Adeno-associated virus (AAV) vectors are currently the leading candidates for virus-based gene therapy because of their broad tissue tropism, non-pathogenic nature and low immunogenicity1. They have been successfully used in clinical trials to treat hereditary diseases such as hemophilia B2 and have been approved for treatment of lipoprotein lipase deficiency in Europe3. Considerable efforts are made to engineer AAV variants with novel and biomedically valuable cell tropisms to allow efficacious systemic administration1,4, yet basic aspects of AAV cellular entry are still poorly understood. In particular, the protein receptor(s) required for AAV entry subsequent to cell attachment, remains enigmatic. Here we use an unbiased, haploid genetic screen to identify critical players in AAV serotype 2 (AAV2) infection. The most significantly enriched gene of the screen encoded an uncharacterized type-I transmembrane protein, KIAA0319L (hereafter termed AAV receptor – AAVR). We characterize AAVR as a protein capable of rapidly endocytosing from the plasma membrane and trafficking to the trans-Golgi network. We show that AAVR directly binds to AAV2 particles and that anti-AAVR antibodies efficiently block AAV2 infection. Moreover, genetic ablation of AAVR renders a wide range of mammalian cell types highly resistant to AAV2 infection. Strikingly, AAVR serves as a critical host factor for all tested AAV serotypes. The importance of AAVR for in vivo gene delivery is further underscored by the robust resistance of AAVR−/− mice to AAV infection. Collectively, the data indicate that AAVR is a universal receptor involved in AAV infection.
AAV2, the most commonly studied AAV serotype, attaches to cells using heparan sulphate proteoglycan (HSPG)5. For several other non-enveloped viruses, initial attachment is followed by engagement of a protein receptor, which dictates entry into the cytoplasm. Whether AAV also requires such a protein receptor is unclear. Surface proteins including human fibroblast growth factor receptor-1 (FGFR1) and hepatocyte growth factor receptor (c-MET) have been reported as putative AAV2 co-receptors6,7. Using isogenic knockout cell lines (Extended Data Fig. 1a, b) however, we observed no significant effect on AAV2 infection in cells lacking FGFR1, and only a minimal consequence of c-MET loss (Extended Data Fig. 1c), suggesting a modest role in AAV2 infection for these proteins. To identify host factors critical for AAV2 infection, we used an unbiased, genome-wide screening approach based on insertional mutagenesis in haploid human cells (HAP1)8. A library of mutagenized cells, carrying knockouts in virtually all non-essential genes, was infected with an AAV2 vector that expresses red fluorescent protein (RFP) (Extended Data Fig. 2a). Mutant cells refractory to AAV2 infection were isolated through iterative cycles of fluorescence-activated cell sorting (Extended Data Fig. 2b). The screen yielded 46 significant hits (Fig. 1a, Supplementary Table 1), many of which were implicated in HSPG biosynthesis (depicted in blue). AAV2 hijacks endosomal pathways to travel from the cell surface to the nucleus, and several endosomal trafficking genes (depicted in green) were prominently identified in the screen, specifically members of the retromer (VPS29, VPS35) and GARP complexes (VPS51, VPS52, VPS53, VPS54). These proteins are involved in retrograde transport from the endosomes to the Golgi9,10, but have not been specifically associated with AAV2 infection before now. The most significantly enriched gene of the screen was KIAA0319L (AAVR), with 570 independent mutations identified. This gene encodes a poorly characterized transmembrane protein. Little is known about the cellular function of AAVR, but it has been linked to dyslexia, with a potential role in neuronal migration11.
Fig. 1. An unbiased, haploid genetic screen identifies KIAA0319L (AAVR), an essential host factor for AAV2 infection.
a, Bubble plot illustrating significance of enrichment of gene-trap insertions within identified genes (relative to unselected control population). Bubbles represent genes with width proportional to number of independent gene trap insertions. Top forty significant genes (p ≤ 0.001) are colored and grouped by function. b, AAV2-RFP infection in wild-type (WT) cells and AAVR knock-out (AAVRKO) cells, evaluated in AAV2-susceptible human and mouse cell lines. c, AAV2-RFP infection of poorly permissive human and murine cell lines with and without AAVR overexpression. Data depicts mean with s.d. error bars for triplicate infections. Infections were performed using MOI 20,000 vg/cell for 24 hrs. The P value was determined using an unpaired, parametric, two-sided student t-test, with a Welch post-correction, where * - P <0.05, ** - P <0.01, *** - P <0.001.
To validate AAVR’s role in AAV2 infection, we used CRISPR/Cas9 genome engineering to generate isogenic AAVR knock-out cell lines (AAVRKO) in a panel of cell types representing various human and murine tissues (Extended Data Table 1). In all eight cell types, AAVR knockout rendered cells highly resistant to AAV2 infection (20,000 viral genomes (vg) per cell) (Fig. 1b). At a multiplicity of infection as high as 100,000 vg/cell, AAVRKO cells still remained poorly susceptible to infection by an AAV2-luciferase vector (Extended Data Fig. 3a). This also held true for wild-type AAV2 infection, where AAV2 replication was negligible in AAVRKO cells (Extended Data Fig. 3b). Notably, c-MET and FGFR1 knock-outs demonstrated no significant effect on infection in multiple cell types (Extended Data Fig. 3e). Genetic complementation of AAVR in AAVRKO cells (Extended Data Fig. 3c) restored susceptibility to AAV2 in all cell types assessed, confirming that the resistance phenotype observed in AAVRKO cells was caused by loss of AAVR expression (Extended Data Fig. 3d). To further examine if AAVR expression is capable of limiting AAV2 infection, we overexpressed AAVR in four cell lines previously identified as poorly permissive to AAV212,13. We observed an increase in susceptibility to AAV2 in all AAVR-overexpressing cell lines compared to wild-type cells, emphasizing AAVR’s critical role in AAV2 infection (Fig. 1c).
