ABSTRACT
The packaging RNA (pRNA) found in the phi29 family of bacteriophage is an essential component of a powerful molecular motor used to package the phage's DNA genome into the capsid. The pRNA forms homomultimers mediated by intermolecular "kissing-loop" interactions, thus it is an example of the unusual phenomenon of a self-associating RNA that can form symmetric higher-order multimers. Previous research showed the pRNAs from phi29 family phages have diverse self-association properties and the kissing-loop interaction is not the sole structural element dictating multimerization. We found that a 3-way junction (3wj) within each pRNA, despite not making direct intermolecular contacts, plays important roles in stabilizing the intermolecular interactions and dictating the size of the multimer formed (dimer, trimer, etc.). Specifically, the 3wj in the pRNA from phage M2 appears to favor a different conformation compared to the 3wj in the phi29 pRNA, and the M2 junction facilitates formation of a higher-order multimer that is more thermostable. This behavior provides insights into the fundamental principles of RNA self-association, and additionally may be useful to engineer fine-tuned properties into pRNAs for nanotechnology.
KEYWORDS: Packaging RNA (pRNA), RNA self-association, RNA structure, 3-way junction (3wj)
Abbreviations
- RNA
ribonucleic acid
- pRNA
packaging RNA
- 3wj
3-way junction
- nt
nucleotide
- SHAPE
selective 2'-hydroxyl acylation analyzed by primer extension
- FRET
Förster resonance energy transfer
- EPR
electron paramagnetic resonance
Introduction
The packaging RNA (pRNA) found in the phi29 family of bacteriophages is an essential component of the motor that packages the genomic DNA directly into the phage capsid.1-12 Within this motor, multiple copies of the pRNA self-associate to form a ring structure that interacts with multiple copies of the gp10 connector protein and gp16 ATPase protein as well as with the empty capsid.4,12-14 Each motor contains multiple copies of the pRNA within this ring, with each copy contacting adjacent partners through intermolecular “kissing-loop” or “hand-in-hand” base-pair interactions (Fig. 1A & B).1,3,15 The pRNAs can also self-associate when free in solution,4,10,13,15-19 thus they are unusual naturally occurring RNA molecules that can form higher-order multimers through intermolecular interaction of identical molecules and without requiring proteins. The most-studied pRNA is from phage phi29,1,2,20-24 but phylogenetically related strains of the phi29 family all contain pRNAs that form similar secondary structures containing several stem-loops, including the loops that make the intermolecular kissing-loop interactions and a 3-way junction.17,19,21,25,26 All pRNAs likely form pentamers or hexamers in the context of the mature packaging motor,4,10,18,27 whereas in solution, without associated proteins, they show variability in their self-association properties in terms of thermostability and stoichiometry.13,16,17,19 Although phi29 pRNA hexamers have been reported free in solution,24 the most readily obtained forms are dimers and trimers.13,16,17,19 The higher-order forms are probably stabilized by their association with the packaging motor proteins. The ability to form these smaller multimers is important as they have been reported to be the “building blocks” for forming the larger ring structures,16,17,28,29 and they can provide a toolbox to construct nanodevices of various sizes and shapes.30-43 By exploring how very similar RNAs are able to self-associate into homomultimers of different sizes, we can better understand the basic determinants of programmed RNA self-association to guide ongoing efforts to engineer artificial self-associating RNAs for use in nanotechnology44,45 and can also aid in understanding the assembly of higher-order RNA structures in general.
Figure 1.

The 3wj affects the stoichiometry of pRNA self-association. (A) Cartoon of pRNA self-association. The pentamer form that is thought to occur within the phage packaging motor is shown (although some reports favor a hexamer form).4,10,18,20 Under most solution conditions, including those explored here, the pRNAs form smaller homomultimers (dimers, trimers) that are the building blocks for larger structures, are useful for nanotechnology applications, and are the forms studied here. The kissing-loop interactions between adjacent copies of the RNA (red boxes) are the known sites of intermolecular contact. (B) Cartoon of the secondary structure of the entire pRNA transcribed during phage infection. The boxed region is sufficient for self-association. The intermolecular kissing-loop interaction sequences are red. The grey region appears to be cleaved off during phage infection and is not functionally necessary. The 3wj is indicated with the dashed cyan box. (C) Schematic of the 3 families of 3wjs assigned by Lescoute and Westhof.46 White numbers on the left-most schematic show the identity of the 3 helices according to their nomenclature. (D) Secondary structures of the minimal self-association domains from M2, M2_phi29 and phi29 pRNAs. M2_phi29 contains the M2 pRNA with the phi29 3-way junction inserted (cyan). The intermolecular base-pairing sequences are in red. (E) Native gel electrophoresis of the pRNAs from panel (D) at 4°C and with 5 mM MgCl2. Assignment of dimer and trimer multimeric states was done in previous biophysical analyses.17
The self-association ability of the pRNA clearly depends on the kissing-loop interactions,21 but the sequences involved in the kissing-loop interactions between copies of self-associated pRNAs differ between strains.13,17,19 This suggests that the kissing-loop interactions might confer different behaviors. However, while the kissing-loop interactions are necessary for self-association, they are not the only determinant of the variability of pRNA multimer thermostability and stoichiometry in solution.17,19 For example, a variety of biophysical experiments have shown that under most conditions, the pRNA from the M2 strain forms trimers in solution that are more thermostable than the dimers formed by pRNAs from the prototype phi29.17 However, swapping the kissing-loop interaction sequences from M2 to phi29 does not transfer this behavior.17 This and other data suggest that the structural features that govern pRNA multimerization extend beyond the intermolecular base-pairing in the kissing-loop interaction and thus, to exploit the pRNAs as a tool for RNA engineering, we must better understand the other elements that modulate their self-association properties.
One element within the pRNAs that could alter self-association is the 3-way junction (3wj). Like the sequence variability of the kissing-loop interactions, the 3wj present in all pRNAs is variable in sequence,17,19,21 suggesting it may play a role in different pRNA self-association properties. It is well-established that RNA 3wjs can facilitate formation of long-range tertiary interactions within larger folded RNAs.46,47 Thus, they may also play a role in promoting distal interactions in homomultimer-forming self-associating RNAs, perhaps by spatially prepositioning the elements that make intermolecular contacts. Based on structures of many folded RNAs, Lescoute and Westhof classified diverse RNA 3wjs into 3 different families depending on the relative lengths of the single-stranded segments within the junction (Fig. 1C).46 Based on this analysis the 3wjs of different pRNAs can be predicted to be in different families, suggesting that the emerging helices are oriented differently relative to one another. Thus, we hypothesized that the 3wj element in pRNA is a key determinant of the stoichiometry of self-association in solution. To test this hypothesis, we used the M2 and phi29 pRNAs as representative models to study different self-association properties. These two were chosen as they have clearly different yet robust multimerization behaviors. We probed the folding characteristics of the 3wjs and used Förster resonance energy transfer (FRET) to query the relative angles between helices emerging from the junctions. We found that the 2 3wjs appear to favor different conformations with different interhelical angles, consistent with their different multimer behaviors. The analyses of several chimeric pRNAs support the conclusion that self-association is affected by the 3wj, and thus the 3wj is a valid RNA module for continued pRNA engineering.
