Abstract
Chromosome breakage elicits transient silencing of ribosomal RNA synthesis, but the mechanisms involved remained elusive. Here we discover an in-trans signaling mechanism that triggers pan-nuclear silencing of rRNA transcription in response to DNA damage. This is associated with transient recruitment of the Nijmegen breakage syndrome protein 1 (NBS1), a central regulator of DNA damage responses, into the nucleoli. We further identified TCOF1-Treacle, a nucleolar factor implicated in ribosome biogenesis and mutated in Treacher Collins syndrome, as an interaction partner of NBS1, and demonstrate that NBS1 translocation and accumulation in the nucleoli is Treacle-dependent. Finally, we provide evidence that Treacle-mediated NBS1 recruitment into the nucleoli regulates rRNA silencing in-trans in the presence of distant chromosome breaks.
The inability to properly respond to DNA damage leads to various disorders and enhanced rates of tumor development in mammals. To avoid genome instability, cells respond to DNA damage by activating a complex network of signaling pathways, collectively termed the DNA damage response (DDR), which is designed for detecting, signaling and repairing of aberrant DNA structures 1.
A central player in the DDR is the ATM protein, a large Serine-Threonine kinase that is activated by DNA double-strand breaks (DSBs) and is mutated in the genome instability syndrome Ataxia telangiectasia (AT) 2. Several proteins are implicated in efficient recruitment of ATM to sites of DSBs and its subsequent activation: NBS1 for example, which is a subunit of the conserved MRE11-RAD50-NBS1 (MRN) complex, directly interacts with ATM, and the MRN complex is involved in the efficient activation of ATM in response to DSBs 3. NBS1 is mutated in Nijmegen breakage syndrome (NBS), a rare autosomal recessive congenital disorder that is associated with short stature, cranio-facial abnormalities including microcephaly, mild mental retardation, immunodeficiency and a strong predisposition to lymphoid malignancy 4.
Other proteins such as the phosphorylated form of the histone variant H2AX (termed γH2AX) and the adaptor MDC1 help to establish a specialized chromatin environment in the regions flanking DSBs, a process believed to enhance and maintain the ATM-dependent DNA damage signaling pathways 5. Recruitment and accumulation of the MRN complex at sites of DSBs requires the direct interaction between a unique divalent Forkhead-associated (FHA)-tandem BRCA1 C-terminal (BRCT) phosphopeptide-binding architecture located at the N-terminus of NBS1, with a constitutively phosphorylated acidic repeat region in MDC1 (SDT repeat region; 6–11).
Induction of DSBs triggers transient inhibition of cell cycle progression (so-called cell cycle checkpoints), especially during S-phase and at the G2-M border. Whether the transcriptional machinery is similarly affected by the presence of DSBs remains a long-standing question. Recent evidence suggests that ATM-dependent signaling cascades can also suppress transcription. For example, an ATM-dependent transcriptional silencing mechanism prevented RNA polymerase II (Pol II) elongation-dependent chromatin decondensation at regions distal to DSBs 12. In addition, ATM-dependent signaling was shown to inhibit local RNA polymerase I (Pol I) transcription in the nucleoli in response to chromosome breaks 13. In both cases, transcriptional silencing seemed to be limited to chromatin regions that are closely located to the sites of breaks and a pan-nuclear transcriptional silencing mechanism in response to DNA damage has so far not been described.
Here we describe an ATM-dependent in-trans signaling mechanism that triggers pannuclear silencing of Pol I transcription in response to DNA breaks. Furthermore we present evidence that in-trans inhibition of rRNA synthesis is regulated by rapid and transient DNA damage-induced nucleolar translocation and accumulation of NBS1, a process that is regulated by ATM and is dependent on direct interaction between NBS1 and the nucleolar factor TCOF1-Treacle. In summary, our findings reveal an unexpected trans-compartmental communication between sites of DNA damage and Pol I mediated transcription in the nucleoli.
Results
In-trans inhibition of Pol I transcription in response to DNA damage is associated with NBS1 recruitment into the nucleoli
It was previously shown that laser micro-irradiation of an individual nucleolus leads to transient silencing of rRNA synthesis within this nucleolus, while transcription in the other nucleoli of the same cell was unaffected 13.
In order to test whether induction of DSBs outside of the nculeolar compartment had the capacity to trigger rRNA silencing in trans we established an imaging-based assay that combined laser micro-irradiation with 5-EU RNA labeling followed by Click-iT chemistry. Surprisingly, we found that irradiation of any sub-nuclear region had a pronounced effect on rRNA transcription in all of the nucleoli of the irradiated cells irrespective of whether or not they were directly hit by the laser beam (Fig. 1a and Supplementary Fig. 1a). This suggests the existence of an as yet unidentified in-trans signaling mechanism that transduces the signal generated by the presence of chromosome breaks into the nucleoli. Further support of such an in-trans signaling mechanism came from the observation that low doses of IR gave a very similar degree of rRNA inhibition as compared to high-dose treatment (Supplementary Fig. 2a).
Figure 1. In-trans inhibition of Pol I transcription in response to DNA damage is associated with NBS1 recruitment into the nucleoli.
(a) Measurement of rRNA synthesis after laser micro-irradiation by 5-EU incorporation. Location of the nucleoli is highlighted (2 independent experiments, n > 30 cells irradiated) (b) NBS1 mobilization into the nucleoli after laser micro-irradiation. Quantification of the percentage of cells with NBS1-GFP positive nucleoli is displayed in the right hand panel. Error bars represent standard error of the mean (S.E.M; 5 independent experiments, n > 60 cells irradiated per experiment). (c) Recruitment of endogenous NBS1 but not MRE11 in the nucleoli after DNA damage. U2OS cells were irradiated with 5 Gy, fixed after 10 min. and stained by IF with the indicated antibodies (2 independent experiments) (d) Quantification of NBS1-GFP translocation was measured as GFP-intensity in laser microirradiation tracks and as increase in standard deviation in the nucleoli. Error bars represent S.E.M (n = 8 nucleoli from 5 cells; 5 independent experiments). (e) Kinetics of rRNA inhibition after IR measured by 5-EU incorporation. Cells were incubated with 5-EU at indicated timepoints after 5 Gy of IR (n > 8000 nucleoli; 1 representative of 2 independent experiments). Scale bars in all panels: 10 µm.
Since Pol I silencing in-cis after exposure of cells to IR was shown to be dependent on ATM signaling and on the presence of the two DDR mediator-adaptor proteins MDC1 and NBS1 13, we asked if these factors were also involved in this trans-compartmental signaling mechanism. While in our hands MDC1 depletion had only a minor effect we found that rRNA repression was dependent on ATM signaling and on NBS1, consistent with previous findings 13 (Supplementary Fig. 3).