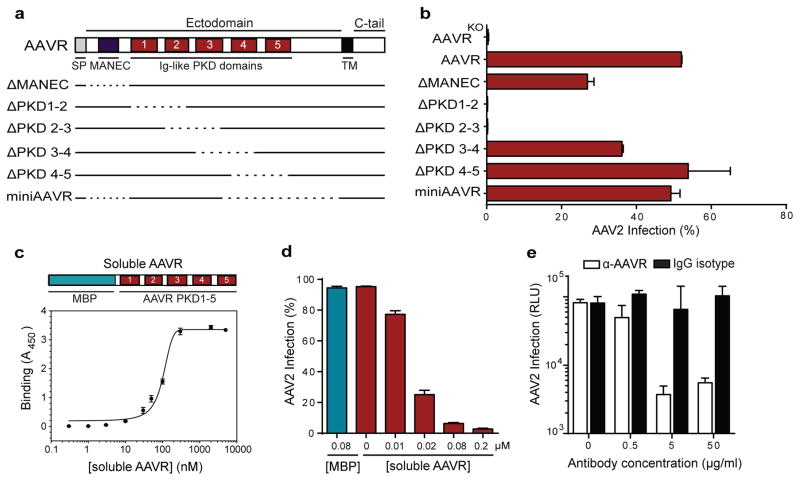
AAVR is a predicted type I transmembrane protein with five Ig-like domains in its ectodomain, referred to as polycystic kidney disease (PKD) domains14. Ig-like domains mediate cell-cell adhesion and are present in various well-characterized virus receptors15, including those for poliovirus, measles virus and reovirus. Based on this similarity to other receptors, and the extreme dependence of AAV2 infection on AAVR, we hypothesized that AAVR acts as an AAV2 receptor. We first determined whether AAVR PKD domains are responsible for mediating AAV2 infection by creating a series of AAVR deletion mutants and expressing each in AAVRKO cells (Fig. 2a). Simultaneous deletion of AAVR PKD domains 1 and 2, or 2 and 3, abrogated its role in AAV2 infection, whereas deletions in other regions were tolerated (Fig. 2b). An AAVR minimal mutant (miniAAVR) comprised of PKD domains 1–3 in its ectodomain, efficiently rescued AAV2 infection, highlighting the significance of the first three PKD domains for infection. Importantly, soluble AAVR (an E.coli-expressed recombinant protein comprising a fusion between maltose-binding protein and AAVR PKD 1–5), but not MBP alone, bound directly to AAV2 particles (Fig. 2c, Extended Data Figure 4a) with a KD of ~150 nM (measured using surface plasmon resonance - Extended Data Fig. 4b). We next investigated whether AAV2 infection could be neutralized in the presence of soluble AAVR. Indeed, infection efficiency was inhibited in a concentration-dependent manner when soluble AAVR was included during infection (Fig. 2d, Extended Data Fig. 4c). Consistent with this inhibition assay, antibodies directed against AAVR were capable of potently blocking AAV2 infection by more than 10-fold when incubated with cells prior to infection, in contrast to control IgG antibodies (Fig. 2e). This suggests that blocking viral access to AAVR on the cell surface substantially limits infection.
Fig. 2. AAVR binds specifically to AAV2 via its Ig-like PKD domains.
a, Schematic of AAVR domains and deletion mutants; dotted line represents deletions. b, AAV2-RFP infection of HAP1 AAVRKO cells expressing AAVR deletion mutants (MOI 20,000 vg/cell). c, ELISA showing binding to AAV2 particles of soluble AAVR (fusion protein between MBP and AAVR PKD 1–5). d, AAV2 neutralization assay incubating cells with soluble AAVR or MBP during AAV2-GFP infection, (MOI 7,500 vg/cell). e, Antibody inhibition assay incubating wild-type HeLa cells with anti-AAVR or IgG isotype control antibodies (at respective concentrations) at 4°C before AAV2-luciferase infection (MOI 1,000 vg/cell). Data depicts mean with s.d. error bars for triplicate infections; transgene expression measured after 24 hrs. SP: signal peptide, MANEC: motif at N-terminus with eight cysteines, PKD: polycystic kidney disease, TM: transmembrane, C-tail: C-terminal cytoplasmic tail, MBP: maltose binding protein, RLU: relative light units.
Characterization of the subcellular localization of AAVR revealed a distinct perinuclear localization, demonstrating a strong association with the cis-medial Golgi marker (giantin), and complete co-localization with the trans-Golgi network (TGN) marker (TGN46) (Fig. 3a). Many TGN proteins are dynamically recycled from the plasma membrane (PM) through motifs in their C-terminal cytoplasmic tail (C-tail) that direct endocytosis and intracellular trafficking (reviewed in16,17). To determine if AAVR is such a recycling receptor, we specifically labeled the cell surface pool of AAVR by incubating live AAVR-complement cells with anti-AAVR antibodies under cold conditions. Cells were warmed to initiate endocytosis and fixed at defined time points. Labeled-AAVR gradually moved from the surface into the cell, and concentrated in a perinuclear location associated with the Golgi marker (Fig. 3b). This rapid endocytosis may explain why we did not observe AAVR at the cell surface in steady state (Fig. 3a). As a control, AAVRKO cells were labeled similarly to AAVR-expressing cells, but no AAVR was detected on these cells (Extended Data Fig. 5a). Interestingly, the intracellular trafficking route of AAVR mapped here is remarkably similar to that of AAV particles, trafficking from the PM to the Golgi18. To determine if AAVR endocytosis contributes to mediating AAV2 infection, we removed the C-tail of AAVR (encoding its endocytic motifs). C-tail deletion (ΔC-tail) led to increased cell surface expression of AAVR (Fig. 3c, Extended Data Fig. 5b) and prevented endocytosis (Extended Data Fig. 5c). Importantly, ΔC-tail was incapable of mediating AAV2 infection upon complementation in AAVRKO cells (Fig. 3d), suggesting that AAVR endocytosis is required for AAV2 infection. We further investigated whether AAVR requires intracellular trafficking all the way to the TGN to mediate infection, by replacing the C-tail of miniAAVR with those of cellular receptors with well-characterized endocytic motifs (Extended Data Fig. 6a). These included the cation-independent mannose 6-phosphate receptor (Ci-MPR), which is the prototypical receptor that traffics from the PM to the TGN19. We also included the low density lipoprotein receptor (LDLR)20 and poliovirus receptor (PVR)21, which both endocytose and traffic between the PM and endosomes but are not reported to travel to the TGN. Each of the fusion constructs displayed cellular localization patterns comparable to their parent receptors, with PVR-tail and LDLR-tail detectable on the cell surface, and MPR-tail co-localized with a TGN marker and displaying a dispersed pattern in the cytoplasm (Extended Data Fig. 6b). Remarkably, all fusion constructs rescued AAV2 infection, albeit to different degrees (Extended Data Fig. 6c). Compared to the restored infection in miniAAVR-expressing cells, the LDLR and PVR fusion constructs yielded 2-fold and 4-fold reduced infectivity, respectively. Conversely, routing AAVR to the TGN using Ci-MPR endocytosis signals resulted in infection rates on par with miniAAVR-expressing cells. Thus, trafficking to the TGN appears to increase AAV infection efficiency, but varying rates of endocytosis between the constructs may also contribute to infectivity differences. Nonetheless, all fusion constructs rescued infectivity to an extent, even those that are not reported to traffic to the TGN, indicating that trafficking to the TGN may not be a strict requirement. Collectively, these data suggest a model where AAVR interacts with AAV at the cell surface and facilitates trafficking to the TGN, but do not exclude the possibility that the interaction is initiated in early/late endosomes and/or the TGN.
