ABSTRACT
Structural centers of motion (pivot points) in the ribosome have recently been identified by measurement of conformational changes in rRNA resulting from EF-G GTP hydrolysis. This series of measurements is extended here to the ribosome's interactions with the cofactor EF-Tu. Four recent EF-Tu bound ribosome structures were compared to unbound structures. A total of 16 pivots were identified, of which 4 are unique to the EF-Tu interaction. Pivots in the GTPase associated center and the sarcin-ricin loop omitted previously, are found to be mobile in response to both EF-Tu and EF-G binding. Pivots in the intersubunit bridge rRNAs are found to be cofactor specific. Head swiveling motions in the small subunit are observed in the EF-Tu bound structures that were trapped post GTP hydrolysis. As in the case of pivots associated with EF-G, the additional pivots described here are associated with weak points in the rRNA structures such as non-canonical pairs and bulge loops. The combined set of pivots should be regarded as a minimal set. Only several states available to the ribosome have been presented in this work. Future, precise crystal structures in conjunction with experimental data will likely show additional functional pivoting elements in the rRNA.
KEYWORDS: EF-G, EF-Tu, intersubunit rotation, non standard base pairs, pivot points, ribosome dynamics, sarcin Ricin loop, translation
Abbreviations
- ASF
A-site finger
- GAC
GTPase Associated Center
- LSU
large subunit
- rRNA
Rrna
- SRL
sarcin ricin loop
- SSU-
small subunit
Introduction
The ribosome is responsible for the dynamic process of translation.1 It is comprised of 2 subunits, each consisting of RNA and protein. In Bacteria, the major co-factors that facilitate this process are the elongation factors EF-Tu and EF-G, initiation factor IF-2, and the release factor RF-3.2 EF-G is thought to coordinate and hasten accommodation and translation of the tRNA by cycles of conformational rigidity and relaxation before and after GTP hydrolysis.3 EF-Tu is believed to control tRNA mobility by disallowing incorrect codon-anticodon interactions.4 With these cofactors and others, the process of translation exhibits multiple motions including tRNA translocation, intersubunit ratcheting, and small subunit head swivel.3,5-7
The motions of tRNA during the various stages of translation, including accommodation, are largely associated with reorientations of a structurally weak pivoting element.5,8-10 Motion also exists in the mechanisms of intersubunit ratcheting and 30S head swiveling, which have previously been analyzed using high-resolution crystal structures,6,11 cryo-EM structures,12 and computational studies.7,13-15 Recent high-resolution crystal structures of EF-G16,17 and EF-Tu18-21 associated ribosomes now allow further characterization of the cofactor dependent elements in the rRNA core.
Major pivoting elements associated with EF-G functionality were reported previously. 22 Herein, this effort is extended to identify pivoting positions associated with EF-Tu function in Thermus thermophilus. This is accomplished by comparison of 4 high resolution crystal structures of ribosome subunits bound and unbound to the cofactor EF-Tu.18-21,23 In 2 bound structures the GTP is not hydrolyzed, while in the other 2 it is. The motions made obvious by alignment of the different structures are tabulated by the resulting greatest interhelix distance in Angstroms. When combined with the earlier EF-G results, a set of elements allowing large scale motion is identified in the rRNAs of T. thermophilus. The differences in the mobility of the described set of rRNAs hint at previously unreported functional differences between the 2 cofactors.
Results
Consistent with earlier studies,22,24 partial overlap exists between pivoting elements associated with EF-Tu binding and those previously found to be associated with EF-G. Three categories of pivots were recognized. This includes those that were active with both EF-Tu and EF-G, those that are only mobile in the presence of EF-Tu, and those that are associated only with EF-G. Tables 1 and 2 summarize the average results for 4 individual comparisons. Detailed results for the individual comparisons are provided in the Supplemental materials as Tables S1-S8. Individual results are tabulated for pre-and post-GTP hydrolysis in Table S9 for 16S rRNA and Table S10 for 23S rRNA.
Table 1.