Results
The three-way junction contributes to pRNA self-association properties
We chose to study the pRNAs from phi29 and M2 phages as models because our previous work showed they have different self-multimerization properties, and thus they can serve as good examples to understand what elements favor dimerization versus trimerization.17 Previously, we used both sedimentation equilibrium and sedimentation velocity analytical ultracentrifugation to establish the propensity of different pRNAs to form multimers of specific size, and we correlated this with measurements by native gel electrophoresis.17 This established native gel electrophoresis as a convenient way to assess the stoichiometry of different pRNA multimers. To determine if the 2 different 3wjs directly influence pRNA self-association, we generated a chimeric construct M2_phi29, in which the phi29 3wj was embedded within the context of the M2 pRNA (Fig. 1D). We used a ~75-nucleotide sequence of the pRNA, which is sufficient for multimerization. The migration patterns on the native gel match the published multimerization properties; at 4°C, M2 forms a trimer, and phi29 forms a dimer.17 Analysis of the chimeric M2_phi29 using this established native gel system showed that it travels at a rate consistent with formation of a dimer, near the mobility of phi29 pRNA but not M2 pRNA (Fig. 1E). We attempted to generate a chimeric pRNA with the M2 pRNA 3wj in the context of the phi29 pRNA, but this RNA formed an aberrant secondary structure (as assayed by chemical probing, data not shown), precluding further analysis. Nonetheless, the result from M2_phi29 suggests that the 3wj is a critical element determining pRNA multimerization properties.
M2 and phi29 pRNA 3-way junctions do not show differences in magnesium-induced folding
Having observed that the pRNA 3wj contributes to establishing the multimer state, we explored why the M2 and phi29 junctions have different properties. The phi29 pRNA 3wj has been proposed to be flexible or conformationally dynamic.14 In comparison, structural information regarding the M2 pRNA 3wj is limited. One hypothesis is that there are fundamental differences in the folding characteristics of the 2 junctions in terms of their conformational dynamics or their ability to form a stable folded structure. If so, the M2 pRNA 3wj might be less dynamic or its structure more stably folded than the phi29 3wj, which could promote the formation of higher-order, more stable multimers. To interrogate the folds of the 2 3wjs, we probed the RNAs with hydroxyl radicals, which cleave RNA backbones independent of sequence or secondary structure. Cleavage by the radicals is related to the solvent accessibility of each ribose.48,49 The method is a powerful way of assessing the formation of locally packed structure in an RNA since it is sensitive to the local protection of the backbone; it has been effective in detecting compact local packing in other RNA junctions.50 Because RNA requires cations to induce folding, hydroxyl radical probing is often performed in the absence and presence of Mg2+ and the resultant patterns are compared to detect regions of tight backbone packing induced by folding. We used this strategy here, reasoning that if the phi29 and M2 pRNA exhibited different Mg2+-induced hydroxyl radical cleavage patterns, this would provide evidence of different folding characteristics that may lead to different multimerization properties.
We performed hydroxyl radical probing of the full-length M2 and phi29 pRNAs (Fig. 2A) at 3 different temperatures (4°C, 25°C, 37°C), in both the absence and presence of Mg2+. As stated above, the Mg2+ may be needed to fold the junctions, and in the case of pRNAs is also necessary to form multimers in solution. Based on our previous biophysical analyses, the pRNA is exchanging between multimeric and monomeric forms; at 4°C both can form multimers, at 25°C M2 pRNA can multimerize while phi29 pRNA multimers are destabilized, and at 37°C both are destabilized,17 hence these various species are likely present in the experiment.
Figure 2.
Hydroxyl radical probing of pRNAs. (A) Secondary structures of the complete, fully functional pRNAs from phi29 and M2; each are ~120 nt long. The shaded region is the minimal self-association domain. The nucleotides in the 3-way junction are labeled and highlighted with the arrows. (B) Hydroxyl radical probing of the full-length phi29 and M2 RNAs. Reactions without Mg2+ are marked as “-“, reactions with Mg2+ are marked as “+”. Temperatures (°C) are indicated as “4”, “25”, and “37”. An RNase T1 sequencing ladder (indicating G bases) and hydrolysis ladder (OH) are included and labeled. The nucleotides in the 3-way junction are labeled and highlighted with the arrows. Asterisk marks a location in the gel where salt-induced effects result in a compaction of a few bands. (C) Quantified and graphed data of the probing gel shown in panel 2B. Black: cleavage pattern without Mg2+. Green: cleavage pattern with Mg2+. Temperatures (°C) are indicated as “4”, “25”, and “37”. The trace of the RNase T1 sequencing ladder with a few locations marked is in red. The approximate location of nucleotides in the 3-way junction is shaded gray. Asterisk in the M2 pRNA graphs marks a location in the gel where salt-induced effects result in a compaction of a few bands and an apparent increase in cleavage at a few nucleotides.
A visual examination of the gel shows that the addition of magnesium resulted in little change in the probing pattern, and also temperature did not induce changes in the patterns (Fig. 2B). However, to more rigorously assess the probing patterns, we quantitated each lane and analyzed the data in a manner similar to our previous studies on other RNAs (Fig. 2C).49 Consistent with the visual examination, at 4°C and 25°C the traces in the absence and presence of Mg2+ are essentially identical for both phi29 and M2 pRNAs. Likewise, the traces overall between these 2 temperatures are very similar for both pRNAs. Within this fairly consistent cleavage pattern, it is interesting to note that some of the nucleotides that are in the junction are somewhat protected from cleavage relative to their neighbors (Fig. 2B & C). Thus, the junction appears to adopt a specific structure in both pRNAs but this structure is not dependent on Mg2+. Most importantly, at 4°C and 25°C there is no evidence that one junction has radically different folding characteristics than the other, or that multimerization induces a structural compaction. Interestingly, in both pRNAs there are some differences in the patterns with and without Mg2+ at 37°C, most notably at the top of the gel. At the increased temperature and without Mg2+, it appears that both pRNAs become less structured in the junction region, consistent with the junction beginning to denature. Not surprisingly, this effect is reversed upon addition of Mg2+, which is known to stabilize RNA folds. In this case the Mg2+ is stabilizing a fold that does not require Mg2+ at lower temperatures, thus likely acting by general charge neutralization. Overall, the folding characteristics of the 2 pRNAs are quite similar, thus it does not appear that one junction has a compact fold that is measurably more stable than the other. This is consistent with previously published selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) probing of both pRNAs, which showed the 3wjs were equally modified.17 These results suggest that the different multimerization properties of the M2 and phi29 pRNAs are not due to gross differences in the folding characteristics of the junctions.
