ABSTRACT
The microRNA-26 (miR-26) family is known to control adipogenesis in non-ruminants. The genomic loci of miR-26a and miR-26b have been localized in the introns of genes encoding for the proteins of the C-terminal domain RNA polymerase II polypeptide A small phosphatase (CTDSP) family. Insulin-induced gene 1 (INSIG1) encodes a protein with a key role in the regulation of lipogenesis in rodent liver. In the present study, we investigated the synergistic function of the miR-26 family and their host genes in goat mammary epithelial cells (GMEC). Downregulation of miR-26a/b and their host genes in GMEC decreased the expression of genes relate to fatty acid synthesis (PPARG, LXRA, SREBF1, FASN, ACACA, GPAM, LPIN1, DGAT1 and SCD1), triacylglycerol accumulation and unsaturated fatty acid synthesis. Luciferase reporter assays confirmed INSIG1 as a direct target of miR-26a/b. Furthermore, inhibition of the CTDSP family also downregulated the expression of INSIG1. Taken together, our findings highlight a functional association of miR-26a/b, their host genes and INSIG1, and provide new insights into the regulatory network controlling milk fat synthesis in GMEC. The data indicate that targeting this network via nutrition might be important for regulating milk fat synthesis in ruminants.
KEYWORDS: CTDSP family, INSIG1, microRNA-26, TAG accumulation
Introduction
The goat (capra hircus) is one of the most important livestock species and the oldest to be domesticated.35,43 Goats have long been used for their milk, meat, hair and skin.11 Milk fat is one of the main components in goat dairy products, and provides an important source of nutrition for humans.2 Ninety-eight percent of goat milk fat is composed of triacylglycerol (TAG),25 which is mainly stored in cytoplasmic lipid droplets and secreted from mammary epithelial cells (MEC).16 Milk fat quality and production have recently become important concerns in the dairy industry. Therefore, a better understanding of the regulation of lipid metabolism in goat mammary epithelial cells (GMEC) will facilitate development of strategies for optimizing goat milk fat quality to help drive the profitability of the dairy industry.
Lipid metabolism in MEC involves various metabolic processes including de novo fatty acid synthesis, fatty acid uptake, and fat droplet formation and secretion. For instance, fatty acids (FA) are synthesized de novo via fatty acid synthase (FASN) and acetyl-CoA carboxylase α (ACACA);7 some of these fatty acids are then unsaturated by stearoyl-CoA desaturase 1 (SCD1) and processed into TAG by diacylglycerol acyltransferase1 (DGAT1) in the endoplasmic reticulum (ER).7 Glycerol-3-phosphate acyltransferase mitochondrial (GPAM), 1-acylglycerol-3-phosphate O-acyltransferase 6 (AGPAT6), and lipin 1 (LPIN1) also are involved in TAG synthesis;32,42,7 Transport of FA from outside the cell occurs primarily via thrombospondin receptor (CD36) and fatty acid-binding protein 4 (FABP4) transports the incoming FA for incorporation into TAG.7 Perilipin 2 (PLIN2), perilipin 3 (PLIN3) and patatin-like-phospholipase domain containing 2 (PNPLA2) are involved in lipid droplet formation.34,28 In addition, peroxisome proliferator-activated receptor gamma (PPARG),47 sterol regulatory element binding transcription factor 1 (SREBF1),18 and insulin-induced gene 1 (INSIG1)54 are key transcription regulators controlling the expression of genes related to FA and TAG synthesis.
INSIG1 is a membrane-spanning (polytopic) protein of the ER that regulates lipid synthesis by retaining sterol regulatory element binding transcription factors (SREBFs) in the ER and preventing their proteolytic activation in the Golgi apparatus.54,15 When INSIG1 is expressed at a low level, SREBF cleavage-activating protein (SCAP) escort SREBF to the Golgi, where they are processed and released into the cytosol on their way to the nucleus to activate transcription of lipogenic genes.13,44
MicroRNA (miRNA) are small, non-coding RNA molecules that regulate mRNA expression at the post-transcriptional level by binding to the 3’- untranslated regions (3’-UTR) of target mRNA to either direct mRNA translation or disrupt mRNA degradation.3 Several miRNA have been reported to modify cell lipid metabolism by regulating lipid-related gene expression. For instance, the miR-26 family is involved in adipocyte development and the acquisition of brown adipocyte characteristics in humans.22 The genomic loci of miR-26a and miR-26b are localized in the introns of genes encoding the protein family C-terminal domain RNA polymerase Ⅱpolypeptide A small phosphatase (CTDSP). Transcription of miR-26a and miR-26b occurs via three genomic loci, miR-26a-1, miR-26a-2 and miR-26b, which reside in the introns of genes coding for CTDSPL, CTDSP2 and CTDSP1, respectively. Analysis of 175 human miRNA across 24 different human tissues revealed that the expression of intronic miRNA coincides with the transcription of their host transcriptional units,5 indicating that the intronic miRNA and its host gene may be co-regulated and generated from a common precursor transcript.
The mechanistic implications of altering a transcription factor or a miRNA need to be considered carefully as their down-stream effects are multifold. Furthermore, their mutual interaction might be essential for the equilibrium of the cellular microenvironment. Previous studies in non-ruminants revealed that activation of PPARG, LXRA and SREBF altered miRNA expression profiles,9,10 hence, revealing a novel and physiologically-important network that could control lipogenesis.
The specific objective of this study was to evaluate the function of miR-26a/b and their host genes on GMEC lipid metabolism. To achieve this objective, we measured their expression profile in goat mammary gland tissue, and utilized an overexpression and downregulation approach to uncover their role on TAG accumulation in goat MEC (GMEC).
Results
miR-26a/b and their host ghenes are dynamically expressed in goat mammary gland tissue
To explore the involvement of miRNA in milk fat synthesis in goat mammary gland, RT-PCR analysis at various time points during lactation cycle revealed several dynamically regulated miRNA (unpublished observations), among which were miR-26a and -26b. As depicted in Fig. 1A, a significant upregulation of both miRNA was observed in early, peak and mid-lactation relative to the dry period. MiR-26a and miR-26b are transcribed from three genomic loci, miR-26a-1, miR-26a-2 and miR-26b that reside in the intros of genes encoding for the CTDSPL, CTDSP2 and CTDSP1 proteins, respectively. Based on these findings, the expression profile of CTDSP1/2/L during the lactation cycle also was evaluated. We observed that CTDSP1/2/L displayed a marked upregulation in early, peak and mid-lactation relative to the dry period, and the expression of CTDSPL was not altered between peak and mid-lactation (Fig. 1B). Similarly, we observed that PPARG, SREBF1 and LXRA, the well-known transcription factors involved in lipid metabolism, exhibited a significant increase during peak-lactation.7,47 Thus, it was hypothesized that induction of miR-26a/b could be a critical event for milk fat synthesis in goat mammary gland which may be regulated by PPARG, SREBF1 and LXRA.
Figure 1.