Large subunit helices mobile with respect to EF-Tu ribosome contact. Tabulated are (columns from left to right) the helix number, average motion measured across all 4 EF-Tu bound structures, aligned stem sequences, pivoting residues, average motion in EF-Tu bound ribosomes pre-GTP hydrolysis, average motion in EF-Tu bound ribosomes post-GTP hydrolysis and the final residues the motion of which was measured. The average of 4 crystal structure measurements is reported in Angstroms under A1 for GTP non-hydrolyzed structures 2XQE, 2WRO and under A2 for post GTP hydrolysis structures 4ABS, 2Y11. Finally the table is separated into three groups of measurements: helices mobile as a result of either cofactor binding are shown in “Both” section, helices mobile as a result of individual cofactor binding are labelled under EF-Tu and EF-G.
|
Both | ||||||
| Helix |
Average |
Stem |
Pivot |
A1 |
A2 |
Final Residue |
| 38 | 4.2 | 868-870+ | 871U-G | 4.8 | 3.6 | 883G |
| 911-913 | ||||||
| 42 | 7.6 | 1030-1031+ | 1032A-G | 7.8 | 7.5 | 1046A |
| 1123-1124 | ||||||
| 69 | 3.7 | 1906+1924 | 1907 G-U | 2.3 | 5.2 | 1914C |
| 76 | 8.8 | 2093-2095+ | 2096U-G | 11.3 | 6.2 | 2116G |
| 2194-2196 | ||||||
| 89 | 4.9 | 2455-2456+ | 2457U-G | 4.8 | 5.0 | 2473U |
| 2495-2496 | ||||||
| 95 | 9.3 | 2646+2674 | 2647U-G | 8.7 | 9.8 | 2661A |
| EF-Tu | ||||||
| 10 | 4.4 | 148+178 | 149 A-G | 4.0 | 4.8 | 171G |
| 59 | 4.8 | 1527+1544 | 1528 Bulge | 5.3 | 4.6 | 1531C |
| EF-G | ||||||
| 34 | 1.7 | 700-702+ 730-732 | 703 Bulge | 1.6 | 1.9 | 715G |
| 84 | 1.2 | 2996-2997+2321-2322 | 2298 Bulge | 1.1 | 1.4 | 2307G |
Table 2.
Small subunit helices mobile with respect to EF-Tu ribosome contact. Tabulated are (columns from left to right) the helix number, average motion measured across all 4 EF-Tu bound structures, aligned stem sequences, pivoting residues, average motion in EF-Tu bound ribosomes pre-GTP hydrolysis, average motion in EF-Tu bound ribosomes post-GTP hydrolysis and the final residues the motion of which was measured The average of 4 crystal structure measurements is reported in Angstroms under A1 for GTP unhydrolyzed structures 2XQD and 2WRN and under A2 for post GTP hydrolysis structures 4ABR and 2Y10. Finally, the table is separated into three groups of measurements: helices mobile as a result of either cofactor binding are shown in “Both” section, helices mobile as a result of individual cofactor binding are labelled under EF-Tu and EF-G.
|
Both | ||||||
|
Helix |
Average |
Stem |
Pivot |
A1 |
A2 |
Final Residue |
| 6 | 6.1 | 62+106 | 63 U-G | 6.7 | 5.6 | 82 |
| 8 | 2.6 | 144-147+175-178 | 152 A-C | 3.1 | 2.2 | 160 |
| 33 | 3.8 | 984-990+ 1215-1221 | 1036 bulge | 4.8 | 2.9 | 1030b |
| 39 | 3.5 | 1118-1124+ 1149-1155 | 1130 A-G | 4.0 | 3.0 | 1137 |
| 40 | 3.2 | 1158+1178 | 1159 U-G | 3.5 | 2.9 | 1167 |
| 44 | 8.0 | 1401+1501 | 1402 C-A | 8.7 | 7.3 | 1447 |
| EF-Tu | ||||||
| 10 | 2.0 | 199-200+217-218 | 201 G-U | 2.2 | 1.8 | 202 |
| 14 | 3.0 | 339-342+347-350 | Unclear | 3.0 | 3.0 | 346 |
| 17 | 3.1 | 438-440+494-496 | 441 A-A | 2.8 | 3.6 | 461 |
| EF-G | ||||||
| 21 | 1.8 | 586-592+647-651 | 593 G-U | 2.0 | 1.6 | 619 |
| 26 | 1.4 | 829-830+856-857 | 831 U-G | 1.8 | 1.0 | 839 |
| 31 | 0.8 | 954-955+1225-1226 | 956 bulge | 0.6 | 1.0 | 968 |
| 36 | 0.5 | 1068-1073+ 1102-1107 | 1074 G-U | 0.5 | 0.5 | 1092 |
| 37 | 0.8 | 1068-1073+1102-1107 | 1074 G-U | 0.7 | 1.0 | 1078 |
| 41 | 0.9 | 1241-1246+1291-1296 | 1242 U-G | 0.4 | 1.5 | 1267 |
| 42 | 1.9 | 1303+1334 | 1304 Bulge | 1.5 | 2.2 | 1317 |
| 43 | 1.7 | 1350+1372 | 1351 G-U | 1.7 | 1.8 | 1361 |
A total of 12 pivots are mobile in both sets of structural comparisons. In the SSU, these are helices h6, h8, h33, h39, h40, and h44- all of which are associated with intersubunit bridges. In the large subunit, these are the A- site finger H38, the b/L12 stalk H42, bridge b1a H69, the uL1 stalk H76, as well as GTPase associated center (GAC), helix H89, and sarcin ricin loop (SRL) helix H95. This set of elements is involved in cofactor binding and the tRNA translocation process.
