Abstract
The impact of different culture conditions on biology of primary cancer cells is not always addressed. Here conditional reprogramming (CRC) was compared with mammary-optimized EpiCult™-B (EpiC) for primary mammary epithelial cell isolation and propagation, allograft generation and genome-wide transcriptional consequences using cancer and non-cancer mammary tissue from mice with different dosages of Brca1 and p53. Selective comparison to DMEM was included. Primary cultures were established with all three media but CRC was most efficient for initial isolation (p<0.05). Allograft development was faster using cells grown in EpiC as compared to CRC (p<0.05). Transcriptome comparison of paired CRC and EpiC cultures revealed 1700 differentially expressed genes by passage 20. CRC promoted Trp53 gene family up-regulation and increased expression of epithelial differentiation genes while EpiC elevated expression of epithelial-mesenchymal-transition genes. Differences did not persist in allografts where both methods yielded allografts with relatively similar transcriptomes. Restricting passage (<7) reduced numbers of differentially expressed genes below 50. In conclusion CRC was most efficient for initial cell isolation but EpiC was quicker for allograft generation. The extensive culture-specific gene expression patterns that emerged with longer passage could be limited by reducing passage number when both culture transcriptomes were equally similar to that of the primary tissue. Defining impact of culture condition and passage on the transcriptome of primary cells could assist experimental design and interpretation. For example differences that appear with passage and culture condition are potentially exploitable for comparative studies targeting specific biological networks in different transcriptional environments.
Keywords: primary cell culture, mammary cancer, Brca1, Genetically engineered mouse models, transcriptome
Introduction
Isolation of primary tumor cells from patient samples is a first step for many types of genetic, biochemical and pharmacological experiments relevant to personalized cancer treatment (Mitra et al., 2013). Retention of genetic characteristics and progenitor cells from the original cancer are important goals for culture methodology. Genetically engineered mouse models (GEMM) of breast cancer serve as preclinical translational analogs for the study of breast cancer pathophysiology and technology development for precision medicine. Here we used Brca1 deficient (L. P. Jones et al., 2005; L. Jones et al., 2008; Nakles et al., 2013) and Brca1 replete (Tilli et al., 2003; Díaz-Cruz et al., 2011) mouse models of breast cancer to compare transcriptional consequences and cancer progenitor cell retention of two different primary cancer epithelial cell culture methodologies: the mammary epithelial cell-optimized EpiCult (B) system (Stemcell Technologies, Vancouver, BC, Canada) (Yamaji et al., 2009) and the epithelial-selective conditionally reprogrammed culture (CRC) technology (Liu et al., 2012; Suprynowicz et al., 2012; Palechor-Ceron et al., 2013; Chapman et al., 2014; Saenz et al., 2014; Ligaba et al., 2015). Previously we employed time-lapse microscopy to show that mammary epithelial cells from wild-type (WT) and Brca1 deficient GEMM undergo morphological changes consistent with epithelial-mesenchymal transition (EMT) within four days of initial culture in EpiCult (B) (EpiC) (Nakles et al., 2013). The experiments presented here were initiated because CRC was reported to retain mammary epithelial cell cuboidal architecture (Saenz et al., 2014). However mandatory inclusion of the irradiated fibroblast feeder layer or conditioned media burdens the technique and the required Rho kinase (ROCK) inhibitor Y-27632 has the potential to alter a number of cellular processes (Olson, 2008) For that reason we thought it sensible to directly compare biological and transcriptional consequences of these two prominent choices for initial isolation and propagation of primary mammary epithelial cells under 2D adherent conditions to further define the advantages and disadvantages of each approach. Previous work reported significant gene expression differences in 3D cultures of undifferentiated human mammary epithelial cells grown as mammospheres as compared to growth under differentiating conditions on collagen (Dontu et al., 2003).
Epithelial-mesenchymal transition (EMT) is observed in many 2D epithelial cell culture conditions and is associated with specific changes in cell morphology and gene expression including up-regulation of Transforming Growth Factor beta (TGFB) signaling pathways (Moreno-Bueno et al., 2009) and dedifferentiation, including loss of Estrogen Receptor alpha (ERa) expression (Novaro et al., 2003). In cancer biology, EMT has been mechanistically linked to metastasis, chemotherapy resistance, and preservation of cancer progenitor cells (Drasin et al., 2011; Luo et al., 2015)
CRC reduces TGFB signaling (Ligaba et al., 2015) and promotes p63 expression, a gene linked to ‘stemness’ in some tissues, but nevertheless expressed in fully differentiated mammary myoepithelial cells (Suprynowicz et al., 2012; Assefnia et al., 2014; Chapman et al., 2014; Melino, Memmi, Pelicci, & Bernassola, 2015). Differentiation of skin keratinocytes is suppressed in CRC with lower levels of keratin gene expression and increased levels of hTERT (Liu et al., 2012; Palechor-Ceron et al., 2013; Chapman et al., 2014). Removal from CRC and placement into alternative media allows the cells to differentiate with increased levels of keratin expression illustrating that some genetic changes imposed by CRC can be reversible (Suprynowicz et al., 2012). Treatment of breast cancer cells with the ROCK inhibitor used for CRC, Y-27632, is known to decrease actomyosin interaction (Bhandary et al., 2015). Actin cytoskeleton changes are integral to EMT (Haynes et al., 2011).
The natural history of GEMM utilized for this study includes development of a range of triple negative mammary cancers from moderately well-differentiated to undifferentiated adenocarcinomas and more EMT-like spindloid/sarcomatoid carcinomas (Diaz-Cruz et al., 2010; Nakles et al., 2013). The same spectrum of histopathology is found in human triple negative breast cancers (Lehmann et al., 2011). One of the human mesenchymal-stem like (MSL) triple negative histologies demonstrates TGFBR3 over-expression (Jovanović et al., 2014).
Here we show that both methodologies can be effectively used for isolation and propagation of primary non-cancer and cancer mammary epithelial cells. The major advantage of the CRC approach was a significantly higher success rate for initial culture (95%). Benefits of EpiC include a less burdensome culture methodology and faster allograft development. Both culture conditions were associated with characteristic transcriptional changes upon prolonged passage (>20). Limiting passage (<7) reduced the magnitude of these changes equivalently for both conditions. Differences found in the paired cells after prolonged culture, for example, variances in allograft formation, up-regulation of p53 pathways under CRC, up-regulation of TGFBR3 under EpiC, could be exploited in targeted studies of cancer biology and therapy.
Materials and Methods
Mouse models
Primary mammary epithelial cells were isolated from cancer and non-cancer mammary tissue from six GEMMs and parental strain wildtype (WT) C57Bl/6 mice (30–46 gm): Brca1floxed (fl)11/wild-type (WT)/Mouse Mammary Tumor Virus (MMTV)-Cre/Trp53−/+ (Brca1−/+/p53−/+) n=6/cancer, n=3/non-cancer; Brca1fl11/fl11/MMTV-Cre/Trp53−/+ (Brca1−/−/p53−/+) n=2/cancer, n=1/non-cancer; tet-op-CYP19A1MMTV-reverse tetracycline-controlled TransActivator(rtTA)/Trp53−/+/MMTV-Cre (CYP19A1p53−/+) n=1/cancer; Brca1fl11/WT/MMTV-Cre/tet-op-CYP19A1/Trp53−/+I− (Brca1−/+/CYP19A1/p53−/+) n=4/cancer; Brca1fl11/fl11/MMTV-Cre/Trp53−/+/MMTV-rtTA/tet-op-Esr1 (Brca1−/−/p53−/+/rtTA/Esr1) n=1/cancer, n=1/non-cancer; tet-op-Simian Virus 40 T AntigenMMTV-tetracycline TransActivator (tTA)/tet-op-Esr1 (TagtTA/Esr1) n=1/non-cancer; WT n=1/non-cancer. GEMMs were generated at Georgetown University, weaned before puberty, and genotyped using tail DNA (Transnetyx, Inc., Cordova, TN). For allograft experiments six-week-old athymic nude (NU (NCr)-Foxn1nu) female mice (n=13) (Harlan Laboratories, Inc., Frederick, MD) (26–30 gm) were acclimatized one week prior to primary cell injection into thoracic and inguinal mammary fatpads through a skin incision under isoflurane anesthesia (4 sites/mouse/106 mammary epithelial cancer cells/site in 25μl PBS/25μl Matrigel) (BD Biosciences, Franklin Lakes, NJ)/1cc syringe/27-gauge needle). All mice were housed in barrier zones in single-sex sterilized ventilated cages with corn-cob bedding (1–4 mice per cage) with ad libitum access to water/irradiated Picolab rodent diet 20 (5053) (Labdiet, St Louis, MO) under 12 h dark/light cycles at Georgetown University with exception of doxycycline-containing diet (Bio-Serv, Frenchtown, NJ) for CYP19A1p53−/+ mice. Mice were monitored weekly with measurement of palpable tumors and euthanized by CO2 inhalation followed by cervical dislocation when mice reached 6 months of age (allograft experiments), 12 months of age (GEMM) or palpable tumors > 1cm3, whichever occurred first. Cancer and non-cancer mammary tissues were removed at necropsy and divided for isolation of viable mammary epithelial cells, formalin fixation, or frozen at −80°C.
Primary mammary epithelial cell culture
Cells were isolated using EpiCult-B Mouse Medium Kit (Stemcell Technologies, Vancouver, BC) (Nakles et al., 2013) and divided for culture under CRC (F medium containing 25% Ham’s F-12 nutrient mix (Thermo Fisher Scientific, Carlsbad, CA) supplemented with 25ng/mL hydrocortisone, 5 μg/mL insulin, 0,1nmol/L cholera toxin (Sigma-Aldrich, St Louis, MO), 0.125 ng/mL epidermal growth factor, 10 μg/mL gentamicin (Thermo Fisher Scientific), 250ng/mL Fungizone (Thermo Fisher Scientific), and 5 μmol/L ROCK inhibitor Y-27632 (Enzo Life Sciences, Farmingdale, NY) in the presence of irradiated Swiss 3T3-J2 mouse fibroblast feeder cells (Chapman et al., 2010; Liu et al., 2012) or EpiC (EpiCult®-B Basal medium (Mouse) supplemented with EpiCult®-B Basal Proliferation Supplements (Mouse), 10ng/mL recombinant human epidermal growth factor (rhEGF), 10% Fetal bovine serum (FBS) (Stemcell Technologies) and 100μg/mL streptomycin/penicillin (Thermo Fisher Scientific) and/or and complete Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 100 μg/mL penicillin, 100 μg/mL streptomycin, and 100 μg/mL glutamine (Thermo Fisher Scientific) at 37°C with 5% CO2. For EpiC culture, fibroblast contamination was reduced by incubating cells in a first flask to allow fibroblasts to adhere (3–4h) and then transferring the epithelial cell enriched non-adherent cells to a second flask for expansion (Darcy et al., 2000). Cells were viably frozen at different passage numbers and re-cultured when needed. Digital images were taken using EVOS™ XL Core Cell Imaging System or Olympus IX+71 microscope/Olympus DP70 camera/DP Controller v3.2.1.276 software (Olympus America Inc., Center Valley, PA).
