouring wild-type TERT, respectively (Figure 3B). Analyses of telomere length using a PCR-based approach [38], Southern blot [40] and fluorescence in situ hybridization, however, did not reveal significant differences between PTs with altered or wild-type TERT (Figures 2C and 2D, Supplementary Figure S6), in agreement with the observations made in clear cell renal cell carcinoma [54]. Given the significantly higher frequency of the TERT promoter hotspot mutation and TERT gene amplification in borderline PTs and malignant PTs, it is plausible that these alterations may play a role in the progression from benign to malignant PTs.
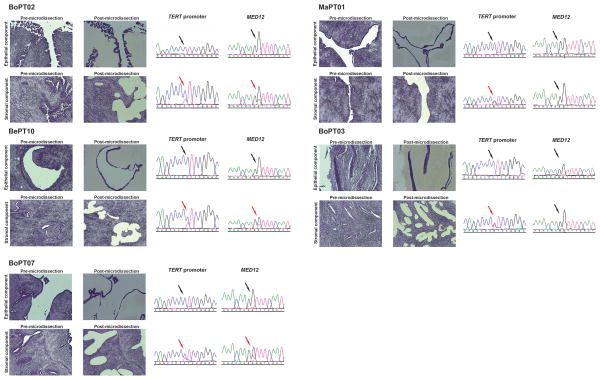
MED12 somatic mutations have been shown to be restricted to the stromal components of fibroadenomas [55]. Based on the similarities between fibroadenomas and PTs, we reasoned that the TERT promoter hotspot mutations would be present in the stromal cells, but absent in the epithelial cells of PTs. Sequencing of MED12 exon 2 and TERT promoter in DNA extracted from the epithelial and stromal components of five laser capture microdissected PTs revealed that both mutations were restricted to the stromal component of these tumours (Figure 4), supporting the latter as the neoplastic component of PTs.
Figure 4. TERT and MED12 somatic genetic alterations are present in the stromal but not in the epithelial components of phyllodes tumours of the breast.
Micrographs of laser capture microdissected stromal and epithelial cells from five phyllodes tumours, and the respective Sanger sequencing traces of the TERT promoter region encompassing the −124 locus and MED12 exon 2 in the separately microdissected stromal and epithelial components. To avoid contamination, a thorough two-step laser-capture microdissection was performed: the component not to be sequenced was first ablated with a laser; after the complete removal of the component not to be analyzed, the component to be analyzed was retrieved using laser-capture microdissection. The presence or absence of the mutations interrogated by Sanger sequencing is indicated by red and black arrows, respectively.
Given that PTs may be difficult to distinguish from fibroadenomas, particularly in the limited tissue sample of a diagnostic core biopsy, and that clinical management of all PTs requires complete excision of the lesion with clear surgical margins to minimize the risk of relapse and distant metastases whereas fibroadenomas may be followed-up clinically, we sought to define whether the TERT promoter −124 C>T hotspot mutation and/or gene amplification would be present in fibroadenomas. No TERT somatic genetic alterations were identified in a series of 100 consecutive fibroadenomas (Supplementary Table S8). Furthermore, we found that analysis of the TERT −124 C>T promoter hotspot mutation coupled with TERT gene copy number differentiates PTs from fibroadenomas with 100.00% (95% CI 95.38–100.00%) specificity and 100.00% (95% CI 85.86–100.00%) positive predictive value (PPV; Table 1), and benign PTs and fibroadenomas with 100.00% (95.38–100.00%) and 100.00% (56.09–100.00%; Table 1) specificity and PPV, respectively. These results suggest that TERT gene promoter sequencing and gene copy number analysis could be used as ancillary tests to differentiate PTs from fibroadenomas.
Table 1.