The phi29 and M2 pRNA 3wjs favor different helical arrangements in solution
Although the M2 and phi29 pRNA 3wjs have similar folding characteristics, they may have different conformations that favor different interhelical angles, placing the kissing-loop interaction elements in positions to affect the multimerization properties. To directly assess the different solution-state interhelical orientation of the helices, we used Förster resonance energy transfer (FRET), which has been used to study RNA junctions.51-55 With previous FRET studies of RNA 3wjs as a guide, we made 3 RNA strands that anneal to form the 3wjs of both the M2 and phi29 pRNAs. This design was chosen to allow us to study the inherent conformation of the 3wj independent from other parts of the pRNA and without potential changes to the structure of the junction induced by multimerization (Fig. 3A). We verified that the 3 RNA oligomers hybridized to form a single product using native gel electrophoresis (Fig. 3B). Samples containing all 3 strands of both phi29 and M2 pRNAs had different migration patterns compared to other combinations, indicating robust and essentially quantitative formation of the 3wj from 3 RNA strands.
Figure 3.
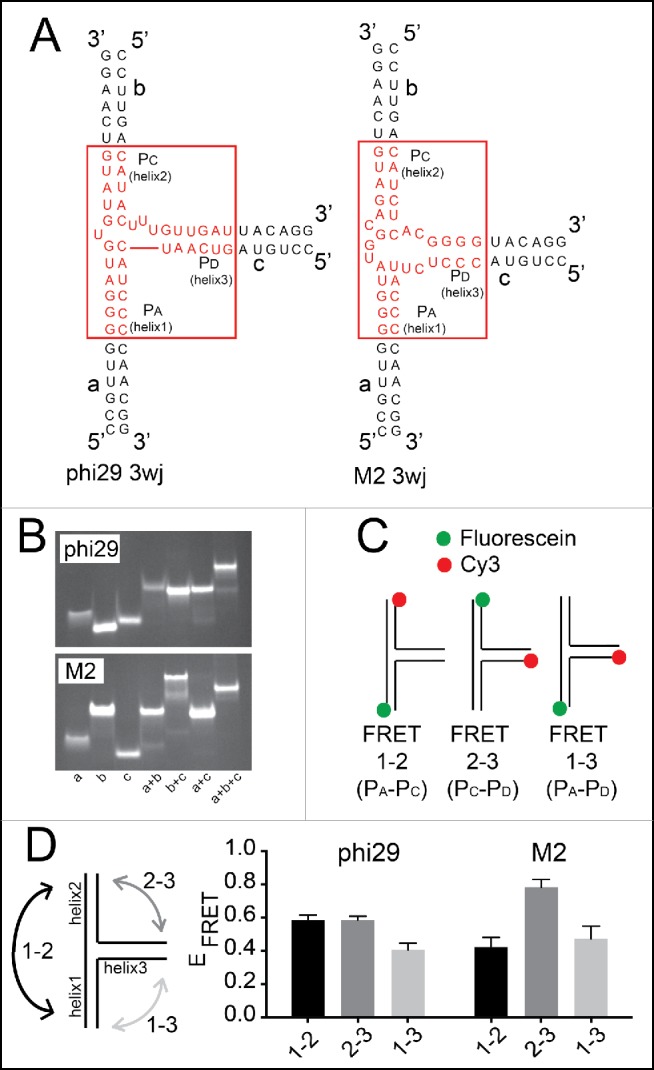
Förster resonance energy transfer (FRET) examination of 3wj conformation. (A) Diagrams of 3wj RNAs used for FRET experiments. These RNAs were comprised of 3 RNA strands (a, b, and c) that anneal to form the 3wj. The 3wjs derived from phi29 and M2 are shown in red. (B) Native gel electrophoresis of combinations of the 3 strands for each pRNA 3wj. The a+b+c combination (1μg each) results in a band of unique mobility, indicating 3wj formation. The native gels contained 5 mM Mg2+ and were run at 25°C. (C) Dye labeling and FRET strategy to examine the relative distances between the ends of helices that emerge from the 3wjs. The fluorophores in FRET 1-2 are sensitive to the distance between the ends of helix 1 (H1) and helix 2 (H2). FRET 2-3 measures the distance between the ends of helix 2 (H2) and helix 3 (H3). FRET 1-3 measures the distance between the ends of helix 1 (H1) and helix 3 (H3). The donor fluorophore (fluorescein) is green and the acceptor fluorophore (Cy3) is red. (D) Left: diagram of the FRET measurements. Right: FRET efficiencies for phi29 and M2 pRNA 3wjs for each label pair. The diagram to the left illustrates the FRET pair for each measurement.
To perform the FRET measurements, the 5’ end of each RNA strand was labeled with a fluorescein or Cy3 dye, and then one unlabeled strand and 2 labeled strands were mixed in different combinations such that each combination had a FRET donor on the 5’ end of one strand and an acceptor on the 5’ end of another (Fig. 3C). This yielded 3 samples; each is able to report on the distance between the 5’ ends of 2 different helices. We measured the FRET efficiency of each combination for both the phi29 and M2 pRNA 3wjs at 25°C (Fig. 3D). For the phi29 construct, “FRET 1-2” and “FRET 2-3” had similar FRET efficiencies, both higher than “FRET 1-3”. This suggests that the distances between the ends of helices 1 and 2, and helices 2 and 3 are, on average, similar. By extension, the distance between the ends of helices 1 and 3 is greater. For the M2 construct, the efficiencies of “FRET 1-2” and “FRET 1-3” were similar, suggesting similar distances between the ends of helices 1 and 2 and 1 and 3. Interestingly, the M2 pRNA 3wj “FRET 2-3” had the highest FRET efficiency, indicating the average distance between the ends of helices 2 and 3 is the closest of all measured. The different patterns in the FRET data from the phi29 and M2 pRNA 3wjs support the hypothesis that the 2 junctions have different interhelical angles. Because the FRET signal reflects the populationally weighted average of conformations in solution, even if the junctions are conformationally dynamic, the data show that they sample somewhat different states or spend different amounts of time in each state.
Higher-order self-association depends on cooperation between favorable junctions
The differing properties of the phi29 and M2 pRNA 3wjs raised the question of whether a population of pRNAs containing a mixture of the 2 junctions but able to form identical kissing-loop interactions would favor dimer or trimer formation. To test this, we combined M2_phi29 and M2 in 2 experiments. When M2 was added to radiolabeled M2_phi29, the M2_phi29 maintained a dimer migration pattern on a native gel; it was not forced into trimers although there is some upward smearing perhaps indicative of very transient higher-order multimers (Fig. 4A). Likewise, when M2_phi29 was added to radiolabeled M2 pRNA, trimer formation was inhibited (Fig. 4B). Because the kissing-loop interactions were identical, this further indicates that the 3wj is a key determinant of self-association behavior and also suggests that the junction facilitates the specific and correct alignment of intermolecular interaction surfaces (kissing-loop base-pairs) within the higher-order structure of the multimer.