Immunohistochemistry and pathological examination
Tissue and cell pellets were fixed in 10% buffered formalin (Fisher Scientific, Hampton, NH) overnight at 4°C, embedded in paraffin and five μm sections stained with hematoxylin and eosin (H&E) or used for immunohistochemistry (IHC) following antigen retrieval with citrate buffer. Primary antibodies: Cytokeratin, Wide Spectrum Screening (CK), Z0622 (Dako, Carpinteria, CA) 1:625, cell pellets, 1:120, tissue; p53 Protein (CM5) Novocastra, NCL-p53-CM5p 1:200, Ki67 NCL+L+Ki67+MM 1:120 (Leica Biosystems Inc., Buffalo Grove, IL); anti-p63 (4A4) Mouse Monoclonal Primary Antibody, 790-4509 (Ventana Medical Systems, Inc. Tucson, AZ) 1:1, Transforming Growth Factor, Beta Receptor III (TGFBR3) sc6199 1:300 (Santa Cruz Biotechnology Inc., Dallas, TX), and Cluster of Differentiation (CD)140A/Platelet-Derived Growth Factor Receptor, Alpha Polypeptide (PDGFRA) APA5 1:100 (BioLegend, San Diego, CA) to label murine fibroblasts (Gawade et al., 2016). A board-certified academic pathologist (B.V.S.K.) blinded to sample identity read histology. Digital images were taken using Nikon Eclipse E800 Microscope/NIS-Elements BR 4.30.02 64-bit software (Nikon Instruments Inc., Melville, USA).
Polymerase Chain Reaction (PCR) and Reverse Transcriptase-PCR (RT-PCR)
DNA was extracted from cell pellets using PureLink® Genomic DNA Mini Kit (Thermo Fisher Scientific) or formalin-fixed, paraffin-embedded (FFPE) allograft sections employing QuickExtract™ FFPE DNA Extraction Kit (Epicenter, Madison, WI) and PureLink® Genomic DNA Mini Kit (Thermo Fisher Scientific) or RNA extracted using an RNeasy Mini Kit (QIAGEN, Gaithersburg, MD), quantified (Nanodrop, Thermo Fisher Scientific) and 1 μg total RNA used to prepare complement DNA (cDNA) (qScript™ cDNA Synthesis Kit, Quanta Biosciences Inc., Gaithersburg, MD) for detection of BRCA1 exon 11 deletion (forward: 5′ GGGTAGTTTGTAAGCATCCA 3′, reverse: 5′ GCCTGTTCCTCCCCTTGTATA 3′), tet-op-CYP19A1 (forward: 5′ CGAGCTCGGTACCCGGGTCG 3′, reverse: 5′ CAGGCATGGCTTCAGGCACGA 3′) (Díaz-Cruz et al., 2011), MMTV-Cre (forward: 5′ GTGAACGTGCAAAACAGGCT 3′, reverse: 5′CGGTGCTAACCAGCGTTTTC 3′), and Actb (beta actin) (forward: 5′ ATCGTGGGCCGCCCTAGGCA 3′, reverse: TGGCCTTAGGGTTCAGAGGG 3′) (GoTaq® Green Master Mix, Promega, Madison, WI). PCR (30 cycles: 1m: 95°C, 1m: 58°C, 3m: 72°C) and RT-PCR (30 cycles: 1m: 95°C, 1m: 60°C, 3m: 72°C) products were visualized by ethidium bromide staining after electrophoresis on agarose gels (Bio-Rad Universal Hood II, Bio-Rad, Hercules, CA).
RNAseq
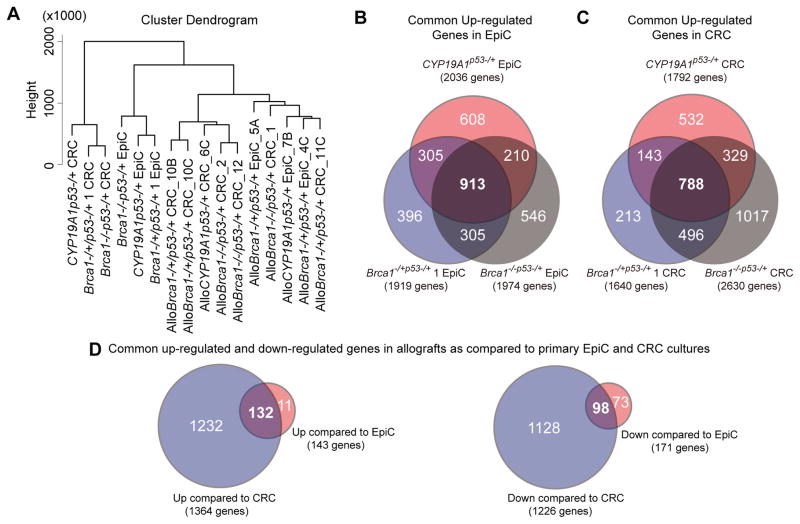
Total RNA was extracted from cell pellets (n=12), mammary cancers (n=2) and allografts (n=10) using Trizol (Thermo Fisher Scientific) followed by phase separation and precipitation using chloroform and isopropanol (Thermo Fisher Scientific), purified (RNeasy Plus Mini Kits, Qiagen, Valencia, CA), analyzed (Nanodrop, Agilent Bioanalyzer 2100, Agilent Technologies, Santa Clara, CA), converted to cDNA (SuperScript II, Invitrogen), sequencing libraries prepared (TruSeq RNA Sample Preparation Kit, Illumina, San Diego, USA), and paired- 5481 (P36), 9668 (P20) and 9740 (P21) n=6, or single- 5504C/D (P7) n=4, and 3189 (P3) n=2, end sequencing performed (HiSeq 2000,Illumina) with read quality determined (FastQC, http://www.bioinformatics.babraham.ac.uk/project/fastqc), contaminated adaptor portions trimmed (Trim Galore, http://www.bioinformatics.babraham.ac.uk/project/trim_galore) [See Gene Expression Omnibus (GEO) database number GSE81212], and alignment to mouse reference genome (mm9) performed (Kim et al., 2013). Transcriptomes were assembled, transcript abundance estimated (FPKM: fragments per kilobase of transcript per million mapped reads), DEGs defined (statistically significant 2-fold up- or 0.5 downregulated expression) (Cufflinks and Cuffdiff (Trapnell et al., 2010, 2012), and mapped reads visualized (Integrative Genomics Viewer (IGV), Robinson et al., 2011; Thorvaldsdóttir et al., 2013). Statistically significant DEGs between individual pairs were identified to define genes that were commonly upregulated in EpiC (n=913) versus CRC (n=788) conditions. Allograft transcriptomes were compared to their respective cell culture transcriptome to identify significant DEGs. Genes that were commonly up- or downregulated in allografts as compared to CRC (up: n=1364, down: n=1226) and EpiC (up: n=143, down: n=171) primary cultures were then compared to identify the subset of genes commonly up- or downregulated irrespective of primary culture/genotype (up: n=132, down n=98). Samples were clustered according to their gene expression profiles using the hclust function in R (R Development Core Team (2008). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org).
Gene ontology, network analyses and motif analysis
Gene ontology (GO) was performed using Partek Genomics Suite 6.6, applying Fisher’s Exact test and restricting analyses to groups with more that two genes employing the default mapping method. Top 19 enrichment score categories were extracted for presentation. Hallmark gene sets (Molecular Signatures Database (MSigDB),http://www.broadinstitute.org/gsea/msigdb/index.jsp) corresponding to commonly up- or downregulated DEGs were further analyzed by Pathway Studio to identify expression targets. DNA sequences in promoter regions (−450bp – TSS – +50bp) of DEGs upregulated in EpiC versus CRC were analyzed to identify transcription factor binding site (TFBS) motifs. Significantly enriched motifs (p-value < 0.001) were recognized utilizing the JASPAR motif database (http://jaspar.genereg.net/) (Kang et al., 2013). A second search algorithm, PSCAN (Zambelli et al., 2009) was used for further validation and results were visualized as a heatmap. Venn diagram were designed by BioVenn (http://www.cmbi.ru.nl/cdd/biovenn/) (Hulsen et al., 2008)
Statistics
Two-tailed Fisher’s exact test was used to compare probabilities of primary culture establishment, allograft development and correlations between genotypes and allograft histologies, logrank test was used to test the identity of allograft development timecourse curves, and Kruskal-Wallis test with Dunn’s multiple comparisons test was used to compare proportion of fibroblasts in the allograft tissues (GraphPad Software, Inc. La Jolla, CA).
Results
Conditional Reprogramming Culture (CRC) conditions were more successful for initial establishment of primary cultures than EpiCult™-B or DMEM
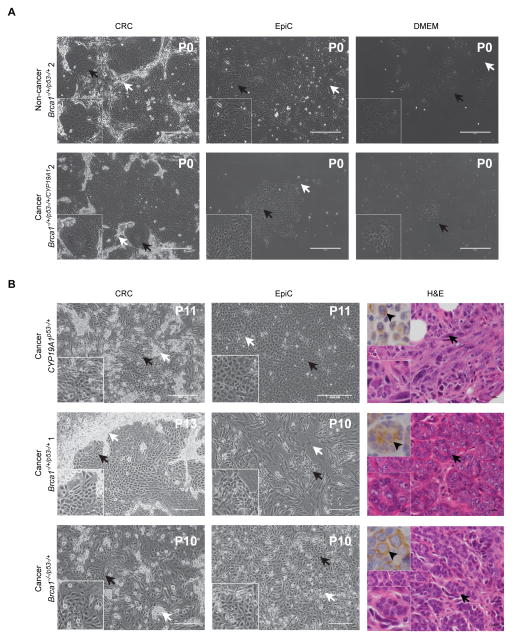
CRC methodology was associated with a significantly higher likelihood of establishing primary cultures of mouse mammary epithelial cells (95%) in comparison to EpiCult™-B (EpiC) (60%, p=0.02) and DMEM (35%, p=0.0003) (Table 1). Doubling times were faster for CRC and EpiC compared to DMEM for both cancer and non-cancer cells (Figure 1A). Brca1 gene dosage did not impact likelihood of prolonged passage of mammary cancer cells for either CRC or EpiC (Figure 1B, Table 1) (Groeneveld, 2012). Eight of nine cancer cell cultures initially established in either CRC or EpiC were successfully moved to DMEM (Table 1).
Table 1.
