Abstract
We show a logical aggregation method that, combined with propositionalization methods, can construct novel structured biological features from gene expression data. We do this to gain understanding of pathway mechanisms, for instance, those associated with a particular disease. We illustrate this method on the task of distinguishing between two types of lung cancer; Squamous Cell Carcinoma (SCC) and Adenocarcinoma (AC). We identify pathway activation patterns in pathways previously implicated in the development of cancers. Our method identified a model with comparable predictive performance to the winning algorithm of a recent challenge, while providing biologically relevant explanations that may be useful to a biologist.
Keywords: Biological pathways, Warmr, TreeLiker, Reactome, Barcode, Logical aggregation
1. Introduction and Background
In the field of Systems Biology researchers are often interested in identifying perturbations within a biological system that are different across experimental conditions. Biological systems consist of complex relationships between a number of different types of entities, of which much is already known [1]. An Inductive Logic Programming (ILP) approach may therefore be effective for this task, as it can represent the relationships of such a system as background knowledge, and use this knowledge to learn potential reasons for differences in a particular condition. We demonstrate a propositionalization-based ILP approach, and apply this to the example of identifying differences in perturbations between two types of lung cancer; Squamous Cell Carcinoma (SCC) and Adenocarcinoma (AC).
A recent large competition run by the SBV Improver organisation, called the Diagnostic Signature Challenge, tasked competitors with finding a highly predictive model distinguishing between these two lung cancer types [2]. The challenge was motivated by the many studies that have also worked on similar tasks, with the aim to find a model with the best predictive performance. The winning method from this competition is a pipeline that exemplifies the classification approaches used for this task [3].
The typical pipeline has three distinct stages. The first stage uses technology such as microarrays or RNAseq, to measure gene expressions across the genome in a number of samples from each of the experimental conditions. The second stage identifies a subset of genes whose expression values differ across conditions. This stage is commonly achieved by performing differential expression analysis and ranking genes by a statistic such as fold change values [4]. A statistical test is then used to identify the set of genes to take forward to stage 3. Alternatively for stage 2, researchers may train a model using machine learning to classify samples into experimental conditions, often using an attribute-value representation where the features are a vector of gene expression values (as performed by the winning SBV Improver model). This approach has the advantage that the constructed model may have found dependencies between genes that would not have been identified otherwise. Researchers use the ‘top’ features from the model to identify the set of genes to take forward to stage 3.
In stage 3 researchers look for connections between these genes by, for example, performing a Gene Set Enrichment Analysis (GSEA) [5]. Here, the set of genes are compared with predefined sets of genes, that each indicate a known relation. For example, a gene set may have a related function, exist in the same location in the cell, or take part in the same pathway.
To bring background knowledge of relations into the model building process, past ILP research integrated stage 2 (finding differentially expressed genes) and stage 3 (GSEA), into a single step [6]. This was achieved using Relational Subgroup Discovery, which has the advantage of being able to construct novel sets by sharing variables across predicates that define the sets. For example, a set could be defined as the genes annotated with two Gene Ontology terms.
Other ways researchers have tried to integrate the use of known relations includes adapting the classification approach of stage 2. New features are built by aggregating across a predefined set of genes. For example, an aggregation may calculate the average expression value for a pathway [7].
A major limitation of current classification approaches is that the models are constructed from either genes or crude aggregates of sets of genes, and so ignore the detailed relations between entities in a pathway. In order to incorporate more complex relations an appropriate network representation is needed, such that biological relations are adequately represented. For example, a simple directed network of genes and proteins does not represent all the complexities of biochemical pathways, such as the dependencies of biochemical reactions. To do this bipartite graphs or hypergraphs can be used [8].
One way to incorporate more complex relations is by creating topologically defined sets, where a property of the network is used to group nodes into related sets. One method to generate these sets is Community Detection [9]. However, this approach can create crude clusters of genes, that do not account for important biological concepts. Biologists may be interested in complex biological interactions rather than just sets of genes.
Network motif and frequent subgraph mining are methods that can look for structured patterns in biological networks [10]. However, in these approaches the patterns are often described in a language which is not as expressive as first order logic. This means they are unable to find patterns with uninstantiated variables, or with relational concepts such as paths or loops.
To our knowledge only one previous work has used ILP for this task [11]. Here the authors propose identifying features consisting of the longest possible chain of nodes in which non-zero node activation implies a certain (non-zero) activation in its successors, which they call a Fully Coupled Flux. Their work is preliminary, with limited evaluation of the performance of this method.
