ABSTRACT
Last finding indicates that post-transcriptional processes are significant in low-oxygen conditions, but their nature is poorly understood. Here, we localized poly(A) RNA and mRNA coding proteins involved and not involved with resistance to hypoxia in Lupinus luteus and Arabidopsis thaliana during submergence and after recovery of aerobic conditions. We showed a strong nuclear accumulation of poly(A) RNA and 6 of 7 studied mRNAs with a concurrent strong reduction in RNA polymerase II transcription during hypoxia. In this study, the nucleus did not accumulate mRNA of the ADH1 (alcohol dehydrogenase 1) gene, which is a core hypoxia gene. The RNA accumulation in the nucleus is among the mechanisms of post-transcriptional gene regulation that prevents translation. However re-aeration was accompanied by a strong increase in the amount of the mRNAs in the cytoplasm and a simultaneous decrease in nuclear mRNAs. This finding indicates that the nucleus is a storage site for those of mRNAs which are not involved in the response to hypoxia for use by the plants after the hypoxic stress. In this study, the highest intensity of RNA accumulation occurred in Cajal bodies (CBs); the intensity of accumulation was inversely correlated with transcription. Under hypoxia, ncb-1 mutants of Arabidopsis thaliana with a complete absence of CBs died sooner than wild type (WT), accompanied by a strong reduction in the level of poly(A) RNA in the nucleus. These results suggest that the CBs not only participate in the storage of the nuclear RNA, but they also could take part in its stabilization under low-oxygen conditions.
KEYWORDS: Cajal bodies, hypoxia, mRNA, post-transcriptional gene regulation, retention of poly(A) RNA
Introduction
The second half of the 20th century saw a significant rise in the number and severity of floods in all continents, affecting crop yields and accordingly the general condition of the agricultural sector in those areas.1 The damage to terrestrial plants during flooding results from hypoxia/anoxia, as the diffusion of O2, CO2 and ethylene in water is nearly 104 times lower than in air.2 The decline in O2 availability reduces the energy produced by plants, as ATP synthesis is less efficient during anaerobic respiration. Under hypoxic conditions, plant cell survival is determined by the levels of carbohydrate available for oxidative phosphorylation. Plants can increase their carbohydrate levels by starch catabolism and glycolysis. Typically, the increase in glycolytic flux is coupled with the regeneration of NAD+ by the fermentation of pyruvate to ethanol.3 Long-term, however, the ethanol produced has damaging consequences for cell integrity and survival.4 In addition, ethanol diffuses out of cells, depleting their carbon reserves. Therefore, many plants have developed an alternative response to low-oxygen conditions: the production of alanine from pyruvate by alanine aminotransferase (AlaAT).5,6 This process includes the generation of 2-oxoglutarate as a co-product that can be further metabolized to succinate via the TCA cycle, thereby providing additional ATP per molecule of sucrose metabolized.7
Plants are able to respond quickly to reduced levels of oxygen in the environment. In cotton, Arabidopsis and poplar, 84 homologous genes are known to be up-regulated and 38 down-regulated in the early response to hypoxia.8 Genes with increased transcription levels participate in anaerobic fermentation, sucrose transport, and reactive-oxygen species regulation, and include ethylene response factor-associated genes. Down-regulation is observed for genes involved in cell wall synthesis and genes that participate in signaling or secondary metabolism. Despite the conservative character of the hypoxia deregulation genes, half of the low-oxygen-induced genes of Arabidopsis encode PUFs (proteins of unknown function). Eleven of the 16 homozygous Arabidopsis mutants of PUFs significantly modify the tolerance to prolonged oxygen deprivation.9,10
Low O2 conditions may result in a strong inhibition of protein synthesis by restricting ribosome association with the start codon of mRNA.11 Branco-Price et al.12 show that this restriction applies to approximately 70% of cellular mRNA. This reduction in energy-consuming protein synthesis saves ATP, which is reduced by 50% after two hours of hypoxia. The translationally repressed mRNAs are highly enriched for proteins associated with translation, including 138 of 219 cytosolic ribosomal proteins.11 Of the 16% of the genes transcripts increased in a cell exposed to 9 h of hypoxic stress, only 4% underwent increased translation. A decrease in translation without a concurrent decrease in the number of transcripts occurred in approximately 50%.12 The presence in the cytoplasm of stress granules (SG) rich in poly(A) RNA, might be the cellular expression of the inhibition of protein synthesis. The stress granules did not exhibit the presence of the translation initiation factors eIF2a; however, the presence of the 40S ribosomal subunit was shown in some of them.13,14 Recently, it has been shown that the UBP1C protein, appearing in SG, binds mRNAs with non-U-rich 3 UTRs. Only a small amount of the transcript gene AHD1 (core hypoxia gene) is related to UBP1C. Mutation or downregulation of UBP1C interferes with seedling establishment and reduces survival because of low-oxygen stress.15
After re-oxygenation, a significant increase of the process of translation was observed, which had no reflection in the change of the transcriptome. The substantial lack of correlation between the changes among particular mRNAs within the total amount of the mRNAs and the mRNA connected with ribosomes, indicates the participation of post-transcriptional regulation of gene expression (PTGR) during hypoxia and re-aeration.16,17,18
Despite advancement in the molecular study of hypoxia, processes such as alternative splicing, edition or interference have not yet been fully understood. It is known that hypoxia increases intron retention events in a number of mRNAs. These events occurred in a number of the mRNAs encoding the splicing machinery. For example, there was enhanced retention of intron 6 of the small nuclear RNP 70K (U1-70K), which led to the production of a truncated and nonfunctional protein. There are also examples of stress-regulated intron skipping. Notably, it also has been shown that 5% of the mRNAs associated with ribosomes contain introns.11
PTGR also includes localization, stabilization or storage of mRNA. However, due to the lack of sufficient research regarding the spatial regulation of gene expression during hypoxia, except for stress granules (SGs), these processes require explanation. We have recently proven the presence of poly(A) RNA in Cajal bodies (CBs), also during hypoxia.19 However, the role of these structures in the metabolism of RNA during the process of hypoxia in plants remains unknown. Cajal bodies are multifunctional domains present within the nuclei of plant and animal cells involved in maturation of different types of RNA. The best known function of CBs is their participation in the process of snRNP biogenesis. This process includes the assembly of snRNP-specific proteins, snRNA methylation and pseudouridylation by scaRNPs, and formation of a complex consisting of U4, U5, and U6 snRNPs.20,21 Thus, the marker for Cajal bodies in plants, except for coilin, are proteins associated with snRNA, such as Sm or U2B’.22,23,24
Accordingly, in this work, we determined the subcellular localisations and analyzed the quantity of poly(A) RNA, along with a few transcripts of protein coding genes, in cells before and during hypoxia and after the removal of stress conditions in Lupinus luteus and Arabidopsis thaliana. We also established the levels of the elongation form of RNA polymerase II and the distribution of splicing factors. The results showed a considerable response of the investigated molecules to hypoxia and during recovery from stress. Lupine is a fodder plant grown in Europe. It is extremely convenient for microscopy research because of the much larger nuclei and Cajal bodies compared with A. thaliana.