Figure 4.
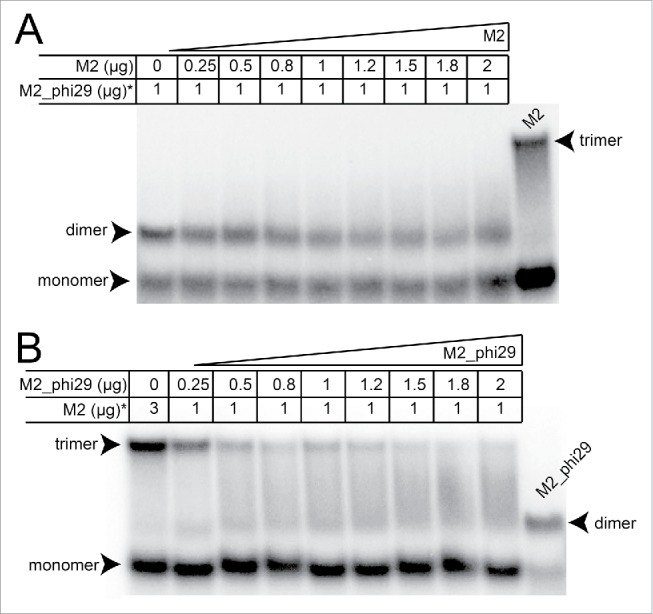
Native gel electrophoresis of M2_phi29 and M2 combinations. (A) Native gel in which increasing amounts of M2 pRNA (Fig. 1D) was added to radiolabeled M2_phi29 (Fig. 1D). The total amount of each pRNA loaded is shown above the lanes. Radiolabeled M2 pRNA (3 μg) is included for reference. The locations of trimers, dimers, and monomers are indicated. Under these conditions, both pRNAs exist as a mix of monomer and multimer. Addition of M2 pRNA does not induce M2_phi29 to form trimers, although some smearing of the dimer band “upwards” is seen. (B) Native gel of unlabeled M2_phi29 added in increasing amounts of radiolabeled M2 pRNA. Lanes are labeled as in panel (A). Radiolabeled M2_phi29 is included for reference. The ability of M2 pRNA to form trimers is disrupted by addition of M2_phi29, resulting in a smear between the two. The native gels contained 5 mM Mg2+ and were run at 4°C.
Discussion
The pRNAs from the phi29 family of bacteriophage are naturally occurring self-associating RNAs that form homomultimers of diverse sizes through kissing-loop base-pairing and thus serve not only as models for this type of RNA behavior but also as scaffolds to engineer RNAs for nanotechnology. Although the pRNAs from different members of the phi29 family have similar secondary structures, recent studies have shown that their inherent self-association behaviors when free in solution differ both in terms of stability and size of the resultant multimers.17,19 This variation provides a way to explore the basic rules of RNA self-association and identify new strategies for pRNA engineering. Two pRNAs that were previously shown to differ in their self-association characteristics are those from the phi29 and M2 phage strains.17 Here, we explored the idea that this difference may depend in part on an element other than the intermolecular kissing-loop base-pair interaction at the interface between identical pRNA molecules. Using a biochemical and biophysical approach, we found that the 3wj in these pRNAs is a key element in driving multimerization behavior. Our data suggest that this is due to differences in the angles favored by the RNA helices that emerge from each junction, which likely spatially position the kissing-loop base-pair elements to affect RNA self-association and the formation of higher-order multimers. Hence, efforts to use the pRNAs for nanotechnology or other uses should take into account the diverse behavior of these 3wjs to create self-associating RNAs with diverse programmed characteristics.
3wjs are common in RNA structures where they spatially position emerging helices, facilitating long-range intramolecular interactions.46,47 In the case of pRNAs, there is no evidence of distal intramolecular interactions that are affected by the 3wjs; rather the junctions may act to alter intermolecular interactions. If true, self-association should be affected by the characteristics of each junction. In fact, the phi29 and M2 pRNA 3wjs are predicted to have different characteristics based on analyses of other 3wjs. Specifically, diverse RNA 3wjs have been classified by Lescoute & Westhof based on crystal structures of a large number of 3wj-containing RNAs.46 They divided the 3wjs into 3 families depending on the relative lengths of the single-stranded segments within the junction that link the helices, finding that this correlates with different helical angles. Briefly, by counting the number of unpaired nucleotides that link each helical element, the junction can be assigned to its likely class. Based on this analysis, the phi29 pRNA 3wj is predicted to be in family A; in this family there is no predicted preference for the arrangements of the 3 helices. However, the M2 pRNA 3wj likely belongs to family B, which has a preferred helical arrangement with 2 emerging helices at an acute angle. Neither 3wj has a pattern and number of unpaired bases that match family C. This classification system predicted inherent differences in the preferred conformations of the phi29 and M2 pRNA 3wjs, supporting a hypothesis regarding different characteristics of these 2 3wjs.
Although the conformation of the M2 pRNA 3wj has not been previously explored, the phi29 pRNA 3wj has been studied by several methods, including pulsed electron paramagnetic resonance (EPR) and X-ray crystallography.14,54,56,57 EPR studies suggested that the PA and PD helices emerge from the junction at an acute angle, similar to the angle predicted in a model based on FRET measurements between 2 pRNAs within a dimer.56 In contrast, the crystal structure of a version of this 3wj RNA that lacked any ability to form intermolecular kissing-loop base-pairs showed these helices to be stacked coaxially, with the third helix (PC) emerging at an acute angle relative to PA (Fig. 5A).54 Furthermore, the crystal structure of the phi29 pRNA 3wj in a tetrameric state showed a different set of angles, with PC at a more acute angle to PD than in the monomeric form (Fig. 5A).14 These observations support the idea that the 3wj from the phi29 pRNA is a conformationally dynamic or flexible element. In fact, this characteristic has been proposed to be important for multimerization.17,56 These data also suggest that the angles between the helices must change as the pRNA goes from a monomeric to a multimeric form (Fig. 5B).
Figure 5.