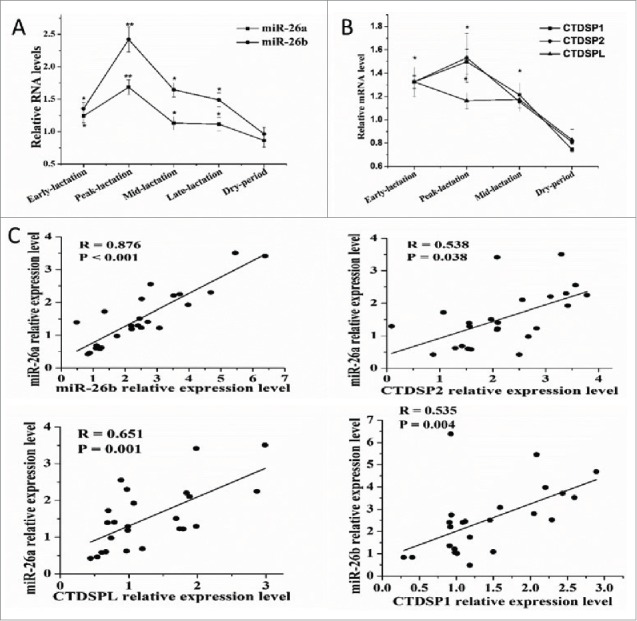
MiR-26a/b are expressed concomitantly with CTDSP1/2/L during lactation. (A) Analysis of miR-26a/b expression in dairy goat mammary gland tissue during different stages of lactation stages. (B) Analysis of CTDSP1/2/L expression in dairy goat mammary gland tissue during different stages of lactation stages. (A) and (B) were performed in quintuplicate and repeated 3 times (n = 15). (C) The correlation between expression levels of miR-26a/b and their host genes in 24 mid-lactation mammary gland of goats. Values are presented as means + standard error of the means, *, P < 0.05; **, P < 0.01.
miR-26a/b are expressed concomitantly with their host genes in mid-lactation mammary gland tissue
To better evaluate the relationship between miR-26a/b and CTDSP1/2/L, data on mRNA expression in mammary gland tissue from goats at mid lactation (n = 24) was used for correlation analysis. There was a strong correlation between miR-26a and miR-26b expression (R = 0.876, P < 0.001, Fig. 1C), and between miR-26a/b and their respective host genes (Fig. 1C) in mid-lactation mammary gland. These results indicated that miR-26a/b and their host genes may be co-regulated in mid-lactation mammary gland.
PPARG, SREBF1 and LXRA modify miR-26 family and their host genes expression profile in GMEC
Because PPARG,47 SREBF112 and LXRA51,1 appear to be key regulators in ruminant mammary gland lipid synthesis, the use of Ad-PPARG, Ad-SREBF1, and the LXRA agonist-T0901317 (T09) to study the expression profile of the miR-26 and their host genes would provide evidence of their link with milk fat synthesis. Compared with the control, as shown in Fig. 2, the expression of miR-26a was increased by ~1.76 (P < 0.01), ~2.28 (P < 0.01) and ~4.99 fold (P < 0.01), respectively (Fig. 2A), in cultures with Ad-PPARG/Ad-SREBF1 and the LXRA agonist-T09. Similar to miR-26a, there was an upregulation compared with the controls of miR-26b in cultures with all the Ad-PPARG/Ad-SREBF1 and T09 (Fig. 2A). The expression of miR-26a/b host genes also was upregulated by all these treatments (Fig. 2B). These results suggested that the miR-26 family and the expression of their host genes are regulated at least in part by the transcription factors PPARG, SREBF1 and LXRA and may be involved in milk fat synthesis in goat mammary gland.
Figure 2.
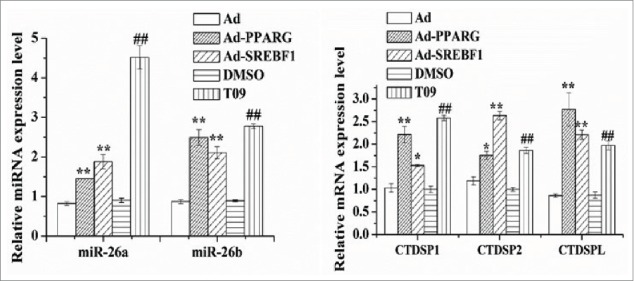
Induction of miR-26a/b and host genes expression by transcription factors. Cells were treated with Ad-PPARG/Ad-SREBF1 and T0901317 (T09), and then the miR-26a/b and their host genes expression levels was quantified by quantitative real-time PCR. Each treatment was carried out in triplicate and repeated 3 times (n = 9). Values are presented as mean ± SEM; *P < 0.05, **P < 0.01 vs Ad, #P < 0.05, ##P < 0.01 vs DMSO.
miR-26a/b and their host genes are functional on the key genes related to lipid metabolism
To investigate the effect of the miR-26 family on the expression of key genes involved in fatty acid metabolism, we transfected GMEC with a miR-26 family inhibitor or mimic. For each inhibitor, RT-PCR detected a greater than 90% decrease of corresponding miRNA levels (Fig. S1A), which reflects a degradation or sequestration of miRNAs by the inhibitor. As assayed by real-time PCR, miR-26 family knockdown decreased the expression of PPARG, LXRA and SREBF1, de novo FA synthesis genes (FASN and ACACA), genes related to TAG synthesis including GPAM, LPIN1 and DGAT1, and SCD1 (Table 1).
Table 1.
miR-26a/b function synergistically with their host genes to regulate FA synthesis gene expression.
| Gene symbol | anti-miR-C/NC | anti-miR-26a/NC | anti-miR-26b/NC | anti-miR-26a/b | siCTDSP1/anti-miR-C | siCTDSP2/anti-miR-C | siCTDSPL/anti-miR-C | anti-miR-26a/siCTDSP2 | anti-miR-26a/siCTDSPL | anti-miR-26b/siCTDSP1 |
|---|---|---|---|---|---|---|---|---|---|---|
| ACACA | 1.30 + 0.08 | 0.83 + 0.01* | 0.89 + 0.03* | 0.79 + 0.02* | 1.00 + 0.03 | 0.81 + 0.04* | 1.00 + 0.07 | 0.76 + 0.02# | 0.76 + 0.04 | 0.86 + 0.10 |
| AGPAT6 | 1.06 + 0.05 | 0.95 + 0.03 | 1.04 + 0.09 | 0.94 + 0.08 | 1.00 + 0.10 | 0.91 + 0.06 | 1.07 + 0.05 | 1.02 + 0.02 | 1.24 + 0.05 | 1.05 + 0.06 |
| CD36 | 0.82 + 0.01 | 0.71 + 0.06 | 0.59 + 0.03* | 0.55 + 0.06* | 1.00 + 0.05 | 0.95 + 0.05 | 0.86 + 0.04 | 0.78 + 0.05$ | 0.46 + 0.06#,$ | 0.54 + 0.04$ |
| DGAT1 | 1.48 + 0.09 | 0.99 + 0.01* | 0.87 + 0.03* | 0.73 + 0.02** | 1.65 + 0.07 | 1.08 + 0.08* | 1.00 + 0.07* | 1.05 + 0.05 | 0.95 + 0.01 | 0.84 + 0.02$ |
| FASN | 1.76 + 0.09 | 1.42 + 0.10 | 0.90 + 0.03** | 0.82 + 0.02** | 1.10 + 0.04* | 1.49 + 0.11 | 1.00 + 0.10* | 1.03 + 0.05#,$ | 1.23 + 0.04 | 0.81 + 0.05$ |
| GPAM | 1.40 + 0.06 | 0.73 + 0.04** | 0.89 + 0.01* | 0.96 + 0.03* | 1.00 + 0.09 | 0.81 + 0.02* | 1.00 + 0.06 | 0.78 + 0.09 | 0.71 + 0.03 | 0.73 + 0.03# |
| INSIG1 | 1.12 + 0.13 | 2.33 + 0.11* | 3.50 + 0.03** | 3.94 + 0.16** | 3.13 + 0.04 | 2.29 + 0.05 | 2.54 + 0.08** | 2.70 + 0.10$ | 2.97 + 0.13 | 3.15 + 0.08 |
| LPIN1 | 1.30 + 0.05 | 0.85 + 0.09* | 0.84 + 0.04* | 0.84 + 0.04* | 1.00 + 0.01 | 1.16 + 0.09 | 1.19 + 0.02 | 0.74 + 0.01$ | 0.86 + 0.03 | 0.84 + 0.03 |
| LXRA | 1.19 + 0.06 | 0.78 + 0.02* | 0.81 + 0.03* | 0.76 + 0.03* | 0.84 + 0.09 | 0.80 + 0.06* | 1.00 + 0.01 | 0.74 + 0.02 | 0.77 + 0.03$ | 0.69 + 0.03$ |
| PLIN2 | 1.16 + 0.07 | 1.00 + 0.09 | 1.05 + 0.04 | 0.83 + 0.01* | 0.90 + 0.06 | 0.77 + 0.06* | 0.81 + 0.06* | 1.05 + 0.09 | 0.71 + 0.01 | 0.79 + 0 09 |
| PNPLA2 | 1.24 + 0.08 | 1.06 + 0.11 | 0.99 + 0.04 | 0.90 + 0.04* | 0.82 + 0.09 | 2.81 + 0.16** | 1.69 + 0.09 | 1.51 + 0.11# | 2.56 + 0.10#,$ | 1.70 + 0.04#,$ |
| PPARG | 0.82 + 0.10 | 0.61 + 0.06** | 0.54 + 0.03** | 0.60 + 0.02** | 0.61 + 0.03* | 0.98 + 0.02 | 0.86 + 0.07 | 0.56 + 0.02$ | 0.69 + 0.07 | 0.48 + 0.06 |
| SCD1 | 1.24 + 0.07 | 0.64 + 0.05** | 0.75 + 0.01* | 0.57 + 0.03** | 1.00 + 0.05 | 0.73 + 0.01** | 1.13 + 0.07 | 0.87 + 0.07 | 0.87 + 0.03 | 0.89 + 0.06 |
| SREBF1 | 1.62 + 0.03 | 1.45 + 0.07 | 0.87 + 0.08** | 0.71 + 0.03** | 0.96 + 0.06** | 1.03 + 0.07* | 1.00 + 0.02* | 0.91 + 0.03# | 0.94 + 0.09# | 0.71 + 0.03 |
GMEC were co-transfected with 120 nM oligonucleotides (60 nM each) and after 48 h post-transfection, the ACACA, AGPAT6, CD36, DGAT1, FASN, GPAM, INSIG1, LPIN1, LXRA, PLIN2, PNPLA2, PPARG, SCD1 and SREBF1 expression levels were quantified by quantitative real-time PCR. All experiments were performed in triplicate and repeated 3 times (n = 9). Values are presented as mean ± SEM;
P < 0.05,
P < 0.01 vs anti-miR-C/NC,
P < 0.05 vs anti-miR-26a/NC or anti-miR-26b/NC,
P < 0.05 vs siCTDSP1/anti-miR-C or siCTDSP2/ anti-miR-C or siCTDSPL/ anti-miR-C.