Four new pivoting elements specific to EF-Tu binding were found. These were helices h14 and h17 in the small subunit and helices H10 and H59 in the large subunit A number of pivots were active in the EF-G bound structures are inactive in the EF-Tu structures. In the LSU, these are intersubunit bridge helices H34, H69, and H84. In the SSU, EF-G specific pivots are in helices- h21, h26, h28, h31, h32, h36, h37, h41, h42, and h43. These include pivots associated with the head swiveling motion initiated through EF-G-GTP hydrolysis and intersubunit ratcheting.
Helices H69 and h28 both showed dependency on GTP hydrolysis by EF-Tu. Large scale motion was observed in structures stalled by a GTP analog in the pre-GTP hydrolysis state,19 while only smaller motions were observed in structures stalled by kirromycin.18,20,21 Structures 2WRN, 2WRO (4V5G)18 were trapped in the EF-Tu-GTP hydrolysis transition state with kirromycin and paramomycin. Structures 2XQD, 2XQE and (4V5L)19 were captured by GDPCP in the pre-hydrolysed state. Structures 2Y10, 2Y11 (4V5R, now superseded by 4V5S)20 and 4ABR, 4ABS (4V8Q)21 were captured in the post- GTP hydrolysis state with kirromycin. In the small subunit, h33 moves with respect to helix h28 alignment in pre-GTP hydrolysis structures 2WRN and 2XQD. The motion measured is 5.4 Angstroms and 5.8 Angstroms respectively. A ∼4Angstrom full head swivel/ is seen as a result of the same alignment in the post-GTP-hydrolysis structures, 2Y10 and 4ABR. Helix H69 moved by 2.2 and 2.3 Angstroms in the post GTP hydrolyzed structures 2Y11, and 4ABS and more dramatically by 4.3, and 6 Angstroms before GTP hydrolysis in structures 2XQE and 2WRO.
Helices h10 and h42 both met the cutoff only once and with an overall average below 2.5 Angstroms were not considered to be mobile by the criteria used here. Helix h8 failed to meet the cut off in only one comparison and based on overall average is included as mobile. Finally, helices h43 and h17 in 2 cases exhibited modest mobility, while in the other 2 cases they showed essentially none. Figs. 1 and 2 show the location of these 3 categories of pivots in the context of the bacterial rRNA secondary structure. The local context of the unique EF-Tu pivots in the 50S subunit is shown as an insert on Fig. 1 and in higher resolution in Fig. S1.
Figure 1.
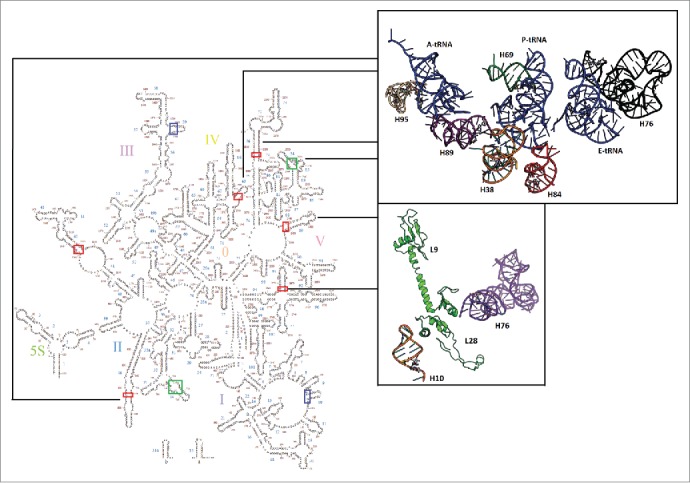
Large subunit map with pivoting positions highlighted. Mobile pivots are highlighted: EF-Tu alone (blue), EF-G alone (green), both (red). The upper structural insert shows a selection of pivoting helices proximal to the tRNA as it moves toward the exit site. The lower insert shows H10 (orange) positioned to interact with proteins bL9 and bL28, which in turn contact the highly mobile H76/ uL1 stalk.