In support of a functional implication of NBS1 in rRNA silencing in response to DNA damage, we observed a focal pattern of GFP-tagged NBS1 in the nucleus that increased after laser micro-irradiation (Fig. 1b). These distinct NBS1-GFP foci colocalized with nucleolar Fibrillarin, suggesting bona fide nucleolar relocalization (Supplementary Fig. 1b). Interestingly, GFP-tagged MRE11 did not show this intercompartmental dynamics in response to DNA damage (Supplementary Fig. 1d). We observed the same focal pattern of endogenous NBS1 in the nucleoli 10 min. after 5 Gy of IR, although the endogenous nucleolar NBS1 foci were smaller than the NBS1-GFP-decorated structures. Again, no signal associated with endogenous MRE11could be detected, thus suggesting an MRE11-independent function of NBS1 (Fig. 1c). Consistently, we also found that rRNA silencing in response to DNA damage was NBS1-dependent but MRE11-independent (Supplementary Fig. 2b).
We then combined laser micro-irradiation with real-time imaging (reviewed in 14) and observed a rapid accumulation of NBS1 around the DSB-containing subnuclear tracks and with a small temporal delay, the focal NBS1-GFP pattern appearing in the nucleoli (Supplementary Fig. 1c). While NBS1-GFP stably associated at sites of laser damage for the entire duration of the experiment (70 min; see Fig. 1d) the nucleolar NBS1-GFP signal reached a maximum at around 15 min post irradiation and gradually decreased, reaching background levels at around 40 min. The kinetics of NBS1 recruitment into the nucleoli perfectly matched the kinetics of rRNA silencing in response to DNA damage (compare Fig. 1d to e), thus suggesting that these two events are functionally linked. In summary, these data reveal that DNA damage-induced in-trans silencing of Pol I transcription is associated with relocalization of a fraction of NBS1 into the nucleoli.
NBS1 is enriched at sites of rRNA transcription in response to DNA damage
Immunofluorescence analysis in laser micro-irradiated cells showed that NBS1-GFP nucleolar foci co-localized with Pol I in the dense fibrillar components of the nucleoli where active transcription of rDNA takes place (Fig. 2a). To directly show NBS1 enrichment within transcribed regions of the nucleoli we used chromatin immunoprecipitation (ChIP) followed by quantitative real time PCR (qRT-PCR) in NBS1-GFP expressing cell lines. The primer pairs for the qRT-PCR included a promoter region 5’ of the transcription start site (H0), a segment inside the coding region of the primary transcript (H1) and a region within the intergenic spacer (H18) of rDNA. A primer pair from the GAPDH gene served as a negative control (see Fig. 2b). ChIP data showed that similar to Pol I, NBS1-GFP is present at the rDNA promoter (H0) and more abundant within the coding region (H1) (Fig. 2c,d).
Figure 2. NBS1 is enriched at sites of rRNA transcription in response to DNA damage.
(a) U2OS cells stably transfected with NBS1-GFP were exposed to laser micro-irradiation. Cells were immunostained with antibodies against Pol I. Quantification of signal intensities was done in ImageJ by applying a single ROI to multiple color channels in the same image and extracting the plot profile (2 independent experiments, one representative image shown) Scale bars: 10 µm. (b) Schematic overview of part of the rDNA repeat with the transcribed region and the first part of the intergenic region. Primer pairs used in the ChIP experiment in (c) and (d) are indicated at the promoter (H0), within the transcriped region (H1) and in the intergenic region (H18). (c) ChIP experiment with antibodies against mouse IgG or Pol I in U2OS cells. GAPDH was used as a nonspecific target. Error bars represent S.E.M (n = 3 independent experiments). (d) ChIP experiment with antibodies against mouse IgG or NBS1 in stably transfected U2OS NBS1-GFP cells. GAPDH was used as a nonspecific target. Error bars represent S.E.M. (n = 4 independent experiments).
Moreover, Pol I localization at the transcriptional start site and within the coding region was not dramatically affected upon IR treatment (Fig. 2c), while NBS1-GFP association with transcribed regions of the rDNA increased after IR, thus suggesting a DNA damage-induced recruitment of NBS1 into actively transcribed regions within the nucleoli (Fig. 2d).
The NBS1 FHA domain mediates nucleolar accumulation of NBS1
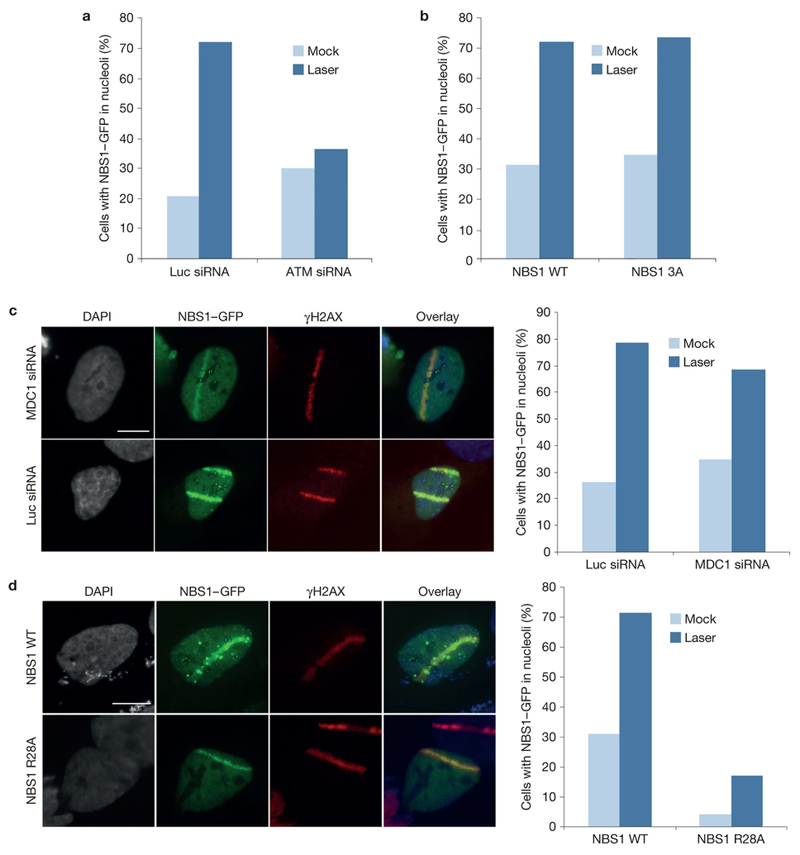
Consistent with Pol I silencing in response to DNA damage being ATM-dependent (Supplementary Fig. 3a; 13), we observed a significant reduction of the percentage of laser micro-irradiated cells with nucleolar NBS1 upon depletion of ATM by RNA interference (Fig. 3a). NBS1 itself is an ATM target but a mutant derivative of NBS1 lacking the three ATM phosphorylation sites (3A mutant) was still efficiently forming nucleolar foci upon laser micro-irradiation, indicating that NBS1 phosphorylation by ATM is not required for its nucleolar translocation (Fig. 3b).
Figure 3. The NBS1 FHA domain mediates ATM-dependent nucleolar accumulation of NBS1.