The aim of this paper is to illustrate how we can identify pathway activation patterns, that differ between biological samples of different classes. A pathway activation pattern is a pattern of active reactions on a pathway. Our novel approach uses known relations between entities in a pathway, and important biological concepts as background knowledge. These patterns may give a biologist different information than models built from simple gene features. We seek to build models that are of comparative predictive performance to those of previous work, while also providing potentially useful explanations.
In this work we take a propositionalization-based ILP approach, where we represent the biological systems as a Prolog knowledge base (composed of first-order rules and facts), and then reduce this to an attribute-value representation (a set of propositions), before using standard machine learning algorithms on this data. We begin with an overview of propositionalization, and a discussion of why it is appropriate for this task.
2. Overview of propositionalization
Propositionalization is a method that transforms data represented in first-order logic to a set of propositions, i.e. a single table representation where each example is represented by a fixed length vector. This is called a reduction. It is possible to make a proper reduction of the representation using Goedel numbering or well-ordering arguments [12]. However, these will have limited practical value as useful structure can be lost or encoded inefficiently, leading to poor inductive ability. Heuristic-based propositionalization methods allow specification of a language bias and a heuristic, in order to search for a subset of potential features which are useful for the learning task.
We have four reasons for adopting a propositionalization-based approach, rather than directly applying an ILP learner. First, separating the feature construction from the model construction means that we have an interesting output in the middle of the process, which we would lose if they were coupled together. For example, the features constructed can represent useful domain knowledge in their own right, as they can describe subgroups of the data which have a different class distribution, or frequent item sets or queries on the data.
Second, propositionalization can be seen as a limited form of predicate invention, where the predicate refers to a property of an individual, or relationships amongst properties of the individual. This means that, when building a model, the features may correspond to complex relationships between the original properties of an individual. In our case they correspond to potentially interesting pathway activation patterns. Hence, we can understand predictions in terms of these higher order concepts, which may give important insights to a biologist.
Third, propositionalization can impose an individual-centred learning approach [12, 13]. This limits predicates to only refer to relationships between properties of an individual – we cannot have a predicate which relates individuals. This strong inductive bias is appropriate for our case, as we do not wish to consider relationships between the individuals. The fourth reason is that we can perform many other learning tasks on the transformed data, with the vast array of algorithms available for attribute-values datasets.
In this work we use query-based propositionalization methods, and now describe some key algorithms. A review of some publicly available propositionalization methods was recently performed by Lavrač et al. [14]. These include Linus, RSD, TreeLiker (HiFi and RelF algorithms), RELAGGS, Stochastic Propositionalization, and Wordification, alongside the more general ILP toolkit, Aleph. Other methods that were not mentioned in that review include Warmr [15], Cardinalisation [16], ACORA [17] and CILP++ [18]. There has also been work on creating propositionalization methods especially for linked open data, both in an automatic way [19], and in a way where manual SPARQL queries are made [20]. The methods in these papers are not appropriate for our work because our data is not entirely made up of linked open data, and we wish to include background rules encoding additional biological knowledge. It is also worth noting that certain kernel methods can be thought of as propositionalization [12].
Wordification treats relational data as documents and constructs word-like features. These are not be appropriate for our task, as they do not correspond to the kind of patterns we are looking for, i.e. features with uninstantiated variables. Stochastic propositionalization performs a randomised evolutionary search for features. This approach may be interesting to consider for future work. CILP++ is a method for fast bottom-clause construction, defined as the most specific clause that covers each example. This method is primarily designed to facilitate the learning of neural networks, and has been reported to perform no better than RSD when used with a rule-based model [18].
ACORA, Cardinalisation and RELAGGS are database inspired methods of propositionalization. They are primarily designed to perform aggregation across a secondary table, with respect to a primary table. ACORA is designed to create aggregate operators for categorical data, whereas RELAGGS performs standard aggregation functions (summation, maximum, average etc.) suitable for numeric data. Cardinalisation is designed to use complex aggregates, where conditions are added to an aggregation. In our work we manually design an aggregation method, described in Section 3.2. These aggregation systems are not appropriate for graph-based datasets, because representing the graph as two tables (denoting edges and nodes) and aggregating on paths through the graph would require many self joins on the edge table. Relational databases are not optimised for this task, such that the resulting queries would be inelegant and inefficient.
