Abstract
Papillary carcinomas are a special histological type of breast cancer, and have a relatively good outcome. We characterised the genomic and phenotypic characteristics of papillary carcinomas, and to determine whether they would constitute an entity distinct from grade- and oestrogen receptor (ER)-matched invasive ductal carcinomas of no special type (IDC-NSTs). The phenotype of 63 papillary carcinomas of the breast and grade- and ER-matched IDC-NSTs was determined by immunohistochemistry. DNA of sufficient quality was extracted from 49 microdissected papillary carcinomas and 49 microdissected grade- and ER-matched IDC-NSTs. These samples were subjected to high-resolution microarray-based comparative genomic hybridisation (aCGH) and MassARRAY Sequenom sequencing analysis of 19 known oncogenes. Papillary carcinomas were predominantly of low histological grade, expressed immunohistochemical markers consistent with a luminal phenotype, and a lower rate of lymph node metastasis and p53 expression than grade- and ER-matched IDC-NSTs. Papillary carcinomas displayed less genomic aberrations than grade- and ER-matched IDC-NSTs; however the patterns of gene copy number aberrations found in papillary carcinomas were similar to those of ER- and grade-matched IDC-NSTs, including 16q losses. Furthermore, PIK3CA mutations were found in 43% and 29% of papillary carcinomas and grade- and ER-matched IDC-NSTs respectively. The genomic profiles of encapsulated, solid and invasive papillary carcinomas, the three morphological subtypes, were remarkably similar. Our results demonstrate that papillary carcinomas are a homogeneous special histological type of breast cancer. The similarities in the genomic profiles of papillary carcinomas and grade- and ER-matched IDC-NSTs suggest that papillary carcinomas may be best positioned as part of the spectrum of ER-positive breast cancers rather than as a distinct entity. Furthermore, the good prognosis of papillary carcinomas may stem from the low rates of lymph node metastasis and p53 expression, low number of gene copy number aberrations, and high prevalence of PIK3CA mutations.
Keywords: breast cancer, papillary carcinoma, comparative genomic hybridisation, immunohistochemistry, mutation, tissue microarrays, in situ hybridisation
Introduction
Breast cancer has been traditionally classified into different histological subtypes based on their cytological and architectural features. The multiple classification systems have been consolidated in the 2003 World Health Organisation classification[1]. This taxonomy recognises the existence of invasive ductal carcinomas of no special type (IDC-NST) and 17 special histological types of breast cancer[1]. Over the last decade, high throughput gene expression profiling and microarray-based comparative genomic hybridisation (aCGH) studies have demonstrated that oestrogen receptor (ER)-positive and ER-negative IDC-NSTs are fundamentally different at the molecular level. Furthermore, IDC-NSTs have been shown to be heterogeneous in terms of their transcriptome and patterns of genomic aberrations, and could be classified into several molecular subtypes[2–5].
Gene expression[4, 6–11] and aCGH[8, 11–14] studies of special histological types of breast cancer have revealed that at the genomic and transcriptomic level, tumours from each of the special histological types of breast cancer are more homogeneous amongst themselves than IDC-NSTs. In addition, some special histological types appear to be almost exclusively ER-positive (e.g. micropapillary, mucinous, tubular/ cribriform, lobular and papillary carcinomas), whereas others (e.g. adenoid cystic, secretory, and metaplastic breast cancers) are uniformly ER-negative[4, 5, 15, 16].
Genotypic-phenotypic correlations between specific genetic aberrations and special histological types of breast cancer have emerged. For instance, adenoid cystic carcinomas and secretory carcinomas are underpinned by the recurrent fusion genes MYB-NFIB[17] and ETV6-NTRK3[18], respectively, and lobular carcinomas are underpinned by loss of function of E-cadherin[3, 5, 7, 11, 15]. Furthermore, micropapillary[13, 14] and mucinous[12] carcinomas have been shown to display distinct constellations of gene copy number aberrations when compared to grade- and ER-matched IDC-NSTs.
Papillary carcinoma of the breast is a histological special type that accounts for approximately 1% of all invasive breast cancers. These tumours usually affect post-menopausal patients and have an overall favourable outcome[1, 19–22]. Histologically, they are characterised by arborescent fibrovascular stalks lined by a layer of neoplastic epithelial cells devoid of an intervening myoepithelial cell layer, a feature that distinguishes them from benign intraductal papillomas[1, 23, 24]. The spectrum of papillary carcinomas currently encompasses encapsulated papillary carcinomas (EPC), solid papillary carcinomas (SPC), and invasive papillary carcinomas (IPC)[21, 22]. EPCs are characterised by a well circumscribed nodule of papillary carcinoma surrounded by a thick fibrous capsule. Although initially perceived as a variant of in situ papillary carcinoma, recent studies have demonstrated that EPCs consistently lack a myoepithelial cell layer surrounding the tumour nodules[22–25]. SPCs are composed of nodules, sheets or coalescent papillae of ovoid-to-spindle-shaped cells growing in a solid pattern, and may display neuorendocrine features[26]. IPCs are characterised by a papillary carcinoma frankly invading surrounding tissue[21].
Limited information is available on the molecular features of papillary carcinomas of the breast[27, 28]. The repertoire of gene copy number aberrations harboured by these tumours is yet to be characterised. We sought to investigate whether papillary carcinomas would constitute an entity distinct from IDC-NSTs at the genomic level or if they would merely constitute a morphological variant of ER-positive IDC-NSTs. We have used an array of methods in order i) to characterise breast papillary carcinomas using immunohistochemistry, aCGH and targeted mutational analysis, and ii) to determine whether they would be distinct from grade- and ER-matched IDC-NSTs in terms of their immunophenotype and genomic aberrations. As a further aim, we investigated whether EPC, SPC and IPC would constitute distinct entities at the immunohistochemical and genomic levels.
Materials and Methods
Cases
Power calculations are described in the Supplementary Methods; assumptions were made based on results from previous studies on special histological types of breast cancer[12–14, 29] and IDC-NSTs[30–34]. Sixty-three papillary carcinomas of the breast were retrieved from the hospital files of The Royal Marsden Foundation Trust (from 2001 to 2008), London, UK, Institut Curie (from 1995 to 2009), Paris, France, The Bergonié Institut (from 1997 to 2005), Bordeaux, France, the Centre Hospitalier Universitaire (from 2003 to 2009), Tours, France, and the Centre Hospitalier Régional (from 2007 to -2010), Orléans, France. Only patients diagnosed and managed in the institution above, whose tumours were <5cm and with no clinical and/ or radiological evidence of distant metastases were included in this study. Exclusion criteria were i) patients with multiple tumours, either ipsi- or contra-lateral; ii) patients who received neoadjuvant chemotherapy; iii) patients for which all histological slides and blocks were not available for review; and iv) tumours not consistent with the diagnosis of EPC, SPC or IPC (see below). Samples were anonymised prior to analysis and the study approved by local research ethics committees of the authors’ institutions. All cases were reviewed by at least three pathologists (RD, MLT, GMG, AVS and JRF) independently. A diagnosis of papillary carcinoma was confirmed in all cases, and subtyping and histological grading were performed by two pathologists (AVS and JRF) on a multi-headed microscope. The tumours were categorised into one of the morphological subtypes (i.e. EPC, SPC and IPC), according to current histological criteria[1, 24, 25, 35], and histological grading was determined using Nottingham grading system[36]. The presence of lympho-vascular invasion and lymph node metastasis was also assessed.
