ABSTRACT
In the last few decades, small regulatory RNA (sRNA) molecules emerged as key regulators in every kingdom of life. Resolving the full targetome of sRNAs has however remained a challenge. To address this, we used an in vivo tagging MS2-affinity purification protocol coupled with RNA sequencing technology, namely MAPS, to assemble full bacterial small RNAs targetomes. The impressive potential of MAPS has been supported by a number of reports. Here, we concisely overview RNA-tagging history that preceded the development of the MAPS assay and expose the range of possible uses of this technology.
keywords: Affinity purification, antisense, MS2, RNA purification, RNA sponge, RNA tag, RybB, RyhB, sRNA, tRNA
The first description of protein tags by Munro and Pelham in 1984 had an immense impact on research.1 Since this seminal work, biologists have been able to pull-down a specific protein of interest and characterize its interacting partners. Following the report of Munro and Pelham, many tags (e.g. Arg-Tag,2 FLAG-Tag3 or His-Tag4) were added to the list of powerful tools available to study protein biology. Over the years, it became clear that the field of RNA biology also needed a similar system. A tagging system specific for RNA would certainly help describe the function of regulatory RNAs such as micro-RNAs or bacterial small RNAs (sRNAs) that have been established as major modulators of gene expression. Hence, it is imperative to understand how these molecules fulfill their functions and to identify any necessary interacting protein or RNA partners.
Artificial RNA tags were first developed and used for the pull-down of specific RNAs. Bachler et. al. (1999) synthesized artificial RNA tags that bound specifically and strongly to the antibiotic streptomycin.5 Named Strepto-Tag, this RNA is structured as a long stem-loop in which nucleotides mainly present in 2 bulges are responsible for the interaction with the antibiotic (Fig. 1A).6 After tagging an RNA of interest, the hybrid RNA is incubated with crude cell extracts to allow in vitro RNA-protein complex assembly. Then, samples are passed through a sepharose column on which streptomycin has been immobilized. Elution of complexes is carried out by the addition of free streptomycin. It is noteworthy to mention that free streptomycin will bind the RNA tag and therefore be present in output samples which could interfere with subsequent analysis. In the same vein, Srisawat and Engelke (2001) also developed RNA tags (termed aptamers) with high specificity for streptavidin.7 Similar to the Strepto-Tag, the structure of the streptavidin tag presents itself as a stem-loop with a bulge. In this case, the nucleotides in the loop, in the bulge and in the stem play a role in the binding to streptavidin7 (Fig. 1B). Srisawat and Engelke demonstrated that the tagged RNA could be expressed in vivo and purified either in its native form or in complex with proteins. Elution is then carried out by addition of free biotin, which breaks the bond between the tag and the affinity column. A downside of this technique is that avidin needs to be added to cell extracts to sequester cellular biotin or biotinylated proteins and prevent them from binding the streptavidin-agarose chromatography column. Streptavidin aptamers have since then been used and improved by many research groups. For example, Leppek and Stoecklin (2013) modified one of the original aptamer by creating perfect complementarity between the basal and terminal stem, and by increasing stem length8 (Fig. 1C). This modification increased the binding to streptavidin efficiency by 3 to 4-fold.8
Figure 1.