Fig. 3. AAVR traffics from the plasma membrane to the trans-Golgi network, and its endocytosis is necessary for AAV2 infection.
a, Endogenous AAVR localization in wild-type HeLa cells shown with markers for cis-medial Golgi (giantin) and trans-Golgi network (TGN46). b,Tracking AAVR endocytosis using anti-AAVR antibodies. AAVR-complement cells were incubated with anti-AAVR antibodies for 1 hr at 4°C, washed and then transferred to 37°C. At respective time points, cells were fixed and anti-AAVR antibodies were visualized to depict the trafficking of surface AAVR. c, AAVR surface expression on AAVRKO cells with and without overexpression of full-length AAVR and ΔC-tail (depicted in schematic). d, AAV2-RFP infection (MOI 20,000 vg/cell; measured after 24 hrs) in AAVRKO cells stably expressing constructs depicted in c. Data depicts the mean with s.d. error bars for triplicate infections. Scale bars represent 10 μm.
To test whether other naturally occurring AAV serotypes are also dependent on AAVR, we infected AAVRKO cells with a panel of AAV serotypes including AAV1, 2, 3B, 5, 6, 8 and 9 (expressing GFP or RFP). We also infected cells with an adenovirus 5 vector expressing RFP (rAd5). AAVRKO cells displayed a robust resistance to all AAV serotypes (Fig. 4a), irrespective of the different glycan attachment factors utilized by each serotype. AAV susceptibility was also restored in AAVR-complemented cells, as previously observed with AAV2. Moreover, there was no significant difference in rAd5 infection amongst the three cell lines tested. The role of AAVR in infection for the tested viruses is therefore specific to AAV and is ubiquitously required for a variety of human and simian-derived AAV serotypes.
Fig. 4. AAVR is a critical host factor for the infection of a wide array of naturally-occurring AAV serotypes, and is essential for AAV infection in vivo.
a, Infection of wild-type HeLa cells, AAVR knock-out (AAVRKO) cells, and AAVRKO cells overexpressing AAVR (AAVR complement), using AAV vectors of different serotypes (MOI 105 vg/cell; RFP/GFP expression measured at 24 hrs). b, Bioluminescence of AAV9-infected wild-type (AAVR+/+), heterozygous (AAVR+/−) and AAVRKO (AAVR−/−) FVB mice over 14 days; representative mice from each group are shown with a radiance range of 5×105 – 1×107 p/s/cm2/sr. c, AAV9-luciferase infection for AAVR+/+, AAVR+/−, and AAVR−/− groups (measured as average radiance) at the respective days post infection. d, AAV9-luciferase infection of mice at Day 7. Data depicts the mean (with s.d. error bars in a and c). The P value was determined using an unpaired, two-sided Mann-Whitney t-test where ** - P <0.01, NS – not significant.
Finally, we tested the contribution of AAVR to in vivo gene delivery. We generated AAVR-KO mice (AAVR−/−) using transcription activator-like effector nuclease (TALEN)-mediated gene targeting. AAVR−/− mice did not display any apparent developmental or physical phenotype. Wild-type (AAVR+/+), heterozygous (AAVR+/−) and AAVR−/− FVB mice (genotypes depicted in Extended Data Fig. 7a) were injected intraperitoneally with AAV9-luciferase, chosen because of its high transduction efficiency in vivo compared to AAV222. Bioluminescence (a measure of luciferase expression) was strongest in the lower abdomen of AAVR+/+ mice, intensifying over 14 days (Fig. 4b, c, Extended Data Fig. 7b). AAVR heterozygosity did not significantly reduce AAV9 infection in vivo; however, AAVR−/− mice displayed a pronounced reduction in bioluminescence, comparable to background levels obtained in uninfected wild-type mice (Fig. 4d, Extended Data Fig. 7c).
Overall, this study identifies AAVR as a key host receptor for AAV infection in vitro and in vivo, using an unbiased and comprehensive genetic screening approach. AAV vector usage for gene therapy is rapidly growing, and recent advances in genome editing23 and vectored immunoprophylaxis24 are expected to further expand its utility. Exploiting AAVR as a tool to improve AAV-based applications may enhance its efficacy in basic research and clinical settings. An understanding of AAVR’s tissue distribution will be of importance to determine the contribution of AAVR to AAV tropism, although additional factors25 including glycan usage, AAV nuclear import and other genes identified in our screen could also influence tropism. Lastly, AAV vectors are commonly used in experimental mouse models; hence expression of AAVR under specific promoters (e.g. for cells in the substantia nigra) in an AAVR−/− background may aid in developing better mouse models for human diseases such as those for neurological disorders.