Control cases - IDC-NSTs
Histological grade- and ER-matched IDC-NSTs were retrieved from the files of The Royal Marsden Foundation Trust from a consecutive cohort of patients accrued between 1994 and 2004. The 63 papillary carcinomas were matched according to histological grade- and ER-status with 63 IDC- NSTs, and their immunohistochemical profiles were compared. aCGH and Sequenom MassARRAY analyses were successfully performed in 49 papillary carcinomas, and these were 1:1 matched with IDC-NSTs according to histological grade- and ER expression. The rationale for matching case and control samples according to grade- and ER status stems from the several lines of evidence suggesting that grade- and ER are strongly associated with the pattern of genomic changes in breast cancer[33, 34, 37, 38].
Immunohistochemistry
To assist the histological review and reclassification of the papillary carcinomas included in this study, five immunohistochemical markers were assessed on full sections to confirm the diagnosis, namely p63, smooth muscle actin, cytokeratin (CK) 5/6, chromogranin and synaptophysin. These markers were employed to demonstrate the lack of myoepithelial cells within papillary fronds and to rule out overt neuroendocrine differentiation (i.e. >50% of neoplastic cells expressing chromogranin and/ or synaptophysin, which would be consistent with a diagnosis of neuroendocrine carcinoma according to the WHO classification[1]). All cases lacked myoepithelial cells and showed positive results in the internal controls (i.e. myoepithelial cells in surrounding non-neoplastic terminal ductal-lobular units or ducts) and failed to display overt neuroendocrine differentiation. The approach for the classification of papillary carcinomas and exclusion of papillary ductal carcinoma in situ (DCIS) and neuroendocrine carcinomas is illustrated in Supplementary Figure 1.
Tissue microarrays (TMAs) containing 63 samples of papillary carcinomas were constructed from paraffin blocks with triplicate 0.6 mm tumour cores as previously described[12]. Immunohistochemical profiles of the included cases of papillary carcinomas (n=63) and grade- and ER-matched IDC-NSTs were assessed on 3μm thick TMA sections, using a panel of antibodies against ER, progesterone receptor (PR), HER2, Ki67, Bcl2, p53, EGFR, CK14, CK17, nestin, caveolin 1 (CAV1), caveolin 2 (CAV2), E-cadherin, and Cyclin D1,as previously described[14, 39–49]. Positive and negative controls were included in each experiment. The results of immunohistochemical analysis were interpreted by two pathologists (RD, MLT), blinded to the results of aCGH, CISH/FISH and MassARRAY analysis. Antibody clones, dilutions, antigen retrieval methods, scoring systems and cut-offs used are described in Supplementary Table 1 and Supplementary Methods. Tumours were classified into molecular subtypes using the immunohistochemical surrogate described by Nielsen et al[50]. Cases with mixed histology (i.e. mixed EPC and SPC) were excluded from supervised analyses of the immunohistochemical and genomic profiles between subtypes of papillary carcinomas.
Microdissection and DNA extraction
Ten representative 10μm-thick sections of papillary carcinomas and controls were cut and stained with nuclear fast red. Microdissection was performed with a sterile needle under a stereomicroscope (Olympus SZ61, Tokyo, Japan) to ensure a percentage of tumour cells greater than 90%, as previously described[13, 14, 51]. DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hamburg, Germany). Double-stranded DNA concentration and DNA quality were measured using the Picogreen® assay and four primer sets in a multiplex PCR, respectively, as previously described[12–14]. Out of 63 microdissected papillary carcinomas, 49 yielded DNA of sufficient quantity and quality for aCGH analysis and mutation profiling using Sequenom MassArray analysis.
Microarray comparative genomic hybridisation
The aCGH platform used for this study comprises ~32,000 BAC clones tiled across the genome, which has been shown to be as robust as, and to have comparable resolution with, high-density oligonucleotide arrays[52–54]. DNA labelling, array hybridisation, and image acquisition were performed as previously described[34, 51]. aCGH data were pre-processed and analysed using an the Base.R script in R version 2.9.0, as previously described (Supplementary Methods)[55, 56]. A categorical analysis was applied to the BACs after classifying them as representing amplification (>0.45), gain (>0.08 and ≤0.45), loss (<-0.08), or no change according to their cbs-smoothed log2 ratio values[34, 57]. Threshold values were determined and validated as previously described[34, 58]. Categorical data were subjected to a multi-Fisher’s exact test with adjustment for multiple-testing using the step-down permutation procedure maxT, which provides strong control of the family-wise type I error rate (FWER), as previously described[12–14, 51], to identify statistically significant differences between the genomic profiles of i) papillary carcinomas and grade- and ER-matched IDC-NSTs, and ii) EPCs, SPCs and IPCs. Unsupervised hierarchical clustering analysis was performed as previously described[12, 13]. Briefly, categorical aCGH states (i.e. gains, losses, and amplifications) were used for clustering, employing Wards clustering algorithm and Euclidean distance. In parallel, 49 grade- and ER-matched IDC-NSTs were selected as controls for the genomic study and subjected to aCGH. Data and the analysis history, script and code are available at http://rock.icr.ac.uk/collaborations/Mackay/Papillary.aCGH/.
Chromogenic/ fluorescence in situ hybridisation (CISH/FISH)
CISH and/or FISH were used to validate selected regions of amplification identified by aCGH (Supplementary Methods), with probes for CCND1 (ZytoVision, Bremerhaven, Germany), HER2 (Vysis, Illinois, USA), and 7q11.23 (in-house generated), as previously described[59, 60]. Full details of CISH/FISH analysis and interpretation are available in the Supplementary Methods.
Sequenom OncoCarta
Forty-nine papillary breast cancers were subjected to mutation screening using the OncoCarta Panel v1.0 (Sequenom Inc., San Diego, CA) detecting 238 mutations in 19 oncogenes as previously described[61, 62]. Details of the analyses are described in the Supplementary Methods. Mutations identified by Sequenom OncoCarta were validated using Sanger sequencing, with primers designed using Primer3 (http://frodo.wi.mit.edu/primer3/) (Supplementary Table 2).
Statistical Analysis
The SPSS statistical software package version 11.5 (SPSS Inc, an IBM Company Headquarters, Chicago, IL, USA) was used for statistical analyses. Correlations between categorical variables were performed using chi-square test and Fisher’s exact test. All p values were two-tailed and 95% confidence intervals were adopted. A p value < 0.05 was considered significant.
Results
Papillary carcinomas are preferentially grade I/ grade II, ER-positive, luminal breast cancers
In this large series of papillary carcinomas, 42 (66.7%), 15 (23.8%) and 6 (9.5%) cases were classified as of histological grades I, II and III, respectively (Table 1), and 39 (61.9%), 9 (14.3%) and 12 (19.0%) cases were subtyped as EPCs, SPCs or IPCs, respectively. Three cases (4.8%) were classified as of mixed encapsulated and solid patterns. The median mitotic count was 14.6 (range, 1-93) mitoses per ten high power fields (Supplementary Table 3). Immunohistochemical analysis revealed that papillary carcinomas were consistently ER-positive and HER2-negative, and displayed a luminal phenotype (Figure 1). Nuclear expression of p53 was found in 1.6% of cases, high levels of Cyclin D1 expression in 84.2% of cases, and expression of basal markers was found in 3 cases (4.8%).