(a) Quantification of the percentage of cells with NBS1-GFP positive nucleoli after laser micro-irradiation. Cells were treated with control or ATM siRNA for 72 h (2 independent experiments; one representative exp. shown; n = number of cells analyzed; Mock: n = 184, 73; Laser: n = 115, 60). (b) Cells were transiently transfected with NBS1-GFP and NBS1-3A-GFP and NBS1-GFP positive nucleoli were quantified after laser micro-irradiation (2 independent experiments; one representative exp. shown; n = number of cells analyzed; Mock: n = 13, 48; Laser: n = 7, 44). (c) As (a) but cells were transfection with siRNA against Luciferase (siLuc) or MDC1 (siMDC1). (2 independent experiments; one representative experiment shown; n = number of cells analyzed; Mock: n = 118, 103; Laser: n = 214, 121). (d) as (c) but cells were transiently transfected with NBS1-GFP or NBS1-R28A-GFP (2 independent experiments; one representative experiment shown; n = number of cells analyzed; Mock: n = 13, 48; Laser: n = 7, 35). Scale bars in all panels: 10 µm.
NBS1 recruitment into γH2AX-marked chromatin regions at sites of DSBs is mediated by MDC1. However, MDC1 depletion by siRNA did not provoke a reduction in nucleolar NBS1-GFP accumulation, even though NBS1 recruitment to sites of DSBs was significantly reduced under these conditions (Fig. 3c). Interestingly though, a mutation in the N-terminal FHA-BRCT region of NBS1 that abrogates its ability to interact with phosphorylated proteins (R28A10), led to a complete loss of nucleolar residence and DNA damage-induced nucleolar enrichment of NBS1-GFP (Fig. 3d). These data strongly suggest that an as yet unidentified phosphorylation-dependent interaction partner of NBS1 that is distinct from MDC1 mediates its nucleolar recruitment in response to DNA damage.
Treacle is a NBS1 FHA-BRCT-interacting protein
In order to identify NBS1 FHA-BRCT-interacting proteins that may mediate trans-compartmental enrichment of NBS1 in the nucleoli we performed an unbiased proteomic screen (see Methods for details). The first N-terminal 382 amino acids of human NBS1 were used as bait and were C-terminally modified with two affinity tags (Strep, HA; see Fig. 4a). One construct was wild type, the other carried the R28A point mutation in the FHA domain, which completely abrogated nucleolar residence of NBS1 (see above). Protein complexes were isolated from untreated cells and from cells treated with 10 Gy of IR by double affinity purification. Retained proteins were then identified by mass spectrometry.
Figure 4. TCOF1-Treacle is an interaction partner of the NBS1 FHA-BRCT region.
(a) Overview of the screening procedure used to identify interaction partners of the NBS1 FHA-BRCT-domain. (b) A selection of interaction partners of NBS1 FHA-BRCT identified in the screen. (c) Protein pull down from Hela nuclear extracts with GST-tagged BRCT (MDC1), N-terminal (1-382) NBS1 WT and an FHA-BRCT-domain mutant (R28A, K160M=DM; (2 independent experiments; one representative image shown). (d) Co-immunoprecipitation of ectopically expressed Strep-tagged Treacle and endogenous NBS1 in 293T cells (2 independent experiments; one representative image shown). (e) Co-immunoprecipitation of ectopically expressed N-terminal fragments of HA-flag-NBS1 and endogenous Treacle in 293T cells. Mutations were introduced to abrogate the FHA- (R28A), BRCT- (K160M) and FHA-BRCT-domain (DM) (2 independent experiments; one representative image shown). (f) Co-immunoprecipitation of endogenous Treacle and endogenous NBS1 +/- 10 Gy of IR in 293T cells (2 independent experiments; one representative image shown).
As expected, unique NBS1 bait peptides were identified in all four purifications (Fig. 4b). MDC1 was only co-purified with wild type NBS1 FHA-BRCT and the interaction was not altered upon IR in agreement with published results. 6,7,15,16. Interestingly, several nucleolar proteins co-purified with the NBS1 FHA-BRCT region. Most pronounced according to the number of unique peptides was TCOF1 (Fig. 4b). The TCOF1 gene encodes a low complexity nucleolar phosphoprotein often referred to as Treacle. TCOF1 is mutated in Treacher-Collins syndrome (TCS), one of the most severe autosomal dominant congenital disorders of craniofacial development. Functionally, TCOF1-Treacle has been implicated in the spatiotemporal control of ribosome biosynthesis, most notably in rRNA transcription and pre-ribosomal RNA processing (reviewed in 17).
We first confirmed the interaction between NBS1 and Treacle biochemically by GST pull down experiments with bacterially purified NBS1 N-terminal fragments. Wild type NBS1-GST (amino acids 1-382) but not an NBS1 FHA-BRCT double mutant (DM: R28A, K160M) pulled down detectable amounts of Treacle from the extract, indicating that interaction of the NBS1 FHA-BRCT region with Treacle requires intact FHA and BRCT phosphopeptide-binding functions (Fig. 4c). Co-immunoprecipitation experiments with over expressed tagged proteins showed that full-length Strep-tagged Treacle could retrieve endogenous NBS1 from cell extracts (Fig. 4d). Moreover, a wild type Flag-HA-tagged N-terminal NBS1 fragment efficiently co-immunoprecipitated with endogenous Treacle, while the FHA mutated fragment (R28A) and the double-mutant fragment (DM: R28A, K160M) did not. The BRCT mutated fragment (K160M) showed residual binding activity, albeit significantly weaker than the wild type (Fig. 4e). This indicates that the NBS1 FHA domain is the predominant interaction partner of Treacle in vitro and that the NBS1 BRCT domains also partially contribute to the interaction.
Endogenous Treacle also co-immunoprecipitated with endogenous NBS1 (Fig. 4f) and, as already shown in the mass spectrometry analysis, Treacle association with NBS1 was not dependent on DNA damage (Fig. 4e,f).
NBS1 binds to conserved phosphorylated SDT-like motifs in Treacle
Treacle consists of three structurally distinct regions: an N-terminal region, a central region consisting of 10 consecutive acidic and Ser-rich sequence stretches and a C-terminal region containing nuclear and nucleolar localization signals 18 (Fig. 5a). Treacle interacts with CK2 19 and its primary sequence contains numerous potential CK2 phosphorylation sites. We found that bacterially purified Treacle fragments are heavily phosphorylated by CK2 in vitro, especially within the repeat region (Supplementary Fig. 4). In order to map the NBS1 interaction site we systematically deleted the individual domains of Treacle in order to assess their impact on the interaction with NBS1 (Fig. 5b). These experiments suggested that the major NBS1 interaction site is located within the N-terminal 225 amino acids of Treacle with a potential minor contribution of the C-terminal region.
Figure 5. Conserved acidic SDT-like phospho-motifs in Treacle mediate the binding to NBS1.
(a) Schematic overview of the protein structure of Treacle. Treacle contains a low complexity N-terminal region, 10 central repeats as well as a low complexity C-terminal region. (b) Five HA-tagged deletion constructs of Treacle, lacking either the N-terminal region (F1), both the N-terminal and the central repeat region (F2), the central repeat region (F3), both the central repeat and the C-terminal region (F4) and the C-terminal region, respectively (F5) were used in co-immunoprecipitation experiments with HA affinity resin after transfection of 293T cells (2 independent experiments; one representative image shown). (c) GST-pulldown with purified GST-Treacle T-2 fragment (amino acids 109 – 330). GST-MDC1 F3 fragment 6 was used as a positive control. Note the band shift induced by CK2 phosphorylation. (d) Alignment of potential NBS1 binding motifs located in the N-terminus of Treacle. (e) ITC data for Treacle SDET, SETE, and SDSE phosphopeptides (2 independent experiments, one representative experiment shown).