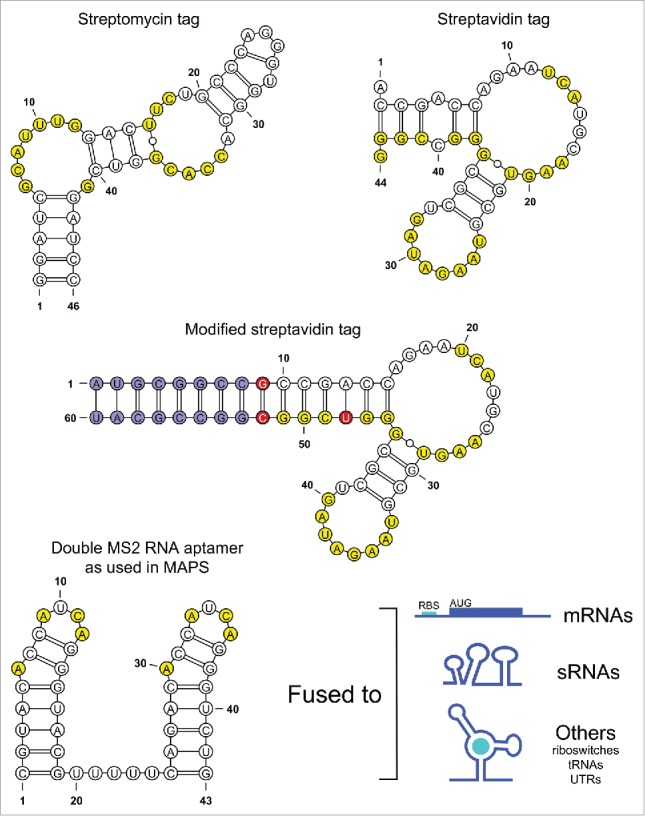
Overview of RNA tags. Nucleotides implicated in the interaction with specific partners are highlighted in yellow. (A) Structure of the Strepto-Tag. (B) Main region of a class I streptavidin aptamer. (C) Structure of a modified class I streptavidin aptamer. Nucleotides highlighted in red are mutations of the original sequence. Nucleotides in blue have been added to the original sequence. (D) Structure of the double MS2 aptamer as used in MAPS. The tag can ben fused to a wide range of RNA molecules to perform MAPS assays.
In the early 2000s, an alternative system was described, exploiting a naturally strong RNA-protein interaction.9 This system is founded on the property of the bacteriophage MS2 coat protein to interact with high specificity with a short RNA, namely the MS2 stem-loop aptamer (for reviews, see refs.10,11,12). During viral infection of Escherichia coli by the MS2 bacteriophage, the MS2 coat protein acts as a threshold signal, dictating its own binding to the MS2 RNA.13,14 This interaction is crucial for induction of the assembly step of the phage’s life cycle, accurate packaging of phage RNA and massive production of functional bacteriophages. In 1998, Bertrand et al. described a fluorescence technique exploiting the MS2 protein-MS2 RNA interaction allowing tracking of a specific mRNA in living cells.15,16 Later, the Vogel group developed an in vivo method to pull-down specific sRNAs and identify their protein partners.17,18 After production of a MS2-tagged sRNA, cell lysates are applied to an amylose resin on which a MBP-MS2 protein (Maltose Binding Protein fused with the MS2 coat protein) has been immobilized. Following elution by addition of a competitor for binding to the MBP, samples were analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS). This technique was revolutionary as it was performed in vivo and didn’t require addition of external proteins or molecules to achieve adequate purification.17,18 Notably, this method allowed purification and identification of a protein found in most sRNA-protein complexes, the RNA chaperone Hfq. The protein Hfq is known to interact with sRNAs to stabilize them in vivo and to facilitate sRNA-mRNA complexes formation.19,20 An opposite in vivo high-throughput technology able to identify ribonucleic partners of specific proteins also exists and was first introduced in 2010, the RIP-seq.21 Two major steps are required to complete a RIP-seq experiment. First, a native co-immunoprecipitation of a protein complexed to its RNA partners is performed. Then, following output collecting, samples are analyzed by RNA sequencing. Both these high-throughput sequencing technologies allow identification of RNA partners of either a specific RNA or protein, respectively.