Table 1. Histopathological and immunohistochemical features of 63 papillary carcinomas and 63 grade- and ER-matched IDC-NSTs.
| N | Papillary carcinomas (n=63) |
IDC-NSTs (n=63) |
p value | |
|---|---|---|---|---|
| Histological grade | 126 | 1** | ||
| I | 42 (66.7%) | 42 (66.7%) | ||
| II | 15 (23.8%) | 15 (23.8%) | ||
| III | 6 (9.5%) | 6 (9.5%) | ||
| Lympho-vascular invasion | 126 | 0.023* | ||
| Present | 10 (15.9%) | 22 (34.9%) | ||
| Absent | 53 (84.1%) | 41 (65.1%) | ||
| Lymph node metastasis | 87 | 0.002* | ||
| Present | 4 (13.8%) | 28 (48.3%) | ||
| Absent | 25 (86.2%) | 30 (51.7%) | ||
| ER | 126 | NP | ||
| Positive | 63 (100%) | 63 (100%) | ||
| Negative | 0 (0%) | 0 (0%) | ||
| PR | 126 | 0.491* | ||
| Positive | 57 (90.5%) | 60 (95.2%) | ||
| Negative | 6 (9.5%) | 3 (4.8%) | ||
| HER2 | 126 | 0.058 | ||
| Negative | 63 (100%) | 58 (92.1%) | ||
| Positive | 0 (0%) | 5 (7.9%) | ||
| Ki67 | 126 | 0.683** | ||
| Low (<10%) | 51 (81.0%) | 49 (77.8%) | ||
| Intermediate (10–30%) | 10 (15.8%) | 13 (20.6%) | ||
| High (>30%) | 2 (3.2%) | 1 (1.6%) | ||
| p53 | 126 | 0.017* | ||
| Positive | 1 (1.6%) | 9 (14.3%) | ||
| Negative | 62 (98.4%) | 54 (85.7%) | ||
| Bcl2 | 123 | 0.109* | ||
| Positive | 62 (98.4%) | 55 (91.7%) | ||
| Negative | 1 (1.6%) | 5 (8.3%) | ||
| Cyclin D1 | 126 | 0.004** | ||
| Low (Allred score 0–3) | 5 (7.9%) | 8 (12.7%) | ||
| Intermediate (Allred score 4–5) | 5 (7.9%) | 18 (28.6%) | ||
| High (Allred score 6–8) | 53 (84.2%) | 37 (58.7%) | ||
| E-cadherin | 124 | 0.614** | ||
| Normal | 55 (90.2%) | 56 (88.9%) | ||
| Reduced | 6 (9.8%) | 6 (9.5%) | ||
| Negative | 0 | 1 (1.6%) | ||
| Cytokeratin 5/6 | 126 | 0.244* | ||
| Positive | 3 (4.8%) | 0 | ||
| Negative | 60 (95.2%) | 63 (100%) | ||
| Cytokeratin 14 | 126 | 1* | ||
| Positive | 1 (1.6%) | 0 | ||
| Negative | 62 (98.4%) | 63 (100%) | ||
| Cytokeratin 17 | 125 | 1* | ||
| Positive | 1 (1.6%) | 1 (1.6%) | ||
| Negative | 61 (98.4%) | 62 (98.4%) | ||
| Basal cytokeratins | 126 | 0.619* | ||
| Positive | 3 (4.8%) | 1 (1.6%) | ||
| Negative | 60 (95.2%) | 62 (98.4%) | ||
| EGFR | 126 | 1 | ||
| Positive | 1 (1.6%) | 0 | ||
| Negative | 62 (98.4%) | 63 (100%) | ||
| Caveolin 1 | 125 | 0.496* | ||
| Positive | 1 (1.6%) | 0 | ||
| Negative | 61 (98.4%) | 63 (100%) | ||
| Caveolin 2 | 125 | 0.496* | ||
| Positive | 1 (1.6%) | 0 | ||
| Negative | 61 (98.4%) | 63 (100%) | ||
| Nestin | 122 | 0.496* | ||
| Positive | 2 (3.2%) | 0 | ||
| Negative | 60 (96.8%) | 60 (100%) | ||
| Chromogranin | 95 | 1* | ||
| Positive | 1 (1.6%) | 0 | ||
| Negative | 62 (98.4%) | 32 (100%) | ||
| Synaptophysin | 95 | 0.548* | ||
| Positive | 2 (3.2%) | 0 | ||
| Negative | 61 (96.8%) | 32 (100%) | ||
| Any basal marker | 126 | 0.619* | ||
| Positive | 3 (4.7%) | 1 (1.6%) | ||
| Negative | 60 (95.3%) | 62 (98.4%) | ||
| Molecular phenotype# | 126 | 0.0576 | ||
| Luminal | 63 (100%) | 58 (92.1%) | ||
| Basal-like | 0 (0%) | 0 (0%) | ||
| HER2 | 0 (0%) | 5 (7.9%) |
Fisher’s exact test
Chi-squared test
molecular phenotype classification according to Nielsen et al.[50];
ER: oestrogen receptor; IDC-NSTs: invasive ductal carcinomas of no special type; NP: not performed (no statistics computed as the value is constant); PR: progesterone receptor.
Figure 1. Histological and immunohistochemical features of papillary carcinomas of the breast.
Representative micrographs of the histological variants of papillary carcinomas: encapsulated papillary carcinoma (A), solid papillary carcinoma (B) and invasive papillary carcinoma (C). Note that regardless of the histological variants, these tumours displayed similar phenotypes with expression of oestrogen receptor (D, E and F) and progesterone receptor (G, H and I), lack of HER2 expression (J, K and L), and high levels of Cyclin D1 expression (M, N and O).
Papillary carcinomas of the breast display relatively simple genomic profiles
aCGH analysis of papillary carcinomas revealed a relative paucity of copy number aberrations, with a median of 12.1% (range 3.23%-34.1%) of BACs showing either gains, losses or amplifications. Regions of recurrent gains and losses occurring in the 49 papillary carcinomas are illustrated in Figure 2 and described in Supplementary Table 4. Consistent with their ER-status and predominance of histological grades I and II, the papillary carcinomas analysed here often displayed the reported genomic features of low-grade, ER-positive breast cancers (i.e. 16q losses, 16p gains and 1q gains)[6, 33, 34].
Figure 2. Frequency plots of chromosomal gains, losses and amplifications observed in 49 papillary carcinomas of the breast.
Frequency plot of copy number gains and losses in 49 papillary carcinomas of the breast (A). The proportion of tumours in which each bacterial artificial chromosome (BAC) clone is gained (green bars) or lost (red bars) is plotted (Y axis) for each BAC clone according to its genomic position (X axis). Frequency plot of amplifications in 49 papillary carcinomas of the breast (B). The proportion of tumours in which each bacterial artificial chromosome (BAC) clone is amplified (green bars) is plotted (Y axis) for each BAC clone according to its genomic position (X axis).
After exclusion of regions mapping to known copy number polymorphisms (http://projects.tcag.ca/variation/), papillary carcinomas were shown to harbour relatively few amplifications. The genomic region most frequently amplified (12% of the cases) mapped to 11q13.3, encompassing CCND1 (Supplementary Table 5). Amplification of this locus was confirmed by CISH using a CCND1 probe in all cases (Supplementary Figures 2A and 2B). These cases consistently displayed high levels of Cyclin D1 expression (Supplementary Figure 2C); as expected, however, Cyclin D1 expression was more prevalent than CCND1 gene amplification (84% vs. 12%) in the papillary carcinomas studied[63]. 7q11.23 amplification was detected by aCGH in two cases and confirmed by CISH using an in-house probe mapping to this genomic region (Supplementary Figures 2D and 2E). Furthermore, consistent with the results of aCGH and immunohistochemical analysis, HER2 gene amplification was not observed in any of the papillary carcinomas studied by means of FISH (data not shown).