A purified GST fragment of this region was efficiently phosphorylated by CK2 in vitro (Supplementary Fig. 4; T-2) and this fragment pulled down the MRN complex from Hela nuclear extracts only when previously phosphorylated by CK2 (Fig. 5c). Notably, this N-terminal region of Treacle features three motifs that are highly reminiscent of the previously described MDC1 SDT motifs 6–9 and form CK2 consensus sites (Fig. 5d). The first motif consists of a Ser residue followed by a Glu and Thr, flanked by three consecutive Glu residues (SETE amino acids 171-174). Further downstream two additional sequence stretches show similarity to the MDC1 SDT motifs: a Ser residue followed by two acidic amino acids and by a Thr residue (SEDT, amino acid 200-203 and SDET, amino acid 207-210, respectively).
To fine-map the NBS1 interaction site in Treacle we assessed the binding affinity of phosphopeptides derived from this region by isothermal titration calorimetry (ITC) (Fig. 5e). The SDET motif around Thr 210 showed the highest binding affinity with a Kd around 1 µM.
Phosphorylated Thr 210 in Treacle is the major interaction site for NBS1
We next mutated Thr 210 to Ala in the context of full-length HA-tagged Treacle and analyzed its binding to Flag-tagged N-terminal NBS1 in 293T cell extracts by Flag immunoprecipitation (Fig. 6a). The T210A mutant was completely deficient for NBS1 interaction and we did not observe a further decrease by additional Ser-Thr substitutions. Mutation of the SETE motif led to a significant reduction in NBS1 binding but did not abrogate it (Fig. 6b), thus suggesting that sequence around Thr 210 is the predominant NBS1-interaction site in Treacle, with a minor contribution of the region around Thr 173.
Figure 6. Phosphorylated Thr 210 in Treacle is the major interaction site for NBS1.
(a) Co-immunoprecipitation of Flag-tagged N-terminal fragments of NBS1 (amino acids 1-382) with HA-tagged Treacle in 293T cells. Binding affinities of wt Treacle, T210A and STTT (S171A, T173A, T203A, T210A) were examined (2 independent experiments; one representative image shown). (b) Mutation of the SETE motif in Treacle leads to reduced NBS1 binding. Co-immunoprecipitation of N-term fragments of NBS1 (amino acids 1-382) with HA-tagged Treacle in 293T cells. Binding affinity of wt Treacle and S171A, T173A Treacle was examined. R28A, K160M (DM) NBS1 was used as a positive control (2 independent experiments; one representative image shown). (c) Schematic representation of the mass spectrometry approach used to identify phosphorylated Thr 210 in vivo (d) Tandem mass spectrum (MS-MS) of the Thr210 phosphorylated peptide sequence DEpTDVEGKPSVK from Treacle. The y and b series indicate fragments at amide bonds of the peptide, unambiguously identifying the phosphorylated Thr210. (e) Isotope cluster of the modified peptide sequence DEpTDVEGKPSVK from Treacle. Using high-resolution mass spectrometry ensures that the phosphorylated peptide is identified with parts permillion (ppm) accuracy (TPM = theoretical peptide mass; MPM = measured peptide mass; MD = mass deviation).
Next we sought to confirm that Thr 210 is phosphorylated in vivo. Proteins extracted from GFP-Treacle-expressing 293T cells were sequentially digested with Trypsin and Asp-N, followed by an enrichment of CK2 phosphopeptides through an affinity resin (Fig. 6c; see Methods for detail). This approach led to the unambiguous identification of a short peptide sequence harboring Thr 210 of Treacle (Fig. 6d), and the mass of this peptide reveals Thr 210 as being phosphorylated (Fig. 6e). Collectively, these data strongly indicate that Thr 210 is indeed phosphorylated by CK2 in vivo, and that this modification constitutes the major interaction site for NBS1.
Treacle mediates NBS1 nucleolar translocation in response to DNA damage and a small fraction of it is transiently recruited to sites of DNA damage
Treacle has originally been described as a nucleolar protein that interacts with upstream binding factor (UBF), an RNA Pol I transcription factor. Indeed, we confirmed nucleolar localization of Treacle and co-localization with UBF by immunofluorescence (Supplementary Fig. 5a). Treacle staining is most pronounced in the dense fibrillar components of the nucleolus where the rDNA transcription takes place, and Treacle and Fibrillarin foci co-localize (Fig. 7a 19).
Figure 7. Treacle mediates NBS1 nucleolar translocation in response to DNA damage and a fraction is transiently recruited to sites of DNA damage.
(a) U2OS cells were immunostained with antibodies against Treacle and Fibrillarin. Quantification of signal intensities using ImageJ plot profile (2 independent experiments; one representative image shown). (b) U2OS cells stably transfected with NBS1-GFP were exposed to laser micro-irradiation. Cells were immunostained with antibodies against Treacle. Quantification of signal intensities using ImageJ plot profile (2 independent experiments; one representative image shown). (c) U2OS cells stably transfected with NBS1-GFP were treated with Treacle siRNAs for 72 h and exposed to laser micro-irradiation. Cells were immunostained with antibodies against γH2AX. A quantification of the percentage of cells with NBS1-GFP positive nucleoli is displayed in the right hand panel (2 independent experiments; one representative image shown, >90 cells irradiated per condition). (d) U2OS cells stably transfected with GFP-Treacle were exposed to laser micro-irradiation (2 independent experiments; one representative image shown). (e) Hela cells were exposed to laser micro-irradiation and cells were immunostained with antibodies against Treacle and γH2AX (2 independent experiments; one representative image shown). (f) U2OS cells stably transfected with GFP-Treacle-c were exposed to laser micro-irradiation (2 independent experiments; one representative image shown). (g) U2OS cells stably expressing GFP-Treacle-c were pre-treated with ATMi (KU55933) and PARPi (ABT888), respectively and exposed to laser micro-irradiation. Cells with increased GFP-Treacle-c signal within the irradiated compartment were quantified (2 independent experiments; both results displayed in graph; n = number of cells analyzed: Mock: n = 39, 31; ATMi: n = 45, 36; PARPi: n = 45, 40). Scale bars in all panels: 10 µm.
Interestingly, upon induction of DSBs by laser micro-irradiation, NBS1-GFP nucleolar foci co-localized with Treacle, indicating that they occupied the same fibrillar structures after DNA damage (Fig. 7b). ChIP experiments in stable GFP-Treacle-expressing cell lines confirmed that similar to Pol I, Treacle mainly occupies the promoter and coding regions of the rRNA gene arrays (Supplementary Fig. 5b). Significantly, depletion of Treacle by siRNA led to a complete loss of NBS1-GFP nucleolar localization and accumulation in response to DNA damage, thus revealing that Treacle acts as a mediator for DNA damage-induced recruitment of NBS1 in the nucleoli (Fig. 7c).