Comparison of cancer and non-cancer primary cell culture outcomes.
| Mouse/Tissue ID | Genotype | Cancer | Non-cancer | Growth | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DMEM | EpiC | CRC | Allografts | ||||||||
| Initial | After EpiC | After CRC | Initial | Initial | EpiC | CRC | |||||
| 1 | 9668 | Brca1 fl11/wt/MMTV-Cre/p53+/− | C | - | ND | Yes | Yes | Yes | Yes | Yes | Yes |
| 2 | 9669 | Brca1 fl11/wt/MMTV-Cre/p53+/− | C | - | ND | ND | Yes | No | Yes | ND | No |
| 3 | 9875 | Brca1 fl11/wt/MMTV-Cre/p53+/− | C | - | Yes | ND | ND | Yes | Yes | ND | ND |
| 4 | 9875 | Brca1 fl11/wt/MMTV-Cre/p53+/− | - | N | Yes | ND | ND | Yes | Yes | ND | ND |
| 5 | 9642 | Brca1 fl11/fl11/MMTV-Cre/p53+/− | C | - | No | ND | No | No | Yes | ND | No |
| 6 | 9642 | Brca1 fl11/fl11/MMTV-Cre/p53+/− | - | N | No | ND | No | No | Yes | ND | ND |
| 7 | 9740 | Brca1 fl11/fl11/MMTV-Cre/p53+/− | C | - | ND | Yes | Yes | Yes | Yes | Yes | Yes |
| 8 | 5481 | CYP19A1 MMTV-rtTA/p53+/−/MMTV-Cre | C | - | ND | Yes | Yes | Yes | Yes | Yes | Yes |
| 9 | 5504C | Brca1 fl11/wt/MMTV-Cre/tet-op-CYP19A1/p53+/− | C | - | Yes | ND | ND | Yes | Yes | ND | ND |
| 10 | 5504D | Brca1 fl11/wt/MMTV-Cre/tet-op-CYP19A1/p53+/− | C | - | Yes | ND | ND | Yes | Yes | ND | ND |
| 11 | 5505C | Brca1 fl11/wt/MMTV-Cre/tet-op-CYP19A1/p53+/− | C | - | No | ND | ND | No | No | ND | ND |
| 12 | 5505D | Brca1 fl11/wt/MMTV-Cre/tet-op-CYP19A1/p53+/− | C | - | No | ND | ND | NO | Yes | ND | ND |
| 13 | 15235 | Brca1fl11/fl11/MMTV-Cre/p53+/−/MMTV-rtTA/tet-op-Esr1 | - | N | ND | ND | No | No | Yes | ND | ND |
| 14 | 15235 | Brca1fl11/fl11/MMTV-Cre/p53+/−/MMTV-rtTA/tet-op-Esr1 | C | - | ND | ND | Yes | No | Yes | ND | Yes |
| 15 | 1135 | MMTV-tTA/Tet-op-Tag/Tet-op-ESR1 | - | N | Yes | ND | ND | Yes | Yes | ND | ND |
| 16 | 9969 | Brca1 fl11/wt/MMTV-Cre/p53+/− | C | - | No | ND | ND | No | Yes | ND | ND |
| 17 | 9969 | Brca1 fl11/wt/MMTV-Cre/p53+/− | - | N | No | ND | ND | Yes | Yes | ND | ND |
| 18 | 9977 | Brca1 fl11/wt/MMTV-Cre/p53+/− | C | - | No | ND | ND | Yes | Yes | ND | ND |
| 19 | 9977 | Brca1 fl11/wt/MMTV-Cre/p53+/− | - | N | No | ND | ND | Yes | Yes | ND | ND |
| 20 | 3189 | WT | - | N | No | ND | ND | Yes | Yes | ND | ND |
| Total | 13 | 7 | 14 | 3 | 8 | 20 | 20 | 3 | 6 | ||
| Percentage of growth | - | - | 35%* | 100% | 62% | 60%* | 95%* | 100% | 66% | ||
ID: Identification. C=Cancer. N=Non-cancer. DMEM: Dulbecco’s Modified Eagle Medium. EpiC: EpiCult™-B Mouse Medium. CRC: Conditionally Reprogrammed Cell culture. ND=Not done.
p=0.02 EpiC vs. CRC, p=0.0003 DMEM vs. CRC, Fisher’s exact, two-tailed.
Figure 1. Cultured primary mammary epithelial cells were derived from mammary cancers and non-cancerous mammary tissue of genetically engineered mice.
(A) Phase contrast images of primary cells cultured in CRC, EpiC and DMEM five days following initial plating without passage. Cells derived from non-cancerous and cancerous mammary gland tissue from the same Brca1−/+/p53−/+ mouse. Bottom left inset shows magnified image of area indicated by black arrows on larger images. (B) Phase contrast images of CYP19A1p53−/+, Brca1−/+/p53−/+ 1 and Brca1−/−/p53−/+ primary cells cultured in CRC and EpiC and H&E images of the mammary cancers from which the cells were derived. Bottom left inset shows magnified image of area indicated by black arrow on larger phase contrast and H&E images. Top left shows representative image of pancytokeratin IHC (arrowheads). Passage number indicated top right (phase contrast images). Black arrows indicate mammary epithelial cells. White arrows indicate fibroblast or feeder cells. Phase contrast images taken at 10X. Size bars= 400μM. H&E images taken at 40X. Size bars= 10μM. H&E: Hematoxylin and Eosin. P: Passage. CRC: Conditionally Reprogrammed Cells. EpiC: EpiCult™B.
Allografts derived from cells cultured under EpiC conditions formed more quickly than those from CRC cultured cells
Allograft formation was investigated using three paired sequentially passaged (>20) CRC and EpiC primary triple negative epithelial cancer cell cultures with different Brca1 gene dosages coupled with p53 haploinsufficiency. Palpable allografts developed significantly faster from EpiC as compared to CRC cultures (p<0.05, logrank, Figure 2A). Loss of both Brca1 alleles significantly increased the probability of allograft formation (EpiC, p=0.007; CRC, p=0.02, Fisher’s Exact, Figure 2B) and appearance of uniform spindloid/sarcomatoid histology (p= 0.04, Fisher’s Exact, Figure 2C, Table 2). Tumorigenicity occurred earlier and was more variable from EpiC as compared to CRC cultures (Figure 2D). Spindloid/sarcomatoid cancers demonstrated cytokeratin expression, consistent with epithelial derivation (Figure 2C, insets), studies of nucleic acids extracted from allograft tissue and/or secondarily cultured cells confirmed presence of GEMM modifications (Figure 2E–I), and the proportion of fibroblasts as compared to cancer cells was low in allografts (CYP19A1p53−/+ CRC: 1.5±0, CYP19A1p53−/+ EpiC: 1.0±0.5, Brca1−/+/p53−/+ CRC:1.8±0.8, Brca1−/+/p53−/+ EpiC: 2.3±0.6*, Brca1−/−/p53−/+ CRC: 0.9±0.4, Brca1−/−/p53−/+ EpiC: 0.6±0.1* CD140A/PDGFRA positive fibroblasts per 40X high power field, *p<0.05, Kruskal-Wallis test, Dunn’s multiple comparisons test) (Figure 2J,K).
Figure 2. Allograft generation from cells grown under EpiC and CRC conditions.
(A) Timecourse of percentage of injected sites developing into palpable allografts from EpiC (black) and CRC (grey) cell cultures. *p<0.05, logrank. (B) Bar graphs illustrating the percentage of injected sites developing into allografts from CYP19A1p53−/+, Brca1−/+/p53−/+ 1, and Brca1−/−/p53−/+ EpiC (black) and CRC (grey) cell cultures. *EpiC, p=0.007; **CRC, p=0.02, CYP19A1p53−/+ compared to Brca1−/−/p53−/+, Fisher’s Exact. (C) Representative H&E images of allografts that developed from CYP19A1p53−/+, Brca1−/+/p53−/+ 1 and Brca1−/−/p53−/+ cell cultures. Top left inset shows representative image of pancytokeratin IHC (arrowheads). Bottom left inset shows representative magnification of H&E image. (D) Timecourse of tumor size development for all injected sites that developed into allografts from EpiC (top, black lines) and CRC (bottom, grey lines) cell cultures. (E) Images of Cre (150 bp) and Actb (Actin) (250 bp) detection by RT-PCR (AlloBrca1−/−/p53−/+ CRC_1B). Markers (M) and NTC: No template negative control shown from same gels. (F) Images showing presence of Brca1 with exon 11 deletion product (600 bp) in AlloBrca1−/−/p53−/+ CRC_2A but absent in WT and AlloCYP19A1p53−/+ CRC_6C negative controls and Actb (Actin) (250 bp) detection by RT-PCR from the same gels with Markers (M). (G) Images of Cre (150 bp) and Actb (Actin) (250 bp) detection by PCR of DNA from mammary epithelial cells secondarily grown in EpiC from AlloBrca1−/−/p53−/+ EpiC. Markers (M) shown from the same gels. (H) Images of CYP19A1 (150 bp) and Actb (Actin) (250 bp) detection by PCR of DNA from AlloCYP19A1p53−/+ CRC_6C with WT negative control. Markers (M) shown from the same gels. Thin lines between gel images indicate lanes taken from same gel. (I) Image showing normalized read coverage across the MMTV-Cre transgene viewed through the Integrative Genomics Viewer (IGV) from AlloBrca−/+/p53−/+ EpiC_5A compared to background negative control WT mammary gland. (J) Representative IHC images of NIH3T3 fibroblasts as positive (arrowhead) and negative (no primary antibody) controls for CD140A/PDGFRA antibody. (K) Representative IHC images of allografts showing detection of CD140A/PDGFRA positive fibroblasts (arrowheads). H&E and IHC images taken at 40_X. Size bars= 10μM.
Table 2.
Allograft development and histology.