All the miR-26a and/or -26b mimic tested could effectively upregulate the expression of the miR-26 family (Fig. S1B). As a response to miR-26 downregulation, cells co-transfected with miR-26a and miR-26b mimic displayed marked up-regulation of genes responsible for de novo FA synthesis (FASN and ACACA), TAG synthesis (GPAM and DGAT1), FA desaturation (SCD1) and lipid droplet formation (PLIN2) (Fig. S2). A dramatic upregulation also was observed for the expression of PPARG and SREBF1 (Fig. S2). Furthermore, the effect of overexpression or knockdown of both miRNA was more pronounced in the expression of some lipid related genessuch as FASN, SREBF1 and LPIN1 (Table 1 and Fig. S2).
Although the miR-26 family and their host genes were co-expressed in goat mammary gland, and miR-26 seem to elicit a function effect on key genes related to lipid metabolism, the effect of CTDSP family members on the expression of genes related to lipid metabolism is still unknown. Therefore, we examined whether all host genes of miR-26a/b play a regulatory role in lipid metabolism. Silencing either of CTDSP family mRNA downregulated the expression of genes related to lipid metabolism in GMEC (Table 1). We further determined whether a synergistic effect exists between miR-26a/b and their host genes in terms of altering the expression of genes related to lipid metabolism. A more pronounced downregulation in the expression of FASN, CD36 and LPIN1 was observed when miR-26a and CTDSP2/L were co-suppressed, and the downregulation of FASN, ACACA, DGAT1, SREBF1 and CD36 was more evident when miR-26a/b and CTDSP1/2/L were co-suppressed (Table 1). Collectively, these data implicated that miR-26a/b may function synergistically with their host genes to regulate lipid metabolism in GMEC.
mir-26a/b function synergistically with their host genes to regulate FA synthesis
To evaluate the potential role of miR-26a/b and CTDSP family in the regulation of FA synthesis and further determine the functional significance of their association, we used 3 indices to analyze TAG accumulation: appearance of fat droplets, TAG content and unsaturated fatty acid (UFA) content. Inhibition of both miR-26a and miR-26b resulted in a marked downregulation of lipid droplet formation (Fig. 3A), TAG content (Fig. 3B) and UFA (Table 2), indicating a significant role of miR-26a/b in milk fat synthesis. To confirm the results from the loss-of-function study, a gain-of-function assay was performed with miR-26a/b mimic. Compared with the control, the miR-26a/b mimic caused a significant increase of the 3 indices (Fig S3 and Table S1). It is noteworthy that the 3 indices all were downregulated upon the knockdown of the CTDSP family (Fig. 3 and Table 2), and the downregulation effect of lipid droplet formation was more evident when miR-26a/b and their host gene family (CTDSP1/2/L) were co-suppressed (Fig. 3A).
Figure 3.
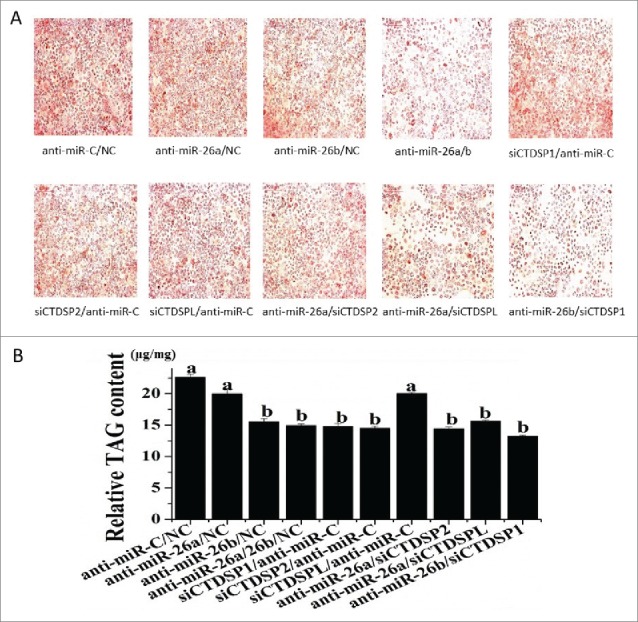
Changes in lipid formation and TAG content in GMEC caused by miR-26a/b and CTDSP family inhibition. GMEC were co-transfected with 120 nM oligonucleotides (60 nM each), and the lipid droplet and TAG level were measured at 48 h posttransfection. (A) Lipid droplet formation after oil red O treatment, scale bar: 200 μm. (B) Cellular TAG content. All experiments were performed in triplicate and repeated 3 times (n = 9). Values are presented as means ± SEM; a, b: significant difference among treatments in anti-miRNAs group (P < 0.05).