Methods
Cell lines and viruses
All cells were grown in media supplemented with 10% fetal calf serum (FCS) (Sigma, St. Louis), 100 IU/ml penicillin/streptomycin (Sigma, St. Louis) and 2 mM L-glutamine (Sigma, St. Louis), and grown in a humidified incubator at 37°C with 5% CO2. HAP18 cells and K562 cells (American Type Culture Collection (ATCC), Manassas, Virginia, USA) were cultured in complete IMDM media. HT29, U2OS (both obtained from ATCC), Caco-2, A549 (both generous gifts from Dr. Lauren Popov, Stanford University, CA), HEK-293T (from Thermo-scientific, USA), H1-HeLa (from ATCC), HuH7 (generous gift from Dr. Peter Sarnow, Stanford University, CA), MEF (generous gift from Dr. Kelly Storek, Stanford University, CA) and NIH3T3 cells (generous gift from Dr. William Kaiser, Emory University, Atlanta, GA) were all cultured in complete DMEM media. Raji cells (expressing DC-SIGN) (generous gift from Dr. Eva Harris, UC Berkeley, CA) were cultured in complete RPMI media. All isogenic knock-out clones were grown in the same media as parent cell lines. HAP1 cells were utilized for haploid genetic screens (see below). Purified, titred stocks of adeno-associated virus (AAV) serotypes 1, 2, 3B, 5, 6, 8 and 9 were purchased from University of North Carolina Chapel Hill Gene Therapy Center Vector Core. These were all self-complementary AAV vectors encoding a reporter fluorescent gene (either GFP or RFP). Purified, titred stocks of AAV9-luciferase were also purchased from this core facility to perform mouse experiments. Adenovirus type 5 vector carrying the mCherry (rAd5-RFP) was constructed by cloning the mCherry cDNA in the pAd/CMV/V5-DEST gateway vector (Invitrogen) according to manufacturer’s protocol.
Antibodies
The following antibodies were used in this study: mouse polyclonal anti-KIAA0319L (ab105385), and rabbit polyclonal anti-giantin (ab24586) were purchased from Abcam (Cambridge, CA); rabbit polyclonal anti-TGN46 antibody (NBP1-49643) was purchased from Novus Biologicals (Littleton, CO); mouse monoclonal anti-GAPDH (GT239) was purchased from Genetex (Irvine, CA); rabbit polyclonal anti-FGFR1 (D8E4) and rabbit IgG2a isotype control were purchased from Cell Signaling Technology (Danvers, MA); mouse monoclonal phycoerythrin-conjugated anti-c-MET antibody (95106) and phycoerythrin-conjugated mouse IgG1 isotype control were purchased from R&D systems Inc. (Minneapolis, MN). A high-affinity F-actin, fluorescently labeled probe (Alexa fluor-660 phalloidin) was used to visualize the cell interior and periphery (Life Technologies, Carlsbad, CA).
Virus infections
Cells were seeded at 10,000 cells/well (96-well plate) overnight. They were then infected with AAV at a multiplicity of infection (MOI) of 20,000 viral genomes/cell (unless otherwise specified) in complete DMEM. Virus infectivity was determined 24 hours post infection by measuring transgene expression (RFP, GFP or luciferase) using flow cytometry or bioluminescence. In the case of wild-type AAV2 infection, HeLa WT or AAVRKO cells were seeded overnight, then infected with wild-type AAV2 (MOI 1,000) in the presence of wild-type adenovirus-5 (helper virus). Twenty-four hrs post infection, RNA was harvested using the Ambion Cell-to-Ct kit (Thermo-Scientific, USA) and the generated cDNA was used to perform quantitative reverse-transcriptase PCR (qRT-PCR). Rep68 mRNA levels was measured (as a means to detect viral replication) and normalized to 18S ribosomal RNA. Primers against Rep68 cDNA included: 5′-CCAATTACTTGCTCCCCAAA-3′ and 5′-CGTTTACGCTCCGTGAGATT- 3′. Primers against 18S rRNA included: 5′-AGAAACGGCTACCACATCCA -3′ and 5′-CACCAGACTTGCCCTCCA- 3′. Recombinant adenovirus expressing RFP (rAd-RFP) was used to infect cells to obtain 50–60% transduction (Fig. 4A), and flow cytometry was used to measure RFP expression. All infections were performed in triplicate, and all data presented is representative of at least two independent experiments.
Haploid genetic screen
The haploid genetic screen was performed similar to the protocol described in 8 with minor changes. Briefly, gene-trap virus was used to create a mutagenized HAP1 library. Of this mutagenized library, 100 million cells were infected with AAV2-RFP at MOI 20,000. After 48 hrs, infected cells underwent fluorescent-activated cell sorting, where RFP-negative cells (approximately 4% of the population) were sorted and grown over a period of 4 days. The resulting sorted cells were then infected again with AAV2 as before, and re-sorted to enrich the RFP-negative (AAV-resistant) population. Thirty million cells of the resistant population were used for genomic DNA isolation. Sequence analysis of gene-trap insertion sites was performed, and the significance of enrichment for each gene in the screen was calculated by comparing how often that gene was mutated and how often the gene carried an insertion in the control data set (due to random integration). For each gene, a p-value was calculated using the one-sided Fisher exact test in R. The p-values were corrected for multiple testing according to the Benjamini and Hochberg method (using the R statistical package), to control for false discovery rate 26. In the case of KIAA0319L, the p-value was lower than the software could report. The numerical value was thus set to 1 × 10−307 (smallest nonzero normalized floating-point number R could report).
Generation of isogenic knock-out cell lines
CRISPR/Cas9 gene editing technology was used to generate isogenic knock-out alleles by targeting exonic sequences shared among all protein-coding transcripts of the respective genes as described in27. The targeted sequences are depicted in Extended Table S1, along with the respective mutations. CRISPR sequence targeting oligos were designed using the Zhang Lab CRISPR design tool (crispr.mit.edu). Oligos corresponding to the guide RNA sequences in Extended Table S1 were synthesized (Integrated DNA Technologies). Guide RNA oligos were directly cloned into Zhang lab generated Cas9-expressing plasmid px330 or px458 (obtained from Addgene.org - plasmid # 63712 or 48138). Respective cells were transiently transfected with guide RNA-encoding plasmid (and GFP-expressing pcDNA vector with guide RNA-px330 plasmids) using Fugene (Promega, Madison, WI). After 48 hours, GFP-expressing cells were subcloned using the BD InFlux Cell Sorter at the Stanford Shared FACS facility. They were then expanded over 2 weeks and screened genotypically for the mutated allele by extracting genomic DNA from subclones (using the quick DNA™ universal 96-kit (Zymo research, CA, USA)), amplifying a 500–700 bp region that encompassed the guide RNA targeted site, and sequencing (ElimBio, CA, USA) the resulting PCR product to identify subclones with KO mutations. B3GALT6 isogenic KO clone was generated using TALENs directed against the nucleotide sequence 5′-TGGCCATGCTGGCCTGGCTG-3′, and the reverse complement sequence of 5′-GAGTTCGTGCTCAAGGCGGA-3′ in the only exon of B3GALT6 (transcript ENST00000379198) as described28. One day after transfection, cells were selected with Blasticidin S (30 μg/mL, Invivogen) for 24 hours, then stained using anti-heparan sulphate antibody. Cells displaying low staining intensity were subcloned by fluorescence-activated cell sorting.