Shortly after, our group used MS2 pull-down assays to purify specific sRNAs in order to analyze RNA-RNA interactions. We used a double MS2 RNA aptamer as a tag (Fig. 1D) and expressed the MS2-sRNA constructs in vivo. These experiments allowed us to demonstrate an unexpected regulatory mechanism of sRNAs. Using a MS2-mRNA (MS2-sdhC) construct to perform affinity purification, Desnoyers and Massé (2012) demonstrated that the interaction of Spot42 sRNA with sdhC mRNA serves as a recruitment platform for Hfq protein.22 Indeed, Spot42 brings Hfq near the translational initiation region (TIR) of sdhC transcript, where the protein competes with the 30S ribosomal subunit to inhibit translation. It was the first example of Hfq being the primary actor in sRNA-mediated genetic regulation. In particular, Desnoyers and Massé (2012) used MS2 pull-down assays with sdhC mRNA bearing different mutations at either Spot42 pairing site or Hfq binding site. Through these experiments, they were able to show in vivo that those mutations impaired interactions with their respective partners, confirming MS2 pull-down assay as a powerful technique to study RNA interactions in vivo.
Next, we wanted to expand and adapt the MS2 pull-down assays to characterize the full targetomes of bacterial sRNAs. These short functional RNAs greatly vary in structure and in mechanism of action.23,24 To achieve genetic regulation, sRNAs often imperfectly base-pair with a set of given mRNAs,25 resulting in various outcomes (repression of translation, mRNA degradation or translation enhancement).23 A single sRNA can even adopt different mechanisms of action depending on the mRNA targeted.25,26 All these characteristics prevent straightforward in silico target prediction. Usually, available bioinformatic programs yield a large pool of false positives and true targets may not present the most intuitive base-pairing pattern.27 To solve this problem, we took advantage of high-throughput RNA sequencing technology (RNA sequencing, RNAseq) combined with MS2-based sRNA pull-down assays. We called this new in vivo tool MAPS for MS2-affinity purification coupled with RNA sequencing (schematic representation in Fig. S1 of Lalaouna et. al., 2015).28 After tagging a sRNA with the MS2 aptamer, its regulatory activity is verified on known target mRNAs by Northern blot analysis. Following this validation step, the MS2-sRNA construct is expressed in vivo and then purified by affinity chromatography. Samples are submitted to RNAseq and results are compared to a control experiment (untagged sRNA MAPS).
As reported in Lalaouna et. al. (2015),28 we applied MAPS to RyhB and RybB, 2 well characterized sRNAs as a proof of principle. Notably, this study allowed us to identify numerous known target mRNAs of both sRNAs, independently from their mode of action (negative or positive regulation). Interestingly, we also identified new targets of RyhB and RybB. In the case of RyhB, 2 targets were identified. The first one is erpA mRNA, which was previously shown to be a target during the course of our study.29 The second is the grxD mRNA that is negatively regulated by RyhB. In the case of the other sRNA, RybB, we were able to demonstrate that yifE mRNA is stabilized following expression of RybB, validating yifE as a positive target. Many other putative targets identified in this study are still awaiting validation. Once these putative targets are validated, we expect to get the full targetome of these sRNAs without discrimination of any targets or technical limitations.
Surprisingly, the identification of new sRNA targets was not the only valuable information revealed by MAPS. In fact, MAPS datasets allowed us to identify a new role for bacterial tRNA-derived RNA fragments (tRFs). Functional tRFs have been reported in the literature and have been gaining interest in the last few years as they seem to be implicated in the regulation of various metabolisms (for reviews, see refs.30-32). The tRF we identified that interacts with RyhB and RybB corresponds to the 3′ external transcribed spacer of glyW-cysT-leuZ pre-tRNA (3′ETSleuZ). Our data indicated that the 3′ETSleuZ acts as a sRNA sponge, which prevents sRNA transcriptional noise in non-inducing conditions.28 Indeed, MAPS data allowed us to demonstrate that the 3′ETSleuZ interacts in vivo with RyhB and RybB sRNA. Then, further experiments conceded evidence that 3′ETSleuZ acts as a concentration threshold setter, buffering a certain amount of sRNA corresponding to their transcriptional noise.