Sensitivity, specificity, positive and negative predictive values of TERT promoter hotspot mutation and/or TERT amplification in the differential diagnosis between fibroadenomas and PTs of the breast.
| TERT genetic alteration status | Sensitivity (%, 95% CIs) | Specificity (%, 95% CIs) | Positive predictive value (%, 95% CIs) | Negative predictive value (%, 95% CIs) | |
|---|---|---|---|---|---|
| Distinguishing PTs from fibroadenomas | TERT −124 C>T promoter mutation and/or amplification status | 39.47% (28.65%–51.36%) | 100.00% (95.38%–100.00%) | 100.00% (85.86%–100.00%) | 68.49% (60.21%–75.78%) |
| TERT −124 C>T promoter mutation status | 35.52% (25.11%–47.41%) | 100.00% (95.36%–100.00%) | 100.00% (84.49%–100.00%) | 67.11% (58.87%–74.45%) | |
| TERT amplification status | 3.94% (1.02%–11.80%) | 100.00% (95.38%–100.00%) | 100.00% (30.99%–100.00%) | 57.80% (50.06%–65.19%) | |
| Distinguishing benign PTs from fibroadenomas | TERT −124 C>T promoter mutation and/or amplification status | 17.50% (7.89%–33.36%) | 100.00% (95.38%–100.00%) | 100.00% (56.09%–100.00%) | 75.18% (66.80%–82.08%) |
| TERT −124 C>T promoter mutation | 17.50% (7.89%–33.36%) | 100.00% (95.38%–100.00%) | 100.00% (56.09%–100.00%) | 75.18% (66.80%–82.08%) | |
| TERT amplification status | NA | NA | NA | NA |
CIs, confidence intervals; NA, not applicable, as the feature was not present in any of the fibroadenomas analyzed; 95% confidence intervals were calculated according to the efficient-score method (corrected for continuity) as previously described[41] based on the method outlined by Wilson [42]. Benign PTs, benign phyllodes tumours of the breast; PTs, phyllodes tumours of the breast.
DISCUSSION
Here we demonstrate that the repertoire of actionable somatic genetic alterations in PTs varies with histologic grade, and that genetic profiling of malignant PTs may assist in the identification of targeted therapies for patients with metastatic disease. We found that the TERT promoter −124 C>T hotspot mutation and/or TERT gene amplification increase in frequency according to the histologic grade of the lesions, and result in increased TERT mRNA expression, supporting their potential role in the progression of PTs. Clonal genetic alterations including MED12 exon 2 and TERT promoter mutations were found to be present in the stromal cells but absent in the epithelial cells of PTs, suggesting that PTs are likely mesenchymal neoplasms.
Tan, et al. have recently reported on the landscape of somatic mutations in breast PTs [10]. In the present study, and in Tan, et al. [10], MED12 exon 2 mutations were the most frequent somatic mutations found in benign and borderline PTs (in the current study, 80% and 64% of benign and borderline PTs harbored MED12 exon 2 mutations, respectively, whereas in Tan, et al. [10], these frequencies were 82% and 63%, respectively). On the other hand, and despite the small sample size of malignant PTs in both studies (n=22 in the current study vs n=10 in Tan, et al.[10]), a difference of borderline significance in the prevalence of MED12 exon 2 mutations in malignant PTs was observed (23% vs. 60%, respectively, p=0.0557). In agreement with our observations, Cani, et al. [11] detected MED12 somatic mutations in 80% of benign (4/5) and borderline (4/5) PTs, but in only 40% (2/5) of malignant PTs. Furthermore, a significantly higher frequency of somatic mutations affecting TP53, RB1 and EGFR was observed in the present study than in Tan, et al. [10]; these differences, however, may merely reflect the higher proportion of malignant PTs in our study (22/76, 29%) than in Tan, et al. [10] (10/79, 13%; Supplementary Table S6). In fact, when adjusted for histological grade, no significant differences in the frequency of somatic mutations affecting the 28 genes assessed in both studies were observed. In agreement with Tan, et al. [10], non-synonymous mutations in cancer-associated genes, such as TP53, RB1, EGFR and NF1, were restricted to borderline and malignant PTs, consistent with the notion that the mutational repertoire of PTs may correlate with their histological grades.