Results
Poly(A) RNA, including coding proteins, accumulates in the nucleus and Cajal bodies during hypoxia
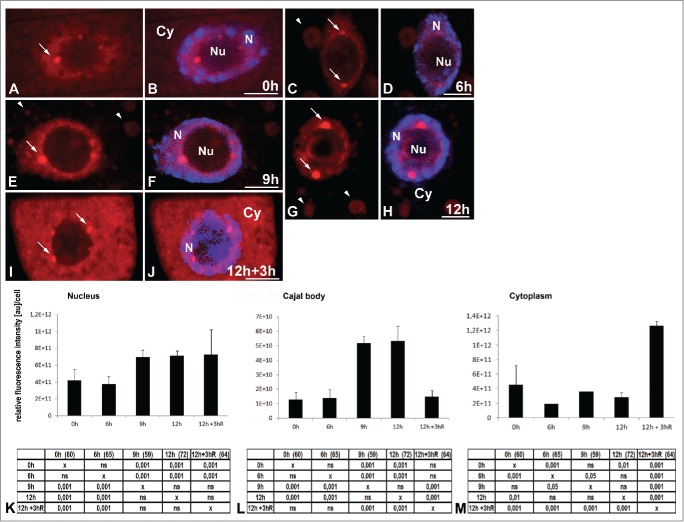
The localization of poly(A) RNA revealed subcellular changes in the distribution and amount of the studied signal in the meristematic root cells of Lupinus luteus under normoxia conditions and during the successive hours of hypoxia (Fig. 1A–M) induced by submerging the seedlings in tap water in the photoperiod condition. In the root cells of lupine under normoxia (at 0 h), the amount of poly(A) RNA in the cytoplasm was similar to the nuclear level (Fig. 1A, B, K, M). In the pictures, the concentration of the signal in the cytoplasm is lower than it is in the nucleus. This is because the cytoplasm volume is larger than that of the cell nucleus. Experimental hypoxia resulted in a strong accumulation of poly(A) RNA in the cell nucleus and a reduction in the cytoplasm (Fig. 1C–H, K, M). The highest level of poly(A) RNA in the nucleus occurred after 12 hours of hypoxia (Fig. 1K). The experiment was not continued because after 15 hours of hypoxia, the seedlings that were transferred back to normoxia exhibited a death rate above 50%. In the nucleus in addition to the homogeneous pool, poly(A) RNA occurred in circular clusters near the nucleolus, during all periods studied (Fig. 1A–J). These nuclear structures rich in poly(A) RNA in the root cells were identified as Cajal bodies in our previous paper.19 We conducted co-localization of the highly conserved eukaryotes core spliceosomal Sm-proteins and poly(A) RNA in the roots after the hypoxia treatment. All the rich poly(A) RNA round clusters were co-localized with the Sm-proteins, which means that they are Cajal bodies (Fig. 2A–I). The amount of poly(A) RNA in the Cajal bodies increased more intensively than it did in the nucleus during hypoxia. At 12 h of hypoxia, we observed an approximately 1.6 times higher nuclear level of poly(A) RNA (Fig. 1K) compared to normoxia, while in the CBs, it had increased by 4.2 times (Fig. 1L). The increase in the quantity of poly(A) RNA correlated with the increase in the diameter of the Cajal bodies (Fig. S1A). In the cytoplasm, the poly(A) RNA level had already decreased after 6 h of hypoxia (Fig. 1 M), accompanied by the appearance of structures similar to stress granules (SGs) (Fig. 1C–H). These poly(A) RNA-rich structures did not co-localize with the 26S rRNA (Fig. 2J–O). Three hours of re-aeration following the 12 hours of hypoxic stress produced a decrease in poly(A) RNA and size of the CBs to the level present before hypoxia (Fig. 1L, Fig. S1A). The SG-like structures disappeared (Fig. 1J), and cytoplasmic poly(A) RNA increased more than three times compared to the levels observed in hypoxia (at 12 h) (Fig. 1M).
Figure 1.
Localization of poly(A) RNA using Cy3(dT) 30 probe in Lupinus luteus meristematic root cells before stress (0 h) (A,B) and after 6 h (C, D), 9 h (E, F), and 12 h (G, H) hypoxia. Poly(A) RNA accumulates in the nucleus during successive hours of hypoxia. In the nuclei, the signal appears in the nuclear bodies (arrows). Poly(A) RNA signal in the cytoplasm decreases, and stress granules like structures form (arrowheads). After 3 h of re-aeration (I, J), the signal reappears in the cytoplasm, and stress granules are not observed. Merging with DAPI staining (B, D, F, H, J). Bar, 10 µm. N- nucleus, Nu- nucleolus, Cy- cytoplasm. Quantitative analysis of poly(A) RNA in the nucleus (K), Cajal body (L), cytoplasm (M) under normoxia, hypoxia and re-aeration conditions. The tables under each plot present the significant differences between groups, with testing probability below the value shown in the table (P<), α = 0.05. In the column heading, the number of analyzed cells in each stage is presented in brackets. ns - nonsignificant differences between groups.
Figure 2.
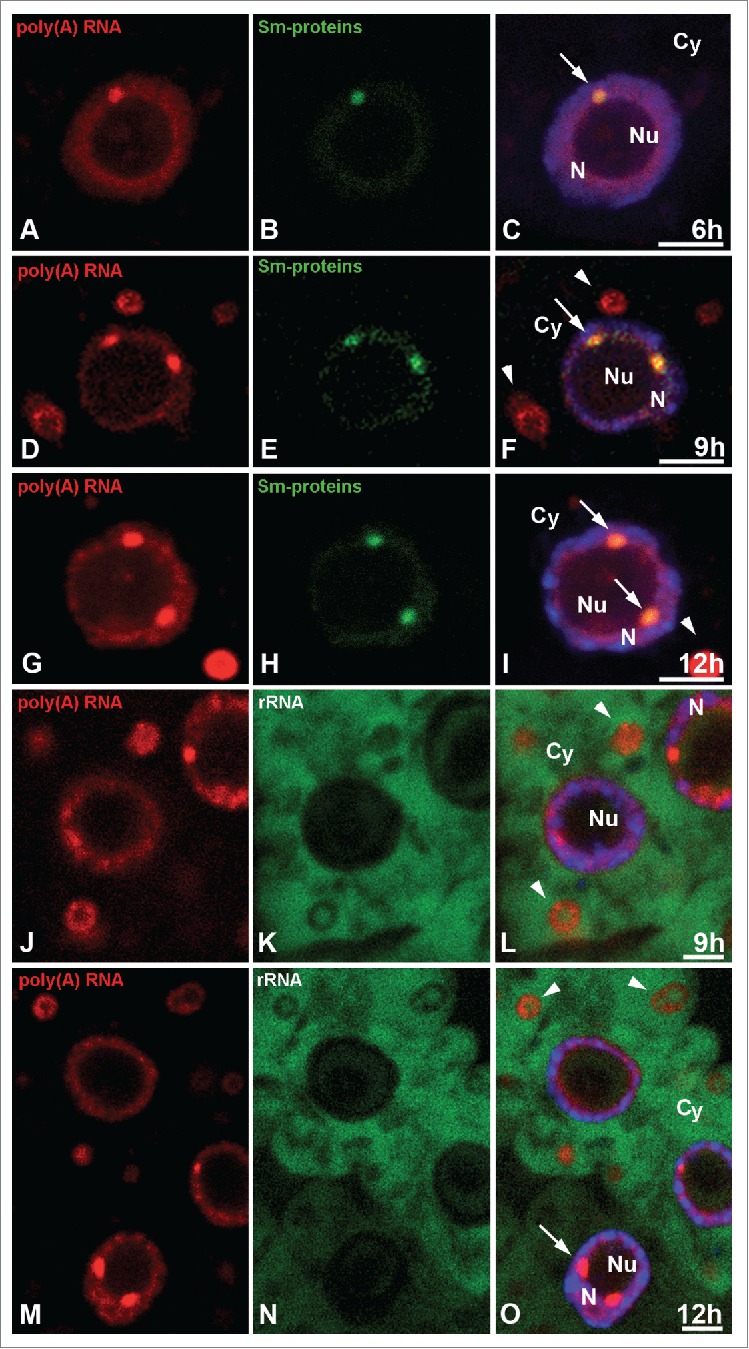
Double labeling of poly(A) RNA (A, D, G) and Sm proteins (B, E, H) in Lupinus luteus meristematic roots after 6 h (A–C), 9 h (D F), and 12 h (G–I) of hypoxia. Strong colocalization of poly(A) RNA in nuclear bodies rich in Sm proteins (arrows) but not in the stress granules (arrowhead). Double in situ hybridization in the 9 h (J, K, L) and 12 h (M, N, O) stress treatment cells of poly(A) RNA (J, M) and 26S rRNA (K, N) indicates that these two RNAs do not colocalize in the stress granules (arrowheads). Merging with DAPI staining (C, F, I, L, O). Bar, 10 µm. N- nucleus, Nu- nucleolus, Cy- cytoplasm, arrows- Cajal bodies.