| Injected Cell Genotype | Mouse ID/site | Growth | Allograft Identifier/RNAseq | Allograft Histopathology | Original Cancer Histopathology |
|---|---|---|---|---|---|
| CYP19A1p53−/+ CRC | B206 MG-A | No | Undifferentiated adenocarcinoma with spindloid/sarcomatoid focus | ||
| B206 MG-C | Yes | AlloCYP19A1p53−/+ CRC_6C | Undifferentiated adenocarcinoma | ||
| B207 MG-A | No | ||||
| B207 MG-C | No | ||||
| B208 MG-A | No | ||||
| B208 MG-C | No | ||||
| B209 MG-A | No | ||||
| B209 MG-C | No | ||||
| CYP19A1p53−/+ EpiC | B206 MG-B | No | |||
| B206 MG-D | No | ||||
| B207 MG-B | Yes | AlloCYP19A1p53−/+ EpiC_7B | Undifferentiated adenocarcinoma with spindloid/sarcomatoid features | ||
| B207 MG-D | No | ||||
| B208 MG-B | No | ||||
| B208 MG-D | No | ||||
| B209 MG-B | Yes | Undifferentiated adenocarcinoma | |||
| B209 MG-D | No | ||||
| Brca1−/+/p53−/+ CRC | B210 MG-A | No | Undifferentiated adenocarcinoma | ||
| B210 MG-B | Yes | AlloBrca1−/+/p53−/+ CRC_10B | Spindloid/sarcomatoid carcinoma with foci of adenocarcinoma | ||
| B210 MG-C | Yes | AlloBrca1−/+/p53−/+ CRC_10C | Spindloid/sarcomatoid carcinoma | ||
| B210 MG-D | No | ||||
| B211 MG-A | No | ||||
| B211 MG-B | No | ||||
| B211 MG-C | Yes | AlloBrca1−/+/p53−/+ CRC_11C | Undifferentiated adenocarcinoma | ||
| B211 MG-D | No | ||||
| Brca1−/+/p53−/+ EpiC | B204 MG-A | No | |||
| B204 MG-B | Yes | Spindloid/sarcomatoid carcinoma with foci of undifferentiated adenocarcinoma | |||
| B204 MG-C | Yes | AlloBrca1−/+/p53−/+ EpiC_4C | Spindloid/sarcomatoid carcinoma with foci of undifferentiated adenocarcinoma | ||
| B204 MG-D | No | ||||
| B205 MG-A | Yes | AlloBrca1−/+/p53−/+ EpiC_5A | Undifferentiated adenocarcinoma with spindloid/sarcomatoid features | ||
| B205 MG-B | Yes | Undifferentiated adenocarcinoma with spindloid/sarcomatoid features | |||
| B205 MG-C | Yes | Undifferentiated adenocarcinoma with spindloid/sarcomatoid features | |||
| B205 MG-D | Yes | Undifferentiated adenocarcinoma with spindloid/sarcomatoid features | |||
| Brca1−/−/p53−/+ CRC | B201 MG-A | Yes | Spindloid/sarcomatoid carcinoma with foci of undifferentiated adenocarcinoma | Undifferentiated adenocarcinoma | |
| B201 MG-B | Yes | AlloBrca1−/−/p53−/+ CRC_1 | Spindloid/sarcomatoid carcinoma | ||
| B201 MG-C | Yes | Spindloid/sarcomatoid carcinoma | |||
| B201 MG-D | No | ||||
| B202 MG-A | Yes | AlloBrca1−/−/p53−/+ CRC_2 | Spindloid/sarcomatoid carcinoma with foci of undifferentiated adenocarcinoma | ||
| B202 MG-B | Yes | Spindloid/sarcomatoid carcinoma | |||
| B202 MG-C | Yes | Spindloid/sarcomatoid carcinoma | |||
| B202 MG-D | No | ||||
| B212 MG-A | No | ||||
| B212 MG-B | Yes | AlloBrca1−/−/p53−/+ CRC_12 | Spindloid/sarcomatoid carcinoma | ||
| B212 MG-C | Yes | Spindloid/sarcomatoid carcinoma | |||
| B212 MG-D | Yes | Spindloid/sarcomatoid carcinoma | |||
| Brca1−/−/p53−/+ EpiC | 689 MG-A | Yes | Spindloid/Sarcomatoid carcinoma | ||
| 689 MG-B | Yes | Spindloid/Sarcomatoid carcinoma | |||
| 689 MG-C | Yes | Spindloid/Sarcomatoid carcinoma | |||
| 689 MG-D | Yes | Spindloid/Sarcomatoid carcinoma | |||
| 692 MG-A | Yes | Spindloid/Sarcomatoid carcinoma | |||
| 692 MG-B | Yes | Spindloid/Sarcomatoid carcinoma | |||
| 692 MG-C | Yes | Spindloid/Sarcomatoid carcinoma | |||
| 692 MG-D | Yes | Spindloid/Sarcomatoid carcinoma |
ID: Identification. EpiC: EpiCult™-B Mouse Medium. CRC: Conditionally Reprogrammed Cell culture.
CRC and EpiC in vitro cultures demonstrated dissimilar gene expression patterns with in vivo allograft gene expression more similar to EpiC than CRC
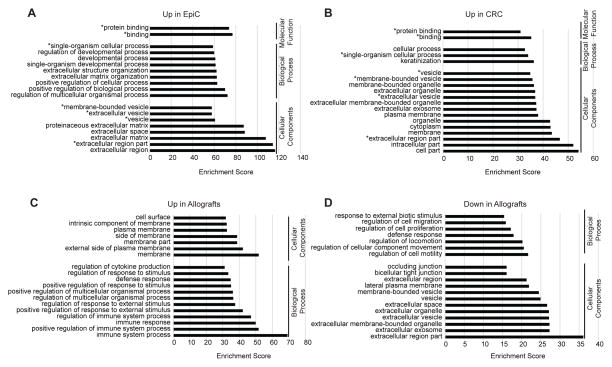
To identify pathways preferentially activated under the different growth conditions, RNAseq was used to characterize transcriptomes of these three paired cell cultures and derived allografts. Hierarchical clustering revealed that culture and growth condition, rather than genetic identity, was the primary determinant of transcriptome similarity. Commonly statistically significantly upregulated DEGs were defined for both EpiC (913) and CRC (788) cultures, independent of genotype (Figure 3, A–C). GO enrichment analyses were performed on these two DEG sets to delineate biological processes and cellular component categories distinctly enriched under the two different culture conditions. More than half (63%) of the top categories identified for each culture condition were unique (Figure 4A, B). Developmental and organismal biological processes were exclusive to EpiC and keratinization to CRC. MSigDB was queried to identify unique Hallmark gene sets associated with the DEGs. Epithelial_Mesenchymal_Transition (EMT) was the most significant Hallmark gene set for upregulated DEGs in EpiC and P53_Pathway for upregulated DEGs in CRC (Table 3A).
Figure 3. Transcriptome characterization of EpiC and CRC cultures and allografts.
(A) Dendrogram illustrating hierarchical clustering of transcriptomes from EpiC and CRC cultures and allografts. (B) Venn diagram illustrating overlaps between commonly upregulated significantly differentially expressed genes in EpiC cultured CYP19A1p53−/+ (red), Brca1−/+/p53−/+ 1 (blue), and Brca1−/−/p53−/+ (grey) primary mammary cancer cells as compared to CRC culture. (C) Venn diagram illustrating the overlap between commonly upregulated significantly differentially expressed genes in CRC cultured CYP19A1p53−/+ (red), Brca1−/+/p53−/+ 1 (blue), and Brca1−/−/p53−/+ (grey) primary mammary cancer cells as compared to EpiC culture. (D) Venn diagrams illustrating the overlap between commonly up-and downregulated significantly differentially expressed genes in allografts as compared to the respective primary cell cultures EpiC (red) and CRC (blue) they developed from.
Figure 4. GO terms associated with commonly significantly differentially expressed genes under different culture and growth conditions.
(A) Top 19 GO or other functional terms associated with the 913 commonly significantly upregulated DEGs in EpiC cultures of mammary cancer cells from CYP19A1p53−/+, Brca1−/+/p53−/+ 1 and Brca1−/−/p53−/+ mice. (B) Top 19 GO or other functional terms associated with the 788 commonly significantly upregulated DEGs in CRC cultures of mammary cancer cells from CYP19A1p53−/+, Brca1−/+/p53−/+ 1 and Brca1−/−/p53−/+ mice. (C) Top 19 GO or other functional terms associated with the 132 commonly significantly upregulated DEGs in allografts of CYP19A1p53−/+, Brca1−/+/p53−/+ 1 and Brca1−/−/p53−/+ mammary cancer cells as compared to the respective EpiC and CRC primary cell cultures they developed from. (D) Top 19 GO or other functional terms associated with the 98 commonly significantly downregulated DEGs in allografts of CYP19A1p53−/+, Brca1−/+/p53−/+ 1 and Brca1−/−/p53−/+ mammary cancer cells as compared to the respective EpiC and CRC primary cell cultures they developed from. GO: Gene Ontology. DEG: Differentially expressed genes.
Table 3.
Hallmark gene sets identified from differentially regulated genes in different culture and growth conditions.
| A. Top 10 Hallmark gene sets identified in DEGs upregulated in EpiC compared to CRC cultures. | |
|---|---|
| EpiC | CRC |
| EPITHELIAL_MESENCHYMAL_TRANSITION | P53_PATHWAY |
| TNFA_SIGNALING_VIA_NFKB | ESTROGEN_RESPONSE_EARLY |
| HYPOXIA | APICAL_JUNCTION |
| INFLAMMATORY_RESPONSE | ESTROGEN_RESPONSE_LATE |
| KRAS_SIGNALING_UP | KRAS_SIGNALING_DN |
| COAGULATION | APICAL_ SURFACE |
| IL6_JAK_STAT3_SINGNALING | KRAS_SIGNALING_UP |
| APOPTOSIS | PI3K_AKT_MTOR_SIGNALING |
| INTERFERON_GAMMA_RESPONSE | MYOGENESIS |
| UV_RESPONSE_DN | APOPTOSIS |
| B. All Hallmark gene sets identified in DEGs upregulated in allografts as compared to EpiC and CRC cultures they were derived from. | |
| EpiC | CRC |
| ALLOGRAFT_ REJECTION | ALLOGRAFT_ REJECTION |
| KRAS_SIGNALING_UP | KRAS_SIGNALING_UP |
| INTERFERON_GAMMA_RESPONSE | INTERFERON_GAMMA_RESPONSE |
| COMPLEMENT | COMPLEMENT |
| INFLAMMATORY_ RESPONSE | INFLAMMATORY_ RESPONSE |
| EPITHELIAL_ MESENCHYMAL_ TRANSITION | |
| C. All Hallmark gene sets identified in DEGs upregulated in original cancers as compared to EpiC and CRC cultures derived from them. | |
| EpiC | CRC |
| KRAS_SIGNALING_UP | ALLOGRAFT_ REJECTION |
| ALLOGRAFT_ REJECTION | KRAS_SIGNALING_UP |
| INFLAMMATORY_ RESPONSE | |
Hallmark gene sets unique to paired comparisons bolded.
Hierarchical clustering demonstrated that EpiC and CRC derived allografts grouped together within the same larger branch as EpiC (Figure 3A). To investigate how allograft gene expression patterns differed from cell culture source, DEGs commonly up- and downregulated in EpiC- and CRC-derived allografts were compared to their parent cultures. CRC-derived allografts showed a log higher number of DEGs (1364 upregulated and 1226 downregulated) as compared to EpiC-derived allografts (143 and 171) (Figure 3D). The majority of up- (92%) and down- (57%) DEGs identified in EpiC-derived allografts were also changed in CRC-derived allografts. GO enrichment analyses demonstrated that all of the top categories identified in the up- and downregulated DEGs were unique (Figure 4C, D). Immune biological processes were exclusive to upregulated DEGs in allografts and motility-related biological processes to the downregulated DEGs. MSigDB analyses demonstrated that the EMT gene set was uniquely identified in DEGs upregulated in CRC-derived allografts (Table 3B) signifying EMT as a process occurring during allograft formation. This was compatible with the spindloid/sarcomatoid histology the majority of the allografts demonstrated, independent of the culture condition they were derived from. Immunologically related gene sets (Allograft_Rejection, Interferon_Gamma_Response, Complement, and Inflammatory_Response) were common to upregulated DEGs for both EpiC- and CRC-derived allografts (Table 3B).