Based on sequence similarity of Treacle with Nopp140, a trafficking nucleolar phosphoprotein that shuttles between the nucleolus, the cytoplasm and Cajal bodies 20, it was speculated that Treacle might also be trafficking to other cellular compartments 19. Indeed, fluorescence recovery after photobleaching (FRAP) experiments in cells expressing a GFP-tagged version of the full-length Treacle cDNA revealed that GFP-Treacle is a mobile protein that shuttles between the nucleoplasm and the nucleoli (Supplementary Fig. 5c). We next sought to determine if nucleoplasmic Treacle was recruited to sites of DNA damage. Laser micro-irradiation followed by fluorescence microscopy revealed weak accumulation of GFP-Treacle and endogenous Treacle within the irradiated subnuclear volume (Fig. 7d and e).
To more robustly demonstrate that a subset of Treacle is transiently recruited to sites of DNA damage we exploited a transcript variant of Treacle (so-called isoform c). This variant uses an alternate terminal exon and lacks ten 3’ exons. The resulting isoform has a significantly shorter and distinct C-terminus as compared to the canonical transcript variant and lacks several putative nuclear and nucleolar localization signals (Supplementary Fig. 6a). This Treacle variant exclusively localized to the nucleoplasm and was efficiently recruited to microlaser-induced DNA breaks, likely due to the lack of competition with the strong nucleolar binding (Fig. 7f). We next sought to determine the requirements for the mobilization of Treacle to DNA lesions. We first tested if NBS1 is needed for Treacle recruitment to sites of DSBs. Surprisingly GFP-Treacle-c was still efficiently recruited to sites of laser-induced DNA damage in NBS1-depleted cells (Supplementary Fig. 6b). However, chemical inhibition of PARP activity significantly reduced Treacle association with DSB sites while an ATM inhibitor did not yield such an effect (Fig. 7g). In addition, PARP inhibition also led to a small but reproducible decrease in the number of cells with NBS1-GFP positive nucleoli (Supplementary Fig. 6c), indicating that Treacle recruitment to sites of DNA damage might be involved in mediating efficient NBS1 nucleolar accumulation.
Together, these data strongly indicate that Treacle mediates DNA damage-induced recruitment of NBS1 in the nucleoli. Moreover, these data also show that the nucleoplasmic pool of Treacle is transiently recruited to sites of DNA damage in a manner that is partly dependent on PARP activity but is independent of ATM activity.
Treacle-dependent NBS1 recruitment into the nucleoli mediates rRNA silencing in response to DNA damage
Having established that in response to DNA damage, Treacle mediates NBS1 recruitment into the nucleoli, we next sought to determine if Treacle is involved in the trans-signaling process that leads to downregulation of rRNA transcription (see above). Treacle itself is an essential factor for efficient rRNA transcription and its inactivation by mutation or its downregulation by RNA interference leads to reduced rates of rRNA synthesis 21,22. Therefore, we carefully compared the reduction in rRNA transcription in response to DNA damage to the reduction in rRNA synthesis upon Treacle depletion. This analysis revealed that rRNA synthesis drops by about 40% in response to DNA damage and upon depletion of Treacle, respectively. However, there was no further reduction of rRNA transcription in response to DNA damage in the absence of Treacle (Fig. 8a). This lack of an additive effect was not due to an exhaustion of rRNA silencing, since rRNA transcription can be reduced further by treatment with Actinomycin D (Fig. 8b). Therefore, these data indicate that the inhibition of rRNA transcription that occurs after Treacle depletion and exposure to DNA damage is conveyed through the same pathway.
Figure 8. Treacle-dependent NBS1 recruitment into the nucleoli mediates rRNA silencing in response to DNA damage.
(a) rRNA transcription was measured by EU labeling after transfection of cells with Treacle shRNA and in the presence of 2 Gy of IR. Error bars represent S.E.M. (3 independent experiments; n represents number of cells analyzed: Con: n =110, 100, 90; shTreacle: n =127, 197, 91; 2 Gy: n = 55, 89, 73; 2 Gy + shTreacle: n = 256, 48, 71). (b) rRNA transcription was measured by EU labeling in the presence of Actinomycin D. Error bars represent S.E.M. (4 independent experiments; n represents number of cells analyzed: Mock: n = 4252, 3848, 3310, 4865; ActD: n = 4113, 3936, 4314, 3975). (c) rRNA transcription was measured by EU labeling after transfection of cells with NBS1-GFP-NOLS. Quantification of signal intensities using ImageJ plot profile (2 independent experiments; one representative image shown). Scale bar: 10 µm. (d) Model of Treacle-NBS1-mediated trans-compartmental communication between sites of DNA breaks and the nucleoli.
However, these data do not provide direct evidence that NBS1 accumulation in the nucleoli is negatively regulating rRNA synthesis in response to DNA damage. Thus, in order to strengthen this conclusion, we fused a nucleolar localization signal (NOLS) to our NBS1-GFP construct and expressed it in Hela cells. Under these conditions recombinant NBS1-GFP-NOLS accumulated in the nucleoli in a subset of cells where it formed a similar focal pattern as observed after DNA damage. Significantly, EU labeling in those cells revealed that accumulation of NBS1 in the nucleoli correlates with low levels of rRNA transcription even in the absence of DNA damage (Fig. 8c). These data support the conclusion that Treacle-mediated NBS1 accumulation in the nucleoli regulates rRNA silencing in response to DNA damage.
Discussion
In this study we identified a signaling pathway that globally inhibits rRNA transcription in response to DSBs via an in-trans mechanism and we present compelling evidence that this Pol I silencing program is associated with rapid and transient recruitment of NBS1 into the nucleoli; a process that depends on the direct interaction between the NBS1 N-terminal FHA-BRCT region and the nucleolar phosphoprotein Treacle. This signaling mode is distinct from the previously reported DSB-induced transcriptional silencing that occurred in the vicinity of the DNA lesions and did not require long-distance signal transduction mechanisms 12,13,23. Specifically, we show that DNA damage induction in randomly chosen nuclear compartments is accompanied by a global rRNA silencing response, which demands trans-compartmental communication between sites of DSBs and the transcription machinery within the nucleoli. Mechanistically, we provide evidence that this trans-compartmental communication is regulated by a genome surveillance complex, consisting of the DDR adaptor protein NBS1 and a nucleolar regulator Treacle (Fig. 8d).
Our findings indicate that the communication between the repair reactions locally at damaged chromosomes and other vital chromatin transaction elsewhere in the nucleus are more widespread than previously thought, and the example of the NBS1-Treacle dynamics might pave the way to dissect the physiological relevance of such intranuclear communication. Our previous work and this study show that nuclear dynamics of NBS1 is coordinated by two distinct ‘adaptor’ proteins, MDC1 and Treacle, respectively. Intriguingly, NBS1 interacts with these adaptors by the same mechanism based on direct interaction of its N-terminal FHA-BRCT region with repeated acidic CK2 phosphorylation sites present both in MDC1 and Treacle. Moreover, the thermodynamic parameters of the association between the NBS1 N-terminal FHA-BRCT region with phosphopeptides derived from the respective interaction sites in MDC1 and Treacle are virtually identical. Thus, it is likely that any given interaction of NBS1 molecules with either MDC1 or Treacle is mutually exclusive and that the relative proportion of MDC1-NBS1 versus Treacle-NBSl complexes only depends on the relative concentration of MDC1 and Treacle in the nucleoplasm. Since the steady state levels of Treacle in the nucleoplasm are very low, we can assume that the bulk of NBS1 associates with MDC1. Nevertheless, as we report in this study, a small fraction of a constitutive NBS1-Treacle complex exists, and can be mobilized after DNA damage.