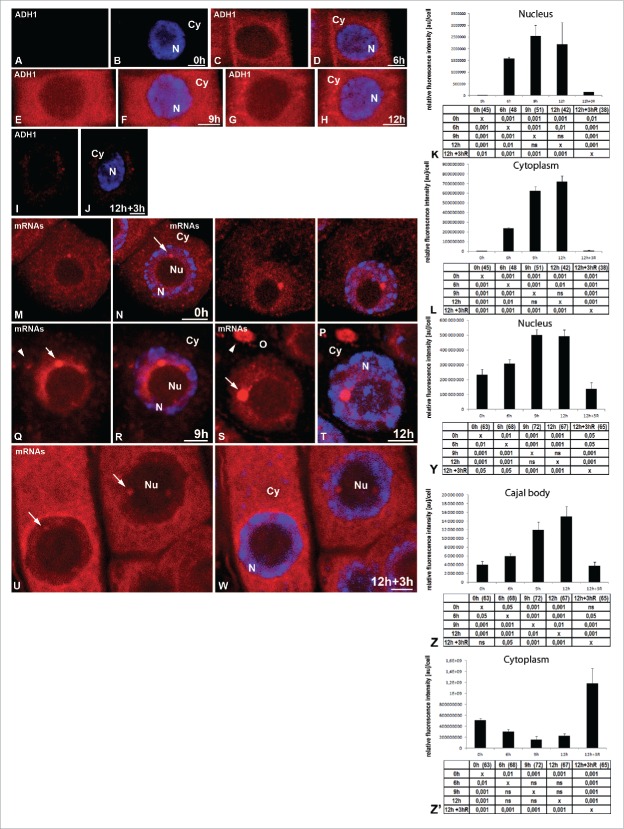
Next, we determined the pattern of the distribution of the RNA coding proteins in the cells under hypoxia and after re-aeration. The mRNA sequences of three housekeeping genes were chosen for this analysis: a cytokinin-specific binding protein, cyclin B1 and pectin methylesterase, which are not involved in the response to hypoxia, and an ortholog of ADH1 (alcohol dehydrogenase 1), which is the core gene involved in the hypoxia stress response. The amount of mRNA ADH1 ortholog was presented at a very low level in the control cells. A strong increase of transcripts during the hours of hypoxia that followed was observed (Fig. 3A–H, K, L), and this confirms hypoxia of the lupin seedlings submerged in tap water. The increase in the amount of the transcript was mainly observed in the cytoplasm (Fig. 3C–H, L). As for the nucleus, there was only a weak increase, approximately 85 times lower than in the cytoplasm, at the 12 h hypoxia treatment, which did not resemble the accumulation observed after the localization of poly(A) RNA. The clusters were not observed in the nucleus. The signal decreased strongly in the cytoplasm and nucleus after three hours of re-aeration (Fig. 3I–L).
Figure 3.
Localization of mRNA ortholog of ADH1. The roots of Lupinus luteus were imaged before (A, B) and after 6 h (C, D), 9 h (E, F), and 12 h (G, H) submerged and after 3 h reoxygenation (I, J). The increase amount of transcript was mainly observed in the cytoplasm under hypoxia condition (C–H). Quantitative analysis of ADH1 in the nucleus (K) and cytoplasm (L) of meristematic roots cells under normoxia, hypoxia and re-aeration conditions. Localization of mixture of three mRNAs (cyclin B1, cytokinin-specific binding protein mRNA and pectin methylesterase) (M, O, Q, S, U) in Lupinus luteus meristematic root cells before stress (0 h) (M, N), after 6 h (O, P), 9 h (Q, R), and 12 h (S, T) hypoxia and after reoxygenation (U, W). Merging with DAPI staining (B, D, F, H, J, N, P, R, T, W). Bar, 10 µm. N- nucleus, Nu- nucleolus, Cy- cytoplasm, arrowhead- stress granules. Histogram showing quantitative analysis of mixture of three mRNAs from all studied stages: Y- nucleus, Z- Cajal body, Z’- cytoplasm. The tables under each plot present the significant differences between groups, with testing probability below the value shown in the table (P<), α = 0.05. In the column heading, the number of analyzed cells in each stage is presented in brackets. ns - nonsignificant differences between groups.
The distribution was similar for the three transcripts that were not responsive to hypoxia stress, but the individual signals were weak in the cells exposed to hypoxic stress. Therefore, we conducted in situ hybridization with a mix of three probes. The localization pattern of mRNAs encoding proteins (Fig. 3M–W) was similar to the distribution of poly(A) RNAs. The accumulation in the nucleus was recorded (Fig. 3Y). Simultaneous localization of the core spliceosomal Sm proteins and three mRNAs in all studied stages revealed that the CBs were also the structures that housed mRNAs encoding proteins (Fig. S1B-M). The amount of mRNA in the CBs increased during the successive hours of hypoxia (Fig. 3Z). Isolated cytoplasmic structures resembling SGs rich in mRNAs were also observed (Fig. 3Q–T). After a 3 h re-aeration, the quantity of transcripts in the cytoplasm increased significantly (Fig. 3U, W, Z’) and was accompanied by a decrease in the amount of mRNAs of the three genes in the nucleus (Fig. 3U, W, Y). Pearson's correlation analysis showed a very high correlation between these events for mRNAs (r= −0.83). After removal of the stress, the amount of mRNA in the CBs returned to pre-hypoxia level (Fig. 3 U, W, Z). This means that among the poly(A) RNA accumulated in the nucleus, there are also those responsible for encoding proteins. The mRNA not involved in the response to hypoxia is transported to the cytoplasm after removal of the stress, whereas the amount of mRNA ADH1 increased during hypoxia is immediately transported to the cytoplasm after transcription.
Transcription reduction during hypoxia
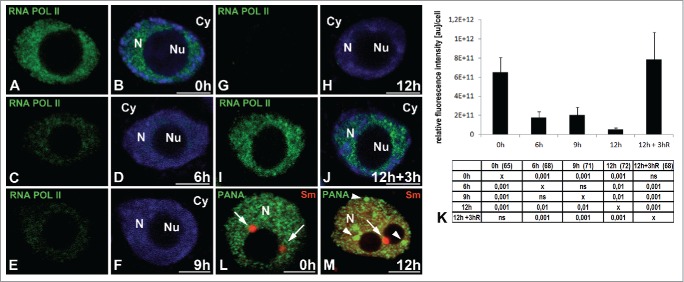
We then examined whether the hypoxia-induced accumulation of poly(A) RNA (including mRNAs) in the cell nucleus was due to intensified transcription. For this purpose, we localized the elongation form of RNA polymerase II with a phosphorylated serine 2 in the CTD domain (Fig. 4A–J). In the nuclei of cells under normoxia, RNA POL II was present homogeneously and at high level in the entire nucleoplasm but not in the nucleolus nor the cytoplasm (Fig. 4A, B). We observed a strong decrease in RNA POL II EF during hypoxia (Fig. 4K). At 6 h of hypoxia (Fig. 4C, D), the fluorescence had decreased more than 3.5 times, while at 12 h (Fig. 4G, H), it had decreased 11 times compared to the period before hypoxia (Fig. 4K). The end of hypoxic stress resulted in an increase in transcription back to normoxia levels (Fig. 4I, J, K). We also localized the PANA antigen, a marker for speckles because cells treated with inhibitors of transcription accumulate splicing factors and enlarge speckles.23,25 The PANA antigen is a component of lupin interchromatin granules representing the splicing speckles observed under the light microscope.23 In the lupine cells, a change in PANA antigen was observed at 12 h hypoxia compared to the control cells, i.e., during the period of the strongest reduction in the amount of the elongated form of RNA POL II. At this stage, in the nucleus, there were many enlarged round speckles that did not co-localize with CBs labeled with Sm antibodies (Fig. 4L, M). The decreasing amount of RNA polymerase II, and the changes in the morphology of the speckles, indicated that a reduction of transcription occurs during hypoxia. Thus, we checked whether only the inhibition of transcription could be responsible for the accumulation of poly (A) RNA in the nucleus. We measured the amount of poly(A) RNA in the nuclei of the root cells treated with transcription inhibitors (actinomicin D, α-amanitin). In the meristematic cells of lupine after using both of the transcription inhibitors, we observed a strong decrease of the poly(A) RNA level in comparison to both the control and the hypoxia-stressed cells (Fig. S1N). In the nucleus, poly(A) RNA occurred as a weak homogeneous pool without clusters, similar to the CBs (Fig. S1Q, R). During hypoxia, there was a strong reduction of the process of transcription; however, this sole phenomenon is not sufficient to account for the accumulation of poly(A) RNA in the nucleus.
Figure 4.