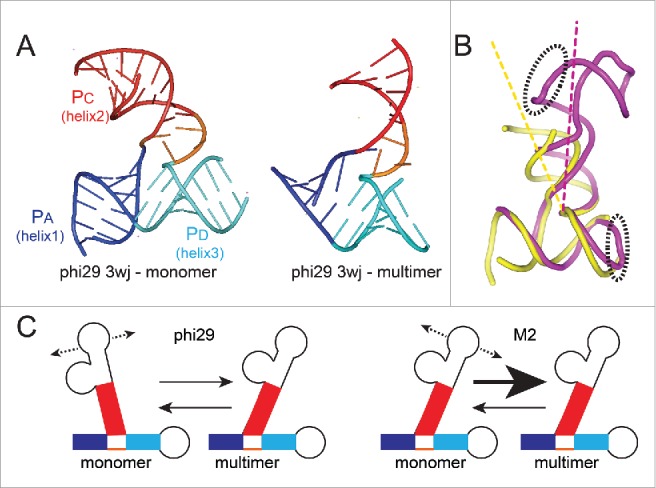
Conformation of 3wjs and putative relationship to multimer formation. (A) Left: Ribbon diagram of the structure of the isolated 3wj from phi29 pRNA, solved by X-ray crystallography (PDB: 4KZ2).54 This RNA did not have the kissing-loop interaction elements and thus was unable to self-associate and is considered to be the structure in monomer form. Right: Ribbon diagram of the structure of the 3wj from phi29 pRNA, within the context of an RNA that was engineered for crystallization but could still self-associate, forming a tetramer in the crystal (PDB: 3R4F).14 For the latter structure, only the 3wj is shown. For both structures, the 3 helices that emerge from the junction are shown in blue, red, and cyan and labeled. (B) Superimposed structures of the 3wj in monomer (yellow) and multimer (magenta) form, using the structures from panel (A). The entire structure is shown, and the elements that form the intermolecular interaction important for self-association are in dashed ellipses. (C) Model for how the 3wj conformation may affect multimer formation. The 3wjs differ in their conformations. If they are conformationally dynamic, we propose the conformational equilibria of sampled states differs. For phi29 pRNA, this conformational equilibrium may be more different from the multimer state than it is in the M2 pRNA.
We used FRET to compare the conformation of the M2 pRNA 3wj to that of the phi29 pRNA, revealing an inherent difference in the average conformations of the 2 junctions in the monomeric state. Specifically, in the M2 pRNA 3wj, helices PC and PD emerge to form a more acute angle than they do in the phi29 pRNA 3wj, while PA and PC are less acute. Both junctions may be conformationally dynamic to some degree; even if this is true, the FRET data show differing conformational landscapes for each pRNA.
When the FRET data are compared to the crystal structures, an interesting correlation is observed: the isolated monomeric phi29 pRNA 3wj has characteristics that more closely resemble the conformation of the phi29 pRNA 3wj monomeric crystal structure. In contrast, the predicted relative angles of the helices emerging from the isolated monomeric M2 pRNA 3wj resemble the phi29 pRNA 3wj in a multimeric state. Most notably, the angle between PC and PD is more acute in the M2 pRNA 3wj in the isolated monomeric fold than it is in the phi29 pRNA 3wj, just as it is in the crystal structure of the phi29 pRNA in a multimer. Based on these data, we propose a model for how the M2 pRNA can form multimers with higher stoichiometry and stability than that of the phi29 pRNA: the 3wj of the M2 pRNA favors a conformation in which the 3 emerging helices are positioned in a way that more closely matches the angle of the helices when they are in a multimer and the kissing-loop interactions are spatially positioned to favor intermolecular interactions (Fig. 5C). This does not indicate that the M2 junction is structurally static; it could be fairly static or conformationally dynamic but on average spends more time in a state conducive to self-association. Thus, the multimerization behavior of diverse pRNAs depends in part on subtle differences in the specific conformational landscape of each 3wj.
In summary, the data presented here show that the 3wj plays a critical role in multimer formation by the naturally occurring self-associating pRNAs, and this is in part through differences in the conformations of the 3wj. Ongoing efforts to engineer artificial RNA nanoparticles into diverse shapes (including nanorings) require careful consideration of the use of diverse RNA elements to achieve self-assembly of the desired structures, to include helical junctions.58-60 Likewise, efforts to engineer the pRNAs for different self-association properties must take into account the different behaviors of 3wjs, to include subtle differences in their conformations and conformational dynamics. These differences may in fact present opportunities for additional engineering, to include efforts to subtly “tune” the behavior of each junction through mutation or alteration of adjoining helices.
Materials and methods
RNA preparation
RNAs for native gels were made using DNA templates generated by PCR with overlapping primers.49 RNAs were generated by in vitro transcription using the PCR products, and purified on 10% polyacrylamide (29:1 acrylamide:bisacrylamide) 7M urea denaturing gels. Bands were identified by UV shadowing, then excised and eluted into RNase-free water overnight at 4°C. The eluted RNA was concentrated using Amicon Ultra filters (Millipore). RNA concentrations were measured by UV absorbance at 260 nm (Nanodrop 2000c, Thermo Scientific).
RNA 5' End labeling
RNA was 5' dephosphorylated using rAPid Alkaline Phosphatase (Roche). One μg of RNA was added to 1X PNK buffer (70 mM Tris-HCl, pH 7.6, 10 mM MgCl2, 5 mM DTT), 20 units PNK enzyme and 32P-γ-ATP (PerkinElmer) in 10 μL, then incubated at 37°C for 1 hour followed by desalting using a Micro Bio-Spin 30 Chromatography Column (BioRad). Labeled RNA was purified on a 10% denaturing polyacrylamide gel. Gels were visualized by autoradiography, and the RNA bands were excised and eluted passively in elution buffer (0.5 M sodium acetate, 0.1% SDS, pH 5.3) overnight. This was followed by ethanol precipitation and the dried pellet was resuspended in RNase-free water prior to use.