Figure 2.
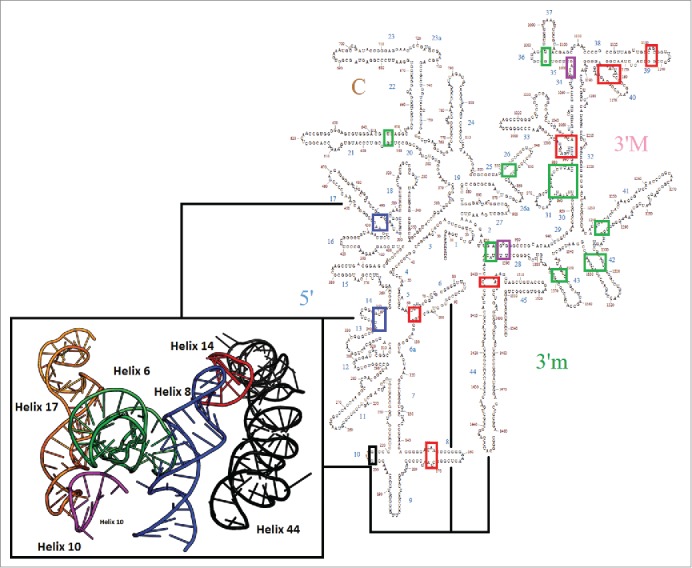
Small subunit secondary structure map with pivoting positions highlighted. Mobile pivots are highlighted: EF-Tu alone (blue), EF-G alone (green), both (red). Two additional pivots proposed previously15 are labeled in purple. Helix h10, which is not considered to be a pivot as discussed in the text is labeled in black. The two EF-Tu specific pivots are in close proximity as shown in the insert. Helix h44 (black), activates bridge B8 helices h8 (blue) and h14 (red), in the small subunit. Mobile helices h6 (green)- the spur, h10 (pink), and h17(orange)-are also proximal to bridge B8.
Discussion
Large subunit
Local motions resulting from cofactor association, intersubunit bridging, and the 30S head swivel play a large role in translation. 3,5-7 With respect to cofactor binding, both EF-G and EF-Tu contact the ribosome primarily at the GTP associated center (GAC), which includes helices H43 and H44.24 Though structurally similar, the 2 cofactors are thought to have a distinct binding mechanism.24 An important feature related to cofactor selectivity in the GAC is the distance between the GAC and the sarcin ricin loop (SRL)24 as well as the incoming cofactor P-loop.25 The measurements obtained here allow up to 10 Angstroms of motion for the SRL which was previously described as immobile.24 Further, the SRL fits the profile of a typical pivoting element, which includes a U-G wobble base pair that closes a 3 way junction. This structure likely allows the SRL the flexibility to accommodate the incoming cofactor.
In response to EF-Tu binding, motion is again seen in the tRNA and a series of pivoting elements around the tRNA extending from the A to the E site (Figs. 1 and 3 and Fig. S1). H76- the uL1 stalk, H38- the A-site finger (ASF) and H42- the bL12 stalk are found to be mobile. The ASF and the uL1 stalk contact the tRNA during translocation while H42 forms a series of functional contacts with the elements of the GAC.25 The E-site- tRNA interaction lies directly upstream of the H82 stem as shown in Fig. S2.
Figure 3.
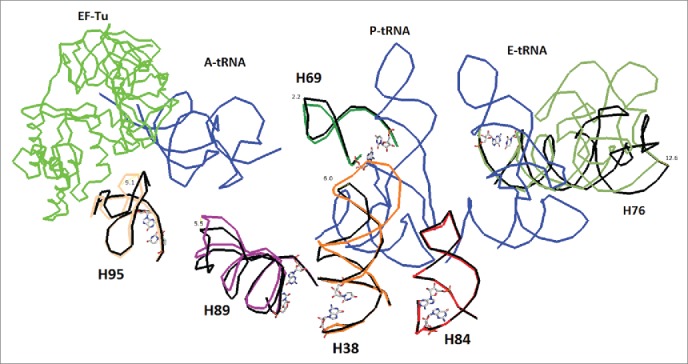
Detailed examination of regions proximal to the tRNA as it moves toward the exit as shown in the upper insert of Fig. 1. The helices may be accommodating the tRNA during translation. Helix 76 is known to guide the tRNA through continuous ionic interaction throughout the process. Helices H84 and H69 are found to be less mobile in EF-Tu bound ribosomes than EF-G bound ribosomes. The A, P, and E site tRNAs are shown in blue. EF-Tu is in green. Pivoting bases are shown as stick models. The structures compared (4V51 black and 4V8Q colored) are pre and post EF-Tu binding.