Papillary carcinomas and grade- and ER-matched invasive ductal carcinomas display a similar immunohistochemical profile
Papillary carcinomas and grade- and ER-matched IDC-NSTs displayed similar immunohistochemical profiles (Table 1). The only differences observed were related to the significantly lower prevalence of p53 nuclear expression (Fisher’s exact test, p = 0.017) and higher levels of Cyclin D1 (chi-squared test, p = 0.004) expression in papillary carcinomas than in grade- and ER-matched IDC-NSTs (Table 1). No differences in terms of expression of HER2, Bcl2, E-cadherin, basal cytokeratins, EGFR, caveolins 1 and 2, and nestin were found. Consistent with their relatively good outcome, papillary carcinomas were significantly less likely to be associated with lympho-vascular invasion and lymph node metastasis at diagnosis than IDC-NSTs (Fisher’s exact tests, p = 0.023 and 0.002, respectively).
Papillary carcinomas are less genomically complex than grade- and ER-matched invasive ductal carcinomas, but display similar patterns of copy number aberrations
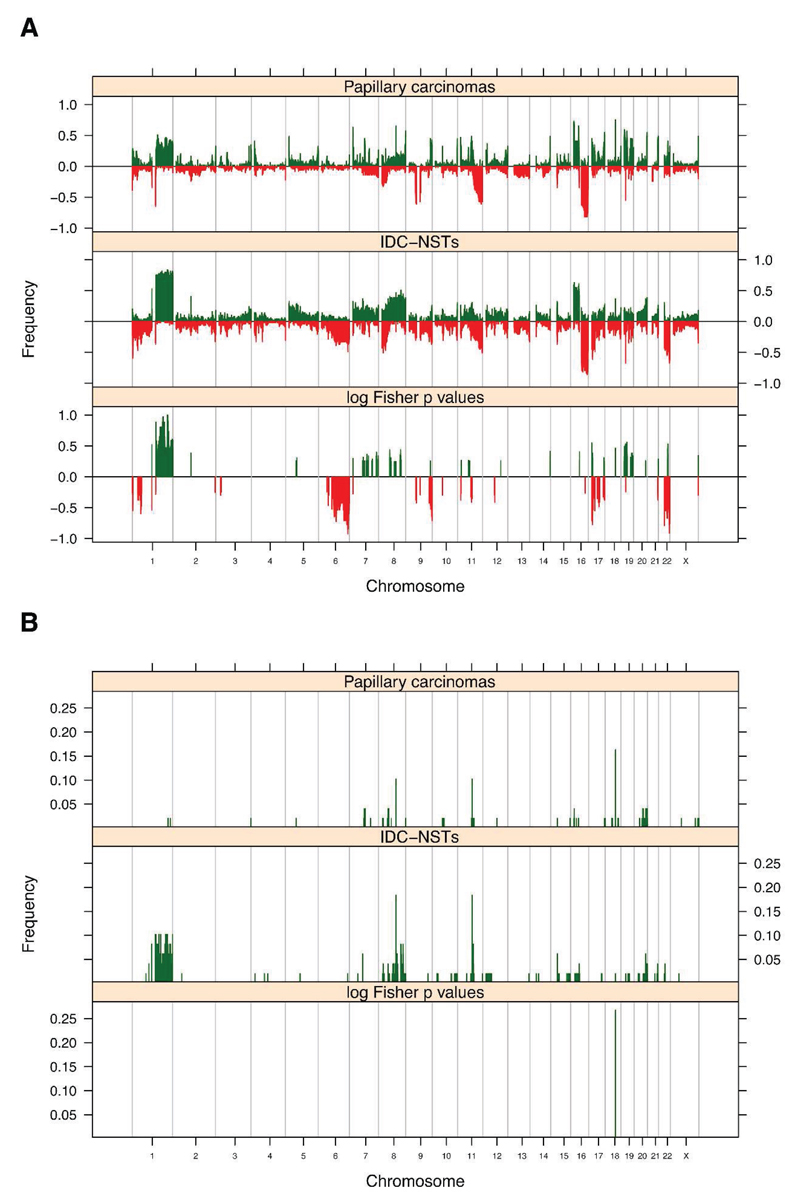
To determine whether papillary carcinomas of the breast are distinct from grade- and ER-matched IDC-NSTs in terms of their number and pattern of genomic aberrations, we compared the genomic profiles of 49 papillary carcinomas with those of 49 grade- and ER-matched IDC-NSTs. This analysis revealed that papillary carcinomas harboured significantly less gene copy number aberrations than grade- and ER-matched IDC-NSTs (median of 12.1% and range of 3.23%-34.1% vs median of 16.9% and range of 6.8%-60.8%, respectively; Mann Whitney U test, p = 0.003). The patterns of copy number gains and losses found in papillary carcinomas were similar to those found in grade- and ER-matched IDC-NSTs, however the prevalence of specific changes was lower in papillary cancers. Significant differences observed included a lower prevalence of 1q whole arm gains and whole arm losses of 6q, 17p, 19p and 22q, and a higher frequency of 19p gains in papillary carcinomas than in grade- and ER-matched IDC-NSTs (multi-Fisher’s exact test p < 0.05; Figure 3A, Supplementary Table 6). After exclusion of regions mapping to known copy number polymorphisms, no significant differences between amplifications observed in papillary carcinomas and grade- and ER- matched IDC-NSTs were detected (Figure 3B). Similar observations were made in a hypothesis generating subgroup analysis of grades I, II or III papillary carcinomas and grade- and ER-matched IDC-NSTs (Supplementary Figures 3A and 3B and data not shown for amplifications).
Figure 3. Comparison of the gene copy number aberrations found in papillary carcinomas and in grade- and ER-matched IDC-NSTs.
Frequency plots of copy number gains and losses (A), and amplifications and deletions (B) observed in 49 papillary carcinomas and 49 grade- and ER-matched IDC-NSTs. In A, the proportion of tumours in which each bacterial artificial chromosome (BAC) clone is gained (green bars) or lost (red bars) is plotted (Y axis) for each BAC clone according to its genomic position (X axis). Inverse Log10 values of the multi-Fisher’s exact test p values are plotted according to genomic position (X axis). In B, the proportion of tumours in which each bacterial artificial chromosome (BAC) clone is amplified (green bars) is plotted (Y axis) for each BAC clone according to its genomic position (X axis). Inverse Log10 values of the multiFisher’s exact test p values are plotted according to genomic position (X axis). ER: oestrogen receptor; IDC-NSTs: invasive ductal carcinomas of no special type.
To determine whether papillary carcinomas would form a discrete group based on their patterns of gene copy number aberrations, we subjected the 49 papillary carcinomas and 49 grade- and ER-matched IDC-NSTs to unsupervised hierarchical clustering. This analysis revealed that although one cluster was significantly enriched for IDC-NSTs (two-tailed Fisher’s exact test, p = 0.0038), papillary carcinomas did not form a separate cluster (Figure 4). Subgroup analyses of grades I and II or grade I tumours only rendered similar findings, with no significant enrichment of IDC-NSTs or papillary carcinomas in any cluster (two-tailed Fisher’s exact test, p = 0.05092 and p = 0.1188; Supplementary Figure 4).
Figure 4. Unsupervised hierarchical clustering analysis of papillary carcinomas and grade- and ER-matched IDC-NSTs.
Hierarchical cluster analysis performed with microarray comparative genomic hybridisation (aCGH) categorical states (i.e. gains, losses and amplifications) using Euclidean distance metric and the Wards algorithm of 49 papillary carcinomas and 49 grade- and ER- matched IDC-NSTs. Amp: amplification; EPC: encapsulated papillary carcinoma; ER: oestrogen receptor; Gain: copy number gain; IDC-NSTs: invasive ductal carcinomas of no special type; IPC: invasive papillary carcinoma; Loss: copy number loss; NC: no copy number change; SPC: solid papillary carcinoma.
Taken together, these observations suggest that papillary carcinomas have less gene copy number aberrations than grade- and ER-matched IDC-NSTs; however, the pattern of genomic aberrations found in papillary carcinomas is similar to that of grade- and ER-matched IDC-NSTs.