Table 2.
miR-26a/b function synergistically with their host genes to regulate FA composition in GMEC.
| Fatty acid | anti-miR-C/anti-miR-C | anti-miR-C/anti-miR-26a | anti-miR-C/anti-miR-26b | anti-miR -26a /-26b | NC | siCTDSP1 | siCTDSP2 | siCTDSPL |
|---|---|---|---|---|---|---|---|---|
| C16:0 (%) | 27.85 + 0.53c | 32.51 + 0.28ab | 31.6 + 0.30b | 34.24 + 0.43a | 28.12 + 0.23B | 32.99 + 0.20A | 31.99 + 0.18A | 29.16 + 0.12AB |
| C16:1 (%) | 4.43 + 0.21a | 3.13 + 0.17b | 2.23 + 0.09c | 1.8 + 0.08d | 4.03 + 0.21A | 2.99 + 0.24B | 3.13 + 0.04B | 3.94 + 0.14A |
| C18:0 (%) | 12.86 + 0.31b | 16.25 + 0.21a | 16.22 + 0.23a | 16.08 + 0.28a | 13.26 + 0.23B | 12.94 + 0.28B | 14.06 + 0.21A | 14.57 + 0.15A |
| C18:1 (%) | 53.01+ 0.56a | 46.43 + 0.39b | 48.66 + 0.63ab | 46.22 + 0.67b | 52.74 + 0.26A | 49.02 + 0.13B | 49.23 + 0.16B | 50.24 + 0.17AB |
| C18:2 (%) | 1.85 + 0.11a | 1.68 + 0.12a | 1.28 + 0.15b | 1.66 + 0.13a | 1.85 + 0.11A | 2.06 + 0.09A | 1.59 + 0.06B | 2.09 + 0.07A |
| SFA* (%) | 40.71 | 48.76 | 47.83 | 50.32 | 41.38 | 45.93 | 46.05 | 43.73 |
| UFA** (%) | 59.29 | 51.24 | 52.17 | 49.68 | 58.62 | 54.07 | 53.95 | 56.27 |
| UFA/SFA | 1.46 | 1.05 | 1.09 | 0.99 | 1.42 | 1.18 | 1.17 | 1.29 |
SFA: saturated fatty acids,
UFA: unsaturated fatty acids.
GMEC were co-transfected with 120 nM anti-miRNA oligonucleotides (60 nM each) or transfected with 60 nM siRNA. Relative fatty acid composition is calculated as percentages relative to the total of these fatty acids identified. All experiments were performed in triplicate and repeated 3 times (n = 9). Values are presented as means ± SEM; a, b: significant difference among treatments in anti-miRNA group (P < 0.05).A, B: significant difference among treatments in siRNA group (P < 0.05).
INSIG1 is a direct miR-26a/b target
Next, we sought to identify possible mechanisms underlying the upregulation of genes encoding the key proteins involved in lipid metabolism in GMEC. Analysis of the 3’-UTR of INSIG1 mRNA revealed one conserved miR-26a/b site. Thus, we hypothesized that miR-26a/b has a pro-adipogenic effect via blunting of the expression of INSIG1. To address the functional relevance between miR-26a/b and INSIG1, we analyzed the expression of miR-26a/b and INSIG1 in 24 mid-lactation goat mammary gland tissue samples. Comparison of INSIG1 level and mRNA levels corresponding to miR-26a and miR-26b resulted in a negative correlation between INSIG1 and miR-26a (R = −0.642, P = 0.001) and miR-26b (R = −0.560, P = 0.004) (Fig. 4A). Consistent with INSIG1 being a miR-26a/b target, the qPCR confirmed the downregulation of INSIG1 mRNA caused by miR-26a/b mimic, while miR-26a/b inhibitor caused a slight but significant upregulation (Fig. 4B and Table 1). Similar results were observed in the protein level of INSIG1 (Fig. 4C).
Figure 4.
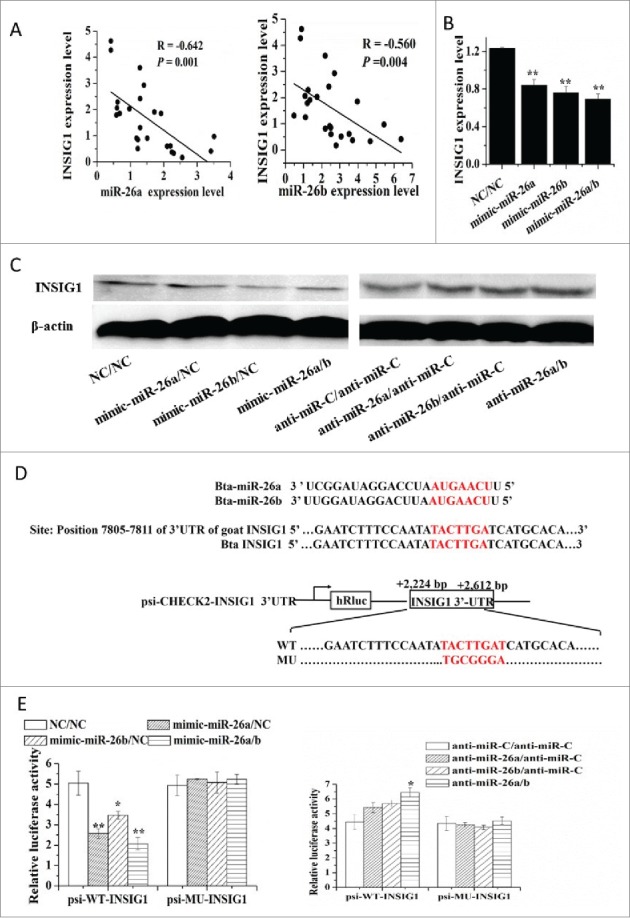
MiR-26 family targets INSIG1. (A) The correlation between expression level of miR-26a/b and INSIG1 in 24 mid-lactation mammary gland tissue samples. (B) Regulation of INSIG1 mRNA level by miR-26a/b mimic. (C) Regulation of INSIG1 protein level by miR-26a/b mimic and inhibitor. (D) MiR-26 family site in INSIG1 3’UTR and the construction of the luciferase (Luc) expression vector fused to the INSIG1 3’UTR. Bta: Bovine; WT, Luc reporter vector with the WT INSIG1 3’UTR (2224 bp to 2612 bp); MU, Luc reporter vector with the mutation at miR-26 family site in INSIG1 3’UTR. (E) Induction or suppression of Luc activity with the WT or MU of the INSIG13’UTR by miR-26 family mimic or inhibitor. GMEC were co-transfected with 120 nM oligonucleotides (60 nM each), WT and MU. The Luc activity was measured at 48 h posttransfection. All experiments were performed in triplicate and repeated 3 times (n = 9). Values are presented as mean + SEM, *, P < 0.05; **, P < 0.01.
To determine whether INSIG1 is a direct target of miR-26a/b, wildtype (INSIG1-WT-3’-UTR) and mutant 3’-UTRs lacking miR-26a/b binding site (INSIG1-MU-3’-UTR) were cloned into psi-CHECK2 luciferase reporter vector downstream of the Renilla luciferase coding region. The relative luciferase activity of INSIG1-WT-3’-UTR in response to the miR-26a/b mimic treatment alone or in combination was reduced by ~50% (P = 0.01), ~31% (P = 0.05) and ~58% (P = 0.01), respectively (Fig. 4E). Such effect did not occur in the INSIG1-MU-3’-UTR or psi-CHECK2 vector alone (Fig. 4E). These data suggested that miR-26a/b cognate site is essential for negative regulation of luciferase expression driven by the INSIG1 3’UTR.
Discussion
Several candidate miRNA have a dynamic expression across different stages of lactation in the mammary gland. For example, miR-135a expression level is associated with lactating stages and directly targets the prolactin receptor to function as a regulator in the goat mammary gland.19 The expression level of miR-27a also was related to lactating stages and seems to play an important role in the regulation of TAG synthesis in GMEC.31 There is evidence that miR-486 plays a key role in bovine mammary gland epithelial cells and promotes milk secretion.27 Several miRNA increased in expression in bovine mammary gland between the dry period and early lactation.50
Although only ~37% of mammalian miRNA are located in the introns of corresponding genes,24 co-regulation of intragenic miRNA and its host genes appears as a common biological phenomenon. Furthermore, evidence from non-ruminant models revealed that intronic miRNA and their host genes always tend to work in parallel, forming regulatory relationships among host genes and intronic miRNA. For example, miR-153 and its host gene islet antigen -2β have opposite functional effects on the insulin and dopamine secretion pathway in mice.53 MiR-33a/b act in concert with SREBF2 to control cholesterol homeostasis.14,17
Unlike other cells or tissues, there is limited information on the relationship between the miR-26 family and their host genes in lactating ruminant mammary gland. The current study revealed a strong positive relationship between the miR-26a/b and their host genes in goat mammary gland tissue, which is consistent with a previous study reporting that the miR-26a/b are expressed concomitantly with their host genes.57 Therefore, the fact that there is a high expression of the miR-26 family and their host genes in goat mammary gland is indicative that these miRNA and their host genes could play a crucial role in the regulation of milk fat synthesis.