H82 is in direct proximity to H68, which has an internal bulge motif that suggests mobility and is known to contact the mobile tRNA in eukaryotes.26 Helix H68 in turn, is in contact with H76- the uL1 stalk, predicted to move as it guides the tRNA toward the exit site. The uL1 stalk contacts the tRNA at residue G2112, but not G2116 as was predicted previously,27 at least not in the 4V5L structure. It appears that the tRNA is accommodated throughout the PTC by a set of highly mobile elements- functionality known for the uL1 stalk but unreported in helices H68 and H82.
Helices H10 and H59 are uniquely mobile in the EF-Tu bound ribosome as shown in the supplemental materials in Fig. S3 and S4. Helix H10 is another mobile element exclusive to the EF-Tu bound ribosome structure. Mobility in H10 can be explained, by its proximity i.e. potential contact with proteins bL9 and bL28, which form a base for the highly mobile H76 that comprises the uL1 stalk bL28, is required for ribosome assembly.28 Helix H59 is exclusively perturbed by EF-Tu. H59 is also a known contact site for the signal recognition particle (SRP).29,30 The Alu domain of the SRP mimics elongation factor structure in the PTC thereby arresting elongation by competition with elongation factor binding on the ribosome.30 The ‘minor-saddle motif’ of the Alu domain is a flexible 3 way junction closed by a base pair mismatch, which-fits the general trend of structures that form pivoting elements.30
Intersubunit bridging
In structures bound to EF-Tu, H69 is activated weakly, primarily in the post GTP-hydrolysis state. This mobility is related to the intersubunit rotation of the ribosome through bridge B2a and h44 of the SSU.22 The combination of the small subunit h28 directed head swiveling motions and h69 mobility as a result of EF-Tu GTP hydrolysis are further evidence for a connected network of cofactor dependent pivoting rRNA elements.
H34 and H84 are also less mobile post EF-Tu binding. This may mean that these bridges are less utilized during the EF-Tu binding event and more heavily used in the intersubunit ratcheting process. Bridge B8 is also found to be highly mobile in the EF-Tu bound structure, while relatively inactive in the EF-G bound ribosome.
Small subunit
Both helices h14 and h8, connected to bridge B8 in the small subunit are found to move ˜3A in the current structure set (Fig. 4). Although h14 is a relatively small helix, it exhibits some conformational freedom in EF-Tu structures as a result. Proximal to this region ( Figs. S5 and S6) are mobile helices h6- the spur, h10, and h17.
Figure 4.
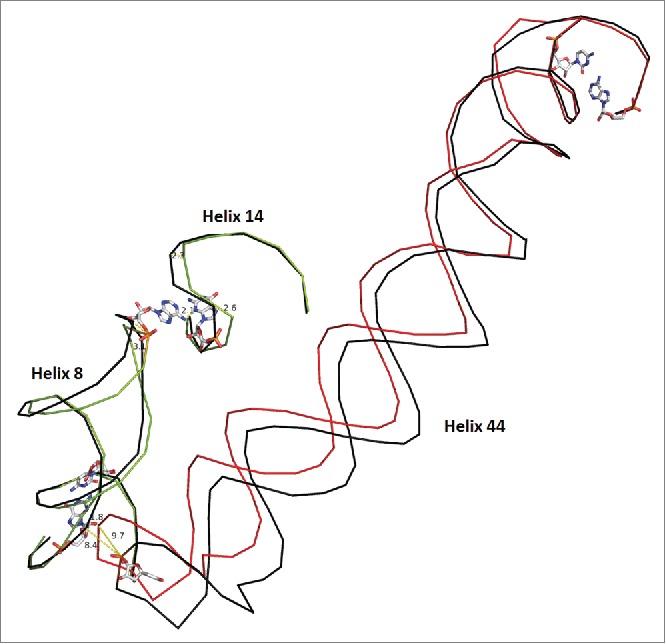
Bridge B8 in the SSU. Helix 44 moves by ∼10 Angstroms in the EF-Tu bound ribosome vs. the unbound molecule. The unbound state is shown in black for each helix with the bound state in a unique color for each protein. Pivoting bases are shown as stick models. Helix h44 contacts h8 and induces a 3.0 Angstrom change in the final residue of h8 which moves 2.1 Angstroms closer to h14. Although h14 is a relatively small helix, it exhibits some conformational freedom, as a result. Helix 10, in proximity to the pivots, displays minor motion and connects h17 to bridge B8 as shown in supplemental Figs. S5 and S6. Overall, the scheme shows a connected network of interactions, which explain partially, the structural consequence of EF-Tu ribosome association. Structure 2J00 is in black and blue. Structure 4ABR is in red and green)
Helix h44 moves by ˜10 Angstroms in the EF-Tu bound ribosome vs. the unbound molecule. h44 contacts h8 and induces a 3.0 Angstrom change in the final residue of h8 which moves 2.1 Angstroms closer to h14 as shown in Fig. 3. As a consequence h14 and h17 now show motion that was not seen in EF-G structures.