Yoshida, et al. [12] have recently identified TERT promoter mutations in 65% of PTs and in 7% of fibroadenomas. Sanger sequencing analysis [12] revealed TERT promoter mutations, mostly −124 C>T but also −146 C>T and −124C>A, in 50%, 87% and 62% of benign, borderline and malignant PTs, and in all but one case, TERT promoter mutations were found in conjunction with MED12 mutations. In addition, 7% of the 58 fibroadenomas analyzed by Yoshida, et al. [12] harboured the TERT promoter−124 C>T hotspot mutation. In accord with Yoshida, et al. [12], our massively parallel sequencing and Sanger sequencing analyses revealed the presence of the TERT promoter−124 C>T hotspot mutation at similar frequencies in borderline and malignant PTs (8/14, 57% vs. 50% in [12] and 12/22, 56% vs. 62% in [12], respectively, p>0.05, Fisher’s exact tests). Our study differed from that of Yoshida, et al. in that the only TERT promoter mutation found in the 76 PTs analyzed here was the −124 C>T hotspot mutation, that the frequency of TERT promoter mutations in benign PTs in this study (7/40, 18%) was significantly lower than that reported by Yoshida, et al. [12] (9/18, 50%, p=0.02, Fisher’s exact test), and that only 52% of the PTs harbouring TERT promoter mutations also displayed MED12 mutations in our study as opposed to 97% of TERT mutant PTs reported by Yoshida, et al. [12] (p<0.0001, Fisher’s exact test). Most importantly, from a diagnostic perspective, the results of our study are fundamentally different from those of Yoshida, et al. [12] in regards to the presence of TERT promoter mutations in fibroadenomas. Whilst four of the 58 (7%) fibroadenomas analyzed by Yoshida, et al. [12] harboured the −124 C>T TERT promoter hotspot mutation, all of the 100 consecutive fibroadenomas analyzed here displayed wild-type TERT. The difference observed in these studies may be attributed to the challenging nature of classifying and grading breast fibroepithelial lesions. In our study, all cases were reviewed by four pathologists with an interest and expertise in breast pathology and the final diagnoses were based on a consensus among the four pathologists. Another potential explanation may stem from the possible differences in the repertoire of somatic mutations in fibroepithelial lesions affecting Asian and North American individuals. Further studies are warranted to define the frequency of TERT promoter mutations in benign PTs and fibroadenomas in patients of different ethnicities.
In our study, PTs harbouring the −124 C>T TERT promoter hotspot mutation or TERT gene amplification were found to consistently display detectable levels of TERT mRNA, and to have significantly higher levels of TERT mRNA than PTs with wild-type TERT. Consistent with the results reported by Hosen, et al. [54], no differences in relative telomere length were observed between PTs with and without TERT promoter mutations or TERT gene amplification. Potential explanations for this observation include that the PTs without TERT mutations may harbour genetic or epigenetic alterations that may activate the alternative lengthening of the telomeres (ALT) mechanism of telomere length maintenance. Interestingly, case MaPT04, one of the TERT wild-type malignant PTs harboured a likely pathogenic mutation affecting ATRX, a gene whose alterations have been shown to result in Alternative Lengthening of Telomeres (ALT) [56,57]. In addition, the association between the −124 C>T TERT promoter hotspot mutation and TERT gene amplification and longer telomeres would presuppose that these alterations happened at the point of telomere crisis. Hence, further studies are warranted to define the exact chronology of the acquisition of these genetic alterations in the evolution of PTs.
Our study has important limitations. Owing to the low recurrence rate (7/72, 10%) and short follow-up (median 27 months) of the patients with PTs analyzed here, it was not possible to address whether TERT somatic genetic alterations are associated with outcome. Defining the impact of TERT somatic genetic alterations on the outcome of patients with benign and borderline PTs remains an important question. Although in our analysis, TERT somatic genetic alterations were found to have a high positive predictive value, its negative predictive value was suboptimal for the distinction between PTs and fibroadenomas. Further studies are warranted to define whether a massively parallel sequencing panel targeting TERT somatic genetic alterations in conjunction with alterations in cancer genes affected in borderline and malignant PTs, including RARA, SETD2, KMT2D, TP53, NF1, RB1, EGFR and RUNX1, would provide an even greater diagnostic accuracy.