Localization of RNA polymerase II with a phosphorylated serine 2 in the CTD domain (elongation form). The roots of Lupinus luteus were imaged before (A, B) and after 6 h (C, D), 9 h (E, F), and 12 h (G, H) submerged and after 3 h reoxygenation (I, J). Decreased signal was observed in successive hours of hypoxia (C–H). After reoxygenation, the signal reappears in the nucleus (I, J). Quantitative analysis of RNA polymerase II elongation form in the nucleus (K) of meristematic root cells under normoxia, hypoxia and re-aeration conditions. The tables under plot present the significant differences between groups, with testing probability below the value shown in the table (P<), α = 0.05. In the column heading, the number of analyzed cells in each stage is presented in brackets. ns - nonsignificant differences between groups. Double immunolocalization in the nuclei cells before (L) and after 12 h hypoxia (M) of PANA antigen (green) and Sm proteins (red). The few large round speckles (arrowheads) not colocalized with Cajal bodies (arrows) occurred in the nucleus after 12 h hypoxia (M). Merging with DAPI staining (B, D, F, H, J). N- nucleus, Nu- nucleolus, Cy- cytoplasm.
The role of the Cajal bodies in the metabolism of poly(A) RNA under hypoxia stress
Because we observed a strong accumulation of poly(A) RNA in Cajal bodies during hypoxia, we attempted to clarify the role of those structures in response to hypoxic stress. For this purpose, we used ncb-1 mutants of Arabidopsis thaliana. Ncb-1 mutants are the only commonly used line with a complete absence of CBs as a result of Atcoilin mutation.26 The 14-day-old seedlings WT and ncb-1 did not display differences in phenotype (Fig. S2A, B). First, differences between ncb-1 and WT plants exposed to hypoxic stress were observed in the morphology and color of leaves after the 4th day. Only 17% of ncb-1 had green leaves, compared to 89% of WT after 5 d of hypoxic stress. The remaining leaves were yellow (Fig. 5A, Fig. S2C, D). This finding indicated that plants without CBs were dying faster under hypoxia than WT plants.
Figure 5.
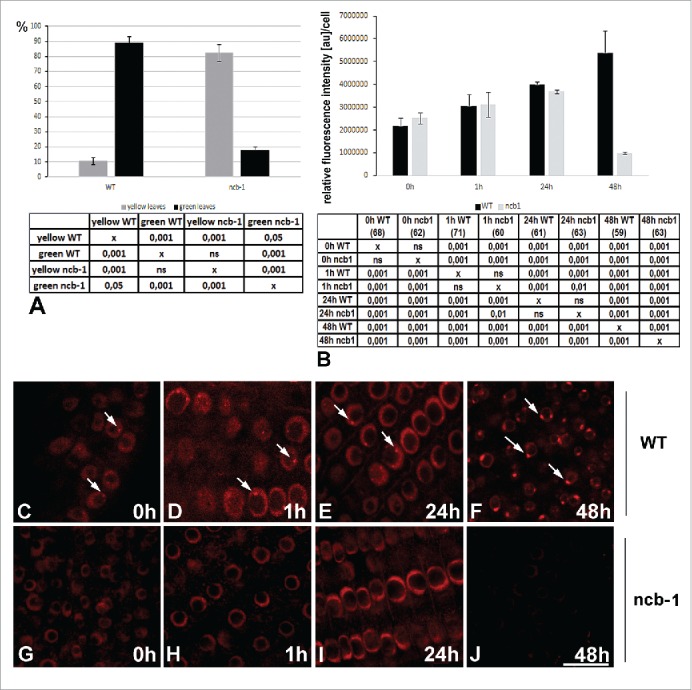
The percentage of Arabidopsis thaliana WT and ncb-1 plants with yellow and green leaves after 5 d of hypoxia (A). The 1602 leaves of 472 WT and 1448 leaves of 424 ncb-1 plants from three different experiments were analyzed. Quantitative analysis of poly(A) RNA in the nuclei of meristematic roots cells of WT and ncb-1 plants (B) under normoxia and after 1 h, 12 h and 48 h hypoxia. Differences in the amount of poly(A) RNA between WT and ncb-1 were observed only after 48 h of stress. The tables under each plot present the significant differences between groups, with testing probability below the value shown in the table (P<), α = 0.05. In the column heading, the number of analyzed cells in each stage is presented in brackets. ns - nonsignificant differences between groups. Localization of poly(A) RNA in root cells WT (C-F) and ncb-1 (G-J) plants under normoxia (C, G) and after 1 h (D, H), 12 h (E, I) and 48 h (F, J) under hypoxia. Arrows indicate Cajal bodies. Bar, 20 µm.
To see if this difference was connected to the participation of CBs in the metabolism of poly(A) RNA, we compared the distribution and amount of poly(A) RNA in WT and ncb-1 plants in normoxia and hypoxia conditions (Fig. 5B–J). In WT plants, similar to lupine, we observed poly(A) RNA accumulated in nuclei and nuclear bodies (Fig. 5B–F). The simultaneous localization of Sm proteins and poly(A) RNA confirmed that they were Cajal bodies (Fig 2E–P). In the roots of plants not exposed to hypoxia, only 2.2% of nuclear poly(A) RNA was present in CBs, increasing to 4.4% at 24 hours of hypoxia and 18% at 48 hours. A decrease in poly(A) RNA was observed in the cytoplasm (Fig S2R). In contrast to WT plants, in ncb-1 mutants, we observed no poly(A) RNA accumulations that could be ascribed to CBs (Fig. 5G–J). However, in the ncb-1 mutants, there was an increase in nuclear poly(A) RNA during hypoxia until 24 h (Fig. 5B, I). At 48 hours, in contrast to WT, there was a significant decrease in poly(A) RNA, and the amount of poly(A) RNA at 48 h of stress was more than five times lower in ncb-1 mutants than in WT plants (Fig. 5B). The CBs increase the tolerance of A. thaliana to hypoxia stress. This may be related to the fact that the CBs affect the level of poly(A) RNA in the cell nucleus during hypoxia.
Different localization of transcripts of ADH1 (alcohol dehydrogenase 1) and the genes that are not responsive to hypoxia
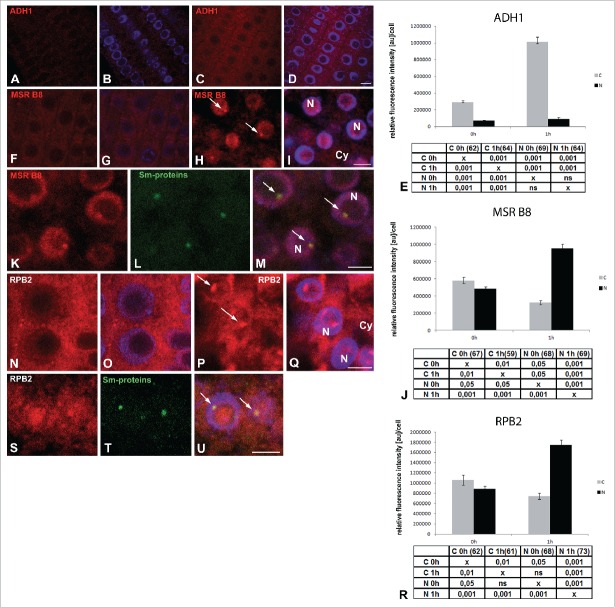
To determine what types of mRNA can be accumulated in the Cajal bodies and nucleus, we decided to conduct in situ hybridization of the transcripts of genes associated with hypoxic stress and housekeeping genes. We localized the transcript of ADH1 (alcohol dehydrogenase 1), the core hypoxia gene. Its amount had increased by more than four times at 1 h of hypoxia (Fig. 6A–D, E). In both normoxia and hypoxia conditions, ADH1 transcripts occurred predominantly in the cytoplasm. In the nuclei, we did not observe clusters resembling nuclear bodies (Fig. 6A–D). We then localized the transcript of the methionine sulfoxide reductase B8 gene (MSR B8) and found it to exhibit nuclear accumulation during hypoxia (Fig. 6F–J). In addition, the mRNA of MSR B8 in 42% of the cells co-localized with the Cajal bodies (Fig. 6K–M). In the cytoplasm, the amount of MSR B8 mRNA decreased (Fig. 6F–J). We also observed nuclear accumulation of the transcripts of RNA polymerase II gene subunit RPB2 (Fig. 6N–R). RPB2 mRNA was present in the Cajal bodies in 36% of the cells (Fig. 6S–U). A similar localization pattern and subcellular change occurred for the transcripts of Sm-like protein (Fig. S3A–E). The results indicate the transcripts that are not responsive to hypoxia accumulated in the nucleus and Cajal bodies.
Figure 6.