Construction of plasmids
To generate the AAVR full length construct and ΔC-tail, Gibson assembly reaction kit (New England Biolabs, UK) was used to insert the gene of interest into a lentiviral-based vector, pLenti-CMV-Puro-DEST (w118-1) (plasmid #17452), digested with EcoRV to remove the DEST cassette (a gift from Eric Campeau) 29. AAVR and derived AAVR genes were amplified from a KIAA0319L cDNA clone (clone ID # 3843301) (purchased from GE Dharmacon, Lafayette, CO), but a single nucleotide polymorphism at position 447 was changed from a ‘T’ to a ‘G’ so that sequence aligned to the annotated human genome. The following primers were used to generate PCR products from the human KIAA0319L cDNA to be cloned directly into pLenti CMV Puro DEST.: AAVR full-length: 5′-ATGTGTGGTGGAATTCTGCAGATACCATGGAGAAGAGGCTGGG – 3′ and 5′-CGGCCGCCACTGTGCTGGATTTACTTATCGTCGTCATCCTTGTAATCCAGGATCTCCTCCCGC - 3′; ΔC-tail: 5′ – GACTCTAGTCCAGTGTGGTG - 3′ and 5′ – CGGCCGCCACTGTGCTGGATTTACTTATCGTCGTCATCCTTGTAATCTCCTTTTTGCCTCTTACAAC- 3′. Note that reverse primer was designed to incorporate a C-terminal 1X FLAG tag sequence.
To generate the AAVR deletion constructs, two or three PCR products were generated using AAVR construct (with FLAG tag) as a template. They were then assembled into the pLenti-CMV-Puro-DEST vector using the Gibson Assembly Reaction. Primers used to amplify the N-terminal fragments for the following constructs were: ΔMANEC: 5′ – GACTCTAGTCCAGTGTGGTG - 3′ and 5′ - CTCACTGGCATCTGTTGAC - 3′, ΔPKD1–2: 5′ – GACTCTAGTCCAGTGTGGTG - 3′ and 5′-CAGTTCCTTTATAACTGGGTATGG - 3′, ΔPKD2–3: 5′ – GACTCTAGTCCAGTGTGGTG - 3′ and 5′ - CTTACGGGGCTCTGGC - 3′, ΔPKD3–4: 5′ – GACTCTAGTCCAGTGTGGTG- 3′ and 5′ – GTAATCCACAGCTTTG TTCAC - 3′, ΔPKD4–5: 5′ – GACTCTAGTCCAGTGTGGTG- 3′ and 5′ – CTTATTGTTTTC AGGTTGCACAAT - 3′, miniAAVR: 5′ – GACTCTAGTCCAGTGTGGTG - 3′ and 5′ - CTCACTGGCATCTGTTGAC - 3′, middle fragment of miniAAVR: 5′ – GTCAACAGATGCCAGTGAGGTATCTGCTGGAGAGAGTGTC - 3′, 5′ - CTTATTGTTTT CAGGTTGCACAAT - 3′.
Primers used to amplify the C-terminal fragments for the following constructs were: ΔMANEC: 5′ – GTCAACAGATGCCAGTGAGACACACTCCTCCAATTCCAT - 3′ and 5′ – ATCCAGAGGTTGATTGTCGAG - 3′; ΔPKD1–2: 5′ – CCATACCCAGTTATAAAGGAACTGCCCCCTGTGGCCAACG- 3′ and 5′ - ATCCAGAGGTTGATTGTCGAG - 3′; ΔPKD2–3: 5′ – GCCAGAGCCCCGTAAGCCTCCTCAGGCAGATGC- 3′ and 5′ – ATCCAGAGGTTGATTGTCGAG - 3′; ΔPKD3–4: 5′ – GTGAACAAAGCTGTGGATT ACCCACCTATAGCCAAGATAACTG- 3′ and 5′ - ATCCAGAGGTTGATTGTCGAG - 3′; ΔPKD4–5: 5′ – ATT GTGCAACCTGAAAACAATAAGAACCTGGTGGAGATCATCTTGGATATC- 3′ and 5′ - ATCCAGAGGTTGATTGTCGAG - 3′; miniAAVR: 5′ – ATTGTGCAACCTGAAAACAATAAGTG TGAGTGGAGCGTGTTATATG - 3′ and 5′ - ATCCAGAGGTTGATTGTCGAG - 3′.
AAVR PKD domains 1–5 (residues 311-787) were expressed in E. coli using the pMAL expression system (New England Biolabs, UK). A bacmid, created from a pFastBac Dual vector containing the cDNA for the KIAA0319L ectodomain fused to a C-terminal influenza hemagglutinin (HA)-tag, was a kind gift from Monique van Oers (Wageningen University, Netherlands), and obtained with the assistance of Mary Waye (The Chinese University of Hong Kong, China)30. cDNA coding for PKD domains 1–5 were cloned out of the pFastBacDual expression vector and inserted into the pMAL-c5X vector, using 5′ - GTATCTGCTGGAGAGAGTGTCC AGATAACC -3′ and 5′ - CAGGTTGTTTTTCCTGCAGGTCACCTGGGATCAGGTTTCAC – 3′, then expressed in NEBexpress cells (New England Biolabs, UK). This resulted in an N-terminally-tagged maltose binding protein (MBP) fusion protein, soluble AAVR. MBP was specifically used as an affinity tag for ease of purification.