Despite these limitations, our findings have possible implications in achieving a more accurate diagnosis of fibroepithelial lesions, given that TERT promoter sequencing and/or TERT gene copy number analysis may assist in the differentiation between fibroadenomas and PTs, thereby improving the clinical management for patients with these tumours. We also demonstrated that a targeted sequencing assay may be employed to identify potentially actionable genetic alterations in malignant PTs, which currently lack options for systemic treatment. Finally, TERT somatic genetic alterations were found to be significantly more prevalent in borderline and malignant PTs than in benign PTs. In conjunction with the observation that borderline and malignant PTs have a higher number of somatic mutations and CNAs than benign PTs, our results would be consistent with the hypothesis that TERT somatic genetic alterations likely play a role in the progression of PTs, potentially by enabling mesenchymal cells of PTs to undergo a greater number of cell divisions and ultimately acquire driver somatic genetic alterations in key cancer genes.
Supplementary Material
Frequency plots illustrating the copy number gains and losses (left) and gene amplifications and homozygous deletions (right) in 25 phyllodes tumours (PTs) subjected to targeted capture massively parallel sequencing. In each panel, the frequency of copy number alterations is presented in the y-axis, with purple bars showing copy number gains or gene amplifications and orange bars showing copy number losses or homozygous deletions accordingly. Genomic positions are arranged along the x-axis.
Each column represents one sample; genes are reported in rows. Only the 227 genes present in all target capture panels are included (Supplementary Methods online). The cancer cell fraction of each mutation is color-coded according to the legend. Clonal mutations are highlighted by an orange box. PT, phyllodes tumour.
Genome plots of the phyllodes tumours with TERT or EGFR amplification, or CDKN2A homozygous deletion. In the genome plots, smoothed Log2 ratios (y-axis) were plotted according to their genomic positions (x-axis).
The relative genomic positions of the TERT transcription (TSS) and translation (ATG) start sites, and the regions targeted by the two hotspot mutations (in red): g.1 295 228 and g.1 295 250, referred to as −124 C>T and 146 C>T, respectively, are illustrated.
Representative Sanger sequencing traces of selected tumour samples and their normal counterparts. Black arrow, wild-type TERT promoter; red arrow, TERT −124 C>T promoter mutation.
DNA was extracted from fresh/frozen phyllodes tumours with (left) and without (right) TERT somatic genetic alterations, and terminal restriction fragments (TRF) were analyzed using the Telo TAGGG Telomere Length Assay[40] (Supplementary Methods online).
List of genes of the MSK-IMPACT targeted capture assays employed in this study.
List of primers used for Sanger sequencing, amplicon targeted re-sequencing and quantitative PCR telomere length assay.
Targeted capture massively parallel sequencing statistics.
List of somatic mutations found in the phyllodes tumours of the breast subjected to targeted capture massively parallel sequencing.
Comparison of the repertoire of non-synonymous somatic mutations in the 25 pyllodes tumours subjected to MSK-IMPACT massively parallel sequencing and the 79 phyllodes tumours of the breast reported by Tan, et al., and between the 76 phyllodes tumours subjected to MED12 exon 2 sequencing analysis (by MSK-IMPACT massively parallel sequencing and/or amplicon re-sequencing and/or Sanger sequencing) and the 79 phyllodes tumours reported by Tan, et al. [10].
Clinico-pathologic characteristics, MED12 exon 2, TERT mutational and amplification status of the phyllodes tumours included in this study, and sequencing analyses performed.
MED12 exon 2 and TERT status of the fibroadenomas included in this study.
Statistical comparison of the mutation rate and the number of genes affected by copy number alterations between phyllodes tumours of different histological grades.
Acknowledgments
SP is funded in part by a Susan G Komen Postdoctoral Fellowship Grant (PDF14298348); GSM is funded by CAPES (#BEX 5714/14-1). Research reported in this publication was supported in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (Grant No. P30CA008748).
Footnotes
Conflict of interest: The authors have no conflicts of interest to declare.