Localization of mRNA in Arabidopsis thaliana meristematic root cells. Localization of mRNA of ADH1 before (A, B) and after 1 h hypoxia (C, D). Localization of mRNA of MSR B8 before (F, G) and after 1 h hypoxia (H, I). Double labeling of MSR B8 transcripts (K) and Sm-proteins (L) in meristematic root cells after 1 h hypoxia (K–M). In situ hybridization to mRNA of RPB2 under normoxia (N–O) and after 1 h hypoxia (P–Q). Simultaneous localization of the mRNA of RPB2 (S) and core spliceosomal Sm proteins (T) in cells after 1 h hypoxia revealed strong colocalization in Cajal bodies (S–U) (arrowheads). Merging with DAPI staining (B, D, G, I, M, O, Q, U). Bar, 20 µm. N- nucleus, Cy- cytoplasm, arrows- Cajal bodies. Histogram shows quantitative analysis of ADH1 (E), MSR B8 (J), RPB2 (U) in the nuclei (N) and cytoplasm (C) under normoxia and hypoxia Arabidopsis thaliana the meristematic roots cells. The tables under plot present the significant differences between groups, with testing probability below the value shown in the table (P<), α = 0.05. In the column heading, the number of analyzed cells in each stage is presented in brackets. ns - nonsignificant differences between groups.
The level of U2 snRNA in the WT and ncb-1 mutant during hypoxia
Cajal bodies are known mainly for their snRNP biogenesis.27 Therefore, we wanted to explore whether the faster death of the ncb-1 mutant compared to that of the WT during hypoxia is also associated with the decreased amount of snRNA. A significant decrease in the amount of splicing factors could impact on processing the mRNA encoding proteins participating in the stress response. In normoxia conditions, we found no statistically significant difference in the amount of U2 snRNA between ncb-1 and WT plants (Fig. 7A–E). The only differences were found at 1 h of hypoxia. The amount of U2 snRNA increased significantly in the WT cell nuclei, while no such increase was observed in ncb-1 (Fig. 7A, F-I). In the subsequent hours of stress, the level of U2 snRNA in WTs decreased at 24 h to the amount present in root cells not exposed to hypoxic stress, and at 48 h, it fell to a level below that value (Fig. 7A, J, K, N, O). The percentage of U2 snRNA in the Cajal bodies in relation to total nuclear levels in WT varied slightly between 3.9–4.2% in the root cells under normoxia and at the first two examined stages of hypoxia (1 h, 24 h). On the second day of hypoxia, the value increased to 7.2%. In ncb-1, no growth was observed at 1 h. At 24 h of hypoxic stress, the amount of U2 snRNA increased, and at 48 hours it decreased below normoxic levels (Fig. 7A, N–Q). A similar level of U2 snRNA in the WT and ncb-1 in the later periods of hypoxia suggests that the mutant did not die faster because of an snRNA deficit.
Figure 7.
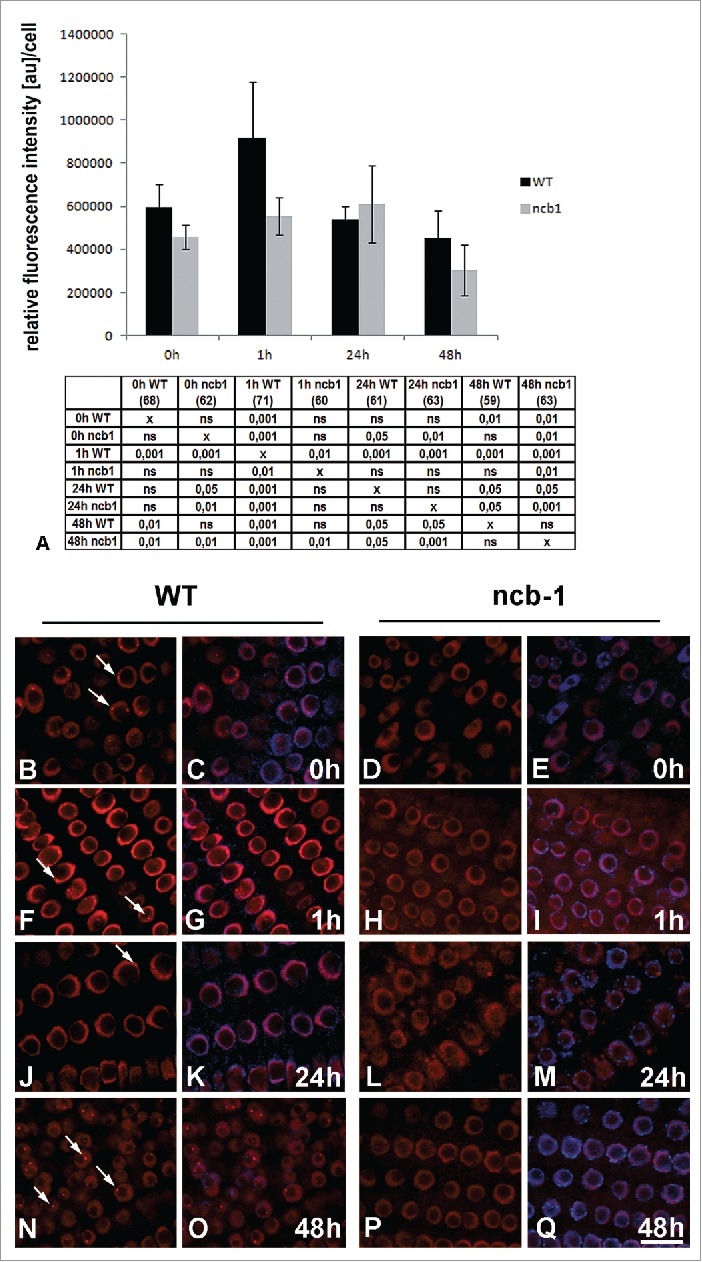
Quantitative analysis of U2 snRNA in Arabidopsis thaliana WT and ncb-1 plant nuclei of the meristematic roots cells under normoxia and hypoxia condition (A). The tables under plot present the significant differences between groups, with testing probability below the value shown in the table (P<), α = 0.05. In the column heading, the number of analyzed cells in each stage is presented in brackets. ns - nonsignificant differences between groups. In column heading, the number of analyzed cells in each stage is presented in brackets. ns - nonsignificant differences between groups. Differences in the amount of U2 snRNA between WT and ncb-1 plants were observed only after 1 h stress. Distribution of U2 snRNA in WT and ncb-1 under normoxia (B–E) and after 1 h (F–I), 12 h (J–M) and 48 h (N–Q) hypoxia. Merging with DAPI staining (C, E, G, I, K, M, O, Q). Bar, 20 µm. Arrows indicate Cajal bodies.
Discussion
We showed dynamic changes in the amount and subcellular location of poly(A) RNA and the mRNA of housekeeping and ADH1 genes in hypoxia stress. The research was conducted in the crop Lupinus luteus due to the easy removal of the stress caused by submergence and the possibility of establishing the sequence of events following recovery. However, due to the low availability of sequences in databases and the lack of mutants of Lupinus luteus, we used A. thaliana in the experiments designed to explain the role of the Cajal bodies.