Papillary carcinomas of the breast harbour recurrent mutations in PIK3CA
Given the genomic similarity between papillary carcinomas of the breast and grade- and ER-matched IDC-NSTs, we sought to determine if papillary carcinomas would be underpinned by genomic aberrations other than gene copy number changes. Given that only formalin-fixed paraffin-embedded material was available, we focused on mutations affecting known oncogenes. To this end, we performed Sequenom mutation profiling using the Oncocarta v1.0 panel on a subset of 49 papillary carcinomas, which revealed 21, two and five mutations in PIK3CA, AKT1, and MET, respectively. All mutations identified by Sequenom analysis were subsequently tested by Sanger sequencing in the index cases. Although all PIK3CA mutations were validated, the two AKT1 mutations and the five MET mutations were shown to be either germline single nucleotide variants or false positive results, respectively. We next sequenced the exons of PIK3CA found to be mutated in papillary carcinomas in the cohort of grade- and ER-matched IDC-NSTs. This analysis revealed a similar prevalence of PIK3CA mutations in both groups: 43% (21/49) of papillary carcinomas and 29% (14/49) of IDC-NSTs (Fisher’s exact test, p = 0.142) (Table 2, Supplementary Table 7).
Table 2. PIK3CA mutations identified in papillary carcinomas and grade- and ER- matched IDC-NSTs using the Oncocarta panel on the Sequenom platform.
| Mutation | Papillary carcinomas (n=49) |
IDC-NSTs (n=49) |
|---|---|---|
| PIK3CA_H1047R | 10 (20.4%) | 13 (26.5%) |
| PIK3CA_E545K | 8 (16.3%) | 1 (2.0%) |
| PIK3CA_N345K | 1 (2.0%) | 0 (0%) |
| PIK3CA_P539R | 1 (2.0%) | 0 (0%) |
| PIK3CA_E542K | 2 (4.1%) | 0 (0%) |
ER: oestrogen receptor; IDC-NSTs: invasive ductal carcinomas of no special type.
Encapsulated, solid and invasive papillary carcinomas are immunophenotypically and genomically similar
As a secondary aim, we sought to define the immunohistochemical and genomic profiles of the three variants of papillary carcinomas (i.e. EPC, SPC and IPC). The only statistically significant phenotypic differences observed between the subtypes were i) that EPCs were significantly more frequently of histological grades I and II (Chi Squared test p<0.0001), ii) that SPCs were more frequently of grades II and III (Chi Squared test p<0.0001), and iii) that SPCs more frequently focally expressed neuroendocrine markers (i.e. <50% of cancer cells) than the other subtypes (Supplementary Table 8). Likewise, the aCGH profiles of EPCs, SPCs and IPCs were similar. Only loss of 16q was significantly more prevalent in EPCs than in both SPCs and IPCs (multi-Fisher’s exact test p < 0.05; Supplementary Table 9 and Figure 5). This finding is likely to stem from the fact that EPCs were significantly more frequently of histological grade I (89.7%) than the two other subtypes (0% and 41.7% in SPCs and IPCs, respectively). Furthermore, after exclusion of regions mapping to known copy number polymorphisms, no differences in amplifications observed in the three morphological subtypes were found (Figure 5). Unsupervised hierarchical clustering of all 49 papillary carcinomas revealed that none of the three morphological variants form discrete clusters (Supplementary Figure 5). Taken together, these immunophenotypic and genomic findings provide circumstantial evidence to suggest that EPC, SPC and IPC may constitute histological variants of the same entity.
Figure 5. Comparative genomic profiling of encapsulated (EPC), solid (SPC) and invasive (IPC) papillary carcinomas.
Frequency plots and multi-Fisher’s exact comparisons of chromosomal gains and losses (A) and amplifications (B) in EPC, SPC and IPC. In A, the proportion of tumours in which each bacterial artificial chromosome (BAC) clone is gained (green bars) or lost (red bars) is plotted (Y axis) for each BAC clone according to its genomic position (X axis). Inverse Log10 values of the multi-Fisher’s exact test p values are plotted according to genomic position (X axis). In B, the proportion of tumours in which each bacterial artificial chromosome (BAC) clone is amplified (green bars) is plotted (Y axis) for each BAC clone according to its genomic position (X axis). Inverse Log10 values of the multi-Fisher’s exact test p values are plotted according to genomic position (X axis).
Discussion
Here we demonstrate that papillary carcinomas of the breast constitute a relatively homogeneous entity and consistently display a luminal phenotype. Their immunohistochemical profiles were remarkably similar to those of grade- and ER-matched IDC-NSTs. The only significant differences observed were the higher prevalence of high Cyclin D1 expression, lower prevalence of p53 expression and a more overt luminal phenotype than IDC-NSTs of similar histological grade.
Papillary carcinomas were also shown to display patterns of gene copy number aberrations that qualitatively are similar to those found in grade- and ER-matched IDC-NSTs[33, 64]. It should be noted, however, that the number of gene copy number aberrations found in papillary carcinomas was significantly lower than those found in grade- and ER-matched IDC-NSTs. Finally, papillary carcinomas were found to harbour PIK3CA mutations in approximately 40% of cases, a feature of ER-positive IDC-NSTs of good prognosis[65].
The observations that papillary carcinomas significantly less frequently displayed lymphovascular invasion or lymph node metastasis at diagnosis, and more frequently harboured a lower prevalence of p53 expression, low number of genomic aberrations and high prevalence of PIK3CA mutations potentially provide a rationale for the relatively good prognosis of papillary carcinomas[19–22]. In fact, previous studies have demonstrated that absence of lymphovascular invasion and lymph node metastasis, lack of p53 expression[66], low number of genomic aberrations[67–69], and presence of PIK3CA mutations[65] are associated with good clinical outcome in patients with ER-positive breast cancers.
Although papillary carcinomas harboured less gene copy number aberrations than grade- and ER-matched IDC-NSTs, the pattern of genomic aberrations found in these two types of breast cancer were similar, in contrast to previous aCGH analyses of other special types of breast cancer, which revealed that micropapillary[13, 14], mucinous[12] and adenoid cystic[29] carcinomas displayed different patterns of genomic aberrations when compared to grade- and ER-matched IDC-NSTs. These observations provide evidence to suggest that although papillary carcinomas are consistently ER-positive, rather than constituting a distinct entity, they belong to the spectrum of ER-positive IDC-NSTs. It is plausible that these tumours may evolve through the same genetic pathways as low grade IDC-NSTs and/ or originate from the same compartment of the normal breast. Consistent with this hypothesis is the observation that the frankly invasive component of papillary carcinomas often loses its characteristic papillary morphology and assumes the histological pattern of IDC-NSTs[1]. Given the distinctive nature of the papillary growth pattern and the fact that no determinant of this phenotype was identified in this study, it is possible that the characteristic architectural features of papillary cancers are underpinned by genetic aberrations other than gene copy number aberrations (i.e. copy number silent loss of heterozygosity events, somatic mutations or fusion genes), epigenetic changes or distinctive tumour-microenvironment interactions. An alternative hypothesis is that the patient’s genetic make-up may predispose to a papillary phenotype in an ER-positive cancer. Finally, another hypothesis is that papillary carcinomas may merely constitute a final stage of development of a DCIS within a pre-existing papilloma, where myoepithelial cells have been completely lost. Given the similarities between the genomic profiles of IDC-NSTs and grade- and ER-matched DCIS[6, 70, 71], this would provide a potential explanation for our findings (i.e. similar patterns of genomic aberrations in papillary carcinomas and grade- and ER-matched IDC-NSTs). In fact, the prevalence of 1q gains, 16q losses, 16p gains and 8q gains in grade- and ER-matched DCIS and IDC-NSTs is strikingly similar[6, 70, 71]; hence, it is probable that a comparison between papillary carcinomas with grade- and ER-matched DCIS would have rendered results similar to those reported in our study. With the advent of massively parallel sequencing, further studies investigating the repertoire of somatic mutations, fusion genes, and epigenetic changes in papillary carcinomas are warranted. It should be noted, however, that there are several lines of evidence from xenograft studies and conditional mouse models driven by distinct genetic aberrations (e.g. RET/PTC3, HPV type 16 E7 protein, BRAF and Ha-Ras expression) that demonstrate that multiple genetic aberrations result in murine cancers that display papillary morphology in several anatomical sites, including thyroid, breast, bladder, pancreas[72–79]. In fact, histological analysis of conditional mouse models of mammary gland cancers revealed that genetic alterations of various pathways, including the ERBB, RAS, WNT, CDK2 and LKB1 pathways, result in tumours with papillary morphology[78, 79]. Therefore, it is likely that papillary carcinomas represent a convergent phenotype[80] rather than a single distinct entity.