AUTHOR CONTRIBUTIONS
EB, BW and JSR-F conceived the study. BW and JSR-F supervised the work. MM, LS, MA and EB provided samples. MM, LS, LN, DSB and JB provided interpreted clinical data. DR, DBS, JB and ML provided interpreted targeted capture sequencing results for a subset of cases. MM, ME, FCG and JSR-F reviewed the cases. FCG, ADP, CM and JSR-F performed the tissue microdissection. SP, GSM, RAI and PKN performed Sanger sequencing and amplicon sequencing. SP, AI, LGM and JP performed the telomerase length assay. Massively parallel sequencing analysis was performed by CKYN, KAB, RSL and IdB. SP performed statistical analyses. SP, CKYN, BW and JSR-F analyzed and interpreted the data. JSR-F wrote the first draft of the manuscript, which was initially reviewed by SP, CKYN and BW. All authors edited and approved the final draft of the manuscript.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Tan PH, Tse G, Lee A, et al. Fibroepithelial tumours. In: Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ, editors. WHO Classification of Tumours of the Breast. 4. IARC press; Lion, France: 2012. [Google Scholar]
- 2.Tan PH, Thike AA, Tan WJ, et al. Predicting clinical behaviour of breast phyllodes tumours: a nomogram based on histological criteria and surgical margins. J Clin Pathol. 2012;65:69–76. doi: 10.1136/jclinpath-2011-200368. [DOI] [PubMed] [Google Scholar]
- 3.Barth RJ., Jr Histologic features predict local recurrence after breast conserving therapy of phyllodes tumors. Breast Cancer Res Treat. 1999;57:291–295. doi: 10.1023/a:1006260225618. [DOI] [PubMed] [Google Scholar]
- 4.Ben Hassouna J, Damak T, Gamoudi A, et al. Phyllodes tumors of the breast: a case series of 106 patients. Am J Surg. 2006;192:141–147. doi: 10.1016/j.amjsurg.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Belkacemi Y, Bousquet G, Marsiglia H, et al. Phyllodes tumor of the breast. Int J Radiat Oncol Biol Phys. 2008;70:492–500. doi: 10.1016/j.ijrobp.2007.06.059. [DOI] [PubMed] [Google Scholar]
- 6.Asoglu O, Ugurlu MM, Blanchard K, et al. Risk factors for recurrence and death after primary surgical treatment of malignant phyllodes tumors. Ann Surg Oncol. 2004;11:1011–1017. doi: 10.1245/ASO.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Mishra SP, Tiwary SK, Mishra M, et al. Phyllodes tumor of breast: a review article. ISRN Surg. 2013;2013:361469. doi: 10.1155/2013/361469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khazai L, Middleton LP, Goktepe N, et al. Breast pathology second review identifies clinically significant discrepancies in over 10% of patients. J Surg Oncol. 2015;111:192–197. doi: 10.1002/jso.23788. [DOI] [PubMed] [Google Scholar]
- 9.Cheng SP, Chang YC, Liu TP, et al. Phyllodes tumor of the breast: the challenge persists. World J Surg. 2006;30:1414–1421. doi: 10.1007/s00268-005-0786-2. [DOI] [PubMed] [Google Scholar]
- 10.Tan J, Ong CK, Lim WK, et al. Genomic landscapes of breast fibroepithelial tumors. Nat Genet. 2015;47:1341–1345. doi: 10.1038/ng.3409. [DOI] [PubMed] [Google Scholar]
- 11.Cani AK, Hovelson DH, McDaniel AS, et al. Next-Gen Sequencing Exposes Frequent MED12 Mutations and Actionable Therapeutic Targets in Phyllodes Tumors. Mol Cancer Res. 2015;13:613–619. doi: 10.1158/1541-7786.MCR-14-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida M, Ogawa R, Yoshida H, et al. TERT promoter mutations are frequent and show association with MED12 mutations in phyllodes tumors of the breast. Br J Cancer. 2015;113:1244–1248. doi: 10.1038/bjc.2015.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinreb I, Piscuoglio S, Martelotto LG, et al. Hotspot activating PRKD1 somatic mutations in polymorphous low-grade adenocarcinomas of the salivary glands. Nat Genet. 2014;46:1166–1169. doi: 10.1038/ng.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saunders CT, Wong WS, Swamy S, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 19.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Mattos-Arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25:1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young SR, Pilarski RT, Donenberg T, et al. The prevalence of BRCA1 mutations among young women with triple-negative breast cancer. BMC Cancer. 2009;9:86. doi: 10.1186/1471-2407-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronglai S, Seshan V. FACETS: Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing. Memorial Sloan-Kettering Cancer Center, Dept of Epidemiology & Biostatistics Working Paper Series; 2015. [Google Scholar]