In both species, the amount of poly(A) RNA (including certain RNAs encoding proteins) in the nucleus increased under hypoxia. In addition, in lupine, the accumulation was accompanied by a strong reduction in RNA polymerase II transcription. However, we know that the inhibition of transcription is not sufficient for the accumulation of poly(A) RNA. Therefore, incomplete or alternative splicing or changes in transport into the cytoplasm might contribute to the accumulation in the nucleus.28,29 For example, nuclei of the fern Marsilea vestita are known to store intron-retaining transcripts during rapid post-transcriptionally controlled spermatogenesis.30 We have also recently observed the accumulation of poly(A) RNA in the cell nucleus following stress induced by removal of the cell wall.31 Together with our observation of the decrease in transcription and increased levels of poly(A) RNA, this finding suggests an enhanced stabilization of RNA that is already present in the cell. Storing a large amount of non-degraded transcript in the nucleus may be one of the causes of the strong decrease in translation in the absence of changes in the amount of mRNA.11,12
Although our research shows a strong accumulation of poly(A) RNA in the cell nucleus, it does not concern all examined transcripts. In A. thaliana and Lupinus luteus, the mRNA of ADH1 did not stored in the cell nucleus during hypoxic stress. ADH1 is core hypoxia gene involved in obtaining ATP by fermentation.10 Our research confirmed that the increase in the amount of ADH1 transcript during hypoxia was located mainly in the cytoplasm. This result suggests that the transcripts encoding proteins necessary for survival in hypoxia are not accumulated in the nucleus but instead are used for translation in the cytoplasm. The A. thaliana genes whose mRNAs are accumulated in the nucleus during hypoxia include MSR B8, RNA polymerase II subunit RPB2 and Sm-like protein. Literature data indicate that MSR B8, like ADH1, is transcribed in low-oxygen conditions, but the efficiency of its translation is significantly lower.11 There are also no reports on the participation of MSR in the response to hypoxic stress. In the case of mRNA of transcript polymerase II subunit RPB2, no changes in the level of translation efficiency were observed at 1 h of hypoxia. However, the level of translation efficiency was much lower than for ADH1.11 Our subcellular measurements of RPB2 mRNA levels showed an increased amount of the transcript and its accumulation in the cell nucleus. At the same time, in lupine, the amount of the active form of that protein was strongly declining during hypoxia. This result suggests a reduction in the translation process, or the accumulation of the enzyme in its inactive form. Similarly, the mRNA of Sm-like protein accumulated in the nucleus of A. thaliana during hypoxia. In the lupine nuclei, we also observed increased amounts of the 3 studied transcripts whose genes were not associated with responses to hypoxic stress. In conclusion, our results show that the nuclei accumulated mRNAs are not core hypoxia genes. This evidence shows that the nucleus is the site of mRNA storage for plants during unfavourable environmental conditions.
Re-aeration caused a significant increase in poly(A) RNA and protein encoding mRNA in the cytoplasm. In the case of the protein coding RNAs, the increase in the cytoplasm was strongly correlated with a decrease in the nucleus, which indicates that the mRNA stored during hypoxia is transported to the cytoplasm and can be translated. The use of the mRNA accumulated in the cell is supported by data showing only a 0.4% increase in total mRNA at 1 h after re-aeration, compared to an increase of more than 50% in mRNAs bound to ribosomes.12 This result suggests that after the removal of stress, transcripts do not change much quantitatively or qualitatively. The strong growth in translation results from molecular processes leading to the transport of mRNA from the site of storage to the site of protein synthesis. There was no strong correlation between the changes in the nuclear and cytoplasmic distribution of poly(A) RNA after re-aeration. However, a significant portion of the poly(A) RNA pool are non-coding RNAs, such as pri-miRNA and long non-coding antisense RNAs that do not leave the cell nucleus.32,33 Thus, the selective storage of mRNA in the cell nucleus, which can then be quickly used after the end of stressful conditions, may be a strategy of plants to survive in hypoxic conditions and recover in normal conditions after stress.
Another site of mRNA storage under stress is in structures resembling stress granules (SGs).34 SGs contain translationally repressed mRNA but also eukaryotic translation initiation factor (eiF4G) or Tudor-SN involved in stress adaptation.35,36 Studies in lupine cells in comparison to A. thaliana allowed better visualization of SGs after the in situ hybridization of poly(A) RNA, including transcripts encoding proteins. We showed that the poly(A) RNA in SG-like structures represented a pool that was not co-localized with ribosomes, which suggests that they do not participate in translation. After removing stress conditions, SGs like structures disappeared after 3 h, indicating their role as a cytoplasmic storage of poly(A) RNA – in addition to the nuclear pool – used after the end of hypoxia.
A recent study shows that the Cajal bodies present in generative and somatic plant cells contain mRNA.19,37 In the two species tested – which suggests the versatility of the process in plants – hypoxia-induced CBs accumulated poly(A) RNA (including RNAs encoding proteins) more intensely than the nucleus overall. The intensity of the accumulation in the CBs was inversely correlated with the level of transcription by RNA polymerase II. It increased when hypoxia reduced mRNA synthesis and decreased with increasing in transcription. This result indicates that the CBs can function as transcript stores. The important role of the CBs in tolerance to low-oxygen conditions was demonstrated by our results in ncb-1 mutants. Plants without CBs died faster under hypoxic conditions. However, death at the 5th day of hypoxia and the a strong reduction in the level of the poly(A) compared to WT indicates that the CBs are involved not only in storage but also in other processes of mRNA metabolism. The CBs may be associated with the stabilization of poly(A) RNA, preventing its degradation in the nucleus. This possibility is backed by the fact that ADH1 mRNA which is translated during hypoxia did not accumulate in the nucleus and did not occur in the nuclear bodies. It has recently been shown that in protoplasts or cells subjected to stress associated with removal of the cell wall, a portion of the transcripts present in the nucleus and having premature termination codons (PTCs) are insensitive to the nonsense-mediated decay (NMD) pathway.38 Some of those transcripts are involved in stress responses.39
On the other hand, it cannot be ruled out that ncb-1 dies sooner because of the disturbances to snRNP assembly. The correct amount of splicing factors is necessary for processing of the transcripts of genes involved in the response to hypoxia. However, differences in the amount of U2 snRNA between ncb-1 and WT were observed only at 1 h of hypoxia. In zebrafish embryos, the lack of CBs may be significant under conditions of increased demand for snRNP.40 The coilin depletion leads to reduced cell proliferation, developmental arrest and cell death in embryos. In our study, a similar amount of U2 snRNA at 24 and 48 h of hypoxia in WT and ncb-1 suggests that the amount of snRNP need not be a limiting factor for survival of hypoxic stress.
In summary, this research on the spatial organization of the transcriptome in both species during hypoxia and then in re-aeration showed the following: (1) as a result of accumulation, the cell nucleus served as a storage for poly(A) RNA and certain mRNAs, preventing their translation; (2) Cajal bodies were connected with the concentration/aggregation and stabilization of poly(A) RNA in the cell nucleus; (3) the accumulation in the cell nucleus was followed by a decrease in RNA polymerase II transcription; (4) cytoplasmic poly(A) RNA, including RNAs encoding proteins, formed stress granules like structure not co-localized with ribosomes; and (5) removal of the hypoxic stress caused a strong increase in cytoplasmic mRNA, which could come from the nucleus and SG like structures.
Materials and methods
Lupinus luteus cv. Sonet (Torseed SA Toruń; Poland) seeds were soaked in 70% (vol/vol) ethanol for 5 min and rinsed five times with sterile, deionized H2O. Seeds were soaked in water for 5 h and subsequently germinated at 21°C for 3 days on water-soaked tissue paper in a growth chamber in the dark. After a three-day germination, the seedlings had 1–2 cm roots and hypocotyls protruding through the seed coat. Hypoxia was imposed by submerging seedlings in tap water in a container at 21°C in a long-day growth chamber. The water level was at least 4 cm above the seedlings. Recovery after hypoxia was conducted under conditions in which the seeds germinated but in a long-day growth chamber.
Arabidopsis thaliana ecotype Columbia-0 and ncb-1 mutant were grown at 22°C in a long-day growth chamber. To impose the hypoxia treatment, 14-day-old seedlings in pots with soil were submerged in plastic containers filled with tap water so that the water level was approximately 4 cm above plants and grown at 22° in the same photoperiod. To ensure a proper submergence of roots, at the beginning the soil in the pots was moved deeply to remove any residual air. To evaluate survival, the color and shape of the leaves were analyzed in 472 WT and 424 ncb-1 plants from three different experiments after 5-day hypoxia. The 1602 and 1448 leaves was respectively analyzed in WT and ncb-1 plants.
Transcription in Lupinus luteus roots was inhibited by actinomycin D and α-amanitin. After 3 d seedlings were transferred on inhibitors-soaked tissue paper in a growth chamber. The roots were treated with 10 μg/ml of α-amanitin and actinomycin D (Sigma) for 12 h before fixation.
In situ specimen preparation
Meristems of Lupinus luteus and Arabidopsis thaliana roots were excised underwater and fixed in 4% formaldehyde (Polyscience) in 50 mM PIPES buffer pH 7.0 for 12 h at 4°C. The fixed roots were washed three times for 15 min in PIPES buffer pH 7.2 and three times for 15 min in PBS buffer pH 7.2. For monolabelling assay, Lupinus luteus 3-4 mm fixed tips of roots were placed in citric acid-buffered digestion solution pH 4,8 containing 5% cellulase (Onuzuka R-10) and 35 U/ml pectinase (Sigma) for 90 min at 35°C. After rinsing with PBS pH 7.2 and distilled water, the root tips were squashed onto slides. For double detection root tips were sectioned underwater into 50 µm thick sections using a Vibratome Leica VT1200. The sections were placed in embryo dishes and treated with 2% cellulase (Onuzuka R-10) and 25 U/ml pectinase (Sigma) in citric buffer pH 4.8 for 25 min at 35°C. Then, the sections were rinsed with PBS pH 7.2 and treated with 0.1% Triton x100 solution in PBS buffer pH 7.2 for 10 min.