To create AAVR fusion constructs, MPR-tail, LDLR-tail, and PVR-tail, the Gibson assembly reaction was used to fuse amplified miniAAVR without its C-terminal to the C-terminal of the respective proteins, and insert it into the pLenti-CMV-Puro-DEST vector. Primers used for amplification and insertion included: miniAAVR without C-terminal and transmembrane domain for MPR-tail: 5′ – GACTCTAGTCCAGTGTGGTG- 3′ and 5′ – CTTATTGTTTTCAGGTTGCACAAT- 3′; MPR C-terminal and transmembrane: 5′ – ATTGTGC AACCTGAAAACAATAAGGCTGTGGGAGCTGTGC- 3′ and 5′ - CGGCCGCCACTGTGC - 3′; miniAAVR without C-terminal and transmembrane domain for LDLR-tail or PVR-tail: 5′ – GACTCTAGTCCAGTGTGGTG- 3′ and 5′ – CTTATTGTTTTCA GGTTGCACAAT - 3′; LDLR or PVR C-terminal and transmembrane: 5′ – ATTGTGCAACCTGAA AACAATAAG- 3′ and 5′ - TAAATCCAGCACAGTGGCGGCCG - 3′.
Generation of stable cell lines
Lentiviral transduction was used to create stable cell lines expressing a selected gene of interest under a CMV promoter. Using Gibson assembly reaction, the respective genes of interest (see construction of plasmids section) were inserted into the pLenti-CMV-Puro-DEST vector, and used as described previously 29. Lentivirus was produced using HEK293 cells and utilized to transduce the respective cell lines overnight. Cells stably expressing the gene of interest were selected by treatment with 1–3 μg/ml puromycin over 2 days (InvivoGen). A lentivirus carrying the mCherry (RFP) gene was used as a control for AAVR complementation in AAVRKO cells.
Flow cytometry
All flow cytometry was performed at the Stanford Shared FACS facility. To perform the haploid genetic screen, fluorescence-activated cell sorting was carried out on a BD FACS Aria flow-cytometer (BD, Franklin Lakes, NewJersey, USA). To measure virus transgene expression (RFP/GFP) in all other experiments, cells were trypsinized 24 hours after infection and a BD LSRII-UV flow cytometer (BD, Franklin Lakes, NewJersey, USA) was used to detect fluorescent cells. For cell surface staining, cells were trypsinized and washed using FACS buffer (1×PBS supplemented with 2% FCS, 1mM EDTA and 0.1% sodium azide). They were subsequently incubated for 40 min at 4°C with the respective primary antibodies at a 1:50 dilution (see Antibody section), washed, and incubated for a further 40 min at 4°C with Alexa488 or Alexa594-conjugated secondary antibodies (1:500 dilution) (if the primary was not conjugated) (Life Technologies, Carlsbad, CA). This was followed by a final wash, and resuspension of cells in FACS buffer before reading fluorescence. All data presented is representative of at least two independent experiments. Data was analyzed and assembled using FlowJo software (TreeStar Inc, Ashland, Oregon, USA).
Immunoblot Analysis
Cell pellets of 2×106 cells were lysed with Laemmli SDS sample buffer containing 5 % β-mercaptoethanol and boiled for 10 minutes at 96°C. Lysates were separated by SDS-PAGE using the Mini-Protean system (Bio-Rad) on 4–15% polyacrylamide gradient gels (Bio-Rad). Proteins were transferred onto PVDF membranes (Bio-Rad) using the Bio-Rad Transblot protein transfer system in a semi-wet preparation. Membranes were blocked by incubating with 1×PBS buffer containing 5% non-fat milk for 1 hr at room temperature (RT). Membranes were subsequently incubated overnight at 4°C with primary antibodies at a dilution of 1:1000 (anti-KIAA0319L antibody) or 1:2000 (anti-GAPDH antibody) in blocking buffer. Membranes were washed 3 times for 5 min using wash buffer (1×PBS buffer with 0.1% Tween-20), and further incubated in HRP-conjugated secondary antibodies (anti-mouse and anti-rabbit- 1:5000 in blocking buffer) (GeneTex) for 1 hr at RT. After another set of three washes, antibody-bound proteins were visualized on film using the West Pico and Extended Duration chemiluminescence peroxide solutions (Thermo-Scientific, USA).
Immunofluorescence
Cells were seeded overnight at 40,000 cells/well onto LabTekII glass chamber slides (Thermo-Scientific, USA). They were washed once with 1×PBS, and either treated or fixed immediately with 4 % paraformaldehyde for 15 minutes. They were washed 3 times with 1×PBS before being incubated for 1 hr at RT with primary antibodies against the respective proteins at a dilution of 1:100 (anti-KIAA0319L and anti-TGN46) or 1:200 (anti-giantin) in IF blocking buffer (PBS with 3% BSA, 1% saponin and 1% Triton X-100). Cells were then washed three times in 1×PBS, and incubated for a further hour in DAPI stain (1:500) and fluorescently-tagged secondary antibodies (Alexa488 anti-mouse and Alexa594 anti-rabbit – Life Technologies) at a dilution of 1:300. Cells were washed a final three times in 1×PBS, and 5 μl of Vectashield (Vector Laboratories Inc, Burlingame, CA) was applied to each slide chamber before a glass cover slip (VWR, USA) was placed over slide to mount samples. Cells were visualized directly with a Zeiss LSM 700 confocal microscope.
Enzyme-linked immunosorbent assay (ELISA)
Purification of the soluble AAVR was achieved through amylose-based MBP affinity chromatography (GE Healthcare). ELISA plates (Corning Costar) were coated overnight at 4°C with 50ul AAV2 virus-like particles (VLPs) at 2.5 μg/ml in 100 mM NaHCO3, pH 9.6. Plates were then washed 2X with TBST (0.05% Tween-20 in TBS) and blocked with 3% BSA in TBST for 1 hr at RT. Subsequent washing was followed by incubation with soluble AAVR or MBP control at the indicated concentrations for 2 hrs at RT. Anti-MBP-HRP (1:500, 1 hr incubation at RT) was used to detect rAAVR1-5 and MBP controls, requiring no secondary antibody. Samples were developed with 1-Step Ultra TMB-ELISA substrate per product instructions (Thermo Scientific, USA) and optical density assayed by microplate reader (Molecular Devices SpectraMax M2e) at 450 nm. Curve fitting was performed in SigmaPlot v12.5 (Systat Software, Inc., USA). All data presented is representative of at least three independent experiments.