The propositionalization methods we use in this work are TreeLiker and Warmr. TreeLiker is a tool that provides a number of algorithms for propositionalization including RelF [21]. RelF searches for relevant features in a block-wise manner, and this means that irrelevant and irreducible features can be discarded during the search. The algorithms in TreeLiker are limited to finding tree-like features where there are no cycles. RelF has been shown to scale much better than previous systems such as RSD, and can learn features with tens of literals. This is important for specifying non-trival pathway activation patterns.
Warmr is a first-order equivalent of frequent item-set mining, where a level-wise search of frequent queries in the knowledge base is performed. Warmr is used as a propositionalization tool by searching for frequent queries in each class. In Warmr it is possible to specify the language bias using conjunctions of literals, rather than just individual literals, and to put constraints on which literals are added. This allows strong control of the set of possible hypotheses that can be considered. Finally, unlike TreeLiker, Warmr can use background knowledge, defined as facts and rules.
3. Methods
An overview of the process we take is shown in Figure 1. First, we extract the reaction graph for each pathway, from Reactome. Second, we infer the instantiated reaction graphs for each instance in the dataset. Third, we identify pathway activation patterns using propositionalization, and then build classification models to predict the lung cancer types. Lastly, we evaluate our models using a hold-out dataset. We begin with a description of the datasets we use in this work.
Fig. 1.
Method overview
3.1. Raw Data
Our approach uses two sources of data: 1) a dataset from Gene Expression Omnibus (GEO) [22] as the set of examples (gene expression values of a set of individuals), and 2) information about biological systems from Reactome.
GEO data Main data
We use a two class lung cancer dataset obtained from GEO, which was previously used in the SBV Improver challenge [2]. This dataset is made up from the following datasets: GSE2109, GSE10245, GSE18842 and GSE29013 (n=174), used as training data, and GSE43580 (n=150), used as hold-out data. We used the examples where the participants were labelled as having either SCC or AC lung cancer. This is the same data organisation as that used in SBV Improver challenge, to allow us to compare our results with the top performing method from this challenge.
This data contains gene expression measurements from across the genome measured by Affymetrix chips. Each example is a vector of 54,614 real numbers. Each value denotes the amount of expression of mRNA of a gene. There is a uniform class distribution of examples, in both the training and holdout dataset.
Reactome- Background Knowledge
We use the Reactome database to provide the background knowledge, describing biological pathways in humans. Reactome [1] is a collection of manually curated peer reviewed pathways. Reactome is made available as an RDF file, which allows for simple passing using SWI-Prolog’s semantic web libraries, and contains 1,351,811 triples. Reactome uses the bipartite network representation of entities and reactions. Entity types include nucleic acids, proteins, protein complexes, protein sets and small molecules. Protein complexes and protein sets can themselves comprise of other complexes or sets. In addition, a reaction may be controlled (activated or inhibited) by particular entities. A reaction is a chemical event, where input entities (known as substrates), facilitated by enzymes, form other entities (known as products).
Figure 2a shows a simple illustration of a Reactome pathway. P nodes denote proteins or protein complexes, R nodes denote reactions, and C nodes denote catalysts. A black arrow illustrates that a protein is an input or output of a reaction. A green arrow illustrates that an entity is an activating control for a reaction. A red arrow illustrates that an entity is an inhibitory control for a reaction. Reaction R1 has 3 protein substrates and 3 protein products, and is controlled by catalyst C. Reactions R3 and R4 both have one protein substrate and one protein product. R3 is inhibited by P2, such that if P2 is present then reaction R3 will not occur. R4 is activated by P3, such that P3 is required for reaction R4 to occur.
Fig. 2.
Reaction Graph illustrations. There are three types of relationships between reactions: follows (black solid lines), activation (green dashed), and inhibition (red dash-dotted).
3.2. Data Processing
Extracting reaction graphs
We reduce the Reactome bipartite graph to a boolean network of reactions. This simplifies the graphs while still adequately encoding the relationships between entities. Previous work has shown that boolean networks are a useful representation of biological systems [23], and unlike gene and protein boolean networks ours encodes the dependencies between reactions.
The boolean networks we create are reaction-centric graphs, where nodes are reactions and directed edges are labelled either as ‘activation’ ,’inhibition’ or ‘follows’ corresponding to how reactions are connected. For example, Figure 2b shows the reaction-centric graph, corresponding the Reactome graph shown in Figure 2a. Reaction R2 follows R1, because in the Reactome graph P1 is an output of R1 and an input to R2. Reaction R1 inhibits R3, because P2 is an output of R1, and it is also an inhibitory control of R3. Reaction R1 activates reaction R4, because P3 is an output of R1, and an activating control of R4.