Arabidopsis thaliana tips of roots were digested in the same solutions as lupine roots in the case of monolabelling. Digestion was performed for 40 min. Then the roots were washed and treated with Triton X100 as described above. All procedures until observation under the microscope were conducted in embryo dishes.
In situ hybridization of poly(A) RNA and double labeling with Sm proteins and 26S rRNA
All double labeling immunocytochemical procedures preceded in situ hybridization procedures. The roots of Arabidopsis thaliana, and Lupine luteus were incubated with antibodies to Sm-proteins (IS2076 CDC Atlanta GA 30333) diluted 1:300 in 1% BSA in PBS pH 7.2 overnight. Then, in situ hybridization was performed according to Niedojadło et al.19 For FISH double labeling (26S rRNA-poly(A) RNA), two probes were applied simultaneously in the hybridization buffer, and the reaction was performed at 28°C. For the detection of 26S rRNA, we used an antisense DNA probe against 26 rRNA 5′ TeT-ATATTAAACTGATAAGAACAGATACTACACTTG. DNA was stained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma). The control reaction was performed in the same manner, except that the hybridization buffer was used without probes, and the primary antibodies were omitted.
Immunocytochemical localization of Sm-proteins, PANA antigen and phosphorylated serine-2 in the CTD domain of RNA POL II
For the double localization of Sm-proteins and PANA antigen, meristematic cells roots of lupine were incubated with mixed antibodies.23 Sm-protein antibodies and anti-PANA antibodies 1:100 in 1% BSA in PBS pH 7.2 overnight at 4°C. For the localization of phosphorylated serine-2 in the CTD domain of RNA POL II, polymerase II antibodies (Chromotek; Germany) diluted 1:100 in 1% BSA in PBS pH 7 overnight at 4°C. DNA was stained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma).
Double labeling of Sm proteins and transcripts in L. luteus and A. thaliana
Immunolocalization of Sm proteins was performed as above, followed by in situ hybridization was used. A. thaliana roots were incubated with proteinase K (Roche) at a concentration of 5 µg/ml for 30 min at 35°C. The following antisense DNA were used for the detection: ADH1 (AT1G77120): 5′-Cy3-TTGCTCCTAACCCAGTAGACAAA CCACAACT; MSR B8 (AT4G21840): 5′-Cy3-TAAACTCTCCTTTGCCTCGTTTATC; PRB2 (AT4G21710): 5′-Cy3 GGGTA ATCTCCTCATCGTCATCGTCTTCTA; Sm-like (AT3G07590): 5′-Cy3-ATTACCCCTCAAGCTAAGATGATCAA GTGTTAC, at concentrations of 50 pmol/ml. The double detection of Sm proteins and transcripts in lupine roots was accomplished according to Niedojadło et al.19 For localization of ADH1 transcripts the sequence identical in 59% to the ADH1 from A. thaliana was found in the database SRA NCBI which typically cover the 5′UTR cis-acting regulatory element essential for the anaerobic induction. We used following antisense DNA probe probe: TATTACAACTAGCGC CTAAGGTTCTTTAAT. For the negative control reactions, the samples were incubated with hybridization buffer, omitting the probes. For fluorescence in situ hybridization of U2 snRNA the following oligonucleotide was using: 5′-Cy3-ATATTAAA CTGATAAGAACAGATACTACACTTG-3′.
Quantitative evaluation of fluorescence signals
To calculate the fluorescence intensity resulting from the in situ hybridization signal and diameter of Cajal bodies, 42-73 cells from three different experiments were analyzed. The numbers of cells used in each experiment are shown in the tables presented under the graphs. For quantitative measurements, each experiment was performed using consistent temperatures, incubation times, and concentrations of probes and antibodies. The results were recorded and analyzed using a Nikon PCM-2000 and Nikon A1R confocal microscope using lasers emitting light at a wavelengths of 405, 488, 543 nm. For NIKON A1R optimized pinhole, long exposure time (400 kHZ) and 60X (numerical aperture, 1.4) Plan Apochromat DIC H oil immersion lens were used. Images were collected sequentially in the blue (DAPI), green (Alexa 488 fluorescence, TeT) and red (Cy3) channels. To minimize bleed-through between fluorescence channels, we employed low laser power (3–10% of maximum power) and single-channel collection. For Nikon PCM-2000 a mid-pinhole, long exposure time (75 μs), and a 100× (numerical aperture, 1.4) Plan Apochromat DIC H oil immersion lens were used. For DAPI staining, an inverted Nikon Eclipse TE 2000 fluorescence microscope equipped with a mercury lamp, a UV-2EC UV narrow-band filter was used. The quantitative analysis was performed using the CeSa Statistical Analyzer (Department of Cell Biology, Nicolaus Copernicus University, Toruń, Poland) software and NIS-Elements AR3.00 (Nikon, Laboratory Imaging). Statistical analysis was performed using PAST41 and Microsoft Excel (Microsoft, Redmond, WA, USA). The non-parametric rank-based Kruskal–Wallis test was used to compare multiple groups, and if significant differences were detected, the Mann–Whitney test with Bonferroni correction was used. Correlation analysis was performed using Pearson's correlation coefficient.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by the Polish Ministry of Science and Higher Education no. NN303 300437. The authors would like to thank Prof. E. Bednarska-Kozakiewicz and Dr D.J. Smoliński (Department of Cell Biology, Nicolaus Copernicus University, Torun, Poland) for critical reading of the manuscript and for constructive comments and M. Świdziński (Department of Cell Biology, Nicolaus Copernicus University, Torun, Poland) for excellent technical assistance. The authors thank Dr P. Glazińskiej (Chair of Plant Physiology and Biotechnology, Nicolaus Copernicus University, Torun, Poland) for helping to generate a probes to ADH1 of Lupinus luteus. We thank Prof. Peter Show for seeds of ncb-1 mutants of Arabidopsis thaliana. This manuscript (1EA3-B545-876A-BD0A-07A8) was edited for proper English language by American Journal Experts.