After these results were obtained by MAPS, we applied the same method to other sRNAs. Results obtained for the sRNA DsrA were particularly exciting.26 DsrA is a well-known effector of positive and negative expression of rpoS and hns mRNA, respectively.33-36 As expected, we first validated that both mRNA targets were co-purified with MS2-DsrA. Surprisingly, MAPS unveiled a new atypical target of DsrA sRNA, the rbsD mRNA. In the case of DsrA-rbsD interaction base-pairing occurs far downstream the TIR, in the open-reading frame (ORF), which is in sharp contrast to most sRNAs that base-pair near the TIR.37 In 1998, Lease et. al. presented in silico predicted base-pairing between DsrA and rbsD at the same location.36 However since then, no report was able to confirm rbsD as a true target of DsrA. DsrA MAPS data prompted us to re-investigate this RNA-RNA interaction further, leading to the confirmation that rbsD was indeed negatively regulated by DsrA, as the sRNA induces mRNA decay following base-pairing.26
Recently, we also applied MAPS to tRNA-derived RNA fragments in order to determine their binding partners. Deriving from pre-tRNA transcript, tRF proved to be a challenge in accomplishing MAPS experiment due to their specific maturation processes. The 3′ETSleuZ was the first tRF candidate to be analyzed by MAPS, which confirmed the in vivo interaction between 3′ETSleuZ and 2 sRNAs (RyhB and RybB) (unpublished data). Likewise, short RNA molecules (˜33 nt) corresponding to the internal transcribed spacers (ITS) of metZ-metW-metV pre-tRNA polycistronic transcript were also successfully tagged and analyzed by MAPS,28,38 leading to the identification of 2 sRNAs (RybB and MicF) interacting with both ITS. Interactions were confirmed by Northern blot analysis after affinity purification.28 While it is still unclear what is the cellular role of the ITS of metZ-metW-metV pre-tRNA, these data suggest a more widespread phenomenon than previously thought.
It is now clear that MAPS is a powerful tool to study interaction partners of various types of RNA molecules whether these partners are mRNA, sRNAs or tRFs (Fig. 1D). Its unique output data set led us to a breakthrough in the bacterial tRFs field, in addition to helping us identify new targets of the well characterized sRNAs RyhB, RybB and DsrA. As sRNAs, tRFs or mRNAs, other type of RNA molecules such as rRNAs and even riboswitches can be tagged with the MS2 aptamer. Therefore, it is not difficult to imagine the diverse application of MAPS.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work has been supported by an operating grant from the Canadian Institutes of Health Research (CIHR) to EM. MCC holds a Ph.D. studentship from the Université de Sherbrooke. EM is a senior scholar from the Fonds de la Recherche en santé du Québec (FRQS).
References
- 1.Munro S, Pelham HR. Use of peptide tagging to detect proteins expressed from cloned genes: deletion mapping functional domains of Drosophila hsp 70. EMBO J 1984; 3:3087-93; PMID:6526011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JC, Derbyshire RB, Cook E, Dunthorne L, Viney J, Brewer SJ, Sassenfeld HM, Bell LD. Chemical synthesis and cloning of a poly(arginine)-coding gene fragment designed to aid polypeptide purification. Gene 1984; 32:321-7; PMID:6335699; http://dx.doi.org/ 10.1016/0378-1119(84)90007-6 [DOI] [PubMed] [Google Scholar]
- 3.Hopp TP, Prickett KS, Price VL, Libby RT, March CJ, Pat Cerretti D, Urdal DL, Conlon PJ. A Short Polypeptide Marker Sequence Useful for Recombinant Protein Identification and Purification. Nat Biotechnol 1988; 6:1204-10; http://dx.doi.org/ 10.1038/nbt1088-1204 [DOI] [Google Scholar]