Surface Plasmon Resonance (SPR) Analysis of binding
SPR was carried out using a BIAcore X instrument (GE Healthcare) using a flow rate of 10 μL/min at 20°C in HBS-P buffer (10 mM HEPES pH 7.5, 150 nM NaCl and 0.005% surfactant P20). His-tagged soluble AAVR (His-tagged MBP fusion with AAVR PKD domains 1–5) at various concentrations was mixed with His-tagged MBP to a total concentration of 0.2 μM in 10 mM sodium acetate buffer pH 4.0 and immobilized on a CM5 sensor chip through amide coupling. MBP at 0.2 μM was sufficient to block non-specific binding to the dextran. For the analysis of binding affinity, all curves were measured in triplicate and were fitted with a Langmuir 1:1 binding model (BIAevaluation software, GE Healthcare).
Antibody inhibition assay
Wild-type HeLa cells were seeded in 96-well plates at 10,000 cells/well overnight. Anti-AAVR antibody (ab105385) or IgG isotype control (both from Abcam, Cambridge, CA) were incubated with cells (at concentrations ranging from 0.5 to 50 μg/ml in DMEM media) for 1 hr at 4°C. Cells were then infected with AAV2-luciferase at MOI 1,000 vg/cell, and left for 24 hrs at 37°C. A luciferase assay kit (#E1500, Promega, Madison, WI) was used to detect bioluminescence, with measurements being taken on the Promega GLOMAX luminometer. Importantly, the storage buffers of both antibodies did not contain preservatives such as azide that could interfere with the assay. All data presented is representative of two independent experiments.
Competitive inhibition assay
HeLa cells were seeded in 96-well plates at 10,000 cells/well overnight. Purified soluble AAVR, or MBP control was then introduced to the medium at the specified concentrations. Cells were transduced with AAV2-GFP at MOI 7,500 vg/cell and incubated for 24 hrs at 37°C. This was followed by trypsinization and measuring transgene expression by flow cytometry. For immunofluorescence imaging, concentration of soluble AAVR and MBP controls was 0.1μM, and transduction was done using 7000 vg/cell. At 24 hrs post-transduction, cells were incubated with 1 μg/ml Hoechst stain (Thermo Scientific) in PBS for 10 min at 37°C, before washing with PBS and subsequent fluorescent imaging (Nikon Eclipse Ti-E). All data presented is representative of two independent experiments.
Tracking surface-bound AAVR using anti-AAVR antibodies
These experiments were performed similarly to Ci-MPR tracking assays, as described in 31. AAVRKO cells with or without overexpression of AAVR or ΔC-tail were incubated at 4°C with anti-AAVR antibodies (approximately 25μg/ml) for 1 hr. Cells were then washed three times with 1xPBS and transferred to 37°C for specific time points (2, 10, 30 and 60 min), at which time they were fixed with 4% PFA for 15 min. Following fixation, immunofluorescence staining (as described above) was performed to visualize AAVR endocytosis. All data presented is representative of two independent experiments.
Ethics statement and animal studies
All the experiments involving animals were conducted in strict accordance with the Institutional Animal Care and Use Committee of Stanford University. Mice were housed in a Stanford University vivarium that is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International. Mice were housed in irradiated disposable caging (Innovive, San Diego, California) with biweekly cage changes. Mice were provided with irradiated food and UV-irradiated, acidified water. Health surveillance was performed via trimester testing of dirty-bedding CD1 sentinels (Charles River Laboratories, Hollister, CA). Sentinels were consistently negative for mouse parvovirus, minute virus of mice, mouse hepatitis virus, rotavirus, murine encephalomyelitis virus, Sendai virus, mouse adenovirus 1 and 2, ectromelia, lymphocytic choriomeningitis virus, pneumonia virus of mice, reovirus 3, Mycoplasma pulmonis, and endo- and ectoparasites. No statistical methods were used to predetermine sample size. In our animal study protocol, we state that the number of animals in each experimental group varies, and is based on similar previous study32. Randomization was not used to allocate animals to experimental groups and the animal studies were not blinded.
AAV infection in mice
TALEN technology was used to create AAVR isogenic knock-out FVB mice (purchased from Cyagen Biosciences, Santa Clara, CA). TALEN targeted sequences were 5′–TGGGAGTCAAGCCAAGTC–3′ and 5′–GCCAGGATATTGTTGGCAGA–3′. Two founder males were mated to FVB/NCrl (Charles River Laboratories, Hollister, CA) females. After 3 rounds of breeding, wild-type (AAVR+/+), heterozygous (AAVR+/−) and homozygous AAVRKO (AAVR−/−) mice were generated, determined by genotyping. All genotypes (wild-type, heterozygous, and knock-out) were obtained in the expected Mendelian ratios after breeding. At 5 weeks of age, animals from each group (AAVR+/+ - n=7 (2 litter mates and 5 purchased FVB mice); AAVR+/− - n=4; and AAVR−/− - n=4) were injected intraperitoneally with 1 × 1011 viral genomes of AAV9-luciferase in 200 μl of 1× phosphate-buffered saline. All the mice recovered from the injection quickly without loss of mobility or interruption of grooming activity. AAVR+/+ and AAVR−/− mice were found to be significantly different in two independent experiments. The second experiment was set up as described above, but groups consisted of 3 mice each, and only AAVR+/+ and AAVR−/− mice were compared.
In vivo bioluminescence imaging
The mice were anesthetized with 2% isofluorane and oxygen. The D-luciferin substrate (Biotium, Hayward, CA) was injected intraperitoneally (3.3 μg per mouse). After 10 min, the mice were then placed in a light-tight chamber, and images were generated using a cryogenically cooled charge-coupling device camera IVIS 100 (Xenogen, Alameda, CA), recording bioluminescence at 1, 10, 60 and 100 sec. The visual output represents the average radiance as the number of photons emitted/second/cm2 as a false color image where the maximum is red and the minimum is dark blue. All animals were imaged on a schedule of 3, 7, 10 and 14 days after AAV vector injection. At each time-point a “region of interest” was designated surrounding each animal in order to quantify the radiance (photons/sec/cm2/radian) being released by luciferase activity. This region was kept the same for each mouse and at each time point. The mean and standard deviation of radiance measurements was determined for each mouse group at each time point.
Statistics
The unpaired, parametric, two-sided student t-test was used for statistical calculations involving two group comparisons in all tissue culture-based experiments (* P < 0.05, ** P < 0.01, *** P < 0.001), with a Welch correction accounting for different standard deviations. An unpaired, two-sided Mann-Whitney t-test was used for statistical calculations involving two group comparisons in in vivo experiments. GraphPad Prism was used for statistical calculations.