In addition to its contact with h8, h44 also contacts the base of h28, which is thought responsible for the majority of the head swivel motion.15,22 Alignment of h28 and h32 in the small subunit results in motion of helix h33, which moves with respect to alignment at both helices in all 4 structures. This is expected as the result of mobility of the B1a bridge-(ASF, S13, S19), which forms contacts with helix 33.31 Head swiveling of approximately 4 Angstroms is seen after alignment of pre GTP hydrolysis EF-Tu bound ribosomes. The swiveling motion is not as robust as that seen in the GTP hydrolyzed EF-G bound ribosome, but is still significant. This is perhaps surprising as only cofactor EF-G is heavily associated with the propagation of intersubunit ratcheting and consequently head swiveling motions.3,22
Conclusions
A network of flexible rRNA interactions described earlier22 proceeds through a series of motions when associated with elongation factors EF-Tu and EF-G. The two cofactors may be producing unique interaction sets. The EF-Tu set involves intersubunit bridge b8 in place of helix H69 of the LSU. Ratcheting and head swiveling motions are activated as a response to EF-G GTP hydrolysis but are weakly activated as a response to EF-Tu binding in agreement with earlier findings.22 Another point of interest is EF-Tu's potential competition with the SRP protein which binds helix H59 in the large subunit, an interaction missing from the EF-G protein. Overall, the results highlight the importance of weak sites in RNA structures in providing function and flexibility. This is likely to occur in other natural or synthetic RNAs. Indeed, the presence of a bulge or non-standard pair is very likely to be a site of significant flexibility in any RNA.
Materials and methods
The PyMOL Molecular Graphics System, Version 1.7.6.2 Schrodinger, LLC (https://www.pymol.org/) was used to measure differences in crystal structures of ribosomal subunits, which are bound and unbound to EF-Tu. All structures were obtained from the PDB32 (http://www.rcsb.org).The pivoting elements were identified by a global structural superposition followed by a local superposition as detailed previously.22 Local sequences, which retained the greatest change after the global alignment, were then manually selected and aligned at the chosen stem sequences. In brief, the local superposition, the ‘align’ command in PyMOL was used on rigid “stem” sequences of mobile rRNA using available scripts.33 Mobility here is defined as large scale motion (at least 2.5 Angstroms) post local alignment of cofactor bound ribosomes to cofactor unbound ribosomes. The cut off range for a pivot is thus 2.5 Angstroms, deemed reasonable against the highest resolution structures compared, All “major pivots” mentioned herein meet this requirement. This method forces a minimal average root mean square distance between all atoms of the stem sequence. Measured motion at the end of the helix is the result of the stem alignment and consequent change at the pivoting position. Single Watson–Crick matches were found suitable as alignments stems as they would yield the superposition of at least 30 atoms- enough to generate reproducible directionality. Though measurements made using this method are relative (choice of alignment affects the magnitude of motion somewhat) the process consistently highlights elements shown to be mobile in experimental studies. Further, because only local stem alignments are used for measurement, observed changes are separated from global conformational changes of the ribosome. However, some flexible helices such as h34 and H68 are not readily detectable because no meaningful stem sequence is adjacent to the pivoting element.