We also show that the three morphological subtypes of papillary carcinomas of the breast (i.e. EPC, SPC and IPC) harbour similar immunohistochemical profiles and patterns of gene copy number aberrations. These observations corroborate the classification of EPC, SPC and IPC as subtypes of papillary carcinomas. Their distinct morphological features may be underpinned by genetic aberrations other than copy number aberrations or by epigenetic changes.
This study has several limitations. First, all analyses were carried out with formalin-fixed paraffin-embedded tissue sections and no germ-line DNA obtained from blood lymphocytes was available from these patients; therefore, massively parallel sequencing for the identification of expressed fusion genes and discovery of novel mutations could not be carried out. Second, the limited sample size of SPCs and IPCs renders the comparisons between subtypes of papillary carcinomas exploratory and hypothesis generating. Third, given the multi-institutional and retrospective nature of the cohort analysed and the incomplete follow-up information available for the patients whose tumours were included in this study, survival analyses could not be performed. Fourth, another potential confounding factor of our study is that the control group of IDC-NSTs was retrieved from The Royal Marsden Foundation Trust, a tertiary hospital; the type of cases managed at that institution may have led to a marginally higher prevalence of lymph node metastasis in the control group than expected in a population-based cohort.
Despite the limitations outlined above, our study demonstrates that papillary carcinomas of the breast are a rather homogeneous special type of breast cancer. In a way akin to good prognosis ER-positive IDC-NSTs, papillary carcinomas are characterised by consistent ER expression, high prevalence of PIK3CA mutations and relatively low rates of p53 expression and gene copy number aberrations. The patterns of gene copy number aberrations found in papillary carcinomas are similar to those found in grade- and ER-matched IDC-NSTs and are unlikely to differ from those reported in grade- and ER-matched DCIS[6, 70, 71], suggesting that they may not constitute a distinct entity but rather a part of the spectrum of ER-positive breast cancers.
Supplementary Material
Acknowledgements
This study was funded by Breakthrough Breast Cancer. JSR-F is one of the recipients of the 2010 CRUK Future Leaders Prize. RD is funded by a grant from the French Higher Education and Research Department. AVS is supported by a grant ‘INTERFACE’ from INSERM. PW is funded by a Wellcome Trust Clinical Fellowship Grant, BW by a CRUK postdoctoral fellowship. We also acknowledge NHS funding to the NIHR Biomedical Research Centre.
Footnotes
Authors’ Contributions
AVS, BW and JSR-F conceived the study, interpreted results and drafted the manuscript. RD, PW, MBL, MLT, AM, RN carried out experiments and analysed data. RA analysed data and provided statistical support. VP, E-PC, FD and EW carried out experiments. GMG, FV and PM provided samples and analysed data. All authors reviewed and approved the submitted version of the manuscript.
Conflict of Interest: None
Microarray comparative genomic hybridisation data, the analysis history, script and code are available at http://rock.icr.ac.uk/collaborations/Mackay/Papillary.aCGH/.
References
- 1.Tavassoli FA, D Pe. Tumours of the breast. International Agency for Research of Cancer (IARC); Lyon: 2003. [Google Scholar]
- 2.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 3.Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010;220:263–280. doi: 10.1002/path.2648. [DOI] [PubMed] [Google Scholar]
- 4.Weigelt B, Horlings HM, Kreike B, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008;216:141–150. doi: 10.1002/path.2407. [DOI] [PubMed] [Google Scholar]
- 5.Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6:718–730. doi: 10.1038/nrclinonc.2009.166. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Garcia MA, Geyer FC, Natrajan R, et al. Transcriptomic analysis of tubular carcinomas of the breast reveals similarities and differences with molecular subtype-matched ductal and lobular carcinomas. J Pathol. 2010;222:64–75. doi: 10.1002/path.2743. [DOI] [PubMed] [Google Scholar]
- 7.Weigelt B, Geyer FC, Natrajan R, et al. The molecular underpinning of lobular histological growth pattern: a genome-wide transcriptomic analysis of invasive lobular carcinomas and grade- and molecular subtype-matched invasive ductal carcinomas of no special type. J Pathol. 2010;220:45–57. doi: 10.1002/path.2629. [DOI] [PubMed] [Google Scholar]
- 8.Vincent-Salomon A, Gruel N, Lucchesi C, et al. Identification of typical medullary breast carcinoma as a genomic sub-group of basal-like carcinomas, a heterogeneous new molecular entity. Breast Cancer Res. 2007;9:R24. doi: 10.1186/bcr1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruel N, Lucchesi C, Raynal V, et al. Lobular invasive carcinoma of the breast is a molecular entity distinct from luminal invasive ductal carcinoma. Eur J Cancer. 2010;46:2399–2407. doi: 10.1016/j.ejca.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Weigelt B, Geyer FC, Horlings HM, et al. Mucinous and neuroendocrine breast carcinomas are transcriptionally distinct from invasive ductal carcinomas of no special type. Mod Pathol. 2009;22:1401–1414. doi: 10.1038/modpathol.2009.112. [DOI] [PubMed] [Google Scholar]
- 11.Bertucci F, Orsetti B, Negre V, et al. Lobular and ductal carcinomas of the breast have distinct genomic and expression profiles. Oncogene. 2008;27:5359–5372. doi: 10.1038/onc.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacroix-Triki M, Suarez PH, Mackay A, et al. Mucinous carcinoma of the breast is genomically distinct from invasive ductal carcinomas of no special type. J Pathol. 2010;222:282–298. doi: 10.1002/path.2763. [DOI] [PubMed] [Google Scholar]
- 13.Marchio C, Iravani M, Natrajan R, et al. Mixed micropapillary-ductal carcinomas of the breast: a genomic and immunohistochemical analysis of morphologically distinct components. J Pathol. 2009;218:301–315. doi: 10.1002/path.2572. [DOI] [PubMed] [Google Scholar]
- 14.Marchio C, Iravani M, Natrajan R, et al. Genomic and immunophenotypical characterization of pure micropapillary carcinomas of the breast. J Pathol. 2008;215:398–410. doi: 10.1002/path.2368. [DOI] [PubMed] [Google Scholar]
- 15.Weigelt B, Geyer FC, Reis-Filho JS. Histological types of breast cancer: how special are they? Mol Oncol. 2010;4:192–208. doi: 10.1016/j.molonc.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reis-Filho JS, Lakhani SR. Breast cancer special types: why bother? J Pathol. 2008;216:394–398. doi: 10.1002/path.2419. [DOI] [PubMed] [Google Scholar]