The basic arrangement of miR-26a-1, miR-26a-2 and miR-26b in the introns of genes encoding the CTDSP protein family remained constant throughout the vertebrate evolution.57 It has been reported that the miR-26 family are involved in human adipogenesis.49,22 More importantly, the miR-26a/b are expressed concomitantly with their host genes in GMEC. Our data revealed a functional significance between miR-26a/b and the CTDSP family in the regulation of lipid metabolism in GMEC, which is consistent with the effect on human adipogenesis22,48 and in cell cycle control.5
The expression of PPARG is essential for fat cell differentiation and normal lipid metabolism in adipose tissue.40 Various long-chain fatty acids20 and chemical compounds23 can bind to and activate PPARG, leading to changes in mRNA expression of target genes.26 Besides PPARG, LXRA and SREBF1 are 2 key transcription factors also involved in lipid metabolism.1 Therefore, the data demonstrating that miR-26a/b overexpression altered PPARG, SREBF1 and LXRA expression profiles and the data showing that cells treated with Ad-PPARG/Ad-SREBF1 and T09 (the agonist of LXRA) up-regulated expression of miR-26 and CTDSP family expression seem to confirm a functional role among the transcription regulators and miR-26a/b. These linkages likely have functional relevance because both the miRNA and transcription factors can influence dozens of target genes. Furthermore, these data reinforce the view that the molecular network of milk fat synthesis is composed of several regulatory interactions. The detailed mechanisms for how transcription factors promote miRNA expression in ruminant mammary cells are worthy of future research efforts.
The increase of UFA concentration by miR-26a/b and their host genes is consistent with the function of SCD1 protein, which is to catalyze the conversion of C16:0 and C18:0 to C16:1 and C18:1.36 Besides the control of UFA synthesis by SCD1, other factors [such as AGPAT6 or DGAT mRNA and protein expression] in coordination with the desaturation process affect the synthesis of TAG. Both DGAT1 and AGPAT6 are key components of the overall pathway leading to synthesis of milk lipid droplets and affect milk fat yield and composition.42,41,33 Although the expression of AGPAT6 did not change, the fact that overexpression of miR-26a/b and their host genes had a significant effect on DGAT1 mRNA expression provides evidence of a mechanistic link between the miRNA and the process of desaturation and esterification
The observation that mimicking or antagonizing miR-26a/b expression in GMEC modified fat droplet, TAG and UFA content is further support of a functional role of miR-26a/b on milk fat synthesis. Among the potential new targets that seems to control goat milk fat synthesis is INSIG1, the present data indicate this is a bona fide target of miR-26a/b. Although the role of INSIG1 in lipid synthesis in non-ruminants has been studied extensively, few studies have addressed the role of the crosstalk among miR-26 family, miR-26 family host genes (CTDSP1/2/L) and miR-26 family target genes (e/g. INSIG1) in the coordination of lipid synthesis in mammary gland.
It is well-established that SCD1, GPAM, FASN and PPARG are mediators of ruminant mammary gland lipid accumulation.7,45,29 Therefore, the data demonstrating upregulation of INSIG1 and downregulation of SREBF1, SCD1, FASN, GPAM and PPARG expression when miR-26a/b and their host genes were knocked down are consistent with a functional relationship. This idea is further supported by earlier reports in mice that overexpression of INSIG1 represses SREBF processing, and in turn, increases the expression of lipogenic genes (Engelking et al., 2004).
Nutrition affects milk composition (influencing its nutritional properties) and modifies the expression of mammary genes, including miRNA expression profiles. For example, out of 30 known or predicted miRNA in the goat mammary gland whose expression seems to be nutritionally regulated,37 22 were differentially expressed in bovine mammary gland in response to a diet containing 5% linseed oil and 5% safflower oil.30 Our data uncovered a functional association of miR-26a/b, their host genes and INSIG1, and provide new insights into the regulatory network controlling milk fat synthesis in GMEC. Further studies are required to address the potential for nutrition to regulate this network.
Overall, the present study revealed the existence of a regulatory loop among miR-26a/b and key lipogenic transcription factors (PPARG, SREBF1 and LXRA) whereby their activation could induce an increase in miRNA expression. Therefore, our data highlight the role of miR-26a/b and their host genes in regulating the synthesis of UFA and TAG in goat mammary cells. Targeting this regulatory network might be crucial for regulating the composition of UFA in goat milk by nutritional means.
Material and methods
Animals and cells
Second-lactation (3-years-old) Xinong Saanen goats of similar weight from the Northwest A&F University herd were used in this study. The Animal Care and Use Committee of the Northwest A&F University (Yangling, Shaanxi, China) approved all the surgical procedures. Twenty-four mid-lactation (120 d after parturition) goats, and five goats each at early-lactation (15 d after parturition), peak-lactation (60 d after parturition) and the dry period (60 d before parturition) were sacrificed for mammary gland tissue sample collection. Total RNA was extracted from the mammary gland tissue using the Trizol reagent (Invitrogen, USA) according the manufacturer’s protocol.
The GMEC were isolated from peak lactation Xinong Saanen goats as described previously.52,46 Briefly, cells were maintained in a basal DMEM/F12 medium (Invitrogen, USA) with 10% of fetal bovine serum (FBS), penicillin/ streptomycin (100 U/mL, Harbin Pharmaceutical Group, China), insulin (5 µg/mL,Sigma, USA), hydrocortisone (1µg/mL, Sigma-Aldrich, USA), epidermal growth factor1 (10 ng/mL, Invitrogen, USA). To induce lactogenesis, GMEC were cultured in a lactogenic medium for 48 h, prior to initial experiments as reported previously.39,20 The lactogenic medium was prepared as the basal medium supplemented with prolactin (2 µg/ mL, Sigma-Aldrich, USA). The details of the GMEC culture were according to our recent publication.28
Cell treatments
The GMEC were seeded into 12-well or 6-well plates and transfection was performed at approximately 80% confluence. Oligonucleotides were obtained from Invitrogen and the details are listed in supplementary Table 2. The final oligonucleotide concentrations were 60 nM for each miR-26a/b mimic (mimic-miRNA), miR-26a/b inhibitor (anti-miRNA) and siRNA. The miRNA mimic is a small chemically-modified double- or single-stranded RNA molecule designed to mimic mature endogenous miRNA after transfected into cells, whereas a miRNA inhibitor is a single-stranded RNA molecule designed to specifically bind to and inhibit endogenous miRNA. We designed a double-stranded RNA as a miR-26 family mimic and single-stranded RNA molecule as miR-26 family inhibitor. The negative control (NC) for the mimic and siRNA treatments was double-stranded and for the inhibitor (anti-miR-C) was single-stranded. The sequences of oligonucleotides are listed in supplementary Table 2. The GMEC were transfected with 60 nM of each oligonucleotide using LipofectamineTM RNAiMAX (Invitrogen, USA) following manufacturer’s instruction. The adenovirus containing green fluorescent protein (Ad-GFP), adenovirus containing peroxisome proliferator-activated receptor-gamma (Ad-PPARG) and adenovirus containing sterol regulatory element binding proteins (Ad-SREBF1) are preserved in our lab. One μM of the LXRA agonist T09 (Sigma, St. Louis, MO, USA) was used for pharmacological treatment of GMEC, and cells were harvested at 48 h for analysis. MiR-26a/b inhibition or overexpression and CTDSP family inhibition were confirmed by real time quantitative PCR (RT-qPCR) as described below.
Isolation and analysis of RNA
Total RNA was obtained from the mammary gland tissue of each goat or GMEC using the Trizol reagent (Invitrogen, USA) according to the manufacturer’s protocol. For mature miRNA analysis, 0.2 µg total RNA were reverse transcribed and the resulting cDNA was amplified by RT-PCR using the S-Poly(T) assay.21,38 the miRNA levels were calculated with the relative quantification method using the 5S rRNA as reference gene. All the primers used for miRNA RT-qPCR are listed in supplementary Table 3.