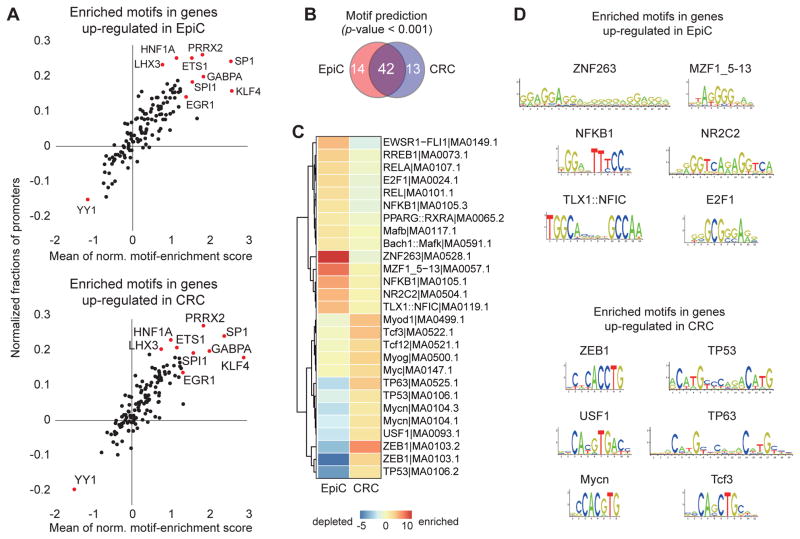
To explore if distinct sets of transcription factors (TF) might be responsible for the significantly different DEGs between EpiC and CRC, transcription factor binding site (TFBS) motifs enriched and depleted in the two datasets were examined using the JASPAR database (Kang et al., 2013) and PSCAN (Zambelli et al., 2009). The JASPAR database demonstrated that the unique sets of DEGs contained many commonly enriched/depleted TFBS (Figure 5A). Similarly, PSCAN demonstrated that the majority of enriched TFBS were shared (61%) but also identified TFBS relatively enriched under the two culture conditions (Fig 5B–D). EpiC enriched TFBS included NFKB1, RELA, and E2F1 sites, TFs positively linked to EMT, the most significant Hallmark gene. However, the TFBS for another EMT-linked gene, ZEB1, was found relatively enriched under CRC so there was no perfect correlation between enriched TFBS and Hallmark gene sets. CRC-enriched unique TFBS included TP53 and TP63, connecting with the P53_Pathway being the most significant Hallmark gene set for CRC.
Figure 5. Transcription Factor Binding Sites associated with commonly significantly differentially expressed genes under different culture conditions.
(A) Scatter plots of Transcription Factor Binding sites (TFBS) enriched motifs identified in DEGs significantly upregulated in EpiC and CRC cultures. All motif occurrences in the DEGs were calculated using the MOODS algorithm with the 130 position-frequency matrices available on the JASPAR website (p value < 0.001). The x-axis and y-axis indicate the mean average of normalized motif enrichment scores and normalized fractions of promoters presented on the Y axis, respectively. Each dot represents a single TFBS and red dots show significantly associated co-TFBSs (normalized proportion > 0.2 and average of normalized motif enrichment score > 1.5). Note that many of the same TFBS are enriched or depleted under both conditions. HNF1A: Hepatocyte Nuclear Factor 1 Homeobox A. PRRX2: Paired Related Homeobox 2. LHX3:.LIM Homeobox 3. ETS1: V-Ets Avian Erythroblastosis Virus E26 Oncogene Homolog 1. SP1: Sp1 Transcription Factor. GABPA: GA Binding Protein Transcription Factor, Alpha Subunit 60kDa. SPI1: Spi-1 Proto-Oncogene. KLF4: Kruppel-Like Factor 4. EGR1: Early Growth Response 1. (B) Venn diagram illustrating the overlap between TFBS significantly enriched in DEGs commonly upregulated in EpiC (red) as compared to CRC (blue) cultures. (C) Heatmap illustrating relative levels of enrichment versus depletion of the 27 TFBS identified as unique to EpiC (14 TFBS) and CRC (13 TFBS) cultures. EWSR1: EWS RNA-Binding Protein1. FLI1: Friend Leukemia Integration1. RREB1:Ras Responsive Element Binding Protein 1. RELA:V-Rel Avian Reticuloendotheliosis Viral Oncogene Homolog A. E2F1: E2F Transcription Factor 1. Rel: V-Rel Avian Reticuloendotheliosis Viral Oncogene Homolog. NFKB1: Nuclear Factor Of Kappa Light Polypeptide Gene Enhancer In B-Cells 1. PPARG: Peroxisome Proliferator-Activated Receptor Gamma. Mafb: V-Maf Avian Musculoaponeurotic Fibrosarcoma Oncogene Homolog B. Bach1: BTB And CNC Homology 1, Basic Leucine Zipper Transcription Factor 1. ZNF: Zinc Finger Protein. MZF1: Myeloid Zinc Finger 1. NR2C2 : Nuclear Receptor Subfamily 2, Group C, Member 2. TLX1: T-Cell Leukemia Homeobox 1. NFICL Nuclear Factor I/C (CCAAT-Binding Transcription Factor). MyoD: Myogenic Differentiation. Tcf3: Transcription Factor 3. Tcf12: Transcription Factor 12. Myog: Myogenin (Myogenic Factor 4). Myc: V-Myc Avian Myelocytomatosis Viral Oncogene Homolog. TP63: Transforming protein 63. TP53: Transforming protein 53. Mycn: V-Myc Avian Myelocytomatosis Viral Oncogene Neuroblastoma Derived Homolog. USF1:Upstream Transcription Factor 1. ZEB1: Zinc Finger E-Box Binding Homeobox 1. (D) Visualization of the six most enriched TFBS motifs identified from DEGs significantly upregulated under EpiC and CRC conditions. TFBS for ZNF263 (also known as ZSCAN12), MZF1_5–13, NFKB1, NR2C2, TLX1::NFIC, and E2F1 were uniquely enriched in genes upregulated under EpiC conditions while TFBS for ZEB1, TP53, USF1, TP63, Mycn, and Tcf3 were uniquely enriched under CRC conditions. Norm: normalized.
Corroboration of p53 pathway changes and variance in EMT-related gene expression
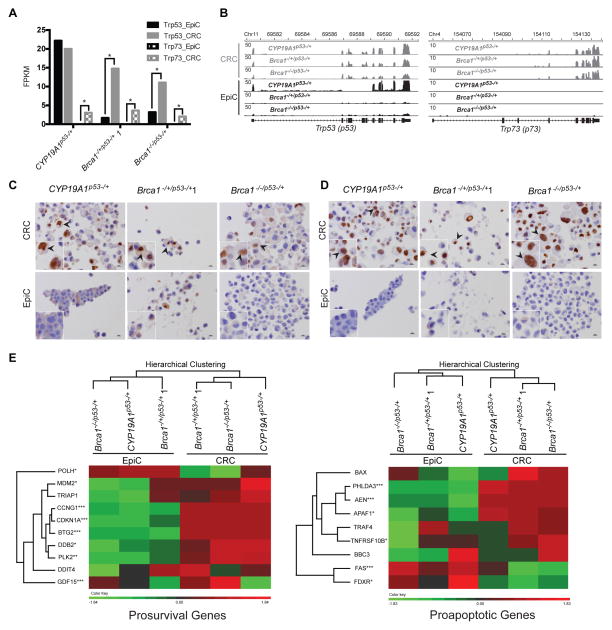
The P53_Pathway and Epithelial_Mesenchymal_Transition were selected for focused study because they were the two most significant Hallmark gene sets and enriched TFBS corresponding to each category were identified. Fragments Per Kilobase of transcript per Million mapped reads (FPKM) were significantly higher for Trp53 and Trp73 under CRC as compared to EpiC conditions (p<0.05) (Figure 6A). Mapped reads were confirmed as corresponding appropriately to the Trp53 and Trp73 exon structures (Figure 6B). Trp63 also demonstrates higher FPKM values under CRC as compared to EpiC in these cells (Assefnia et al., 2014), consistent with reports from others reporting CRC conditions upregulating p63 expression (Suprynowicz et al., 2012; Chapman et al., 2014). Immunohistochemical staining intensity, relative numbers of cells staining, and nuclear localization of p53 and p63 were higher in CRC as compared to EpiC cultured cells (Figure 6C). Hierarchical clustering revealed that samples grouped by culture condition rather than genotype when examining relative expression levels of defined p53 regulated prosurvival and proapoptotic genes (Allen et al., 2014), consistent with higher activation levels of p53 family members under CRC conditions (Figure 6D).
Figure 6. Expression levels of p53 family members were higher under CRC as compared to EpiC culture conditions.
(A) Bar graphs illustrating relative FPKM (Fragments Per Kilobase of transcript per Million mapped) reads of Trp53 and Trp73 in EpiC (black) and CRC (grey) in primary CYP19A1p53−/+, Brca1−/+/p53−/+ 1, and Brca1−/−/p53−/+ cancer cell cultures. *p<0.05, Cuffdiff_pval. (B) Images showing normalized read coverage across the Trp53 and Trp73 loci viewed through the Integrative Genomics Viewer from CYP19A1p53−/+, Brca1−/+/p53−/+ 1, and Brca1−/−/p53−/+ EpiC (black) and CRC (grey) cultures. (C) Representative images of immunohistochemistry (IHC) for p53 in CYP19A1p53−/+, Brca1−/+/p53−/+ 1, and Brca1−/−/p53−/+ cell pellets prepared from EpiC and CRC cultures. Insets bottom left show magnified images of cells indicated by arrows in larger images. Black arrows indicate representative cells with detection of nuclear-localized p53. IHC images taken at 40_X. Size bars= 10μM. (D) Representative images of immunohistochemistry (IHC) for p63 in CYP19A1p53−/+, Brca1−/+/p53−/+ 1, and Brca1−/−/p53−/+ cell pellets prepared from EpiC and CRC cultures. Insets bottom left show magnified images of cells indicated by arrows in larger images. Black arrows indicate representative cells with detection of nuclear-localized p63. IHC images taken at 40_X. Size bars= 10μM. (E) Dendrograms and heatmaps of unsupervised hierarchical clustering of FPKM values of p53-regulated prosurvival and proapoptotic genes from EpiC and CRC CYP19A1p53−/+, Brca1−/+/p53−/+ 1, and Brca1−/−/p53−/+ cultures. POLH: Polymerase (DNA Directed), Eta. MDM2: MDM2 Proto-Oncogene, E3 Ubiquitin Protein Ligase. TRIAP1: TP53 Regulated Inhibitor Of Apoptosis 1. CCNG1: Cyclin G1. CDKN1A: Cyclin-Dependent Kinase Inhibitor 1A (P21, Cip1). BTG2: BTG Family, Member 2. DDB2: Damage-Specific DNA Binding Protein 2, 48kDa. PLK: Polo-Like Kinase. DDIT4: DNA-Damage-Inducible Transcript 4. GDF: Growth Differentiation Factor. BAX: BCL2-Associated X Protein. PHLDA3: Pleckstrin Homology-Like Domain, Family A, Member 3. AEN: Apoptosis Enhancing Nuclease. APAF1: Apoptotic Peptidase Activating Factor 1. TRAF4: Tissue Necrosis Factor Receptor-Associated Factor 4. TNFRSF: Tumor Necrosis Factor Receptor Superfamily. BBC3: BCL2 Binding Component 3. FAS: Fas Cell Surface Death Receptor. FDXR: Ferredoxin Reductase. *One EpiC/CRC pair, **Two EpiC/CRC pairs, ***Three EpiC/CRC pairs, p<0.05, Cuffdiff_pval.