- 17.Persson M, Andren Y, Mark J, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tognon C, Knezevich SR, Huntsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 19.Solorzano CC, Middleton LP, Hunt KK, et al. Treatment and outcome of patients with intracystic papillary carcinoma of the breast. Am J Surg. 2002;184:364–368. doi: 10.1016/s0002-9610(02)00941-8. [DOI] [PubMed] [Google Scholar]
- 20.Grabowski J, Salzstein SL, Sadler GR, et al. Intracystic papillary carcinoma: a review of 917 cases. Cancer. 2008;113:916–920. doi: 10.1002/cncr.23723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pal SK, Lau SK, Kruper L, et al. Papillary carcinoma of the breast: an overview. Breast Cancer Res Treat. 2010;122:637–645. doi: 10.1007/s10549-010-0961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakha EA, Gandhi N, Climent F, et al. Encapsulated papillary carcinoma of the breast: an invasive tumor with excellent prognosis. Am J Surg Pathol. 2011;35:1093–1103. doi: 10.1097/PAS.0b013e31821b3f65. [DOI] [PubMed] [Google Scholar]
- 23.Hill CB, Yeh IT. Myoepithelial cell staining patterns of papillary breast lesions: from intraductal papillomas to invasive papillary carcinomas. Am J Clin Pathol. 2005;123:36–44. doi: 10.1309/xg7tpq16dmjav8p1. [DOI] [PubMed] [Google Scholar]
- 24.Collins LC, Schnitt SJ. Papillary lesions of the breast: selected diagnostic and management issues. Histopathology. 2008;52:20–29. doi: 10.1111/j.1365-2559.2007.02898.x. [DOI] [PubMed] [Google Scholar]
- 25.Collins LC, Carlo VP, Hwang H, et al. Intracystic papillary carcinomas of the breast: a reevaluation using a panel of myoepithelial cell markers. Am J Surg Pathol. 2006;30:1002–1007. doi: 10.1097/00000478-200608000-00011. [DOI] [PubMed] [Google Scholar]
- 26.Nassar H, Qureshi H, Volkanadsay N, et al. Clinicopathologic analysis of solid papillary carcinoma of the breast and associated invasive carcinomas. Am J Surg Pathol. 2006;30:501–507. doi: 10.1097/00000478-200604000-00011. [DOI] [PubMed] [Google Scholar]
- 27.Tsuda H, Uei Y, Fukutomi T, et al. Different incidence of loss of heterozygosity on chromosome 16q between intraductal papilloma and intracystic papillary carcinoma of the breast. Jpn J Cancer Res. 1994;85:992–996. doi: 10.1111/j.1349-7006.1994.tb02895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oikawa M, Nagayasu T, Yano H, et al. Intracystic Papillary Carcinoma of Breast Harbors Significant Genomic Alteration Compared with Intracystic Papilloma: Genome-wide Copy Number and LOH Analysis Using High-Density Single-Nucleotide Polymorphism Microarrays. Breast J. 2011;17:427–430. doi: 10.1111/j.1524-4741.2011.01110.x. [DOI] [PubMed] [Google Scholar]
- 29.Wetterskog D, Lopez-Garcia MA, Lambros MB, et al. Breast adenoid cystic carcinomas constitute a genomically distinct subgroup of triple-negative and basal-like breast cancers. J Pathol. 2011 doi: 10.1002/path.2974. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 30.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Adelaide J, Finetti P, Bekhouche I, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- 32.Andre F, Job B, Dessen P, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15:441–451. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- 33.Natrajan R, Lambros MB, Geyer FC, et al. Loss of 16q in high grade breast cancer is associated with estrogen receptor status: Evidence for progression in tumors with a luminal phenotype? Genes Chromosomes Cancer. 2009;48:351–365. doi: 10.1002/gcc.20646. [DOI] [PubMed] [Google Scholar]
- 34.Natrajan R, Lambros MB, Rodriguez-Pinilla SM, et al. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res. 2009;15:2711–2722. doi: 10.1158/1078-0432.CCR-08-1878. [DOI] [PubMed] [Google Scholar]
- 35.Tavassoli FA, Devilee P. Tumours of the breast. International Agency for Research of Cancer (IARC). IARC Press; Lyon: 2003. [Google Scholar]
- 36.Elston CWEI. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 37.Loo LW, Grove DI, Williams EM, et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64:8541–8549. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- 38.Melchor L, Honrado E, Huang J, et al. Estrogen receptor status could modulate the genomic pattern in familial and sporadic breast cancer. Clin Cancer Res. 2007;13:7305–7313. doi: 10.1158/1078-0432.CCR-07-0711. [DOI] [PubMed] [Google Scholar]
- 39.Harvey JM, Clark GM, Osborne CK, et al. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 40.Reis-Filho JS, Savage K, Lambros MB, et al. Cyclin D1 protein overexpression and CCND1 amplification in breast carcinomas: an immunohistochemical and chromogenic in situ hybridisation analysis. Mod Pathol. 2006;19:999–1009. doi: 10.1038/modpathol.3800621. [DOI] [PubMed] [Google Scholar]
- 41.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 42.Tan DS, Marchio C, Jones RL, et al. Triple negative breast cancer: molecular profiling and prognostic impact in adjuvant anthracycline-treated patients. Breast Cancer Res Treat. 2008;111:27–44. doi: 10.1007/s10549-007-9756-8. [DOI] [PubMed] [Google Scholar]
- 43.Ali HR, Dawson SJ, Blows FM, et al. A Ki67/BCL2 index based on immunohistochemistry is highly prognostic in ER positive breast cancer. J Pathol. 2011 doi: 10.1002/path.2976. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 44.Callagy GM, Pharoah PD, Pinder SE, et al. Bcl-2 is a prognostic marker in breast cancer independently of the Nottingham Prognostic Index. Clin Cancer Res. 2006;12:2468–2475. doi: 10.1158/1078-0432.CCR-05-2719. [DOI] [PubMed] [Google Scholar]
- 45.Dawson SJ, Makretsov N, Blows FM, et al. BCL2 in breast cancer: a favourable prognostic marker across molecular subtypes and independent of adjuvant therapy received. Br J Cancer. 2010;103:668–675. doi: 10.1038/sj.bjc.6605736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdel-Fatah TM, Powe DG, Ball G, et al. Proposal for a modified grading system based on mitotic index and Bcl2 provides objective determination of clinical outcome for patients with breast cancer. J Pathol. 2010;222:388–399. doi: 10.1002/path.2775. [DOI] [PubMed] [Google Scholar]
- 47.Savage K, Lambros MB, Robertson D, et al. Caveolin 1 is overexpressed and amplified in a subset of basal-like and metaplastic breast carcinomas: a morphologic, ultrastructural, immunohistochemical, and in situ hybridization analysis. Clin Cancer Res. 2007;13:90–101. doi: 10.1158/1078-0432.CCR-06-1371. [DOI] [PubMed] [Google Scholar]
- 48.Savage K, Leung S, Todd SK, et al. Distribution and significance of caveolin 2 expression in normal breast and invasive breast cancer: an immunofluorescence and immunohistochemical analysis. Breast Cancer Res Treat. 2008;110:245–256. doi: 10.1007/s10549-007-9718-1. [DOI] [PubMed] [Google Scholar]
- 49.Parry S, Savage K, Marchio C, et al. Nestin is expressed in basal-like and triple negative breast cancers. J Clin Pathol. 2008;61:1045–1050. doi: 10.1136/jcp.2008.058750. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 51.Marchio C, Natrajan R, Shiu KK, et al. The genomic profile of HER2-amplified breast cancers: the influence of ER status. J Pathol. 2008;216:399–407. doi: 10.1002/path.2423. [DOI] [PubMed] [Google Scholar]