Inferring instantiated reaction graphs
Boolean networks [23] are a common abstraction in biological research, but these are normally applied at the gene or protein level not at the reaction level. In order to use a boolean network abstraction on a reaction network, we apply a logical aggregation method that aggregates measured probe values (from the GEO dataset) into reactions. This creates a binary value for each reaction, to create instantiated versions of the reaction-centric graph created in the previous step.
Before we can use this logical aggregation we first transform the original probe values into binary values, an estimated value denoting whether a gene is expressed or not. We do this using Barcode [24], a tool for converting the continuous probe values to binary variables, by applying previously learnt thresholds to microarray data. It is important to note that Barcode makes it possible to compare gene expressions, both within a sample, and between samples that are potentially measured by different arrays.
The logical aggregation process is illustrated in Figure 3. This process takes the binary probe values as input, and uses the structure provided by the Reactome graph, and key biological concepts, to build reaction level features. As we have already described, each reaction has a set of inputs that are required for a particular reaction. We interpret each reaction input as a logical circuit with the following logical rules. The relationship between probes and proteins is treated as an OR gate (matched by Uniprot IDs), because multiple probes can encode for same protein. We are assuming that the measurement from a single probe indicates with high probability whether the protein product is present or not. The formation of a protein complex requires all of its constituent proteins and therefore is treated as an AND gate. A protein set is a set of molecules that are functionally equivalent such that only one is needed for a given reaction, and so this is treated as an OR gate. Inputs to a reaction are treated as an AND gate. A reaction is on if the inputs are on, any activating agents are on, and any inhibitory agents are off. We note that both protein sets and protein complexes can themselves comprise of arbitrarily nested complexes or sets.
Fig. 3.
Illustration of logical aggregation. Known biological mechanisms can be represented as OR or AND gates. The triangular nodes are binary probe values, created using barcode.
Figure 3 illustrates the logical aggregation rules of a single reaction. This reaction has two inputs and one activating control. The two inputs are a protein complex and a protein set, and the values of these are calculated using their own aggregation processes, labelled A and B. The aggregation in process A, starts with the binary probe values, and first infers the values of three proteins. The protein complex is then assigned a value of on because all proteins required for this complex are present (are all on themselves). The aggregation in process B starts by inferring the values of two proteins from the probe values. One protein is on and the other is off. The protein set is assigned the value on because only one protein in this set is required for this protein set to be on. There also exists an activating control for the reaction, a protein whose value is determined by a process labelled C in this figure. This protein is assigned the value on, because both probe values are on, when at least one is required. As all inputs are on and the activating control is also on, the reaction is assigned the value on.
3.3. Searching for Pathway Activation Patterns
In order to identify pathway activation patterns we first find pathways that are most likely to contain these patterns, using the training data. We then use three approaches to identify pathway activation patterns within the ‘top’ pathways, and evaluate the identified activation patterns using the hold-out data.
Identifying predictive pathways
To identify pathways we first run TreeLiker on each pathway. This generates a set of attribute-value features for each instantiated pathway. We use TreeLiker with the RelF algorithm and the following language bias:
set(template,[reaction(–R1,#onoroff),link(+R1,–R2,!T1), reaction(+R2,#onoroff),link(+R2,-R3,!T2),reaction(+R3,#onorff), link(!RA,-R4,!T3),link(+R4,!RB,!T4),link(+R1,-R2,#T1), link(+R2,-R3,#T2),link(!RA,-R4,#T3),link(+R4,!RB,#T4)])
This language bias contains two types of literals; reaction/2 and link/3. The second argument of the reaction literal is always constrained to be a constant depicting if a reaction is on or off. The link literal depicts the relationship between two reactions, where the third argument of the link literal can be a variable or a constant describing the type of relationship - either follows, activates or inhibits. For example, an identified pattern may contain the literal link(r1,r2,follows), specifying that an output entity of reaction r1 is an input to reaction r2.
We then test the performance of the features of each pathway, using 10 fold cross validation. We use the J48 decision tree algorithm (from Weka) because this builds a model that give explanations for the predictions. We calculate the average accuracy across folds, for each pathway, and rank the pathways from highest to lowest accuracy. We then use the top ranked pathways as input to three different methods, to identify predictive pathway activation patterns.