For mRNA, 0.5 µg total RNA was reverse transcribed with the PrimeScript R RT Reagent Kit with gDNA Eraser (Perfect Real time, Takara, Japan) and amplified with SYBR Premix Ex TaqII (Perfect Real Time, Takara, Japan), the mRNA level was calculated using the relative quantification method normalizing with the geometric mean6 of the 3 internal controls (ubiquitously expressed transcript, mitochondrial ribosomal protein L39 and ribosomal protein).6,20,8 After amplification, the products were confirmed by agarose gel electrophoresis and sequencing.
Western blot
Cells were lysed in ice-cold lysis buffer supplemented with RIPA buffer (Solarbio, China) supplemented with PMSF (Pierce, USA). Cell lysates were rotated for 30 minutes at room temperature before the insoluble material was removed by centrifugation at 12,000 × g for 10 min at 4 °C. Total protein was quantified using the PierceTM BCA Protein assay kit (ThermoScientific, Rockford, IL, USA). Equal amounts of protein were separated by SDS/PAGE. Proteins were transferred onto nitrocellulose membranes and membranes were probed with the indicated primary antibodies overnight, washed and incubated with the appropriate HRP-conjugated secondary antibodies. Signals were detected by the chemiluminescent ECL Western blot detection system (Pierce, USA).
A rabbit polyclonal antibody against human INSIG1 was purchased from Bioss (bs-5074R, China, 1:500) and monoclonal mouse anti-βactin was obtained from CWBIO (China, 1:1000). Polyclonal anti-rabbit/ anti-mouse horseradish peroxidase-conjugated IgG (Tiagen, China, 1:1000) were used as secondary antibodies.
Oil red O staining
Cells were washed with phosphate-buffer-solution (PBS), fixed in 10% paraformaldehyde (in PBS) for 1h at 4 °C, washed twice with PBS, stained by incubation with 5% oil red O in isopropanol for 20 min, washed thrice with PBS and then photographed.
Triacylglycerol assay
Cells were washed with PBS (4 °C) and quantification of intracellular TAG was performed using the a colorimetric assay kit (Cayman chemical, USA) according to the manufacturer’s protocol on an XD 811G Biochemistry Analyzer (Shanghai Odin Science & Technology Company, China). Triacylglycerol concentration was normalized to total protein determined by BCA Protein assay kit (Pierce/Thermo, USA) as described previously.4,55
Luciferase reporter assay
The 388 bp segment containing predicted miR-26 family target site in the 3’UTR of INSIG1 (NCBI no. NM_001077909.1) was amplified from GMEC cells cDNA. The forward primer: 5’-ccgctcgagCCCATCTCAGTTTGCTTTAG-3’, the reverse primer: 5’-taagaatgcggccgc ACTTTGGTTCACATACTTGC-3’, the lower-case nucleotides of primers were added with restriction sites Xho I or Not I. The 388 bp products were inserted into the Xho I and Not I restriction sites of the psiCHECK-2 vector (Promega, USA). The wild-type reporter served as template to generate reporter constructs with mutant 3’UTR of INSIG1, primers were designed for overlap PCR as follows, forward primer: 5’- GAATCTTTCCAATAtgcgggaCATGCACAG-3’, the reverse primer: 5’-TGCATGtcccgcaTATTGGAAAGATTC-3’, the lower-case nucleotides of primers were mutant for the miR-26 family target site. Seven nucleotides (TACTTGA) in the target site which were complementary to the miR-26 family seed region were mutated to (TGCGGGA). Correct insertion and successful mutation were validated by sequencing.
The GMEC were seeded in 48-well plates at a density of 50,000 cells per well one day before transfection. Transfections were performed with a total of 0.33 µg of each reporter construct using the X-tremeGENE HP DNA Transfection Reagent (Roche, Switzerland) according to the manufacturer’s protocol. After a 6 h recovery period in medium, cells were co-transfected with 120 nM of oligonucleotides mixture (60 nM each) using LipofectamineTM RNAiMAX according to the manufacturer’s protocol. Cells were harvested 48 h after transfection and assayed for Renilla and firefly luciferase activities using the Dual-Glo luciferase assay system (Promega, USA) by a thermo scientific varioskan flash (Thermo scientific, USA).
Fatty acid extraction and analysis
Total fatty acid was extracted and methylated from approximately 100 mg cells using 2 mL of vitriol oil/methanol (2.5:1, v/v) as previously described.47,56 Methylated samples were analyzed by gas chromatography-mass spectrometry (GC-MS) with HP-5 column (Agilent Technologies, Palo Alto, CA). The relative proportions of fatty acid were determined as percentages of the total peak areas that could be identified.51
Statistical analysis
Data are presented as mean ± SEM. Differences between groups were analyzed by applying Student’s 2-tailed t-test for independent samples. Differences were considered statistically significant at P < 0.05 (*, *P < 0.05;**, **P < 0.01).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank professor D. M. Gou from Shenzhen Key Laboratory of Microbial Genetic Engineering,College of Life Sciences, Shenzhen University, Guangdong, PR. China for providing miRNA analysis kit. This work was jointly supported by the National Natural Science Foundation of China (No. 31372281; Beijing, China), the Special Fund for Agro-scientific Research in the Public Interest (No. 201103038, Beijing, China), and the Transgenic New Species Breeding Program of China (No. 2014ZX08009-051B, Beijing, China).