Mechanistically, the local increase in NBS1 concentration in the nucleoli upon induction of DNA damage is not yet clear. It is possible that posttranslational modifications of Treacle in response to DNA damage may lead to a conformational change that may favor Treacle-NBS1 association or facilitate release of the NBS1-Treacle complex from DSBs. Consistent with this idea, Treacle has been identified as an ATM target 24 and we could confirm these results (see Supplementary Fig. 7).
How does Treacle-dependent NBS1 mobilization in the nucleoli mediate rRNA silencing in response to DNA damage? One simple mechanism may be that NBS1 mediates its effect on rRNA transcription indirectly through Treacle. When in a complex with NBS1, Treacle may simply not be able anymore to promote efficient rRNA transcription.
Both TCS and NBS are associated with craniofacial dysmorphism but only NBS is associated with microcephaly and mental retardation 4. This raises the question if the defect in craniofacial development in TCS and NBS are the consequence of a common functional role of the two affected genes, TCOF1 and NBS1, respectively. However, while the dysmorphic head features of NBS patients derive from a primary developmental defect in the formation of the brain, the cranio-facial abnormalities in TCS patients are likely the result of increased apoptosis in the neuronal crest stem cell population early during development 25. In addition, the recent finding that TCS is also caused by mutations in Pol I and Pol III subunits strongly supports the hypothesis that it is primarily a ribosomopathy 26.
What are the benefits that cells could derive from a global rRNA silencing program triggered by the presence of DSBs? One intriguing possibility is provided by recent observations in yeast, where high rRNA transcription rates are associated with DNA repair defects and genome instability 27. These observations suggest that in response to DNA damage, transient silencing of transcriptional activity within the rDNA repeats may be important to maintain genome integrity.
Methods
Cell culture
U2OS, HeLa and 293T cells were grown in DME containing 10 % fetal bovine serum (Invitrogen), 100 U penicillin, and 100 µg/ml streptomycin. Where indicated, the culture medium was supplied with 50 ng/ml Actinomycin D, 10 µM ATM inhibitor (KU55933) or Veliparib (ABT888) for 1h. U2OS derivative cell lines expressing GFP-Treacle protein in a doxycycline-responsive manner were isolated by cotransfecting U2OS cells with pcDNA6/TR (Invitrogen) and pcDNA4-TO-GFP-Treacle constructs and selecting stably transfected cells with 400 µg/ml Zeocin and 5 µg/ml Blasticidin S (Invitrogen). U2OS GFP-MRE11 and U2OS GFP-Treacle-c stably expressing cells were selected with 400 µg/ml G418. The U2OS-NBS1-GFP2 cell line was previously described 16.
Plasmids and transfections
Transient transfections of Hela and U2OS cells were done using Lipofectamine LTX with plus reagent (Invitrogen) according to manufacturer’s specifications. Transient transfection of 293T cells was done using calcium phosphate. Full length Treacle was synthesized in pcDNA4-TO-strep-HA (Genscript USA inc.) and GFP was inserted to generate pcDNA4-TO-strep-HA-GFP-Treacle. NBS1-GFP2, NBS1-R28A-2GFP and NBS1-3A-2GFP (S278, S343 and S397) were described 16,28. NBS1-GFP-NOLS and GFP-NOLS were generated by digestion of NBS1-2GFP with BsrGI, NotI to remove one GFP and insert the NOLS (QDLWQWRKSL) sequence c-terminally. LpEXPR-IBA103-Nterm-NBS1-NLS-FLAG-HA-Strep-WT, R28A, K160M or DM (R28A and K160M) was generated by insertion of N-term-NBS1 (1-380)-NLS into pEXPR-IBA103-oligo. GFP-Treacle-C construct was generated by insertion of the TCOF1 isoform C into pAcGFP-C1. Treacle GST-fragments were generated by PCR amplification and subcloning into pGEX-4T3. Strep-HA-GFP-Treacle fragments were generated by PCR amplification and religation. Strep-HA-tagged NBS1 for co-purification and mass spectrometry was generated by insertion of an N-terminal NBS1 WT/R28A fragment (1-380) into pTGSH with a C-terminal strep-HA fusion.
RNA interference and site directed mutagenesis
The siRNA oligonucleotides were obtained from Microsynth AG. For annotations and sequences see Supplementary Table 1. siRNA oligonucleotides targeting MRE11 were from Ambion (MRE11 siRNA-59: AMBION 4427038, siRNA ID#:8959; MRE11 siRNA-60, AMBION 4427038, siRNA ID#:8960). The siRNA transfections were performed with 50 or 100 nM siRNA duplexes using Lipofectamine RNAiMAX (Invitrogen). Samples were harvested 72 h after initiation of transfection unless stated otherwise. For mutagenesis primer sequences see Supplementary Table 1. shRNA was generated by insertion of an oligo (see Supplementary Table 1) into the pSUPERIOR.puro vector (Oligoengine). Cells were transfected with Lipofectamine LTX with plus reagent (Invitrogen) according to manufacturer’s specifications and harvested 4-5 days post transfection.
Generation of DNA damage
X-ray irradiation was done with a XYLON.SMART 160E-1.5 device (150kV, 6 mA; YXLON International A/S, Taastrup, Denmark) delivering 11.8 mGy per second. Soft X-rays were largely filtered out with a 3 mm aluminum filter.
5-EU incorporation
In situ detection of nascent rRNA was done with the Click-iT RNA Alexa Fluor 594, 488 Imaging Kit (Invitrogen, Molecular Probes). Briefly, cells were incubated for 20 min in the presence of 5-EU starting at the time of irradiation. Samples were fixed in 4% paraformaldehyde at room temperature for 12 min and permeabilized in 0.25% Triton-X100 for 5 min at room temperature. Samples were then processed according to manufacturer’s recommendation. For high content ScanR analysis a fibrillarin staining was used to create a nucleolar mask for quantification of rRNA transcription. In general, the mean intensity of the 5-EU signal was measured within the nucleolar mask. In the case of ActD treated cells the total intensity of the 5-EU signal was measured in the nucleus.
Microscopy and image analysis
Quantitative image analysis for measurement of fluorescence intensities was done as described previously 29. The images were obtained with a 20x 0,75 NA (UPLSAPO20x) dry objective, a quadruple-band filter set for Dapi, FITC, Cy3 and Cy5 fluorescent dyes, a MT20 Illumination system and a digital monochrome Hamamatsu C9100 EM-CCD camera. Camera resolution is 200nM x 200nM per pixel (binning 1, 40x). Image analysis was performed with Olympus propriety ScanR automated image and data analysis software using standard algorithms for detection of nuclei and sub-objects within nuclei. Typically, 49 images (corresponding to 2000-4000 sub-objects) were acquired under non-saturating conditions for each data point allowing robust measurements of experimental parameters such as intensities.