A series of structures were compared in T. thermophilus using structures 2J01, 2J02 (4V51)23 and 2WDI, 2WDG (4V5C)34 as references. The global alignment of the standard structure sets showed an RMSD of 0.432 for the 16S rRNA and 0.345 for the 23S rRNA after removal of all non-rRNA structures. Standard structures 2J01 and 2J02 were first compared against EF-G bound structure pairs 4JUW, 4JUX (4V9H).16 In this case, the RMSD values were 1.951 for the 16S rRNA and 0.911 for the 23S rRNA far exceeding the background cutoff values as did all the other comparisons undertaken. PDB files 2WRI, 2WRJ (4V5F) were also compared.17 Finally, standard structures 2J00, 2J01 were compared against 4 EF-Tu bound structures 2WRN, 2WRO (4V5G), 4ABR, 4ABS (4V8Q), 2XQD, 2XQE (4V5L), and 2Y10, 2Y11 (4V5R). 18-21 The structure from PDB set 4V5L is trapped in in a state prior to GTP hydrolysis, while structures 4V5R and 4V8Q are trapped in a post-GTP hydrolysis state. Although structure 4V5G is described as immediately after GTP hydrolysis, the EF-Tu domain conformation is thought to be similar to the pre-GTP hydrolysis conformation.18 Results from this structure were thus averaged with those of structure 4V5L.
A potential problem with the method is areas of disorder in the compared crystal structures. Large B-factor regions may very well yield false positives in the identification of mobile rRNAs. However, high B value areas were not discounted as erroneous, because flexible RNA is likely to yield crystals which are inherently disordered. To address this issue, published crystal structures, produced by various crystallization protocols were compared as described above. These consistently showed similar mobility as a result of cofactor binding, and in full agreement with published literature. Finally, it should be noted that the set of pivots is a minimal set representing the major points of flexibility. To this end, an average minimal motion of 2.5 Angstroms in the 4 comparisons considered was required for a pivot to be considered significant. Borderline cases likely exist such as h10 and h42 which were considered here and a change in the criterion would therefore reveal additional prospects.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by the National Aeronautics and Space Administration Astrobiology Center for Ribosome Evolution and Adaptation at the Georgia Institute of Technology (NNA09DA78A) and National Aeronautics and Space Administration Grants NNX14AK36G and NNX14AK16G to GEF.
References
- 1.Frank J, Gonzalez RL Jr. Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem 2010; 79:381-412; PMID:20235828; http://dx.doi.org/ 10.1146/annurev-biochem-060408-173330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helgstrand M, Mandava CS, Mulder FAA, Liljas A, Sanyal S, Akke M. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J Mol Biol 2007; 365:468-79; PMID:17070545; http://dx.doi.org/ 10.1016/j.jmb.2006.10.025 [DOI] [PubMed] [Google Scholar]
- 3.Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science 2013; 340:1235970; PMID:23812721; http://dx.doi.org/ 10.1126/science.1235970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krab IM, Parmeggiani A. Mechanisms of EF-Tu, a pioneer GTPase. Prog Nucleic Acid Res Mol Biol 2002; 71:513-51; PMID:12102560; http://dx.doi.org/ 10.1016/S0079-6603(02)71050-7 [DOI] [PubMed] [Google Scholar]
- 5.Noller HF, Yusupov MM, Yusupova GZ, Baucom A, Cate JH. Translocation of tRNA during protein synthesis. FEBS Lett 2002; 514:11-6; PMID:11904173; http://dx.doi.org/ 10.1016/S0014-5793(02)02327-X [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science 2009; 325:1014-17; PMID:19696352; http://dx.doi.org/ 10.1126/science.1175275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitford PC, Blanchard SC, Cate JH, Sanbonmatsu KY. Connecting the kinetics and energy landscape of tRNA translocation on the ribosome. PLoS Comput Biol 2013; 9:e1003003; PMID:23555233; http://dx.doi.org/ 10.1371/journal.pcbi.1003003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature, 1974; 250:546-51; PMID:4602655; http://dx.doi.org/7029310 10.1038/250546a0 [DOI] [PubMed] [Google Scholar]
- 9.Harvey SC, McCammon JA. Intramolecular flexibility in phenylalanine transfer RNA. Nature 1981; 294:286-7; PMID:7029310; http://dx.doi.org/ 10.1038/294286a0 [DOI] [PubMed] [Google Scholar]
- 10.Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 2003; 10:899-906; PMID:14566331; http://dx.doi.org/ 10.1038/nsb1003 [DOI] [PubMed] [Google Scholar]
- 11.Jin H, Kelley AC, Ramakrishnan V. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci U S A 2011; 108:15798-803; PMID:21903932; http://dx.doi.org/ 10.1073/pnas.1112185108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, et al.. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell 2003; 113:789-801; PMID:12809609; http://dx.doi.org/ 10.1016/S0092-8674(03)00427-6 [DOI] [PubMed] [Google Scholar]