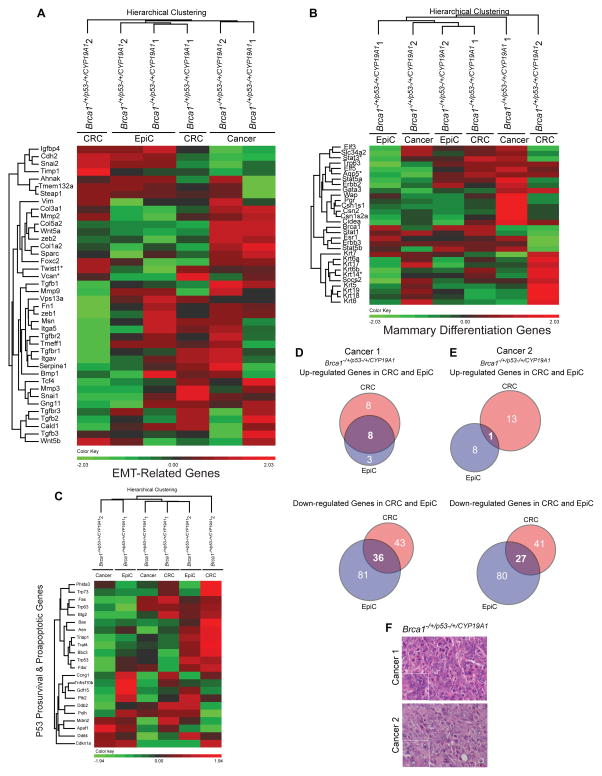
A similar approach was taken for examination of EMT-related genes. Allograft samples were included because Epithelial_Mesenchymal_Transition was a significant Hallmark gene set identified when CRC derived allograft transcriptomes were compared to those of their parent cultures (Table 3B). Hierarchical clustering of defined EMT-related genes separated groups by culture and growth condition as compared to genotype with allografts clustered more closely to EpiC as compared to CRC cultures (Figure 7A). Of note, Zeb1 and Zeb2 FPKM levels, TFs that bind TFBS ZEB1 (Figure 5D) were significantly lower in CRC as compared to EpiC conditions (Figure 7A). Because the TGF-β pathway is a defined activator of EMT (Gonzalez et al., 2014) and previously reported to be downregulated under CRC conditions (Ligaba et al., 2015), FPKM levels of Tgfb1,2,3 and Tgfbr1,2,3 were specifically included (Figure 7A). Tgfbr3 FPKM values were significantly higher under EpiC as compared to CRC conditions in all three paired cultures while Tgfb3 was higher in two pairs and Tgfb2 and Tgfbr1 in one (Figure 7A,B). Tgfbr3 reads mapped appropriately to Tgfbr3 exon structure (Figure 7C). Intensity of membrane-localized staining and numbers of cells stained for TGFBR3 using immunohistochemistry were higher in cells cultured under EpiC as compared to CRC conditions (Figure 7D).
Figure 7. Expression levels of epithelial mesenchymal transition genes were higher under EpiC as compared to CRC culture conditions.
(A) Dendrogram and heatmap of unsupervised hierarchical clustering of FPKM values of epithelial mesenchymal transition (EMT)-related genes from CYP19A1p53−/+, Brca1−/+/p53−/+ 1, and Brca1−/−/p53−/+ EpiC and CRC cancer cell cultures and allografts. *One EpiC/CRC pair, **Two EpiC/CRC pairs, ***Three EpiC/CRC pairs, p<0.05, Cuffdiff_pval. Msn: Moesin. Cdh2: Cadherin 2, Type 1. Cald1: Caldesmon 1. Tgfb3: Transforming growth factor beta 3. Wnt5b: Wnt5b: Wingless-Type MMTV Integration Site Family, Member 5B. Serpine1: Serpin Peptidase Inhibitor, Clade E (Nexin, Plasminogen Activator Inhibitor Type 1), Member 1. Itgav: Integrin, Alpha V. Tmem132a: Transmembrane Protein 132A. Tgfb3: Transforming growth factor beta 3. Steap: Six-Transmembrane Epithelial Antigen Of Prostate 2. Tgfbr1: Transforming growth factor beta receptor 1. Bmp1: Bone Morphogenetic Protein 1. Col5a2: Collagen, Type V, Alpha 2. Col1a2: Collagen, Type I, Alpha 2. Vcan: Versican. Timp1: TIMP Metallopeptidase Inhibitor 1. Wnt5a: Wingless-Type MMTV Integration Site Family, Member 5A. Tgfb1: Transforming growth factor beta 1. Mmp3: Matrix Metallopeptidase 3. Foxc2: Forkhead Box C2. Tgfbr3: Transforming growth factor beta receptor 3. Sparc: Secreted Protein, Acidic, Cysteine-Rich (Osteonectin). Snai1: Snail Family Zinc Finger 1. Zeb2: Zinc Finger E-Box Binding Homeobox 2. Col3a1: Collagen, Type III, Alpha 1. Twist1: Twist Family BHLH Transcription Factor 1. Fn1: Fibronectin 1. Itga5: Integrin, Alpha 5 (Fibronectin Receptor, Alpha Polypeptide). Mmp2: Matrix Metallopeptidase 2. Mmp9: Matrix Metallopeptidase 9. Zeb1: Zinc Finger E-Box Binding Homeobox 1. Gng 1: Guanine Nucleotide Binding Protein (G Protein), Gamma 11. Tcf4: Transcription Factor 4. Vim: Vimentin. Igfbp4: Insulin-Like Growth Factor Binding Protein 4. Tgfbr2: Transforming growth factor beta receptor 2. Snai2: Snail Family Zinc Finger 2. Tmeff1: Transmembrane Protein With EGF-Like And Two Follistatin-Like Domains 1. Ahnak: AHNAK Nucleoprotein. Vps13a: Vacuolar Protein Sorting 13 Homolog A. (B) Bar graphs illustrating relative FPKM (Fragments Per Kilobase of transcript per Million mapped) reads of Tgfbr3 in EpiC (black) and CRC (grey) in primary CYP19A1p53−/+, Brca1−/+/p53−/+ 1, and Brca1−/−/p53−/+ cancer cell cultures. *p<0.05, Cuffdiff_pval. (C) Images showing normalized read coverage across the Tgfbr3 locus viewed through the Integrative Genomics Viewer from CYP19A1p53−/+, Brca1−/+/p53−/+ 1, and Brca1−/−/p53−/+ EpiC (black) and CRC (grey) cultures. (D) Representative images of immunohistochemistry (IHC) for TGFBR3 in CYP19A1p53−/+, Brca1−/+/p53−/+ 1, and Brca1−/−/p53−/+ cell pellets prepared from EpiC and CRC cultures. Insets bottom left show magnified images of cells indicated by arrows in larger images. Black arrows indicate representative cells with detection of membrane-localized TGFBR3. IHC images taken at 40_X. Size bars= 10μM.
Expression of mammary epithelial cell differentiation genes was not retained in allografts
Genes linked to mammary gland differentiation in vivo were selected for specific examination because retention of mammary epithelial cell differentiation-related gene expression in primary culture and allografts is of interest to mammary gland biologists and breast cancer researchers. Hierarchical clustering indicated that for all but one culture (EpiC: Brca1−/−/p53−/+) the number of differentiation-related genes with higher FPKM values was greatest under CRC conditions followed by EpiC and then allografts (Figure 8A). Keratin (K) genes expressed in vivo in human breast cancers (K5,6,7,8,14,18,19) and normal differentiated luminal (K7,8,18,19) and basal (K5, 6,14,17) mammary epithelial cells were included in the analyses (Trask et al., 1990; Taylor-Papadimitriou et al., 1989; Livasy et al., 2006; Grimm et al., 2006; Shao et al., 2012; Kang et al., 2014; Visvader et al., 2014) Overall keratin FPKM levels were highest under CRC conditions, consistent with higher Intensity of membrane-localized staining and numbers of cells staining for pan-cytokeratin (Figure 8B) and identification of keratinization as a unique GO enriched biological process (Figure 4B). Esr1 expression was not significantly altered by culture condition but Erbb3 was higher under CRC in all three pairs (Figure 8A).
Figure 8. Relative expression levels of mammary epithelial cell differentiation genes in EpiC and CRC cultures and allografts.
(A) Dendrogram and heatmap of unsupervised hierarchical clustering of FPKM values of mammary epithelial cell differentiation-related genes from CYP19A1p53−/+, Brca1−/+/p53−/+ 1, and Brca1−/−/p53−/+ EpiC and CRC cancer cell cultures and allografts. *One EpiC/CRC pair, **Two EpiC/CRC pairs, ***Three EpiC/CRC pairs, *p<0.05, Cuffdiff_pval. Brca1: Breast Cancer 1, Early Onset. Gata3: GATA Binding Protein 3. Krt18: Keratin 18, Type I. Krt8: Keratin 8, Type II. Esr1: Estrogen Receptor 1. Aqp5: Aquaporin 5. Stat1: Signal Transducer And Activator Of Transcription 1, 91kDa. Csn2: Casein Beta. Csn1s1: Casein Alpha S1. Csn1s2a: Casein Alpha S2-Like A, Pseudogene. Trp63: Tumor Protein P63. Erbb2: Erb-B2 Receptor Tyrosine Kinase 2. Krt19: Keratin 19, Type I. Krt7: Keratin 7, Type II. Elf3: E74-Like Factor 3 (Ets Domain Transcription Factor, Epithelial-Specific). Erbb3: Erb-B2 Receptor Tyrosine Kinase 3. Elf5: E74-Like Factor 5 (Ets Domain Transcription Factor). Krt6b: Keratin 6B, Type II. Krt17: Keratin 17, Type I. Krt5: Keratin 5, Type II. Krt6a: Keratin 6A, Type II. Slc34a2: Solute Carrier Family 34 (Type II Sodium/Phosphate Cotransporter), Member 2. Krt14: Keratin 14, Type I. Cidea: Cell Death-Inducing DFFA-Like Effector A. Wap: Whey acidic protein. Stat3: Signal Transducer And Activator Of Transcription 3 (Acute-Phase Response Factor). Socs2: Suppressor Of Cytokine Signaling 2. Stat5a: Signal Transducer And Activator Of Transcription 5A. Stat5b: Signal Transducer And Activator Of Transcription 5B. Pgr: Progesterone Receptor. (B) Representative images of immunohistochemistry (IHC) for pancytokeratin in CYP19A1p53−/+, Brca1−/+/p53−/+ 1, and Brca1−/−/p53−/+ cell pellets prepared from EpiC and CRC cultures. Insets bottom right show magnified images of cells indicated by arrows in larger images. Black arrows indicate representative cells showing cytoskeletal-localization. IHC images taken at 40_X. Size bars= 10μM.