Method 1
This approach simply takes a pathway of interest, generates a single model using the J48 algorithm using the training data, and then evaluates this performance on the hold-out data. The decision tree can then be viewed to determine which activation patterns are predictive of lung cancer type. We demonstrate this approach with the top-ranked pathway.
Method 2: Warmr approach
We illustrate using Warmr to generate pathway activation patterns, using one of our identified ‘top’ pathways.
We use Warmr with two particular concepts in the background knowledge. First, we use a predicate longestlen/3, that calculates the longest length of on reactions in an example, for the pathway on which Warmr is being run. The arguments are: 1) the beginning reaction of a path, 2) the end reaction of the path with longest length, and 3) the length of this path. This longest length concept corresponds to the fully coupled flux of a previous work [11].
Second, we use the predicates inhibloop/1 and actloop/1, that depict inhibition and activation loops, where a path of on reactions form a loop and one of the edges is an inhibition or activation edge, respectively. Inhibition and activation loops are common biological regulatory mechanisms [25].
We then use the OneR (Weka) algorithm to identify the single best pathway activation pattern found by Warmr, and then evaluate this pattern on the holdout data.
Method 3: Combined approach
Our combined method takes advantage of the beneficial properties of the two algorithms, by using Warmr to extend the patterns identified by TreeLiker. This effectively switches the search strategy from the block-wise approach of TreeLiker, to the level-wise approach of Warmr. The reason for doing this is to identify any relations between reactions that exist between entities within the TreeLiker feature, that could not be identified in TreeLiker due to its restriction to tree structures. This results in long, cyclical features that neither TreeLiker nor Warmr would be able to find on their own.
While we could use the features generated by method 1, and extend these, in this section we also demonstrate the possibility of using our approach for generating descriptions of subgroups. We identify a subgroup with the CN2SD algorithm [26], using the training data. The activation patterns defining this subgroup are then extended using Warmr. The following code is an example language bias we use in Warmr:
rmode(1: (r(+S,-A,1),link(A,-\B,follows),link(B,-\C,_),r(S,C,0), r(S,B,0), link(B,-\D,_),r(S,D,1),link(A,-\E,_),r(S,E,1))).
rmode(1: link(+A,+B,#)).
The first rmode contains the feature that was previously identified using TreeLiker. The second rmode uses the literal link, to allow Warmr to add new links to the TreeLiker feature. After extending the activation pattern using Warmr, we then evaluate this on the hold-out data.
4. Results
To reiterate, the aim of this work is to build explanatory models that help biologists understand the system perturbations associated with conditions, in this case lung cancer. Therefore, although we give the classification performance of our models in order to make the quantitative performance comparison, we additionally emphasis the form that the classification models take and how these are of interest to biologists. Table 1 shows the top 5 pathways found using our TreeLiker/J48 method, and the size of each reaction graph.
Table 1.
Top 5 pathways identified. Mean accuracy across 10 folds of cross-validation on the training dataset.
| Ranking | Pathway | Accuracy | Number of nodes (reactions) | Number of edges |
|---|---|---|---|---|
| 1 | Hexose uptake | 78.74% | 18 | 25 |
| 2 | Hyaluronan biosynthesis | 77.59% | 18 | 25 |
| 4 | Mitotic G1-G1/phases | 76.74% | 51 | 59 |
| 4 | Creatine metabolism | 78.64% | 6 | 6 |
| 5 | Cell Cycle | 77.59% | 322 | 492 |
4.1. Quantitative evaluation and comparison with SBV Improver model
To provide a quantitative comparison of our models, we compare to the winning classifier of the SBV Improver challenge. We use the area under the ROC curve (AUC) metric, to evaluate the ranking performance of the models. We generate confidence intervals for the AUC using a stratified bootstrapped approach (with 2000 bootstraps) [27]. We also use permutation testing to compare the performance of our models with a random model. We generate 2000 random rankings, with the same class distribution as our data, and calculate the AUC for each of these rankings. We then find the proportion of random rankings with an AUC greater than that of our model. We refer to this as the permutation P value.