References
- 1.Alexis V, Clarles T, Minako I, Thomas G, Amalia T. Liver X receptor activation promotes polyunsaturated fatty acid synthesis in macrophages relevance in the context of atherosclerosis. Arterioscler Thromb Vasc Biol 2015; 35(6):1357-65; PMID:25838428; http://dx.doi.org/ 10.1161/ATVBAHA.115.305539 [DOI] [PubMed] [Google Scholar]
- 2.Anderson SM, Rudolph MC, McManaman JL, Neville MC. Key stages in mammary gland development. Secretory activation in the mammary gland: it's not just about milk protein synthesis! Breast Cancer Res 2007; 9(1):204; PMID:17338830; http://dx.doi.org/ 10.1186/bcr1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136(2):215-33; PMID:19167326; http://dx.doi.org/ 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartholomew SR, Bell EH, Summerfield T, Newman LC, Miller EL, Patterson B, Niday ZP, Ackerman WE, Tansey JT.. Distinct cellular pools of perilipin 5 point to roles in lipid trafficking. Biochimi Biophys Acta 2012; 1821(2):268-78;; PMID:2206371; http://dx.doi.org/15701730 10.1016/j.bbalip.2011.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005; 11(3):241-247; PMID:15701730; http://dx.doi.org/ 10.1261/rna.7240905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bionaz M, Loor JJ. Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol Genomics 2007; 29(3):312-9; PMID:17284669; http://dx.doi.org/ 10.1152/physiolgenomics.00223.2006 [DOI] [PubMed] [Google Scholar]
- 7.Bionaz M. Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics 2008; 9(1):366; PMID:18671863; http://dx.doi.org/ 10.1186/1471-2164-9-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet M, Bernard L, Bes S, Leroux C. Selection of reference genes for quantitative real-time PCR normalisation in adipose tissue, muscle, liver and mammary gland from ruminants. Animal 2013; 7(08):1344-53; PMID:23552195; http://dx.doi.org/ 10.1017/S1751731113000475 [DOI] [PubMed] [Google Scholar]
- 9.Daimiel-Ruiz L, Klett-Mingo M, Konstantinidou V, Mico V, Aranda JF, Garcia B, Martinez-Botas J, Davalos A, Fernandez-Hernando C, Ordovas JM. Dietary lipids modulate the expression of miR-107, an miRNA that regulates the circadian system. Mol Nutr Food Res 2015; 59(3):552-65; PMID:25522185; http://dx.doi.org/ 10.1002/mnfr.201400616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dharap A, Pokrzywa C, Murali S, Kaimal B, Vemuganti R. Mutual induction of transcription factor PPARgamma and microRNAs miR-145 and miR-329. J Neurochem 2015; 135(1):139-46; PMID:26119485; http://dx.doi.org/ 10.1111/jnc.13220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubeuf JP, Morand-Fehr P, Rubino R. Situation, changes and future of goat industry around the world. Small Ruminant Res 2004; 51(2):165-73; http://dx.doi.org/ 10.1016/j.smallrumres.2003.08.007 [DOI] [Google Scholar]
- 12.Eberlé D, Hegarty B, Bossard P, Ferré P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 2004; 86(11):839-48; PMID:15589694; http://dx.doi.org/11111080 10.1016/j.biochi.2004.09.018 [DOI] [PubMed] [Google Scholar]
- 13.Edwards PA, Tabord D, Kast HR, Venkateswaran A. Regulation of gene expression by SREBP and SCAP. Biochim Biophys Acta 2000; 1529(1):103-13; PMID:11111080; http://dx.doi.org/ 10.1016/S1388-1981(00)00140-2 [DOI] [PubMed] [Google Scholar]
- 14.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem 2010; 285(44):33652-61; PMID:20732877; http://dx.doi.org/ 10.1074/jbc.M110.152090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong Y, Lee JN, Lee PC, Goldstein JL, Brown MS, Ye J. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metab 2006; 3(1):15-24; PMID:16399501; http://dx.doi.org/ 10.1016/j.cmet.2005.11.014 [DOI] [PubMed] [Google Scholar]
- 16.Hansen HO, Grunnet I, Knudsen J. Triacylglycerol synthesis in goat mammary gland. The effect of ATP, Mg2+ and glycerol 3-phosphate on the esterification of fatty acids synthesized de novo. Biochem J 1984; 220:513-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horie T, Nishino T, Baba O, Kuwabara Y, Nakao T. MicroRNA-33b knock-in mice for an intron of sterol regulatory element-binding factor 1 (Srebf1) exhibit reduced HDL-C in vivo. Scientific Reports 2014; 4; PMID:24931346; http://dx.doi.org/ 10.1038/srep05312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 2002; 109(9):1125-31; PMID:11994399; http://dx.doi.org/ 10.1172/JCI0215593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji Z, Dong F, Wang G, Hou L, Liu Z, Chao T, Wang J. miR-135a targets and regulates prolactin receptor gene in goat mammary epithelial cells. DNA Cell Biol 2015; 34(8):534-40; PMID:26102062; http://dx.doi.org/ 10.1089/dna.2015.2904 [DOI] [PubMed] [Google Scholar]
- 20.Kadegowda AKG, Bionaz M, Piperova LS, Erdman RA, Loor JJ. Peroxisome proliferator-activated receptor-γ activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents. J Dairy Sci 2009; 92(9):4276-89; PMID:19700688; http://dx.doi.org/ 10.3168/jds.2008-1932 [DOI] [PubMed] [Google Scholar]
- 21.Kang K, Zhang X, Liu H, Wang Z, Zhong J, Huang Z, Peng X, Zeng Y, Wang Y, Yang Y, et al. . A novel real-time PCR assay of microRNAs using S-Poly(T), a specific oligo(dT) reverse transcription primer with excellent sensitivity and specificity. PLoS One 2012; 7(11):e48536; PMID:23152780; http://dx.doi.org/ 10.1371/journal.pone.0048536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karbiener M, Pisani DF, Frontini A, Oberreiter LM, Lang E, Vegiopoulos A, Mössenböck K, Bernhardt GA, Mayr T, Hildner F, et al. . MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells 2014; 32(6):1578-90; PMID:24375761; http://dx.doi.org/ 10.1002/stem.1603 [DOI] [PubMed] [Google Scholar]
- 23.Khandoudi N, Delerive P, Isabelle B-B, Buckingham RE, Staels B, Bril A. Rosiglitazone, a peroxisome proliferator-activated receptor-γ, inhibits the Jun NH2-terminal kinase/activating protein 1 pathway and protects the heart from ischemia/reperfusion injury. Diabetes 2002; 51(5):1507-14; PMID:11978649; http://dx.doi.org/19165215 10.2337/diabetes.51.5.1507 [DOI] [PubMed] [Google Scholar]
- 24.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009; 10(2):126-39; PMID:19165215; http://dx.doi.org/ 10.1038/nrm2632 [DOI] [PubMed] [Google Scholar]
- 25.Kompan D, Komprej A. The Effect of Fatty Acids in Goat Milk on Health. INTECH: Open Access Publisher; 2012 [Google Scholar]
- 26.Lee G, Elwood F, McNally J, Weiszmann J, Lindstrom M, Amaral K, Nakamura M, Miao S, Cao P, Learned RM, et al. . T0070907, a selective ligand for peroxisome proliferator-activated receptor gamma, functions as an antagonist of biochemical and cellular activities. J Biolog Chem 2002; 277(22):19649-57; PMID:11877444; http://dx.doi.org/ 10.1074/jbc.M200743200 [DOI] [PubMed] [Google Scholar]
- 27.Li D, Xie X, Wang J, Bian Y, Li Q, Gao X, Wang C. 2015a. MiR-486 regulates lactation and targets the PTEN gene in cow mammary glands. PLoS One 10(3):e0118284; PMID:25738494; http://dx.doi.org/23537996 10.1371/journal.pone.0118284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Luo J, Wang H, Shi H, Zhu J, Sun Y, Yu K, Yao D. Adipose triglyceride lipase regulates lipid metabolism in dairy goat mammary epithelial cells. Gene 2015b; 554(1):125-30; PMID:25307872; http://dx.doi.org/23537996 10.1016/j.gene.2014.10.020 [DOI] [PubMed] [Google Scholar]
- 29.Li J, Luo J, Xu H, Wang M, Zhu J, Shi H, Haile AB, Wang H, Sun Y. Fatty acid synthase promoter: characterization, and transcriptional regulation by sterol regulatory element binding protein-1 in goat mammary epithelial cells. Gene 2015c; 561(1):157-64; PMID:25688876; http://dx.doi.org/23537996 10.1016/j.gene.2015.02.034 [DOI] [PubMed] [Google Scholar]