Primary cells cultured for a limited number of passages under either condition maintained gene expression patterns that were not significantly different from source tissue
We next determined if shortening passage number (P=7) reduced the number of significantly altered DEGs between the two culture conditions. Cells were isolated from two cancers that developed in the same mouse (Brca−/+/p53−/+/CYP19A1). Transcriptomes were compared to each other as well as to the parent cancer from which the cells were derived. There was no clustering by culture or growth condition for EMT-related, mammary differentiation or p53 pathway genes (Figure 9A–C) and the magnitude of significantly different DEGs between each culture condition and the parent cancer were similar (Figure 9D, E) even given the disparate cancer histologies (Cancer 1: poorly differentiated adenocarcinoma; Cancer 2: spindloid/sarcomatoid carcinoma) (Figure 9F). The few commonly significantly up- or downregulated DEGs mapped to less than three Hallmark gene sets (Allograft_Rejection, Kras_Signaling_up, Inflammatory_Response) (Table 3C). Normal wild-type mammary epithelial cells cultured for three passages before RNAseq analyses showed similar results with few statistically significant DEGs between culture conditions, consistent with the concept of limiting passage number in order to reduce culture induced transcriptome changes (O’Driscoll et al., 2006).
Figure 9. Limiting culture duration restrained significant changes in EMT-related, mammary differentiation-related, and p53 pathway genes.
(A) Dendrogram and heatmap of unsupervised hierarchical clustering of FPKM values of EMT-related genes from paired EpiC and CRC cultures (P7) from two different Brca1−/+/p53−/+/CYP19A1 cancer cell cultures and cancers they were derived from (Cancer 1, Cancer 2). *One EpiC/CRC pair, *p<0.05, Cuffdiff_pval. (B) Dendrogram and heatmap of unsupervised hierarchical clustering of FPKM values of mammary epithelial cell differentiation-related genes from paired EpiC and CRC cultures (P7) from two different Brca1−/+/p53−/+/CYP19A1 cancer cell cultures and cancers they were derived from (Cancer 1, Cancer 2). *One EpiC/CRC pair, *p<0.05, Cuffdiff_pval. (C) Dendrogram and heatmap of unsupervised hierarchical clustering of FPKM values of p53-regulated prosurvival and proapoptotic genes from paired EpiC and CRC cultures (P7) from two different Brca1−/+/p53−/+/CYP19A1 cancer cell cultures and cancers they were derived from (Cancer 1, Cancer 2). *One EpiC/CRC pair, *p<0.05, Cuffdiff_pval. (D) Venn diagrams illustrating the overlap between DEGs significantly up- and downregulated in EpiC (blue) and CRC (red) cultures as compared to the parent Cancer 1. (E) Venn diagrams illustrating the overlap between DEGs significantly up- and downregulated in EpiC (blue) and CRC (red) cultures as compared to the parent Cancer 2. (F) Representative H&E images of Cancer 1 and Cancer 2 that coincidently developed in the same Brca1−/+/p53−/+/CYP19A1 mouse. Magnified images lower left. Images taken at 40X. Size bars= 10μM.
Discussion
The high success rate of CRC for initial primary cell isolation and propagation was consistent with previous reports (Chapman et al 2010; Chapman et al 2014; Ligaba et al, 2015; Liu et al 2012; Saenz et al. 2014). Here we demonstrated that mammary epithelial cells initially isolated in CRC can be transitioned to another culture methodology. An advantage of CRC is that growth of epithelial cells is selectively enhanced under the conditions used here (Liu et al. 2012). Therefore not only did use of CRC for initial isolation rate increase the number of successful cultures, it also reduced fibroblast contamination.
Faster development of allografts with EpiC-cultured cells and acquisition of higher levels of EMT-related gene expression in allografts derived from CRC-cultured cells was consistent with the notion that EMT induces cancer stem cell characteristics in normal and transformed mammary epithelial cells thereby increasing their tumor-initiating capacity (Mani et al., 2008). Higher levels of EMT genes under EpiC as compared to CRC conditions paralleled previous reports of EpiC cultured cells undergoing EMT (Nakles et al., 2013) in comparison to retention of cuboidal architecture (Saenz et al.) and suppression of the EMT-related TGFB pathway (Ligaba et al., 2015) under CRC conditions. Here we documented upregulated TGFBR3 expression, a protein linked to a subtype of triple negative breast cancers in women (Jovanović et al., 2014), under EpiC conditions. Provocatively we found the TFBS for ZEB1, an EMT-linked gene, enriched in CRC-associated DEGs. However FPKM levels for ZEB1 and its paralog ZEB2 were significantly lower in CRC as compared to EpiC conditions. Competitive binding between ZEB1 and alternatively-acting Nkx2.5 has been described in vascular smooth muscle cells (Ponticos et al., 2004). A similar mechanism may act under CRC conditions. A second possibility for the absence of ZEB1 associated EMT under CRC conditions is the reported repressive action of p53 on ZEB1/ZEB2 (Sánchez-Tilló et al., 2012). The significant difference between numbers of injected sites developing into allografts between Brca1 replete and Brca1 deficient cells for both culture conditions may also be linked to EMT. BRCA1 has been described as an EMT suppressor (Bai et al., 2014).
Reports on the CRC technique have emphasized a link between increased p63 expression and retention of stem/progenitor cells (Suprynowicz et al., 2012; Liu et al., 2012; Chapman et al., 2014; Ligaba et al., 2015; Brown et al., 2015). Here we showed that in mammary epithelial cancer cells p63 upregulation is part of a more generalized increase in expression levels of all p53 gene family member genes including Trp53 and Trp73. Faster development of EpiC cultured allografts may have signified that, at least for these cancer cells, higher levels of Trp63 expression was not required for retention of cancer progenitor cells. If the goal for primary mammary epithelial cancer cell culture is allograft generation, the readily applied EpiC culture methodology enhanced allograft development. Presence of Allograft_Rejection as a significant Hallmark gene set present in the transcriptomes of allografts and primary cancers was consistent with the expected presence of immune cells in vivo but not in vitro.
Limiting passage number to reduce culture-induced changes is not a new concept (O’Driscoll et al., 2006). Importantly here we showed that if isolation of primary mammary epithelial cells is primarily for transcriptome characterization, the less cumbersome EpiC culture methodology was acceptable. Moreover, we demonstrated that even though CRC promoted retention of epithelial cell architecture, this did not translate to superior preservation of the parent cancer transcriptome.
We documented differential activation of genetic networks linked to cancer biology in the two culture systems, p53 pathway under CRC conditions and TGFB pathway under EpiC. Therapeutic approaches for both of these pathways have been developed (Connolly et al., 2012; Duffy et al., 2014). A possible future direction would be to compare response of targeted therapeutics in these paired cultures to establish how modulations in a baseline transcriptome might impact in vitro testing results. It is possible this could improve the predictability of in vitro testing for some therapeutics (Hwu et al., 2006). Studies here focused on triple negative cancer cells that lack measurable levels of Estrogen Receptor (ER) alpha protein. A future direction is to explore utilization of these culture methodologies for ER positive breast cancers. Small molecule inhibitors of TGFB can increase maintenance of ER alpha expression in primary mammary epithelial cells (Fridriksdottir et al., 2015). One possibility would be to test how application of TGFB small molecule inhibitors would influence ER alpha expression levels in these cultures.
In conclusion, both EpiC and CRC culture conditions were used to generate and propagate primary mammary epithelial cell cultures. CRC was the more efficient modality for initial generation and EpiC for allograft generation in these experiments. Because the two conditions result in culture-specific transcriptome alterations, the methodology selected for extended culture may be dictated by the experimental questions that are being posed. It is possible to move from either of these two specialized media to more generic culture condition such as DMEM. Either culture condition can be used to isolate primary mammary epithelial cells for transcriptome inspection but passage should be limited for best approximation to intact tissue.
Acknowledgments
Funding. Research performed here supported by NIH NCI RO1 CA112176 (PAF), NIH NCI 5P30CA051008 (Histology and Tissue, Genomics and Epigenomics, and Animal Shared Resources), the Intramural Research Program of NIDDK/NIH, and King Khalid University, Abha, Saudi Arabia (AMA).
We thank Sarah Dabydeen for preparation of the sequencing libraries and Harold Smith, NIDDK, NIH for next generation sequencing.
Footnotes
Declaration of interest.
AMA, KK, SG, WW, XZ, BK, LH, & PAF declare no conflict of interest. Georgetown University has been awarded a patent by the United States Patent Office (9,279,106) for conditional cell reprogramming. This technology has been licensed exclusively to a new biotechnology company, Propagenix, for commercialization. Georgetown University and the inventor on this manuscript (XL) receives payments and potential royalties from Propagenix.
Authorship contributions. AMA (conception and design, data acquisition, analysis and interpretation of data, drafting, revising the manuscript and final approval), KK (bioinformatics conception and design, data acquisition, analysis and interpretation, drafting, revising the manuscript and final approval), SG (design, data acquisition, analysis and interpretation of data, drafting, revising the manuscript and final approval), WW (data acquisition, manuscript revision and final approval), XZ (bioinformatics analyses, manuscript revision and final approval), BK (acquisition of data and interpretation, manuscript revision and final approval), LH (data analysis, manuscript revision and final approval), XL (design, data analysis, manuscript revision and final approval), PAF (conception and design, analysis and interpretation of data, drafting and revising the manuscript and final approval).