Laser micro irradiation experiments were performed as described previously 28 with a PALM micro beam equipped with a 355 nm UV-A laser (Zeiss). Confocal images were obtained on a LSM780 (Zeiss) using a Pln Apo 40x 1.4 Oil DICII objective. All images were taken under non-saturated conditions and images to be compared were acquired with the same settings. Non-confocal images were acquired on a Leica DMI 6000B equipped with a Leica DFC365FX camera using the HCX PL APO 63x1.4 oil. Scoring of cells with NBS1-GFP2 positive nucleoli and GFP-Treacle or GFP-Treacle-c laser stripes was done manually. Cells would be scored as positive or negative based on visual presence of GFP signal within nucleoli/laser tracks. Intensity plots for co-localization were generated with the Plot Profile function in ImageJ. For co-localization one ROI was applied to individual channels of the same image and intensities were measured. Time-lapse microscopy to follow accumulation of Nbs1-GFP2 within laser tracks and in the nucleoli was done using a Axiocam MRm (Zeiss) and Axio Vision software. Images were obtained with a LD Plan Neofluar 40x/0.6 corrM27 dry objective. Quantitative analysis of timelapse images was done in imageJ where time-lapse images were converted into stacks. Stacks were aligned using the StackReg plugin. After alignment a single ROI was applied to the stack and the average intensity or standard deviation was measure for the NBS1-GFP2 within laser tracks or in the nucleoli respectively.
FRAP experiments were performed on a UltraView Vox live cell imaging system (Perkin Elmer) mounted on a Nikon Ti-E inverted microscope and integrated with a Photokinesis unit for photo-bleaching experiments. Cells were grown in glass-bottom Labtek dishes (Nunc, 155361) and kept under physiologic conditions for the duration of the experiment (37°C incubator, custom-made CO2-independent medium, covered by a layer of mineral oil to prevent evaporation). Images were acquired using a Nikon 60x/1.4 APO oil objective, filter sets for GFP, and a Hamamatsu C9100-50 EMCCD camera using propriety Volocity software (Perkin Elmer). GFP-Treacle protein was bleached by a 488 nm laser (100% output, 4 iterations). The bleached region was designed to bleach one nucleolus. Fluorescence recovery in the bleached region was monitored by acquiring 10 images prior to bleaching, 10 frames/sec for the first 3 sec followed by 30 frames/min for 15 minutes An unbleached region was measured in order to allow background subtraction. The average of signal intensities prior to bleaching was set to 1 and the background level to 0 and intermediate values were transformed accordingly.
Real time PCR
RNA was isolated with RNeasy, Quiagen. cDNA synthesis was based on 1 µg RNA using M-MLV Reverse Transcriptase (Promega, M170A) according to Manufacturer’s recommendation with random hexamer primers. Real-time PCR was performed using 10 ng cDNA with Roche SYBR Green I Master (Cat. 04707516001). Samples were analyzed on the LightCycler480 and quantified with LightCycler480 quantification software. For primer sequences see Supplementary Table 1.
Chromatin immunprecipitation
Cells were fixed with 1% formaldehyde for 10 min and the reaction was stopped by addition of 1.25 M glycine. Cells were washed in cold Pbs and suspended in buffer A (100mM TRis-HCL pH8, 10 mM DTT), followed by 15 min incubation on ice and 15 min incubation at 37° C. Cells were pelleted and resuspended in Buffer B (10 mM EDTA, 10 mM EGTA, 10 mM Hepes pH8, 0.25% TritonX-100). Again, cells were pelleted and resuspended in Buffer C (10 mM EDTA, 0.5 mM EGTA, 10 mM Hepes pH8, 200mM NaCL). Finally cells were pelleted and resuspended in lysis Buffer D (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.1). After sonication to yield DNA fragments of 0.5–1 kb, 100 ug chromatin was diluted tenfold with IP buffer (16.7 mM Tris-HCl pH 8.1, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X-100). Proteins of interest were immunoprecipitated with Salmon sperm DNA/Protein A/G Agarose (Millepore, 16-157/16-201) overnight with the respective antibodies. Beads were washed extensively with washing buffer 1A (10 mM Tris-HCL pH8, 150 mM NaCl, 0.1% SDS, 1% TritonX-100), washing buffer 1B (10 mM Tris-HCL pH8, 500 mM NaCl, 0.1% SDS, 1% TritonX-100) and washing buffer 2 (0.25 M LiCl, 0.5% NP-40, 0.5% Sodiumdeoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8) before elution in 100 µl Elution buffer (1% SDS,100 mM NaHCO3). Proteinase K was added to each sample and reversion of crosslinks was done by heating for 1 hour at 37° C followed by over night incubation at 65 °C. QIAquick PCR purification kit (Qiagen) was used to purify eluted DNA. 7% of purified DNA was amplified by 40 cycles using a LightCycler480 (Roche) with the SYBR Green detection system. A standard curve was included for each primer pair in order to compare the abundance at different regions of the rDNA repeat.
Immunological techniques
Total cell lysates were prepared in Laemmli sample buffer (50 mM Tris, pH 6.8, 100 mM DTT, 2% SDS, 0.1% bromophenol blue, and 10% glycerol). Alternatively, cell extracts were prepared by the following buffer (50 mM Tris, pH 7.5, 120 mM NaCl, 0.5% NP-40, and 1 mM EDTA). Extracts were supplemented with 2 mM MgCl2 and benzonase and incubated for 30 min at 4°C. Lysates were cleared by centrifugation at 14000 g for 15 min and protein concentration was measured using Bradford protein assay. Immunoprecipitations were done using Anti-Flag M2 affinity gel (Sigma, A2220), Monoclonal anti-HA agarose (Sigma, A2095), Dynabeads M-280 Streptavidin (Invitrogen, 112.05D) or Treacle antibody (Sigma Life Science, HPA038237), in combination with Protein A agarose.
Other antibodies used in this study were: Mouse γH2AX (Millipore, cloneJBW302, cat. #:05-636), Rabbit RPA194 (Santa Cruz, sc28714), Rabbit NBS1 (Novus Biologicals, NB100-143), Mouse NBS1 (Genetex, Clone 1D7, GTX70224), Rabbit HA (Abcam, ab9110), Rabbit Flag (Sigma, F7425), Mouse Fibrillarin (Abcam, 38F3, ab4566), Rabbit SMC1 (Abcam, ab9262), Rabbit Bid pS61 (Bethyl, A300-527A), Mouse Mcm7 (Santa Cruz, DCS141, sc65469), Mouse UBF (Santa Cruz f-9, sc-13125), Mouse GFP (Roche, 11814460001), Mouse Tubulin (Sigma, DM1A, T6199), Mouse Mre11 (Abcam, 12D7, ab214), Sheep MDC1 3835 (gift from Steve Jackson).
Kinase assays
CK2 phosphorylation assay and pull down were done as previously described 6 Briefly, 100 ng of recombinant CK2a (Milipore) was added to 1 µg of purified GST-fusion protein in CK2 kinase buffer (20 mM MOPS, pH 7.2, 25 mM β-glycerophosphate, 5 mM EGTA, 1 mM sodium orthovanadate, 37.5 mM MgCl2, 1 µM ATP and 10 µCi γ-[32P]ATP and incubated from 10 min at 30°C. In case of GST-pulldown after in vitro phosphorylation, γ-[32P]ATP was excluded. Kinase reactions were inactivated by boiling in SDS sample buffer and were run on SDS polyacrylamide gels.