- 52.Coe BP, Ylstra B, Carvalho B, et al. Resolving the resolution of array CGH. Genomics. 2007;89:647–653. doi: 10.1016/j.ygeno.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 53.Tan DS, Lambros MB, Natrajan R, et al. Getting it right: designing microarray (and not ‘microawry’) comparative genomic hybridization studies for cancer research. Lab Invest. 2007;87:737–754. doi: 10.1038/labinvest.3700593. [DOI] [PubMed] [Google Scholar]
- 54.Gunnarsson R, Staaf J, Jansson M, et al. Screening for copy-number alterations and loss of heterozygosity in chronic lymphocytic leukemia--a comparative study of four differently designed, high resolution microarray platforms. Genes Chromosomes Cancer. 2008;47:697–711. doi: 10.1002/gcc.20575. [DOI] [PubMed] [Google Scholar]
- 55.Mackay A, Tamber N, Fenwick K, et al. A high-resolution integrated analysis of genetic and expression profiles of breast cancer cell lines. Breast Cancer Res Treat. 2009;118:481–498. doi: 10.1007/s10549-008-0296-7. [DOI] [PubMed] [Google Scholar]
- 56.Natrajan R, Weigelt B, Mackay A, et al. An integrative genomic and transcriptomic analysis reveals molecular pathways and networks regulated by copy number aberrations in basal-like, HER2 and luminal cancers. Breast Cancer Res Treat. 2010;121:575–589. doi: 10.1007/s10549-009-0501-3. [DOI] [PubMed] [Google Scholar]
- 57.Reis-Filho JS, Drury S, Lambros MB, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat Genet. 2008;40:809–810. doi: 10.1038/ng0708-809b. author reply 810-802. [DOI] [PubMed] [Google Scholar]
- 58.Geyer FC, Weigelt B, Natrajan R, et al. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J Pathol. 2010;220:562–573. doi: 10.1002/path.2675. [DOI] [PubMed] [Google Scholar]
- 59.Lambros MB, Natrajan R, Reis-Filho JS. Chromogenic and fluorescent in situ hybridization in breast cancer. Hum Pathol. 2007;38:1105–1122. doi: 10.1016/j.humpath.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 60.Lambros MB, Simpson PT, Jones C, et al. Unlocking pathology archives for molecular genetic studies: a reliable method to generate probes for chromogenic and fluorescent in situ hybridization. Lab Invest. 2006;86:398–408. doi: 10.1038/labinvest.3700390. [DOI] [PubMed] [Google Scholar]
- 61.Fumagalli D, Gavin PG, Taniyama Y, et al. A rapid, sensitive, reproducible and cost-effective method for mutation profiling of colon cancer and metastatic lymph nodes. BMC Cancer. 2010;10:101. doi: 10.1186/1471-2407-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lambros MB, Wilkerson PM, Natrajan R, et al. High-throughput detection of fusion genes in cancer using the Sequenom MassARRAY platform. Lab Invest. 2011 doi: 10.1038/labinvest.2011.110. [DOI] [PubMed] [Google Scholar]
- 63.Elsheikh S, Green AR, Aleskandarany MA, et al. CCND1 amplification and cyclin D1 expression in breast cancer and their relation with proteomic subgroups and patient outcome. Breast Cancer Res Treat. 2008;109:325–335. doi: 10.1007/s10549-007-9659-8. [DOI] [PubMed] [Google Scholar]
- 64.Roylance R, Gorman P, Papior T, et al. A comprehensive study of chromosome 16q in invasive ductal and lobular breast carcinoma using array CGH. Oncogene. 2006;25:6544–6553. doi: 10.1038/sj.onc.1209659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Di Cosimo S, Baselga J. Phosphoinositide 3-kinase mutations in breast cancer: a “good” activating mutation? Clin Cancer Res. 2009;15:5017–5019. doi: 10.1158/1078-0432.CCR-09-1173. [DOI] [PubMed] [Google Scholar]
- 66.Abdel-Fatah TM, Powe DG, Agboola J, et al. The biological, clinical and prognostic implications of p53 transcriptional pathways in breast cancers. J Pathol. 2010;220:419–434. doi: 10.1002/path.2663. [DOI] [PubMed] [Google Scholar]
- 67.Gravier E, Pierron G, Vincent-Salomon A, et al. A prognostic DNA signature for T1T2 node-negative breast cancer patients. Genes Chromosomes Cancer. 2010;49:1125–1134. doi: 10.1002/gcc.20820. [DOI] [PubMed] [Google Scholar]
- 68.Jonsson G, Staaf J, Vallon-Christersson J, et al. Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Res. 2010;12:R42. doi: 10.1186/bcr2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Russnes HG, Vollan HK, Lingjaerde OC, et al. Genomic architecture characterizes tumor progression paths and fate in breast cancer patients. Sci Transl Med. 2010;2:38ra47. doi: 10.1126/scitranslmed.3000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vincent-Salomon A, Lucchesi C, Gruel N, et al. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clin Cancer Res. 2008;14:1956–1965. doi: 10.1158/1078-0432.CCR-07-1465. [DOI] [PubMed] [Google Scholar]
- 71.Robanus-Maandag EC, Bosch CA, Kristel PM, et al. Association of C-MYC amplification with progression from the in situ to the invasive stage in C-MYC-amplified breast carcinomas. J Pathol. 2003;201:75–82. doi: 10.1002/path.1385. [DOI] [PubMed] [Google Scholar]
- 72.Powell DJ, Jr, Russell J, Nibu K, et al. The RET/PTC3 oncogene: metastatic solidtype papillary carcinomas in murine thyroids. Cancer Res. 1998;58:5523–5528. [PubMed] [Google Scholar]
- 73.Ledent C, Marcotte A, Dumont JE, et al. Differentiated carcinomas develop as a consequence of the thyroid specific expression of a thyroglobulin-human papillomavirus type 16 E7 transgene. Oncogene. 1995;10:1789–1797. [PubMed] [Google Scholar]
- 74.Coppee F, Gerard AC, Denef JF, et al. Early occurrence of metastatic differentiated thyroid carcinomas in transgenic mice expressing the A2a adenosine receptor gene and the human papillomavirus type 16 E7 oncogene. Oncogene. 1996;13:1471–1482. [PubMed] [Google Scholar]
- 75.Burniat A, Jin L, Detours V, et al. Gene expression in RET/PTC3 and E7 transgenic mouse thyroids: RET/PTC3 but not E7 tumors are partial and transient models of human papillary thyroid cancers. Endocrinology. 2008;149:5107–5117. doi: 10.1210/en.2008-0531. [DOI] [PubMed] [Google Scholar]
- 76.Bumaschny V, Urtreger A, Diament M, et al. Malignant myoepithelial cells are associated with the differentiated papillary structure and metastatic ability of a syngeneic murine mammary adenocarcinoma model. Breast Cancer Res. 2004;6:R116–129. doi: 10.1186/bcr757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang ZT, Pak J, Huang HY, et al. Role of Ha-ras activation in superficial papillary pathway of urothelial tumor formation. Oncogene. 2001;20:1973–1980. doi: 10.1038/sj.onc.1204315. [DOI] [PubMed] [Google Scholar]
- 78.Rosner A, Miyoshi K, Landesman-Bollag E, et al. Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am J Pathol. 2002;161:1087–1097. doi: 10.1016/S0002-9440(10)64269-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McCarthy A, Lord CJ, Savage K, et al. Conditional deletion of the Lkb1 gene in the mouse mammary gland induces tumour formation. J Pathol. 2009;219:306–316. doi: 10.1002/path.2599. [DOI] [PubMed] [Google Scholar]
- 80.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–38. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.