- 30.Li R, Beaudoin F, Ammah AA, Bissonnette N, Benchaar C, Zhao X, Ibeagha-Awemu EM. Deep sequencing shows microRNA involvement in bovine mammary gland adaptation to diets supplemented with linseed oil or safflower oil. BMC Genom 2015d; 16(1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin X-Z, Luo J, Zhang LP, Wang W, Shi H-B, Zhu J-J. miR-27a suppresses triglyceride accumulation and affects gene mRNA expression associated with fat metabolism in dairy goat mammary gland epithelial cells. Gene 2013; 521(1):15-23; PMID:23537996; http://dx.doi.org/ 10.1016/j.gene.2013.03.050 [DOI] [PubMed] [Google Scholar]
- 32.Little JL, Wheeler FB, Fels DR, Koumenis C, Kridel SJ. Inhibition of fatty acid synthase induces endoplasmic reticulum stress in tumor cells. Cancer Res 2007; 67(3):1262-9; PMID:17283163; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-1794 [DOI] [PubMed] [Google Scholar]
- 33.Littlejohn MD, Tiplady K, Lopdell T, Law TA, Scott A, Harland C, Sherlock R, Henty K, Obolonkin V. Expression variants of the lipogenic AGPAT6 gene affect diverse milk composition phenotypes in bos taurus. PLoS One 2014; 9(1):e85757; PMID:24465687; http://dx.doi.org/ 10.1371/journal.pone.0085757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McManaman JL, Russell TD, Schaack J, Orlicky DJ, Robenek H. Molecular determinants of milk lipid secretion. J Mammary Gland Biol Neoplasia 2007; 12(4):259-68; PMID:17999166; http://dx.doi.org/ 10.1007/s10911-007-9053-5 [DOI] [PubMed] [Google Scholar]
- 35.Missohou A, Talaki E, Laminou IM. Diversity and genetic relationships among seven West African goat breeds. Asian Australasian J Anim Sci 2006; 19(9):1245; http://dx.doi.org/ 10.5713/ajas.2006.1245 [DOI] [Google Scholar]
- 36.Miyazakia M, Ntambi JM. Role of stearoyl-coenzyme A desaturase in lipid metabolism. Prostaglandins Leukot Essent Fatty Acids 2003; 68(2):113-21; PMID:12538075; http://dx.doi.org/26473604 10.1016/S0952-3278(02)00261-2 [DOI] [PubMed] [Google Scholar]
- 37.Mobuchon L, Marthey S, Le Guillou S, Laloë D, Le Provost F, Leroux C. Food deprivation affects the miRNome in the lactating goat mammary gland. PloS One 2015; 10(10):e0140111; PMID:26473604; http://dx.doi.org/ 10.1371/journal.pone.0140111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niu Y, Zhang L, Qiu H, Wu Y, Wang Z, Zai Y, Liu L, Qu J, Kang K, Gou D. An improved method for detecting circulating microRNAs with S-Poly(T) Plus real-time PCR. Sci Rep 2015; 5:15100; PMID:26459910; http://dx.doi.org/ 10.1038/srep15100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson DG, Matitashvili EA, Bauman DE. The inhibitory effect of trans-10, cis-12 CLA on lipid synthesis in bovine mammary epithelial cells involves reduced proteolytic activation of the transcription factor SREBP-1. J Nutr 2004; 134(10):2523-7; PMID:15465741 [DOI] [PubMed] [Google Scholar]
- 40.Rosen ED, Spiegelman BM. PPARgamma : a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem 2001; 276(41):37731-34; PMID:11459852; http://dx.doi.org/ 10.1074/jbc.R100034200 [DOI] [PubMed] [Google Scholar]
- 41.Schennink A, Heck JM, Bovenhuis H, Visker MH, van Valenberg HJ, van Arendonk JA. Milk fatty acid unsaturation: genetic parameters and effects of stearoyl-CoA desaturase (SCD1) and acyl CoA: diacylglycerol acyltransferase 1 (DGAT1). J Dairy Sci 2008; 92(5):2135-43; PMID:18420645; http://dx.doi.org/ 10.3168/jds.2007-0825 [DOI] [PubMed] [Google Scholar]
- 42.Schennink A, Stoop WM, Visker MH, Heck JM, Bovenhuis H, van der Poel JJ, van Valenberg HJ, van Arendonk JA. DGAT1 underlies large genetic variation in milk-fat composition of dairy cows. Anim Genet 2007; 38(5):467-73; PMID:17894561; http://dx.doi.org/ 10.1111/j.1365-2052.2007.01635.x [DOI] [PubMed] [Google Scholar]
- 43.Sepehri B, Seyedabadi HR. Molecular analysis of khalkhali goat population based on cytB region of mitochondrial DNA. Biol Forum – An Int J 2012; 7(1):1311-6 [Google Scholar]
- 44.Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell Metab 2012; 16(4):414-9; PMID:23000402; http://dx.doi.org/ 10.1016/j.cmet.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi H, Luo J, Zhu J, Li J, Sun Y, Lin X, Zhang L, Yao D, Shi H. PPARγ regulates genes involved in triacylglycerol synthesis and secretion in mammary gland epithelial cells of dairy goats. PPAR Research 2013a; 2013:1-10; PMID:23710163; http://dx.doi.org/24889218 10.1155/2013/310948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi H, Shi H, Luo J, Wang W, Haile AB, Xu H, Li J. Establishment and characterization of a dairy goat mammary epithelial cell line with human telomerase (hT-MECs). Anim Sci J 2014; 85(7):735-43; PMID:24889218; http://dx.doi.org/ 10.1111/asj.12206 [DOI] [PubMed] [Google Scholar]
- 47.Shi HB, Luo J, Yao DW, Zhu JJ, Xu HF, Shi HP, Loor JJ. Peroxisome proliferator-activated receptor-γ stimulates the synthesis of monounsaturated fatty acids in dairy goat mammary epithelial cells via the control of stearoyl-coenzyme A desaturase. J Dairy Sci 2013b; 96(12):7844-53; PMID:24119817; http://dx.doi.org/24140453 10.3168/jds.2013-7105 [DOI] [PubMed] [Google Scholar]
- 48.Song G, Xu G, Ji C, Shi C, Shen Y, Chen L, Zhu L, Yang L, Zhao Y, Guo X. The role of microRNA-26b in human adipocyte differentiation and proliferation. Gene 2014; 533(2):481-7; PMID:24140453; http://dx.doi.org/ 10.1016/j.gene.2013.10.011 [DOI] [PubMed] [Google Scholar]
- 49.Sun D, Zhang J, Xie J, Wei W, Chen M, Zhao X. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS Letters 2012; 586(10):1472-9; PMID:22673513; http://dx.doi.org/ 10.1016/j.febslet.2012.03.068 [DOI] [PubMed] [Google Scholar]
- 50.Wang M, Moisa S, Khan MJ, Wang J, Bu D, Loor JJ. MicroRNA expression patterns in the bovine mammary gland are affected by stage of lactation. J Dairy Sci 2012a; 95(11):6529-35; PMID:22959945; http://dx.doi.org/21090118 10.3168/jds.2012-5748 [DOI] [PubMed] [Google Scholar]
- 51.Wang W, Luo J, Zhong Y, Lin X-Z, Shi H-B, Zhu J-J, Li J, Sun Y-T, Zhao W-S. Goat liver X receptor α, molecular cloning, functional characterization and regulating fatty acid synthesis in epithelial cells of goat mammary glands. Gene 2012b; 505(1):114-20; PMID:22634102; http://dx.doi.org/21090118 10.1016/j.gene.2012.05.028 [DOI] [PubMed] [Google Scholar]
- 52.Wang Z, Luo J, Wang W. Characterization and culture of isolated primary dairy goat mammary gland epithelial cells Sheng wu gong cheng xue bao. Chin J Biotechnol 2010; 26(8):1123-7; PMID:21090118 [PubMed] [Google Scholar]
- 53.Xu H, Abuhatzira L, Carmona GN, Vadrevu S, Satin LS, Notkins AL. The Ia-2beta intronic miRNA, miR-153, is a negative regulator of insulin and dopamine secretion through its effect on the Cacna1c gene in mice. Diabetologia 2015; 58(10):2298-306; PMID:26141787; http://dx.doi.org/ 10.1007/s00125-015-3683-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Brown MS. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 2002; 110(4):489-500; PMID:12202038; http://dx.doi.org/ 10.1016/S0092-8674(02)00872-3 [DOI] [PubMed] [Google Scholar]
- 55.Yu K, Shi HB, Luo J, Li J, Zhao WS, Tian HB, Shi HP. PPARG modulated lipid accumulation in dairy GMEC via regulation of ADRP gene. J Cell Biochem 2015; 116(1):192-201; PMID:25169669; http://dx.doi.org/ 10.1002/jcb.24958 [DOI] [PubMed] [Google Scholar]
- 56.Zhu JJ, Luo J, Wang W, Yu K, Wang HB, Shi HB, Sun YT, Lin XZ, Li J. Inhibition of FASN reduces the synthesis of medium-chain fatty acids in goat mammary gland. Animal 2014; 8(9):1469-78; PMID:24909980; http://dx.doi.org/ 10.1017/S1751731114001323 [DOI] [PubMed] [Google Scholar]
- 57.Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang JR, Zeng C, Zhuang SM. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucl Acids Res 2011; 40(10):4615-25; PMID:22210897; http://dx.doi.org/ 10.1093/nar/gkr1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