Isothermal titration calorimetry
The affinity and thermodynamic parameters for NBS1 interactions with synthetic phosphopeptides was determined as previously described 10. Briefly, isothermal titration calorimetry was done with a VP-ITC instrument (MicroCal). NBS1 samples were dialyzed extensively into 50 mM HEPES (pH 7.5), 150 mM NaCl, 2 mM β-mercaptoethanol. Prior to use, all peptides were desalted and buffer exchanged using NAP-5 purification columns (GE Healthcare) into the relevant buffer. Peptides (0.5–1.5 mM) were titrated into 0.05 mM NBS1. Data were analyzed with Origin 7.0 software. The peptide sequences were as follows: SETE: TLVpSEpTEEE; SDET: SSSpSDEpTDVE; SDSE: SEEpSDpSEEE.
Mass spectrometry
Double affinity purification and mass spectrometry analysis for identification of Treacle was done by Dualsystems Biotech AG (CaptiVate Shotgun proteomics service). For enrichment and mass spectrometric analysis of CK2-specific phosphopeptides, 293T cells were transfected with GFP-TCOF1 and harvested in high salt RIPA buffer (50 mM Tris pH 7.5, 400 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.1% Na-deoxycholate, 2 mM Na-orthovanadate, 5 mM NaF, 5 mM Glycero-2-phosphate, protease inhibitors (Roche)). Proteins from cleared lysate were precipitated in acetone, dissolved in urea, reduced with dithiothreitol, alkylated with chloroacetamide 30, and digested using Lys-C and modified sequencing grade trypsin (Sigma) followed by digestion with sequencing grade Asp-N (Promega). Protease digestion was terminated by addition of trifluoroacetic acid and peptides were purified using reversed-phase Sep-Pak C18 cartridges (Waters). CK2-specific phosphopeptides were enriched using the PTMScan® Phospho-CK2 Substrate Motif (S*DXE or T*DXE) Kit (Cell Signaling). Mass spectrometric experiments were performed on a nanoscale UHPLC system (EASY-nLC1000 from Proxeon Biosystems) connected to an Orbitrap Q-Exactive equipped with a nanoelectrospray source (Thermo Fisher Scientific). Each peptide fraction was auto-sampled and separated on a 15 cm analytical column (75 µm ID) in-house packed with 1.9 µm C18 beads using a 2 h gradient ranging from 5% to 40% acetonitrile in 0.5% formic acid at a flow rate of 200 nl/min. The Q Exactive was operated in data-dependent acquisition mode and all samples were analyzed using previously described ‘sensitive’ acquisition method 31. All raw data analysis was performed with MaxQuant software suite 32 version 1.3.0.5 supported by the Andromeda search engine 33.
Supplementary Material
Acknowledgements
We thank Steve Jackson for reagents, Raffaella Santoro for technical support in relation to rRNA transcription, Christoffel Dinant for critical discussion of the FRAP data and members of the Department of Gynecology, the Novo Nordisk Foundation Center for Protein Research and the National Institute of Medical Research for helpful discussions. This work was supported by grants from the Swiss National Foundation (3100A0-111818 and 31003A-144284), Promedica Foundation, Lundbeckfonden (R93-A8863), Novo Nordisk Foundation and by the Kanton of Zürich. S.J.S. is supported by the Medical Research Council UK (U117584228).
Footnotes
Author Contribution
D.H.L. designed and performed most of the experiments, analyzed data and contributed to writing the paper; F.H. designed the mass spectrometry screen that identified TCOF1 and performed some of the biochemical analysis; S.J. and M.L.N. designed and performed most of the mass spectrometry analysis, analyzed data and contributed to writing the paper; J.A.C. and S.J.S. designed and performed the ITC experiments, analyzed data and contributed to writing the paper; M.A. provided feedback in experimental design, performed some of the microscopic analysis and revised the written manuscript; L.I.T. provided feedback in experimental design and performed some of the microscopic analysis; K.G. performed some of the ChIP experiments; M.G. purified proteins, generated reagents and performed in vitro phosphorylation assays and pull down analysis; M-B.R. and M.Gr. generated reagents; D.F. supervised the project; C.L. discovered NBS1 nucleolar localization and provided feedback in the design of the microscopic analysis and interpretation of the data; J.L. and M.S. coordinated and supervised the project, designed experiments, analyzed data and wrote the paper.
References
- 1.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 3.Difilippantonio S, Nussenzweig A. The NBS1-ATM connection revisited. Cell Cycle. 2007;6:2366–2370. doi: 10.4161/cc.6.19.4758. [DOI] [PubMed] [Google Scholar]
- 4.Chrzanowska KH, Gregorek H, Dembowska-Bagińska B, Kalina MA, Digweed M. Nijmegen breakage syndrome (NBS) Orphanet Journal of Rare Diseases. 2012;7:13. doi: 10.1186/1750-1172-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair. 2006;5:534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 6.Spycher C, et al. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. The Journal of Cell Biology. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melander F, et al. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. The Journal of Cell Biology. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci USA. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd J, et al. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams RS, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-Dependent Chromatin Changes Silence Transcription In cis to DNA Double-Strand Breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruhlak MJ, et al. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- 14.Lukas C, Bartek J, Lukas J. Imaging of protein movement induced by chromosomal breakage: tiny ‘local’ lesions pose great ‘global’ challenges. Chromosoma. 2005;114:146–154. doi: 10.1007/s00412-005-0011-y. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg M, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 16.Lukas C, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakai D, Trainor PA. Treacher Collins syndrome: unmasking the role of Tcof1/treacle. Int J Biochem Cell Biol. 2009;41:1229–1232. doi: 10.1016/j.biocel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wise CA, et al. TCOF1 gene encodes a putative nucleolar phosphoprotein that exhibits mutations in Treacher Collins Syndrome throughout its coding region. Proc Natl Acad Sci USA. 1997;94:3110–3115. doi: 10.1073/pnas.94.7.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isaac C, et al. Characterization of the nucleolar gene product, treacle, in Treacher Collins syndrome. Mol Biol Cell. 2000;11:3061–3071. doi: 10.1091/mbc.11.9.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- 21.Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci USA. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon J, et al. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci USA. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19:276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 24.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 25.Jones NC, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dauwerse JG, et al. Mutations in genes encodingsubunits of RNA polymerases I and III cause Treacher Collins syndrome. Nature Publishing Group. 2010;43:20–22. doi: 10.1038/ng.724. [DOI] [PubMed] [Google Scholar]
- 27.Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of Ribosomal RNA Gene Copies Maintains Genome Integrity. Science. 2010;327:693–696. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- 28.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol. 2003;5:255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 29.Gudjonsson T, et al. TRIP12 and UBR5 Suppress Spreading of Chromatin Ubiquitylation at Damaged Chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen ML, et al. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat Meth. 2008;5:459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- 31.Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J Proteome Res. 2012;11:3487–3497. doi: 10.1021/pr3000249. [DOI] [PubMed] [Google Scholar]
- 32.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 33.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.