- 4.Hochuli E, Bannwarth W, Döbeli H, Gentz R, Stüber D. Genetic Approach to Facilitate Purification of Recombinant Proteins with a Novel Metal Chelate Adsorbent. Nat Biotechnol 1988; 6:1321-5; http://dx.doi.org/ 10.1038/nbt1188-1321 [DOI] [Google Scholar]
- 5.Bachler M, Schroeder R, von Ahsen U. StreptoTag: a novel method for the isolation of RNA-binding proteins. RNA N Y N 1999; 5:1509-16; http://dx.doi.org/ 10.1017/S1355838299991574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Windbichler N, Schroeder R. Isolation of specific RNA-binding proteins using the streptomycin-binding RNA aptamer. Nat Protoc 2006; 1:637-40; PMID:17406291; http://dx.doi.org/ 10.1038/nprot.2006.95 [DOI] [PubMed] [Google Scholar]
- 7.Srisawat C, Engelke DR. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA N Y N 2001; 7:632-41; http://dx.doi.org/ 10.1017/S135583820100245X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leppek K, Stoecklin G. An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins. Nucleic Acids Res 2014; 42:e13; PMID:24157833; http://dx.doi.org/ 10.1093/nar/gkt956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson HE, Liljas L, Uhlenbeck OC. RNA Recognition by the MS2 Phage Coat Protein. Semin Virol 1997; 8:176-85; http://dx.doi.org/ 10.1006/smvy.1997.0120 [DOI] [Google Scholar]
- 10.Patel DJ. Adaptive recognition in RNA complexes with peptides and protein modules. Curr Opin Struct Biol 1999; 9:74-87; PMID:10047585; http://dx.doi.org/ 10.1016/S0959-440X(99)80010-4 [DOI] [PubMed] [Google Scholar]
- 11.Nagai K. RNA-protein complexes. Curr Opin Struct Biol 1996; 6:53-61; PMID:8696973; http://dx.doi.org/ 10.1016/S0959-440X(96)80095-9 [DOI] [PubMed] [Google Scholar]
- 12.De Guzman RN, Turner RB, Summers MF. Protein-RNA recognition. Biopolymers 1998; 48:181-95; PMID:10333745; http://dx.doi.org/ 10.1002/(SICI)1097-0282(1998)48:2%3c181::AID-BIP7%3e3.0.CO;2-L [DOI] [PubMed] [Google Scholar]
- 13.Beckett D, Wu HN, Uhlenbeck OC. Roles of operator and non-operator RNA sequences in bacteriophage R17 capsid assembly. J Mol Biol 1988; 204:939-47; PMID:3221401; http://dx.doi.org/ 10.1016/0022-2836(88)90053-8 [DOI] [PubMed] [Google Scholar]
- 14.Pickett GG, Peabody DS. Encapsidation of heterologous RNAs by bacteriophage MS2 coat protein. Nucleic Acids Res 1993; 21:4621-6; PMID:8233800; http://dx.doi.org/ 10.1093/nar/21.19.4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell 1998; 2:437-45; PMID:9809065; http://dx.doi.org/ 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- 16.Zenklusen D, Wells AL, Condeelis JS, Singer RH. Imaging real-time gene expression in living yeast. CSH Protoc 2007; 2007:pdb.prot4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Said N, Rieder R, Hurwitz R, Deckert J, Urlaub H, Vogel J. In vivo expression and purification of aptamer-tagged small RNA regulators. Nucleic Acids Res 2009; 37:e133; PMID:19726584; http://dx.doi.org/ 10.1093/nar/gkp719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corcoran CP, Rieder R, Podkaminski D, Hofmann B, Vogel J. Use of aptamer tagging to identify in vivo protein binding partners of small regulatory RNAs. Methods Mol Biol Clifton NJ 2012; 905:177-200; PMID: 22736004; http://dx.doi.org/20980440 10.1007/978-1-61779-949-5_11 [DOI] [PubMed] [Google Scholar]