Fe(II)-EDTA hydroxyl radical probing
32P end-labeled RNA was heat-denatured to 85°C for 15 seconds then cooled to room temperature. Each reaction contained RNA (at least 200,000 cpm), 30 mM HEPES-KOH (pH 7.5), 0.1 μg whole yeast tRNA, 5 mM MgCl2 for Mg samples or 0 mM MgCl2 for control samples, and RNase-free water to a total volume of 7 μL. Samples were then incubated at 4°C, 25°C or 37°C for 15 minutes to reach folding equilibrium. Cleavage was initiated by adding 1 μL Fe(II)-EDTA solution (10 mM (NH4)2Fe(SO4)2 and 18 mM EDTA, pH 8.0), 1 μL 1.5% H2O2, and 1 μL 10 mM NaAscorbate. The reaction was then incubated at 4°C, 25°C or 37°C for 5 minutes. Each reaction was quenched by adding 1 μL of 100 mM thiourea. Samples were precipitated with 10 μL of 3M sodium acetate and 100 μL 100% EtOH, pelleted by centrifugation, and dried. Samples were measured by scintillation counting and normalized based on counts per minute to each other by resuspending in 9 M urea loading buffer before being resolved on a denaturing sequencing gel along with the corresponding RNase T1 ladder and –OH ladder.49 Gels were electrophoresed for 2 hours, transferred to Whatman paper, dried, and observed by phosphorimaging. Each lane was quantified using ImageQuant to yield a trace of intensity vs. position in the gel. Each trace was normalized to total counts in the lane and baseline-corrected using a double polynomial curve fit, then graphed using the program KaleidaGraph.49
RNA synthesis and preparation of 3wjs
RNA oligonucleotides for 3wj formation were chemically synthesized (Integrated DNA Technologies). Three-way junctions were constructed by mixing 3 separate RNA strands (1 μg each) in 90 mM Tris-borate (pH 7.0) and 5 mM MgCl2, and annealed by heating at 85°C for 15 seconds and slowly cooled to 4°C. Sequences:
phi29 a strand: 5' –CCGUUGGGGAUGUGUAUGUCAAGG– 3'
phi29 b strand: 5' –CCUUGACAUACUUUGUUGAUUACAGG– 3'
phi29 c strand: 5' –CCUGUAGUCAAUCAUCCCCAACGG– 3'
M2 a strand: 5' –CCGUUGGGGUAUGGCAGAUGUCAAGG– 3'
M2 b strand: 5' –CCUUGACAUCUCACGGGGUACAGG– 3'
M2 c strand: 5' –CCUGUACCCUCUUACCCCAACGG– 3'
Förster resonance energy transfer (FRET)
Three-way junctions were constructed by mixing one fluorescein-labeled strand, one Cy3-labeled strand and the remaining unlabeled strand (each 10 nM) in 90 mM Tris-borate (pH 7.0) and 5 mM MgCl2, then annealed by heating at 85°C for 15 seconds and slowly cooled to 4°C. Samples were placed in a quartz fluorimeter cell (Starna Cells) and measured using a FluoroMax-3 (Horiba Jobin Yvon) at 25°C. FRET efficiency values (EFRET) were measured and calculated using the acceptor normalization method as described.51,52,61 Briefly, for each 3wj, emission spectra were recorded from 500 nm to 650 nm using 2 excitation wavelengths (490 nm or 550 nm). The spectra were also recorded from reaction buffer and donor-only sample controls. Emission from donor-acceptor samples that were excited at 490 nm had components from direct excitation of donor, direct excitation of acceptor, and acceptor emission due to energy transfer (Equation A and other equations shown below). Emission from donor samples that were excited at 490 nm contained pure donor emission with no energy transfer (Equation B). Emission from samples that were excited at 550 nm contained pure acceptor emission with no energy transfer (Equation C). The fluorescence intensity is a function of the extinction coefficient of the fluorophores (ϵ) and quantum yield (Φ) at the excited wavelength. So, the direct excitation of the donor was subtracted using the spectrum of donor-only sample (Equation A) and donor-acceptor sample (Equation B), resulting in an extracted spectrum that only has components due to acceptor emission (Equation D). The extracted acceptor emission spectrum was combined with the direct excitation of the acceptor emission spectrum to calculate the EFRET (Equation E). Since the ratios of emission values were determined at the same wavelengths, the quantum yield terms reduced to unity. The ratios ϵD(ν490) / ϵA(ν550) and ϵA(ν490) / ϵA(ν550) values measured from the absorption spectra were 0.45 and 0.23 respectively. The (ratio)A is calculated using the measured intensity of the Cy3 peak for the donor-acceptor sample excited at 490 nm over that of the Cy3 peak for the donor-acceptor sample excited at 550 nm.
Equation A (Direct excitation of donor + Direct excitation of acceptor + Acceptor emission due to energy transfer):
Equation B (Direct excitation of donor):
Equation C (Direct excitation of acceptor):
Equation D (Direct excitation of acceptor + Acceptor emission due to energy transfer):
Equation E (Extracted acceptor / direct acceptor excitation) = Equation D / Equation C:
Native gel electrophoresis
Native gel electrophoresis experiments were performed as described.49 Briefly, 3 μg RNA in a total volume of 10 μL was loaded. The pRNA sample was renatured by heating to 85°C for 15 seconds in Mg-Buffer (66 mM Tris, 34 mM HEPES, 5 mM MgCl2, pH 7.4) then snap-cooled on ice. Samples were mixed with equal volume of 2X native gel loading buffer (132 mM Tris, 68 mM HEPES, 10% glycerol, 0.5 mg/mL bromophenol blue, 0.5 mg/mL xylene cyanol, 10 mM MgCl2, pH 7.4) and were run on an 8% polyacrylamide native gel (29:1 acrylamide:bisacrylamide with Mg-Buffer for Mg2+ gels). Both gel and buffer were pre-incubated at desired temperatures and the gels were pre-run for 20 minutes at 5 W. Gels were run at 5 W for 10 hours and then stained with 1% ethidium bromide for visualization.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
The authors thank David Costantino, Dr. Mair Churchill and Dr. Mark Johnston for critical reading of the manuscript and for insightful comments, and current and former Kieft Lab members for thoughtful discussions and technical assistance. The FRET data was collected using the instruments at the Biophysics Core at the University of Colorado, Anschutz Medical Campus. The authors thank Dr. David M. Lilley of the University of Dundee for discussion about FRET. Viral RNA studies in the Kieft Lab are funded by National Institutes of Health Grants GM097333 and GM081346 (and previously by an Early Career Scientist Award from the Howard Hughes Medical Institute). YH was an American Heart Association Predoctoral Fellow (13PRE14500042).