- 25.Schwarz JM, Rödelsperger C, Schuelke M, et al. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 26.Carter H, Chen S, Isik L, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shihab HA, Gough J, Cooper DN, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi Y, Sims GE, Murphy S, et al. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martelotto LG, Ng C, De Filippo MR, et al. Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations. Genome Biol. 2014;15:484. doi: 10.1186/s13059-014-0484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weigelt B, Geyer FC, Natrajan R, et al. The molecular underpinning of lobular histological growth pattern: a genome-wide transcriptomic analysis of invasive lobular carcinomas and grade-and molecular subtype-matched invasive ductal carcinomas of no special type. J Pathol. 2010;220:45–57. doi: 10.1002/path.2629. [DOI] [PubMed] [Google Scholar]
- 37.Arriola E, Marchio C, Tan DS, et al. Genomic analysis of the HER2/TOP2A amplicon in breast cancer and breast cancer cell lines. Lab Invest. 2008;88:491–503. doi: 10.1038/labinvest.2008.19. [DOI] [PubMed] [Google Scholar]
- 38.O’Callaghan NJ, Fenech M. A quantitative PCR method for measuring absolute telomere length. Biol Proced Online. 2011;13:3. doi: 10.1186/1480-9222-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kannan K, Inagaki A, Silber J, et al. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3:1194–1203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu KD, Orme LM, Shaughnessy J, Jr, et al. Telomerase and telomere length in multiple myeloma: correlations with disease heterogeneity, cytogenetic status, and overall survival. Blood. 2003;101:4982–4989. doi: 10.1182/blood-2002-11-3451. [DOI] [PubMed] [Google Scholar]
- 41.Julious SA. Two-sided confidence intervals for the single proportion: comparison of seven methods by Robert G. Newcombe, Statistics in Medicine 1998; 17:857–872. Stat Med. 2005;24:3383–3384. doi: 10.1002/sim.2164. [DOI] [PubMed] [Google Scholar]
- 42.Wilson EB. Probable inference, the law of succession, and statistical inference. Journal of the American Statistical Association. 1927;22:209–212. [Google Scholar]
- 43.Piscuoglio S, Murray M, Fusco N, et al. MED12 somatic mutations in fibroadenomas and phyllodes tumours of the breast. Histopathology. 2015;67:719–729. doi: 10.1111/his.12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinhold N, Jacobsen A, Schultz N, et al. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet. 2014;46:1160–1165. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heidenreich B, Rachakonda PS, Hemminki K, et al. TERT promoter mutations in cancer development. Curr Opin Genet Dev. 2014;24:30–37. doi: 10.1016/j.gde.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 46.Borah S, Xi L, Zaug AJ, et al. Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science. 2015;347:1006–1010. doi: 10.1126/science.1260200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vinagre J, Almeida A, Populo H, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 48.Nault JC, Zucman-Rossi J. TERT promoter mutations in primary liver tumors. Clin Res Hepatol Gastroenterol. 2016;40:9–14. doi: 10.1016/j.clinre.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 49.Li C, Wu S, Wang H, et al. The C228T mutation of TERT promoter frequently occurs in bladder cancer stem cells and contributes to tumorigenesis of bladder cancer. Oncotarget. 2015;6:19542–19551. doi: 10.18632/oncotarget.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heidenreich B, Rachakonda PS, Hosen I, et al. TERT promoter mutations and telomere length in adult malignant gliomas and recurrences. Oncotarget. 2015;6:10617–10633. doi: 10.18632/oncotarget.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Biase D, Gandolfi G, Ragazzi M, et al. TERT Promoter Mutations in Papillary Thyroid Microcarcinomas. Thyroid. 2015;25:1013–1019. doi: 10.1089/thy.2015.0101. [DOI] [PubMed] [Google Scholar]