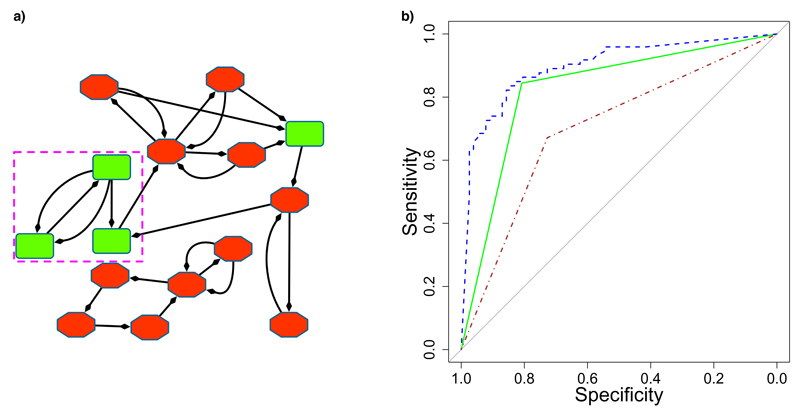
We select the top pathway identified in the training data – hexose uptake. After retraining J48 on the whole training data, evaluation on the hold-out data gives an AUC of 0.820 (95% confidence interval (CI): 0.764-0.890). This model is better than a random model (permutation P value < 0.001). The SBV method, evaluated on the hold-out data has an AUC of 0.913 (95% CI: 0.842-0.947). The confidence intervals overlap such that we cannot find a difference in performance between our model and the SBV model. Figure 4 shows the ROC curves of the SBV and hexose uptake models. Our hexose uptake model is a decision tree with a single feature:
Fig. 4.
Results
(a) Example of the hexose uptake pathway for a particular individual. Green squares: on reactions, red octagons: off reactions. Identified feature of three on reactions shown in pink, dashed box.
(b) ROC curves comparing performance. SBV Improver: blue, dashed; TreeLiker & J48: green, solid; Warmr & OneR: red, dash-dotted.
reaction(A,1), link(A,B,_), link(B,C,_), reaction(C,1), reaction(B,1).
This corresponds to a chain of three on reactions, where the model predicts SSC if this feature exists and AC otherwise. This pathway activation pattern is present in 67 of the 76 individuals with SSC, and 17 of the 74 individuals with AC. In Figure 4a we show an example instantiation of the hexose uptake pathway, for a particular individual. For this individual, the three variables A,B,C in the feature given above, are instantiated to the following reactions:
A. GLUT1 + ATP <=> GLUT1 :ATP.
B. GLUT1 + ATP <=> GLUT1 +ATP.
C. alpha-D-Glucose + ATP => alpha-D-glucose 6-Phosphate + ADP.
4.2. Results for Warmr method
We illustrate the value of our Warmr only method using the cell cycle pathway (ranked fifth in Table 1). The more complex background predicates that we have defined for Warmr are only relevant when the pathway itself contains particular relationships. For example, the activation loop predicate will only be potentially beneficial when a pathway contains an activation edge, that may potentially be identified as the activation within an activation loop. The cell cycle is the top ranked pathway that contains all three kinds of edges; follows, activation and inhibition. The OneR classifier generated with the Warmr features has an AUC of 0.699 (95% CI: 0.625-0.773), on the hold-out data.
While this model performs worse than the SBV Improver model and the hexose uptake pathway TreeLiker/combined model (in terms of AUC), it still has predictive value (Permutation P value < 0.001). The identified rule is complex and potentially interesting to a biologist:
actloop(C),largestlen(E,F,G),greaterthan(G,5),link(E,H,follows),r(H,0)
The rule states that a sample is classified as SCC cancer if there is a self activating loop for a reaction C, and that the longest chain of on reactions is from reaction E to reaction F, which is a chain at least 6 reactions long. Additionally, following reaction E there is also a reaction H that itself is not on.
This suggests that one of the differences between the SCC and AC cancer is that in the cell cycle SCC tumours have a self activating loop, that causes a longer chain of reactions to occur than in the AC tumour types. An instantiation of this feature is shown in Figure 5, for a particular individual. In this example there is a chain of 7 on reactions, and this also contains the self-activating loop.
Fig. 5.
The pattern found by Warmr instantiated for individual GSM1065725. There is a self-activating loop, highlighted by the grey box.