References
- 1.Guo PX, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi 29 DNA. Science 1987; 236:690-4; PMID:3107124; http://dx.doi.org/ 10.1126/science.3107124 [DOI] [PubMed] [Google Scholar]
- 2.Meijer WJ, Horcajadas JA, Salas M. Phi29 family of phages. Microbiol Mol Biol Rev 2001; 65:261-87; second page, table of contents; PMID:11381102; http://dx.doi.org/ 10.1128/MMBR.65.2.261-287.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo PX, Bailey S, Bodley JW, Anderson D. Characterization of the small RNA of the bacteriophage phi 29 DNA packaging machine. Nucl Acid Res 1987; 15:7081-90; PMID:3116499; http://dx.doi.org/ 10.1093/nar/15.17.7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, Anderson DL, Rossmann MG. Structure determination of the head-tail connector of bacteriophage phi29. Acta Crystallogr D Biol Crystallogr 2001; 57:1260-9; PMID:11526317; http://dx.doi.org/ 10.1107/S0907444901010435 [DOI] [PubMed] [Google Scholar]
- 5.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature 2001; 413:748-52; PMID:11607035; http://dx.doi.org/ 10.1038/35099581 [DOI] [PubMed] [Google Scholar]
- 6.Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J Mol Bio 2002; 315:663-76; PMID:11812138; http://dx.doi.org/ 10.1006/jmbi.2001.5278 [DOI] [PubMed] [Google Scholar]
- 7.Serwer P. Models of bacteriophage DNA packaging motors. J Struct Biol 2003; 141:179-88; PMID:12648564; http://dx.doi.org/ 10.1016/S1047-8477(02)00628-7 [DOI] [PubMed] [Google Scholar]
- 8.Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DE. Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi 29. Proc Natl Acad Sci U S A 2007; 104:11245-50; PMID:17556543; http://dx.doi.org/ 10.1073/pnas.0701323104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol 2007; 5:e59; PMID:17311473; http://dx.doi.org/ 10.1371/journal.pbio.0050059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson DL, Rossmann MG. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure 2008; 16:1267-74; PMID:18682228; http://dx.doi.org/ 10.1016/j.str.2008.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Gen 2008; 42:647-81; PMID:18687036; http://dx.doi.org/ 10.1146/annurev.genet.42.110807.091545 [DOI] [PubMed] [Google Scholar]
- 12.Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature 2009; 461:669-73; PMID:19794496; http://dx.doi.org/ 10.1038/nature08443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson MA, Wood JP, Capaldi SA, Baron AJ, Gell C, Smith DA, Stonehouse NJ. Affinity of molecular interactions in the bacteriophage phi29 DNA packaging motor. Nucl Acid Res 2006; 34:2698-709; PMID:16714447; http://dx.doi.org/ 10.1093/nar/gkl318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding F, Lu C, Zhao W, Rajashankar KR, Anderson DL, Jardine PJ, Grimes S, Ke A. Structure and assembly of the essential RNA ring component of a viral DNA packaging motor. Proc Natl Acad Sci U S A 2011; 108:7357-62; PMID:21471452; http://dx.doi.org/ 10.1073/pnas.1016690108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood JPA, Capaldi SA, Robinson MA, Baron AJ, Stonehouse NJ. RNA multimerisation in the DNA packaging motor of bacteriophage ϕ29. J Theoret Med 2005; 6:127-4; http://dx.doi.org/ 10.1080/10273660500149802 [DOI] [Google Scholar]
- 16.Chen C, Sheng S, Shao Z, Guo P. A dimer as a building block in assembling RNA. A hexamer that gears bacterial virus phi29 DNA-translocating machinery. J Biol Chem 2000; 275:17510-6; PMID:10748150; http://dx.doi.org/ 10.1074/jbc.M909662199 [DOI] [PubMed] [Google Scholar]
- 17.Hao Y, Kieft JS. Diverse self-association properties within a family of phage packaging RNAs. RNA 2014; 20:1759-74; PMID:25246655; http://dx.doi.org/ 10.1261/rna.045948.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single-molecule dual-view system. EMBO J 2007; 26:527-37; PMID:17245435; http://dx.doi.org/ 10.1038/sj.emboj.7601506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu X, Schroeder SJ. Different sequences show similar quaternary interaction stabilities in prohead viral RNA self-assembly. J Biol Chem 2011; 286:14419-26; PMID:21349846; http://dx.doi.org/ 10.1074/jbc.M110.191064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C, Guo P. Sequential action of six virus-encoded DNA-packaging RNAs during phage phi29 genomic DNA translocation. J Virol 1997; 71:3864-71; PMID:9094662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, Zhang C, Guo P. Sequence requirement for hand-in-hand interaction in formation of RNA dimers and hexamers to gear phi29 DNA translocation motor. RNA 1999; 5:805-18; PMID:10376879; http://dx.doi.org/ 10.1017/S1355838299990350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garver K, Guo P. Mapping the inter-RNA interaction of bacterial virus phi29 packaging RNA by site-specific photoaffinity cross-linking. J Biol Chem 2000; 275:2817-24; PMID:10644747; http://dx.doi.org/ 10.1074/jbc.275.4.2817 [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Guo P. Magnesium-induced conformational change of packaging RNA for procapsid recognition and binding during phage phi29 DNA encapsidation. J Virol 1997; 71:495-500; PMID:8985376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell 1998; 2:149-55; PMID:9702202; http://dx.doi.org/ 10.1016/S1097-2765(00)80124-0 [DOI] [PubMed] [Google Scholar]
- 25.Bailey S, Wichitwechkarn J, Johnson D, Reilly BE, Anderson DL, Bodley JW. Phylogenetic analysis and secondary structure of the Bacillus subtilis bacteriophage RNA required for DNA packaging. J Biol Chem 1990; 265:22365-70; PMID:2125049 [PubMed] [Google Scholar]
- 26.Zhao W, Saha M, Ke A, Morais MC, Jardine PJ, Grimes S. A three-helix junction is the interface between two functional domains of prohead RNA in 29 DNA packaging. J Virol 2012; 86:11625-32; PMID:22896620; http://dx.doi.org/ 10.1128/JVI.01370-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao F, Zhang H, Guo P. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucl Acid Res 2008; 36:6620-32; PMID:18940870; http://dx.doi.org/ 10.1093/nar/gkn669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mat-Arip Y, Garver K, Chen C, Sheng S, Shao Z, Guo P. Three-dimensional interaction of Phi29 pRNA dimer probed by chemical modification interference, cryo-AFM, and cross-linking. J Biol Chem 2001; 276:32575-84; PMID:11371551; http://dx.doi.org/ 10.1074/jbc.M100045200 [DOI] [PubMed] [Google Scholar]
- 29.Trottier M, Mat-Arip Y, Zhang C, Chen C, Sheng S, Shao Z, Guo P. Probing the structure of monomers and dimers of the bacterial virus phi29 hexamer RNA complex by chemical modification. RNA 2000; 6:1257-66; PMID:10999603; http://dx.doi.org/ 10.1017/S1355838200992501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee TJ, Schwartz C, Guo P. Construction of bacteriophage phi29 DNA packaging motor and its applications in nanotechnology and therapy. Annal Biomed Eng 2009; 37:2064-81; PMID:19495981; http://dx.doi.org/ 10.1007/s10439-009-9723-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye X, Hemida M, Zhang HM, Hanson P, Ye Q, Yang D. Current advances in Phi29 pRNA biology and its application in drug delivery. Wiley Interdiscip Rev RNA 2012; 3:469-81; PMID:22362726; http://dx.doi.org/ 10.1002/wrna.1111 [DOI] [PubMed] [Google Scholar]