- 19.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 2011; 3; PMID:20980440; http://dx.doi.org/ 10.1101/cshperspect.a003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papenfort K, Vanderpool CK. Target activation by regulatory RNAs in bacteria. FEMS Microbiol Rev 2015; 39:362-78; PMID:25934124; http://dx.doi.org/ 10.1093/femsre/fuv016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 2010; 40:939-53; PMID:21172659; http://dx.doi.org/ 10.1016/j.molcel.2010.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desnoyers G, Massé E. Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev 2012; 26:726-39; PMID:22474262; http://dx.doi.org/ 10.1101/gad.182493.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lalaouna D, Simoneau-Roy M, Lafontaine D, Massé E. Regulatory RNAs and target mRNA decay in prokaryotes. Biochim Biophys Acta 2013; 1829:742-7; PMID:23500183; http://dx.doi.org/ 10.1016/j.bbagrm.2013.02.013 [DOI] [PubMed] [Google Scholar]
- 24.Michaux C, Verneuil N, Hartke A, Giard J-C. Physiological roles of small RNA molecules. Microbiology 2014; 160:1007-19; PMID:24694375; http://dx.doi.org/ 10.1099/mic.0.076208-0 [DOI] [PubMed] [Google Scholar]
- 25.Papenfort K, Vogel J. Small RNA functions in carbon metabolism and virulence of enteric pathogens. Front Cell Infect Microbiol 2014; 4:91; PMID:25077072; http://dx.doi.org/ 10.3389/fcimb.2014.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lalaouna D, Morissette A, Carrier M-C, Massé E. DsrA regulatory RNA represses both hns and rbsD mRNAs through distinct mechanisms in Escherichia coli. Mol Microbiol 2015; 98:357-69; PMID:26175201; http://dx.doi.org/ 10.1111/mmi.13129 [DOI] [PubMed] [Google Scholar]
- 27.Li W, Ying X, Lu Q, Chen L. Predicting sRNAs and Their Targets in Bacteria. Genomics Proteomics Bioinformatics 2012; 10:276-84; PMID:23200137; http://dx.doi.org/ 10.1016/j.gpb.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lalaouna D, Carrier M-C, Semsey S, Brouard J-S, Wang J, Wade JT, Massé E. A 3′ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol Cell 2015; 58:393-405; PMID:25891076; http://dx.doi.org/ 10.1016/j.molcel.2015.03.013 [DOI] [PubMed] [Google Scholar]
- 29.Wright PR, Richter AS, Papenfort K, Mann M, Vogel J, Hess WR, Backofen R, Georg J. Comparative genomics boosts target prediction for bacterial small RNAs. Proc Natl Acad Sci U S A 2013; 110:E3487-96; PMID:23980183; http://dx.doi.org/ 10.1073/pnas.1303248110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wegrzyn G, Wegrzyn A. Is tRNA only a translation factor or also a regulator of other processes? J Appl Genet 2008; 49:115-22; PMID:18263978; http://dx.doi.org/ 10.1007/BF03195257 [DOI] [PubMed] [Google Scholar]
- 31.Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet 2014; 5:171; PMID:24966867; http://dx.doi.org/ 10.3389/fgene.2014.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanai A. Disrupted tRNA Genes and tRNA Fragments: A Perspective on tRNA Gene Evolution. Life 2015; 5:321-31; PMID:25629271; http://dx.doi.org/ 10.3390/life5010321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc Natl Acad Sci U S A 1995; 92:2003-7; PMID:7534408; http://dx.doi.org/ 10.1073/pnas.92.6.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J 1996; 15:3993-4000; PMID:8670904 [PMC free article] [PubMed] [Google Scholar]
- 35.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A 1998; 95:12462-7; PMID:9770508; http://dx.doi.org/ 10.1073/pnas.95.21.12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci U S A 1998; 95:12456-61; PMID:9770507; http://dx.doi.org/ 10.1073/pnas.95.21.12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desnoyers G, Bouchard M-P, Massé E. New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet TIG 2013; 29:92-8; PMID:23141721; http://dx.doi.org/ 10.1016/j.tig.2012.10.004 [DOI] [PubMed] [Google Scholar]
- 38.Lalaouna D, Massé E. Identification of sRNA interacting with a transcript of interest using MS2-affinity purification coupled with RNA sequencing (MAPS) technology. Genomics Data 2015; 5:136-8; PMID:26484242; http://dx.doi.org/ 10.1016/j.gdata.2015.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]