4.3. Results for Warmr/TreeLiker combined method
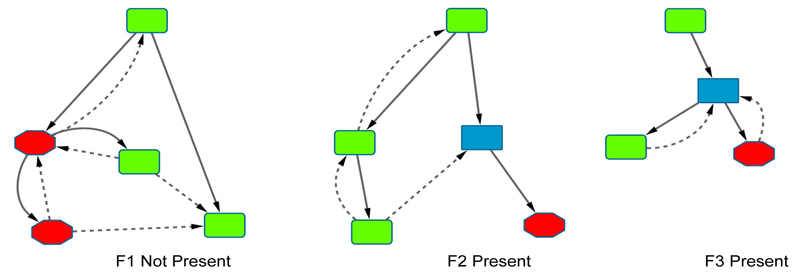
As explained above, the feature used in the top pathway was very simple, and hence we demonstrate the value of our Warmr/TreeLiker combined approach on more complex features, identified from the hyaluronan biosynthesis and export pathway, ranked second in Table 1. Figure 6 shows the three features describing the subgroup identified by this approach. We can see that the additional edges that Warmr finds give a more complete view of the relations between the reactions in these features. This information may be important when a biologist analyses these results. The subgroup described by these three features has 58 true positives and 9 false positives in the hold-out data.
Fig. 6.
The three features in the subgroup description. Solid lines represent the feature found by TreeLiker, dotted lines show the Warmr extensions. on reactions: green, rounded squares; off: red octagons; on or off: blue squares.
5. Conclusions
In this work we have shown the potential of ILP methods for mining the abundance of highly structured biological data. Using this method we have identified differences in pathway activation patterns that go beyond the standard analysis of differentially expressed genes, enrichment analysis, gene feature ranking and pattern mining for common network motifs. We have also demonstrated the use of logical aggregation with a reaction graph, and how this simplifies the search for hypotheses to an extent that searching all pathways is tractable. We have introduced a novel approach that uses Warmr to extend features initially identified with TreeLiker. This makes it possible to search for long cyclical features.
We have identified pathway activation patterns predictive of the lung cancer type, in several pathways. The model we built on the hexose uptake pathway has predictive performance comparable with the top method from a recent challenge, but also provides biologically relevant explanations for its predictions. Each identified activation pattern is evaluated on the hold-out data, such that this should be the expected performance on new, unseen examples. The pathway activation patterns we have found are in clinically relevant pathways [28]. Patterns identified using this method may give diagnostic and clinical insights that biologists can develop into new hypotheses for further investigation.
Acknowledgments
LACM received funding from the Medical Research Council (MC_UU_12013/8).
References
- [1].Croft David, Mundo Antonio Fabregat, Haw Robin, Milacic Marija, Weiser Joel, Wu Guanming, Caudy Michael, Garapati Phani, Gillespie Marc, Kamdar Maulik R, et al. The Reactome pathway knowledgebase. Nucleic acids research. 2014;42(D1):D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rhrissorrakrai Kahn, Rice J Jeremy, Boue Stephanie, Talikka Marja, Bilal Erhan, Martin Florian, Meyer Pablo, Norel Raquel, Xiang Yang, Stolovitzky Gustavo, Hoeng Julia, et al. SBV Improver Diagnostic Signature Challenge: Design and results. Systems Biomedicine. 2013 Sep;1(4):3–14. [Google Scholar]
- [3].Tarca Adi L, Than Nandor Gabor, Romero Roberto. Methodological approach from the best overall team in the SBV Improver Diagnostic Signature Challenge. Systems Biomedicine. 2013;1(4):217–227. [Google Scholar]
- [4].Draghici Sorin. Statistical intelligence: effective analysis of high-density microarray data. Drug discovery today. 2002;7(11):S55–S63. doi: 10.1016/s1359-6446(02)02292-4. [DOI] [PubMed] [Google Scholar]
- [5].Subramanian Aravind, Tamayo Pablo, Mootha Vamsi K, Mukherjee Sayan, Ebert Benjamin L, Gillette Michael A, Paulovich Amanda, Pomeroy Scott L, Golub Todd R, Lander Eric S, Mesirov Jill P. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005 Oct;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gamberger Dragan, Lavrač Nada, Železnỳ Filip, Tolar Jakub. Induction of comprehensible models for gene expression datasets by subgroup Using ILP to Identify Pathway Activation Patterns in Systems Biology 15 discovery methodology. Journal of Biomedical Informatics. 2004 Aug;37(4):269–284. doi: 10.1016/j.jbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- [7].Holec Matĕj, Klma Jiřì, Železnỳ Filip, Tolar Jakub. Comparative evaluation of set-level techniques in predictive classification of gene expression samples. BMC Bioinformatics. 2012 Jun;13(Suppl 10):S15. doi: 10.1186/1471-2105-13-S10-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Whelan Ken, Ray Oliver, King Ross D. Yeast Systems Biology. Springer; 2011. Representation, simulation, and hypothesis generation in graph and logical models of biological networks; pp. 465–482. [DOI] [PubMed] [Google Scholar]