- 13.Whitford PC, Sanbonmatsu KY. Simulating movement of tRNA through the ribosome during hybrid-state formation. J Chem Phys 2013; 139:121919; PMID:24089731; http://dx.doi.org/ 10.1063/1.4817212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanbonmatsu KY. Computational studies of molecular machines: the ribosome. Curr Opin Struct Biol 2012; 22:168-74; PMID:22336622; http://dx.doi.org/ 10.1016/j.sbi.2012.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohan S, Donohue JP, Noller HF. Molecular mechanics of 30S subunit head rotation. Proc Natl Acad Sci U S A 2014; 111:13325-30; PMID:25187561; http://dx.doi.org/ 10.1073/pnas.1413731111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 2013; 340:1235490; PMID:23812720; http://dx.doi.org/ 10.1126/science.1235490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 2009; 326:694-9; PMID:19833919; http://dx.doi.org/ 10.1126/science.1179709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV 4th, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. 2009 Science 2009; 326:688-94; PMID:19833920; http://dx.doi.org/ 10.1126/science.1179700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science 2010; 330:835-8; PMID:21051640; http://dx.doi.org/ 10.1126/science.1194460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmeing TM, Voorhees RM, Kelley AC, Ramakrishnan V. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat Struct Mol Biol 2011; 18:432-6; PMID:21378964; http://dx.doi.org/ 10.1038/nsmb.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neubauer C, Gillet R, Kelley AC, Ramakrishnan V. Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science 2012; 335:1366-9; PMID:22422985; http://dx.doi.org/ 10.1126/science.1217039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paci M, Fox GE. Major centers of motion in the large ribosomal RNAs. Nuc Acids Res 2015; 43:4640-9; PMID:25870411; http://dx.doi.org/16959973 10.1093/nar/gkv289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006; 313:1935-42; PMID:16959973; http://dx.doi.org/ 10.1126/science.1131127 [DOI] [PubMed] [Google Scholar]
- 24.Sergiev PR, Bogdanov AA, Dontsova OA. How can elongation factors EF-G and EF-Tu discriminate the functional state of the ribosome using the same binding site? FEBS Lett 2005; 579:5439-42; PMID:16213500; http://dx.doi.org/ 10.1016/j.febslet.2005.09.010 [DOI] [PubMed] [Google Scholar]
- 25.Lancaster L, Lambert NJ, Maklan EJ, Horan LH, Noller HF. The sarcin–ricin loop of 23S rRNA is essential for assembly of the functional core of the 50S ribosomal subunit. RNA 2008; 14:1999-2012; PMID:18755834; http://dx.doi.org/ 10.1261/rna.1202108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Susorov D, Mikhailova T, Ivanov A, Sokolova E, Alkalaeva E. Stabilization of eukaryotic ribosomal termination complexes by deacylated tRNA. Nucl Acids Res 2015; 43:3332-3343; PMID:25753665; http://dx.doi.org/ 10.1093/nar/gkv171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sergiev PV, Lesnyak DV, Kiparisov SV, Burakovsky DE, Leonov AA, Bogdanov AA, Brimacombe R, Dontsova OA, Function of the ribosomal E-site: a mutagenesis study. Nucl Acids Res 2005; 33:6048-56; PMID:16243787; http://dx.doi.org/ 10.1093/nar/gki910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maguire BA, Wild DG. The roles of proteins L28 and L33 in the assembly and function of Escherichia coli ribosomes in vivo. Mol Microbiol 1997; 23:237-45; PMID:9044258; http://dx.doi.org/ 10.1046/j.1365-2958.1997.2131578.x [DOI] [PubMed] [Google Scholar]
- 29.Halic M, Becker T, Pool MR, Spahn CM, Robert A. Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 2004; 427:808-814; PMID:14985753; http://dx.doi.org/ 10.1038/nature02342 [DOI] [PubMed] [Google Scholar]
- 30.Kempf G, Klemens W, Sinning I. Structure of the complete bacterial SRP Alu domain. Nucl Acids Res 2014; 42:12284-94; PMID:25270875; http://dx.doi.org/ 10.1093/nar/gku883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kietrys AM, Szopa A, Bakowska-Zywicka K. Structure and function of intersubunit bridges in prokaryotic ribosome. Biotechnologia 2009; 1:48-58 [Google Scholar]
- 32.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN. Bourne PE. The protein data bank. Nucleic Acids Res 2000; 28:235-42; PMID:10592235; http://dx.doi.org/ 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. http://apt-browse.org/browse/ubuntu/trusty/universe/amd64/pymol/1.7.0.0-1/file/usr/share/pymol/scripts/metaphorics/alignment.pml [Google Scholar]
- 34.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 2009; 16:528-33; PMID:19363482; http://dx.doi.org/ 10.1038/nsmb.1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