- 52.Chiba K, Johnson JZ, Vogan JM, et al. Cancer-associated TERT promoter mutations abrogate telomerase silencing. Elife. 2015:4. doi: 10.7554/eLife.07918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu T, Brown TC, Juhlin CC, et al. The activating TERT promoter mutation C228T is recurrent in subsets of adrenal tumors. Endocr Relat Cancer. 2014;21:427–434. doi: 10.1530/ERC-14-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hosen I, Rachakonda PS, Heidenreich B, et al. TERT promoter mutations in clear cell renal cell carcinoma. Int J Cancer. 2015;136:2448–2452. doi: 10.1002/ijc.29279. [DOI] [PubMed] [Google Scholar]
- 55.Lim WK, Ong CK, Tan J, et al. Exome sequencing identifies highly recurrent MED12 somatic mutations in breast fibroadenoma. Nat Genet. 2014;46:877–880. doi: 10.1038/ng.3037. [DOI] [PubMed] [Google Scholar]
- 56.Clynes D, Jelinska C, Xella B, et al. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat Commun. 2015;6:7538. doi: 10.1038/ncomms8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Napier CE, Huschtscha LI, Harvey A, et al. ATRX represses alternative lengthening of telomeres. Oncotarget. 2015;6:16543–16558. doi: 10.18632/oncotarget.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frequency plots illustrating the copy number gains and losses (left) and gene amplifications and homozygous deletions (right) in 25 phyllodes tumours (PTs) subjected to targeted capture massively parallel sequencing. In each panel, the frequency of copy number alterations is presented in the y-axis, with purple bars showing copy number gains or gene amplifications and orange bars showing copy number losses or homozygous deletions accordingly. Genomic positions are arranged along the x-axis.
Each column represents one sample; genes are reported in rows. Only the 227 genes present in all target capture panels are included (Supplementary Methods online). The cancer cell fraction of each mutation is color-coded according to the legend. Clonal mutations are highlighted by an orange box. PT, phyllodes tumour.
Genome plots of the phyllodes tumours with TERT or EGFR amplification, or CDKN2A homozygous deletion. In the genome plots, smoothed Log2 ratios (y-axis) were plotted according to their genomic positions (x-axis).
The relative genomic positions of the TERT transcription (TSS) and translation (ATG) start sites, and the regions targeted by the two hotspot mutations (in red): g.1 295 228 and g.1 295 250, referred to as −124 C>T and 146 C>T, respectively, are illustrated.
Representative Sanger sequencing traces of selected tumour samples and their normal counterparts. Black arrow, wild-type TERT promoter; red arrow, TERT −124 C>T promoter mutation.
DNA was extracted from fresh/frozen phyllodes tumours with (left) and without (right) TERT somatic genetic alterations, and terminal restriction fragments (TRF) were analyzed using the Telo TAGGG Telomere Length Assay[40] (Supplementary Methods online).
List of genes of the MSK-IMPACT targeted capture assays employed in this study.
List of primers used for Sanger sequencing, amplicon targeted re-sequencing and quantitative PCR telomere length assay.
Targeted capture massively parallel sequencing statistics.
List of somatic mutations found in the phyllodes tumours of the breast subjected to targeted capture massively parallel sequencing.
Comparison of the repertoire of non-synonymous somatic mutations in the 25 pyllodes tumours subjected to MSK-IMPACT massively parallel sequencing and the 79 phyllodes tumours of the breast reported by Tan, et al., and between the 76 phyllodes tumours subjected to MED12 exon 2 sequencing analysis (by MSK-IMPACT massively parallel sequencing and/or amplicon re-sequencing and/or Sanger sequencing) and the 79 phyllodes tumours reported by Tan, et al. [10].
Clinico-pathologic characteristics, MED12 exon 2, TERT mutational and amplification status of the phyllodes tumours included in this study, and sequencing analyses performed.
MED12 exon 2 and TERT status of the fibroadenomas included in this study.
Statistical comparison of the mutation rate and the number of genes affected by copy number alterations between phyllodes tumours of different histological grades.