- [9].Danon Leon, Diaz-Guilera Albert, Duch Jordi, Arenas Alex. Comparing community structure identification. Journal of Statistical Mechanics: Theory and Experiment. 2005;2005(09):P09008. [Google Scholar]
- [10].Kim Wooyoung, Li Min, Wang Jianxin, Pan Yi. Biological network motif detection and evaluation. BMC Systems Biology. 2011 Dec;5(Suppl 3):S5. doi: 10.1186/1752-0509-5-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Holec Matĕj, Železnỳ Filip, Kléma Jiřì, Svoboda Jiřì, Tolar Jakub. Using bio-pathways in relational learning. Inductive Logic Programming. 2008:50. [Google Scholar]
- [12].Raedt Luc De. Logical and relational learning. Springer Science & Business Media; 2008. [Google Scholar]
- [13].Flach Peter, Lachiche Nicolas. Inductive Logic Programming. Springer; 1999. 1BC: A first-order Bayesian classifier; pp. 92–103. [Google Scholar]
- [14].Lavrač Nada, Vavpetič Anže. Logic Programming and Nonmonotonic Reasoning. Springer; 2015. Relational and semantic data mining; pp. 20–31. [Google Scholar]
- [15].Dehaspe Luc, Raedt Luc De. Mining association rules in multiple relations. In: Lavrač Nada, Džeroski Sašo., editors. Inductive Logic Programming. Springer; Berlin Heidelberg: 1997. Jan, pp. 125–132. number 1297 in Lecture Notes in Computer Science. [Google Scholar]
- [16].Ahmed Chowdhury Farhan, Lachiche Nicolas, Charnay Clément, Jelali Soufiane El, Braud Agnès. Flexible propositionalization of continuous attributes in relational data mining. Expert Systems with Applications. 2015 [Google Scholar]
- [17].Perlich Claudia, Provost Foster. Distribution-based aggregation for relational learning with identifier attributes. Machine Learning. 2006;62(1–2):65–105. [Google Scholar]
- [18].Franç Manoel VM, Zaverucha Gerson, d’Avila Garcez Artur S. Fast relational learning using bottom clause propositionalization with artificial neural networks. Machine Learning. 2014;94(1):81–104. [Google Scholar]
- [19].Ristoski Petar, Paulheim Heiko. A comparison of propositionalization strategies for creating features from linked open data. Linked Data for Knowledge Discovery. 2014:6. [Google Scholar]
- [20].Ristoski Petar. The Semantic Web Latest Advances and New Domains. Springer; 2015. Towards linked open data enabled data mining; pp. 772–782. [Google Scholar]
- [21].Kuželka Ondřej, Železnỳ Filip. Block-wise construction of tree-like relational features with monotone reducibility and redundancy. Machine Learning. 2010 Aug;83(2):163–192. [Google Scholar]
- [22].Edgar Ron, Domrachev Michael, Lash Alex E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang Rui-Sheng, Saadatpour Assieh, Albert Réka. Boolean modeling in systems biology: an overview of methodology and applications. Physical Biology. 2012 Oct;9(5) doi: 10.1088/1478-3975/9/5/055001. 055001. [DOI] [PubMed] [Google Scholar]
- [24].McCall Matthew N, Jaffee Harris A, Zelisko Susan J, Sinha Neeraj, Hooiveld Guido, Irizarry Rafael A, Zilliox Michael J. The Gene Expression Barcode 3.0: improved data processing and mining tools. Nucleic acids research. 2014;42(D1):D938–D943. doi: 10.1093/nar/gkt1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tyson John J, Chen Katherine C, Novak Bela. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Current opinion in cell biology. 2003;15(2):221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- [26].Lavrač Nada, Kavšek Branko, Flach Peter A, Todorovski Ljupčo. Subgroup discovery with CN2-SD. The Journal of Machine Learning Research. 2004;5:153–188. [Google Scholar]
- [27].Robin Xavier, Turck Natacha, Hainard Alexandre, Tiberti Natalia, Lisacek Frédérique, Sanchez Jean-Charles, Müller Markus. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wu Rongrong, Galan-Acosta Lorena, Norberg Erik. Glucose metabolism provide distinct prosurvival benefits to non-small cell lung carcinomas. Biochemical and biophysical research communications. 2015;460(3):572–577. doi: 10.1016/j.bbrc.2015.03.071. [DOI] [PubMed] [Google Scholar]