- 32.Shu D, Moll WD, Deng Z, Mao C, Guo P. Bottom-up Assembly of RNA Arrays and Superstructures as Potential Parts in Nanotechnology. Nano Lett 2004; 4:1717-23; PMID:21171616; http://dx.doi.org/ 10.1021/nl0494497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westhof E, Masquida B, Jaeger L. RNA tectonics: towards RNA design. Fold Des 1996; 1:R78-88; PMID:9079386; http://dx.doi.org/ 10.1016/S1359-0278(96)00037-5 [DOI] [PubMed] [Google Scholar]
- 34.Yan H. Materials science. Nucleic acid nanotechnology. Science 2004; 306:2048-9; PMID:15604395; http://dx.doi.org/ 10.1126/science.1106754 [DOI] [PubMed] [Google Scholar]
- 35.Ko SH, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Synergistic self-assembly of RNA and DNA molecules. Nat Chem 2010; 2:1050-5; PMID:21107369; http://dx.doi.org/ 10.1038/nchem.890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dibrov SM, McLean J, Parsons J, Hermann T. Self-assembling RNA square. Proc Natl Acad Sci U S A 2011; 108:6405-8; PMID:21464284; http://dx.doi.org/ 10.1073/pnas.1017999108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nature materials 2012; 11:316-22; PMID:22367004; http://dx.doi.org/ 10.1038/nmat3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikawa J, Furuta H, Ikawa Y. RNA tectonics (tectoRNA) for RNA nanostructure design and its application in synthetic biology. Wiley Interdiscip Rev RNA 2013; 4:651-64; PMID:23836522; http;//dx.doi.org/ 10.1002/wrna.1185 [DOI] [PubMed] [Google Scholar]
- 39.Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum Gen Ther 2005; 16:1097-109; PMID:16149908; http://dx.doi.org/ 10.1089/hum.2005.16.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoeprich S, Guo P. Computer modeling of three-dimensional structure of DNA-packaging RNA (pRNA) monomer, dimer, and hexamer of Phi29 DNA packaging motor. J Biol Chem 2002; 277:20794-803; PMID:11886855; http://dx.doi.org/ 10.1074/jbc.M112061200 [DOI] [PubMed] [Google Scholar]
- 41.Khisamutdinov EF, Jasinski DL, Guo P. RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano 2014; 8:4771-81; PMID:24694194; http://dx.doi.org/ 10.1021/nn5006254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L, Liu J, Diao Z, Shu D, Guo P, Shen G. Evaluation of specific delivery of chimeric phi29 pRNA/siRNA nanoparticles to multiple tumor cells. Mol Biosyst 2009; 5:1361-8; PMID:19823753; http://dx.doi.org/ 10.1039/b903428e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tarapore P, Shu Y, Guo P, Ho SM. Application of phi29 motor pRNA for targeted therapeutic delivery of siRNA silencing metallothionein-IIA and survivin in ovarian cancers. Mol Ther 2011; 19:386-94; PMID:21063391; http://dx.doi.org/ 10.1038/mt.2010.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo P. Bacterial virus phi29 DNA-packaging motor and its potential applications in gene therapy and nanotechnology. Methods Mol Biol 2005; 300:285-324; PMID:15657489 [DOI] [PubMed] [Google Scholar]
- 45.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat Nanotechnol 2011; 6:658-67; PMID:21909084; http://dx.doi.org/ 10.1038/nnano.2011.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA 2006; 12:83-93; PMID:16373494; http://dx.doi.org/ 10.1261/rna.2208106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de la Pena M, Dufour D, Gallego J. Three-way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold. RNA 2009; 15:1949-64; PMID:19741022; http://dx.doi.org/ 10.1261/rna.1889509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dixon WJ, Hayes JJ, Levin JR, Weidner MF, Dombroski BA, Tullius TD. Hydroxyl radical footprinting. Method Enzymol 1991; 208:380-413; PMID:1664026; http://dx.doi.org/ 10.1016/0076-6879(91)08021-9 [DOI] [PubMed] [Google Scholar]
- 49.Kieft JS, Costantino DA, Filbin ME, Hammond J, Pfingsten JS. Structural methods for studying IRES function. Method Enzymol 2007; 430:333-71; PMID:17913644; http://dx.doi.org/ 10.1016/S0076-6879(07)30013-X [DOI] [PubMed] [Google Scholar]
- 50.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Bio 1999; 292:513-29; PMID:10497018; http://dx.doi.org/ 10.1006/jmbi.1999.3095 [DOI] [PubMed] [Google Scholar]
- 51.Fessl T, Lilley DM. Measurement of the change in twist at a helical junction in RNA using the orientation dependence of FRET. Biophys J 2013; 105:2175-81; PMID:24209863; http://dx.doi.org/ 10.1016/j.bpj.2013.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ouellet J, Melcher S, Iqbal A, Ding Y, Lilley DM. Structure of the three-way helical junction of the hepatitis C virus IRES element. RNA 2010; 16:1597-609; PMID:20581129; http://dx.doi.org/ 10.1261/rna.2158410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.St-Pierre P, McCluskey K, Shaw E, Penedo JC, Lafontaine DA. Fluorescence tools to investigate riboswitch structural dynamics. Biochimica et biophysica acta 2014; 1839:1005-19; PMID:24863161; http://dx.doi.org/ 10.1016/j.bbagrm.2014.05.015 [DOI] [PubMed] [Google Scholar]
- 54.Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, Chi YI. Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA 2013; 19:1226-37; PMID:23884902; http://dx.doi.org/ 10.1261/rna.037077.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savinov A, Perez CF, Block SM. Single-molecule studies of riboswitch folding. Biochimica et biophysica acta 2014; 1839:1030-45; PMID:24727093; http://dx.doi.org/ 10.1016/j.bbagrm.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X, Tung CS, Sowa GZ, Hatmal MM, Haworth IS, Qin PZ. Global structure of a three-way junction in a phi29 packaging RNA dimer determined using site-directed spin labeling. J Am Chem Soc 2012; 134:2644-52; PMID:22229766; http://dx.doi.org/ 10.1021/ja2093647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shu D, Zhang H, Petrenko R, Meller J, Guo P. Dual-channel single-molecule fluorescence resonance energy transfer to establish distance parameters for RNA nanoparticles. ACS Nano 2010; 4:6843-53; PMID:20954698; http://dx.doi.org/ 10.1021/nn1014853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Afonin KA, Grabow WW, Walker FM, Bindewald E, Dobrovolskaia MA, Shapiro BA, Jaeger L. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat Protoc 2011; 6:2022-34; PMID:22134126; http://dx.doi.org/ 10.1038/nprot.2011.418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Afonin KA, Kasprzak WK, Bindewald E, Kireeva M, Viard M, Kashlev M, Shapiro BA. In silico design and enzymatic synthesis of functional RNA nanoparticles. Account Chem Res 2014; 47:1731-41; PMID:24758371; http://dx.doi.org/ 10.1021/ar400329z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Afonin KA, Viard M, Koyfman AY, Martins AN, Kasprzak WK, Panigaj M, Desai R, Santhanam A, Grabow WW, Jaeger L, et al. Multifunctional RNA nanoparticles. Nano letters 2014; 14:5662-71; PMID:25267559; http://dx.doi.org/ 10.1021/nl502385k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol 1992; 211:353-88; PMID:1406315; http://dx.doi.org/ 10.1016/0076-6879(92)11020-J [DOI] [PubMed] [Google Scholar]