References
- 1.Bailey-Serres J, Fukao T, Gibbs DJ, Holdsworth MJ, Lee SC, Licausi F, Perata P, Voesenek LA, van Dongen JT. Making sense of low oxygen sensing. Trends Plant Sci 2012; 17:129–38; PMID:22280796; http://dx.doi.org/ 10.1016/j.tplants.2011.12.004 [DOI] [PubMed] [Google Scholar]
- 2.Armstrong W, Drew MC. Root growth and metabolism under oxygen deficiency. Plant roots. The hidden half (3rd edition), Published by Marcel Dekker, New York & Basel: 2002; 729–76 [Google Scholar]
- 3.Bailey-Serres J, Lee SC, Brinton E. Waterproofing crops: effective flooding survival strategies. Plant Physiol 2012; 160:1698–709; PMID:23093359; http://dx.doi.org/ 10.1104/pp.112.208173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Limami AM, Glévarec G, Ricoult C, Cliquet JB, Planchet E. Concerted modulation of alanine and glutamate metabolism in young Medicago truncatula seedlings under hypoxic stress. J Exp Botany 2008; 59:2325–35; http://dx.doi.org/ 10.1093/jxb/ern102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocha M, Sodek L, Licausi F, Hameed MW, Dornelas MC, van Dongen JT. Analysis of alanine aminotransferase in various organs of soybean (Glycine max) and in dependence of different nitrogen fertilisers during hypoxic stress. Amino Acids 2010; 39:1043–53; PMID:20414691; http://dx.doi.org/ 10.1007/s00726-010-0596-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narsai R, Rocha M, Geigenberger P, Whelan J, van Dongen JT. Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. New Phytol 2011; 190:472–87; PMID:21244431; http://dx.doi.org/ 10.1111/j.1469-8137.2010.03589.x [DOI] [PubMed] [Google Scholar]
- 7.Sweetlove LJ, Beard KF, Nunes-Nesi A, Fernie AR, Ratcliffe RG. Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 2010; 15:462–70; PMID:20554469; http://dx.doi.org/ 10.1016/j.tplants.2010.05.006 [DOI] [PubMed] [Google Scholar]
- 8.Christianson JA, Llewellyn DJ, Dennis ES, Wilson IW. Comparisons of early transcriptome responses to low-oxygen environments in three dicotyledonous plant species. Plant Signal Behav 2010; 5:1006–9; PMID:20724824; http://dx.doi.org/ 10.4161/psb.5.8.12231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LA, Bailey-Serres J. Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol 2011; 190:457–71; PMID:21231933; http://dx.doi.org/ 10.1111/j.1469-8137.2010.03590.x [DOI] [PubMed] [Google Scholar]
- 10.Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, Yaghoubi-Masihi A, Fukao T, Bailey-Serres J. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 2010; 152:1484–500; PMID:20097791; http://dx.doi.org/ 10.1104/pp.109.151845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juntawong P, Girke T, Bazin J, Bailey-Serres J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA 2014a; 111:E203–212; http://dx.doi.org/ 10.1073/pnas.1317811111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 2008; 56:743–55; PMID:18665916; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03642.x [DOI] [PubMed] [Google Scholar]
- 13.Weber C, Nover L, Fauth M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 2008; 56(4):517–30; PMID:18643965; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03623.x [DOI] [PubMed] [Google Scholar]
- 14.Kedersha N, Ivanov P, Anderson P. Stress granules and cell signalling: more than just a passing phase? Trends Biochem Sci 2013; 38(10):494–06; PMID:24029419; http://dx.doi.org/ 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorenson R, Bailey-Serres J. Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 2014; 111:2373–8; PMID:24469793; http://dx.doi.org/ 10.1073/pnas.1314851111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z, Kumari S, Zhang L, Zheng Y, Ware D. Characterization of miRNA in response to short-term waterlogging inthree inbred lines of Zea mays. PLoS One 2012; 7(6):e39786; PMID:22768123; http://dx.doi.org/ 10.1371/journal.pone.0039786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juntawong P, Bailey-Serres J. Dynamic light regulation of translation status in Arabidopsis thaliana. Front Plant Sci 2012; 3:66; PMID:22645595; http://dx.doi.org/ 10.3389/fpls.2012.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mustroph A, Barding GA Jr, Kaiser KA, Larive CK, Bailey-Serres J. Characterization of distinct root and shoot responses to low-oxygen stress in Arabidopsis with a focus on primary C- and N-metabolism. Plant Cell Environ 2014; 37:2366–80; PMID:24450922 [DOI] [PubMed] [Google Scholar]
- 19.Niedojadło J, Kubicka E, Kalich B, Smoliński DJ. Poly(A) RNAs including coding proteins RNAs occur in plant Cajal bodies. PLoS One 2014; 9:e111780; PMID:25369024; http://dx.doi.org/ 10.1371/journal.pone.0111780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J 2002; 21(11):2746–56; PMID:12032087; http://dx.doi.org/ 10.1093/emboj/21.11.2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nesic D, Tanackovic G, Krämer A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J Cell Sci 2004; 117:4423–33; PMID:15316075; http://dx.doi.org/ 10.1242/jcs.01308 [DOI] [PubMed] [Google Scholar]
- 22.Cui P, Moreno Díaz, de la Espina S. Sm and U2B” proteins redistribute to different nuclear domains in dormant and proliferating onion cells. Planta 2003; 217(1):21–31; PMID:12721845 [DOI] [PubMed] [Google Scholar]
- 23.Niedojadło J, Mikulski Z, Dełeńko K, Szmidt-Jaworska A, Smoliński DJ, Epstein AL. The perichromatin region of the plant cell nucleus is the area with the strongest co-localisation of snRNA and SR proteins. Planta 2012; 236(2):715–26; PMID:22526497; http://dx.doi.org/ 10.1007/s00425-012-1640-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smoliński DJ, Wróbel B, Noble A, Zienkiewicz A, Górska-Brylass A. Periodic expression of Sm proteins parallels formation of nuclear Cajal bodies and cytoplasmic snRNP-rich bodies. Histochem Cell Biol 2011; 136(5):527–41; PMID:21904826; http://dx.doi.org/ 10.1007/s00418-011-0861-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ali GS, Golovkin M, Reddy AS. Nuclear localization and in vivo dynamics of a plant-specific serine/arginine-rich protein. Plant J 2003; 36:883–93; PMID:14675452; http://dx.doi.org/ 10.1046/j.1365-313X.2003.01932.x [DOI] [PubMed] [Google Scholar]
- 26.Collier S, Pendle A, Boudonck K, v.Rij T, Dolan L, Shaw P. A distant coilin homologue is required for the formation of Cajal bodies in Arabidopsis. Mol Biol Cell 2006; 17:2942–51; PMID:16624863; http://dx.doi.org/ 10.1091/mbc.E05-12-1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA 2013; 4(1):17–34; PMID:23042601; http://dx.doi.org/ 10.1002/wrna.1139 [DOI] [PubMed] [Google Scholar]
- 28.Muthuswamy S, Meier I. Genetic and environmental changes in SUMO homeostasis lead to nuclear mRNA retention in plants. Planta 2011; 233(1):201–8; PMID:20872268; http://dx.doi.org/ 10.1007/s00425-010-1278-7 [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Zhou T. Dynamical analysis of mCAT2 gene models with CTN-RNA nuclear retention. Phys Biol 2015; 12:016010; PMID:25619276; http://dx.doi.org/ 10.1088/1478-3975/12/1/016010 [DOI] [PubMed] [Google Scholar]
- 30.Boothby TC, Zipper RS, van der Weele CM, Wolniak SM. Removal of retained introns regulates translation in the rapidly developing gametophyte of Marsilea vestita. Dev Cell 2013; 24:517–29; PMID:23434411; http://dx.doi.org/ 10.1016/j.devcel.2013.01.015 [DOI] [PubMed] [Google Scholar]
- 31.Dełeńko K, Niedojadło J, Łabędzka A, Wiśniewska E, Bednarska-Kozakiewicz E. Dedifferentiation of Arabidopsis thaliana cells is accompanied by a strong decrease in RNA polymerase II transcription activity and poly(A+) RNA and 25S rRNA eradication from the cytoplasm. Protoplasma 2015; 252:537–46; PMID:25248757; http://dx.doi.org/ 10.1007/s00709-014-0700-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novikova IV, Hennelly SP, Tung CS, Sanbonmatsu KY. Rise of the RNA machines: exploring the structure of long non-coding RNAs. J Mol Biol 2013; 425:3731–46; PMID:23467124; http://dx.doi.org/ 10.1016/j.jmb.2013.02.030 [DOI] [PubMed] [Google Scholar]
- 33.Arikit S, Zhai J, Meyers BC. Biogenesis and function of rice small RNAs from non-coding RNA precursors. Curr Opin Plant Biol 2013; 16:170–9; PMID:23466255; http://dx.doi.org/ 10.1016/j.pbi.2013.01.006 [DOI] [PubMed] [Google Scholar]
- 34.Adjibade P, Mazroui R. Control of mRNA turnover: implication of cytoplasmic RNA granules. Semin Cell Dev Biol 2014; 34:15–23; PMID:24946962; http://dx.doi.org/ 10.1016/j.semcdb.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 35.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 2009; 10:430–6; PMID:19461665; http://dx.doi.org/ 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- 36.Yan C, Yan Z, Wang Y, Yan X, Han Y. Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J Exp Botany 2014; 65:5933–44; http://dx.doi.org/ 10.1093/jxb/eru334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smoliński DJ, Kołowerzo A. mRNA accumulation in the Cajal bodies of the diplotene larch microsporocyte. Chromosoma 2012; 121:37–48; http://dx.doi.org/ 10.1007/s00412-011-0339-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Göhring J, Jacak J, Barta A. Imaging of endogenous messenger RNA splice variants in living cells reveals nuclear retention of transcripts inaccessible to nonsense-mediated decay in Arabidopsis. Plant Cell 2014; 26:754–64; PMID:24532591; http://dx.doi.org/ 10.1105/tpc.113.118075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, et al.. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 2012; 40:2454–69; PMID:22127866; http://dx.doi.org/ 10.1093/nar/gkr932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strzelecka M, Oates AC, Neugebauer KM. Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus 2010; 1:96–108; PMID:21327108; http://dx.doi.org/ 10.4161/nucl.1.1.10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistic software package for education and data analysis. Palaeontologia Electronica 2001; 4:1–9 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.