References
- Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA, Sullivan KD, Galbraith MD, Luo X, Kraus WL, et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife. 2014;3:e02200. doi: 10.7554/eLife.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assefnia S, Kang K, Groeneveld S, Yamaji D, Dabydeen S, Alamri A, Liu X, Hennighausen L, Furth PA. Trp63 is regulated by STAT5 in mammary tissue and subject to differentiation in cancer. Endocrine-Related Cancer. 2014;21:443–457. doi: 10.1530/ERC-14-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Chan HL, Scott A, Smith MD, Fan C, Herschkowitz JI, Perou CM, Livingston AS, Robbins DJ, Capobianco AJ, Pei XH. BRCA1 suppresses epithelial-to-mesenchymal transition and stem cell dedifferentiation during mammary and tumor development. Cancer Research. 2014;74:6161–6172. doi: 10.1158/0008-5472.CAN-14-1119. [DOI] [PubMed] [Google Scholar]
- Bhandary L, Whipple RA, Vitolo MI, Charpentier MS, Boggs AE, Chakrabarti KR, Thompson KN, Martin SS. ROCK inhibition promotes microtentacles that enhance reattachment of breast cancer cells. Oncotarget. 2015;6:6251–6266. doi: 10.18632/oncotarget.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Dabbs DJ, Lee AV, McGuire KP, Ahrendt GM, Bhargava R, Bhargava R, Davidson NE, Brufsky AM, Johnson RR, Oesterreich S, McAuliffe PF. Developing in vitro models of human ductal carcinoma in situ from primary tissue explants. Breast Cancer Research and Treatment. 2015;153:311–321. doi: 10.1007/s10549-015-3551-8. [DOI] [PubMed] [Google Scholar]
- Chapman S, Liu X, Meyers C, Schlegel R, McBride AA. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. The Journal of Clinical Investigation. 2010;120:2619–2626. doi: 10.1172/JCI42297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S, McDermott DH, Shen K, Jang MK, McBride AA. The effect of Rho kinase inhibition on long-term keratinocyte proliferation is rapid and conditional. Stem Cell Research & Therapy. 2014;5:60. doi: 10.1186/scrt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-β targeted cancer therapy. International Journal of Biological Sciences. 2012;8:964–978. doi: 10.7150/ijbs.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darcy KM, Zangani D, Lee PH, Ip MM. Isolation and Culture of Normal Rat Mammary Epithelial Cells. In: Ip MM, Asch BB, editors. Methods in Mammary Gland Biology and Breast Cancer Research. Springer US; 2000. pp. 163–175. [Google Scholar]
- Diaz-Cruz ES, Cabrera MC, Nakles R, Rutstein BH, Furth PA. BRCA1 deficient mouse models to study pathogenesis and therapy of triple negative breast cancer. Breast Disease. 2010;32:85–97. doi: 10.3233/BD-2010-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Cruz ES, Sugimoto Y, Gallicano GI, Brueggemeier RW, Furth PA. Comparison of increased aromatase versus ERα in the generation of mammary hyperplasia and cancer. Cancer Research. 2011;71:5477–5487. doi: 10.1158/0008-5472.CAN-10-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes & Development. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drasin DJ, Robin TP, Ford HL. Breast cancer epithelial-to-mesenchymal transition: examining the functional consequences of plasticity. Breast Cancer Research: BCR. 2011;13:226. doi: 10.1186/bcr3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MJ, Synnott NC, McGowan PM, Crown J, O’Connor D, Gallagher WM. p53 as a target for the treatment of cancer. Cancer Treatment Reviews. 2014;40:1153–1160. doi: 10.1016/j.ctrv.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Fridriksdottir AJ, Kim J, Villadsen R, Klitgaard MC, Hopkinson BM, Petersen OW, Rønnov-Jessen L. Propagation of oestrogen receptor-positive and oestrogen-responsive normal human breast cells in culture. Nature Communications. 2015;6:8786. doi: 10.1038/ncomms9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawade S, Mayer C, Hafen K, Barthlott T, Krenger W, Szinnai G. Cell growth dynamics in embryonic and adult mouse thyroid revealed by a novel approach to detect thyroid gland subpopulations. Thyroid. 2016;26:591–599. doi: 10.1089/thy.2015.0523. [DOI] [PubMed] [Google Scholar]
- Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Science Signaling. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm SL, Bu W, Longley MA, Roop DR, Li Y, Rosen JM. Keratin 6 is not essential for mammary gland development. Breast Cancer Research: BCR. 2006;8:R29. doi: 10.1186/bcr1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeneveld S. Comparison of two culture techniques for the development of mammary cancer cell cultures from genetically engineered mouse models of human breast cancer. Ludwig-Maximilians-Universität München; München: 2012. [Google Scholar]
- Haynes J. Dynamic actin remodeling during epithelial-mesenchymal transition depends on increased moesin expression. Molecular Biology of the Cell. 2011;22:4750–4764. doi: 10.1091/mbc.E11-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwu P, Bedikian AY, Grimm EA. Challenges of chemosensitivity testing. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2006;12:5258–5259. doi: 10.1158/1078-0432.CCR-06-1656. [DOI] [PubMed] [Google Scholar]
- Jones LP, Li M, Halama ED, Ma Y, Lubet R, Grubbs CJ, Deng CX, Rosen EM, Furth PA. Promotion of mammary cancer development by tamoxifen in a mouse model of Brca1-mutation-related breast cancer. Oncogene. 2005;24:3554–3562. doi: 10.1038/sj.onc.1208426. [DOI] [PubMed] [Google Scholar]
- Jones L, Tilli M, Assefnia S, Torre K, Halama E, Parrish A, Rosen EM, Furth P. Activation of estrogen signaling pathways collaborates with loss of Brca1 to promote development of ERα-negative and ERα-positive mammary preneoplasia and cancer. Oncogene. 2008;27:794–802. doi: 10.1038/sj.onc.1210674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanović B, Beeler JS, Pickup MW, Chytil A, Gorska AE, Ashby WJ, Lehmann BD, Zijstra A, Pietenpol JA, Moses HL. Transforming growth factor beta receptor type III is a tumor promoter in mesenchymal-stem like triple negative breast cancer. Breast Cancer Research: BCR. 2014;16:R69. doi: 10.1186/bcr3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Robinson GW, Hennighausen L. Comprehensive meta-analysis of Signal Transducers and Activators of Transcription (STAT) genomic binding patterns discerns cell-specific cis-regulatory modules. BMC Genomics. 2013;14:4. doi: 10.1186/1471-2164-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Yamaji D, Yoo KH, Robinson GW, Hennighausen L. Mammary-specific gene activation is defined by progressive recruitment of STAT5 during pregnancy and the establishment of H3K4me3 marks. Molecular and Cellular Biology. 2014;34:464–473. doi: 10.1128/MCB.00988-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of Clinical Investigation. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligaba SB, Khurana A, Graham G, Krawczyk E, Jablonski S, Petricoin EF, Glazer RI, Upadhyay G. Multifactorial analysis of conditional reprogramming of human keratinocytes. PloS One. 2015;10:e0116755. doi: 10.1371/journal.pone.0116755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. The American Journal of Pathology. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Modern Pathology: An Official Journal of the United States and Canadian Academy of Pathology, Inc. 2006;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- Luo M, Brooks M, Wicha MS. Epithelial-mesenchymal plasticity of breast cancer stem cells: implications for metastasis and therapeutic resistance. Current Pharmaceutical Design. 2015;21:1301–1310. doi: 10.2174/1381612821666141211120604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino G, Memmi EM, Pelicci PG, Bernassola F. Maintaining epithelial stemness with p63. Science Signaling. 2015;8:re9. doi: 10.1126/scisignal.aaa1033. [DOI] [PubMed] [Google Scholar]
- Mitra A, Mishra L, Li S. Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends in Biotechnology. 2013;31:347–354. doi: 10.1016/j.tibtech.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Bueno G, Peinado H, Molina P, Olmeda D, Cubillo E, Santos V, Palacios J, Portillo F, Cano A. The morphological and molecular features of the epithelial-to-mesenchymal transition. Nature Protocols. 2009;4:1591–1613. doi: 10.1038/nprot.2009.152. [DOI] [PubMed] [Google Scholar]
- Nakles RE, Kallakury BV, Furth PA. The PPARγ agonist efatutazone increases the spectrum of well-differentiated mammary cancer subtypes initiated by loss of full-length BRCA1 in association with TP53 haploinsufficiency. The American Journal of Pathology. 2013;182:1976–1985. doi: 10.1016/j.ajpath.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakles RE, Millman SL, Cabrera MC, Johnson P, Mueller S, Hoppe PS, Schroeder T, Furth PA. Time-lapse imaging of primary preneoplastic mammary epithelial cells derived from genetically engineered mouse models of breast cancer. Journal of Visualized Experiments: JoVE. 2013;(72) doi: 10.3791/50198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novaro V, Roskelley CD, Bissell MJ. Collagen-IV and laminin-1 regulate estrogen receptor alpha expression and function in mouse mammary epithelial cells. Journal of Cell Science. 2003;116:2975–2986. doi: 10.1242/jcs.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll L, Gammell P, McKiernan E, Ryan E, Jeppesen PB, Rani S, Clynes M. Phenotypic and global gene expression profile changes between low passage and high passage MIN-6 cells. The Journal of Endocrinology. 2006;191:665–676. doi: 10.1677/joe.1.06894. [DOI] [PubMed] [Google Scholar]
- Olson MF. Applications for ROCK kinase inhibition. Current Opinion in Cell Biology. 2008;20:242–248. doi: 10.1016/j.ceb.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palechor-Ceron N, Suprynowicz FA, Upadhyay G, Dakic A, Minas T, Simic V, Johnson M, Albanese C, Schlegel L, Liu X. Radiation induces diffusible feeder cell factor(s) that cooperate with ROCK inhibitor to conditionally reprogram and immortalize epithelial cells. The American Journal of Pathology. 2013;183:1862–1870. doi: 10.1016/j.ajpath.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticos M, Partridge T, Black CM, Abraham DJ, Bou-Gharios G. Regulation of collagen type I in vascular smooth muscle cells by competition between Nkx2.5 and deltaEF1/ZEB1. Molecular and Cellular Biology. 2004;24:6151–6161. doi: 10.1128/MCB.24.14.6151-6161.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nature Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz FR, Ory V, AlOtaiby M, Rosenfield S, Furlong M, Cavalli LR, Johnson MD, Liu X, Schlegel R, Wellstein A, Riegel AT. Conditionally reprogrammed normal and transformed mouse mammary epithelial cells display a progenitor-cell-like phenotype. PloS One. 2014;9:e97666. doi: 10.1371/journal.pone.0097666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Tilló E, Liu Y, de Barrios O, Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A, Postigo A. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cellular and Molecular Life Sciences: CMLS. 2012;69:3429–3456. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao MM, Chan SK, Yu AM, Lam CC, Tsang JYS, Lui PC, Law BK, Tan PH, Tse GM. Keratin expression in breast cancers. Virchows Archiv: An International Journal of Pathology. 2012;461:313–322. doi: 10.1007/s00428-012-1289-9. [DOI] [PubMed] [Google Scholar]
- Suprynowicz FA, Upadhyay G, Krawczyk E, Kramer SC, Hebert JD, Liu X, Yuan H, Cheluvaraju C, Clapp PW, Boucher RC, et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20035–20040. doi: 10.1073/pnas.1213241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Papadimitriou J, Stampfer M, Bartek J, Lewis A, Boshell M, Lane EB, Leigh IM. Keratin expression in human mammary epithelial cells cultured from normal and malignant tissue: relation to in vivo phenotypes and influence of medium. Journal of Cell Science. 1989;94:403–413. doi: 10.1242/jcs.94.3.403. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high- performance genomics data visualization and exploration. Briefings in Bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilli MT, Frech MS, Steed ME, Hruska KS, Johnson MD, Flaws JA, Furth PA. Introduction of estrogen receptor-alpha into the tTA/TAg conditional mouse model precipitates the development of estrogen-responsive mammary adenocarcinoma. The American Journal of Pathology. 2003;163:1713–1719. doi: 10.1016/s0002-9440(10)63529-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask DK, Band V, Zajchowski DA, Yaswen P, Suh T, Sager R. Keratins as markers that distinguish normal and tumor-derived mammary epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:2319–2323. doi: 10.1073/pnas.87.6.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes & Development. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji D, Na R, Feuermann Y, Pechhold S, Chen W, Robinson GW, Hennighausen L. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes & Development. 2009;23:2382–2387. doi: 10.1101/gad.1840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Research. 2009;37:W247–252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]