Extended Data
Extended Data Figure 1. Surface molecules, FGFR1 and c-MET, are not essential for AAV2 infection.
a, Region of FGFR1, c-MET, or B3GALT6 genes (previously-identified co-receptors/attachment factors5–7) targeted by CRISPR guide RNA or TALENs in wild-type HAP1 cells, and the resulting genotypes of derived knock-out cell lines. (see full sequence in Extended Data Table 1). All CRISPR- or TALEN-created mutations disrupt the open reading frame of the targeted gene. b, Surface staining for the respective receptors in respective cell lines. Isotype antibodies for the receptor antibodies were used as controls. c, AAV2-RFP infection (MOI 5,000 viral genomes (vg)/cell; measured after 24 hrs) of wild-type and knock-out cell lines. Data depicts the mean with s.d. for triplicate infections. * - p <0.05, *** - p <0.001; analyzed using an unpaired, parametric, two-sided student t-test, with a Welch post-correction. c-MET: hepatocyte growth factor receptor; FGFR1: fibroblast growth factor receptor-1. FITC or PE refer to fluorescently-labeled antibody conjugates used to visualize surface receptors. MOI: multiplicity of infection, RFP: red-fluorescent protein, SSC: side scatter.
Extended Data Figure 2. Haploid, unbiased genetic screen evaluating host factors important for AAV2 infection.
a, A schematic depicting the strategy for the AAV2 genetic screen. A library of mutagenized haploid, HAP1 cells was created with a retroviral gene trap vector, and subsequently infected with AAV2-RFP (MOI 20,000 vg/cell) for 24 hrs. RFP-negative cells were sorted using FACS to isolate those cells with mutations in genes essential for AAV2 infection. These cells were re-infected for a second iteration of selection. DNA was then extracted from this enriched population and sequenced to specifically map where the gene trap insertions occurred that resulted in the mutation. b, The gating strategy for the FACS-based AAV2 screen. FACS: fluorescence-activated cell sorting, RFP: red-fluorescent protein, SSC: side scatter.
Extended Data Figure 3. AAVR is a critical host factor for AAV2 infection.
a, Effect of AAVR isogenic knock-out (AAVRKO) upon AAV2-luciferase infection, evaluated in HAP1 and HeLa cell background from MOI of 100 to 100,000 vg/cell. b, Quantitative RT-PCR to detect wild-type AAV2 infection in wild-type (WT) HeLa or AAVRKO cells. Cells were infected with wild-type AAV2 and adenovirus (helper virus required for AAV2 replication), and AAV2 rep68 mRNA levels were measured to assess AAV2 infection. c, Immunoblot analysis evaluating AAVR expression in WT, AAVRKO and AAVRKO overexpressing AAVR (AAVR Comp.) cell lines of HAP1 and HeLa origin. GAPDH was immunoblotted as a control. AAVR (predicted 115 kDa) appears at 150 kDa due to 6 glycosylation sites. d, AAV2-luciferase infection (MOI 20,000 vg/cell; measured after 24 hrs) in AAVRKO cells stably complemented with AAVR or control lentiviral vector, evaluated in several AAV2-susceptible human and mouse cell lines. e, Comparison of AAV2-RFP infection (MOI 20,000 vg/cell; measured after 24 hrs) in WT, AAVRKO, c-METKO and FGFR1KO cells, evaluated in several AAV2-susceptible human cell lines. RLU: relative light units. Data depicts the mean with s.d. error bars for triplicate infections.
Extended Data Figure 4. AAVR specifically binds to AAV2.
a, ELISA measurement of the binding to AAV2 particles of MBP at concentrations of 0.05 – 2,000 nM. This serves as a control to the ELISA data depicted in Figure 2c. b, Representative surface plasmon resonance sensograms (collected in triplicate), with a ligand (AAVR) concentration of 4nM and an analyte (AAV-2) concentration as indicated, to measure binding of AAV-2 particles to AAVR. c, Simultaneous addition to cells of AAV2-GFP particles with soluble AAVR or MBP (both at 0.1 μM) to evaluate AAVR’s binding effect on AAV2 infection. Fluorescence was imaged 24 hrs post infection. This data complements Figure 2d. Data in a depicts the mean with s.d. error bars for triplicate infections. Scale bars represent 50 μm.
Extended Data Figure 5. AAVR ΔC-tail is detected at the cell surface and does not endocytose to the TGN.
AAVRKO cells (a) or ΔC-tail-expressing cells (c) were incubated with anti-AAVR antibodies for 1 hr at 4°C, washed and then transferred to 37°C. At respective time points, cells were fixed and antibody-bound AAVR was visualized. This data complements Figure 3b. b, Permeabilized and unpermeabilized immunostaining of full-length AAVR and ΔC-tail when expressed in AAVRKO cells. This data complements Figure 3c. Scale bars represent 10 μm.
Extended Data Figure 6. AAVR endocytosis is crucial for AAV2 infection.
a, Schematic of the AAVR minimal construct (miniAAVR) and domain-swapped derivatives probing the localization of AAVR through the swapping of AAVR’s C-tail with that of well-characterized recycling receptors: cation-independent mannose-6-phosphate receptor (Ci-MPR) (traffics from plasma membrane (PM) through endosomes to the TGN), low density lipoprotein receptor (LDLR) and poliovirus receptor (PVR) (both traffic from PM to endosomal compartments but are not reported to traffic to TGN). b, Corresponding permeabilized and unpermeabilized immunofluorescence images of constructs depicted in a when expressed in AAVRKO cells. c, AAV2-RFP infection (MOI 20,000 vg/cell; measured after 24 hrs) in AAVRKO cells stably expressing constructs depicted in a. Data depicts the mean with s.d. for triplicate infections. Scale bars represent 10 μm.
Extended Data Figure 7. AAVR is essential for AAV infection in vivo.
a, Genotypes of FVB mice littermates used to perform in vivo studies. AAVR KO (AAVR−/−) were bred from heterozygous (AAVR+/−) parent mice; AAVR+/− and AAVR−/− mice display frameshift mutations in targeted genes in 1 or 2 alleles respectively. Sequences recognized by the TALENs are displayed in yellow. b, AAV9-luciferase infection (as measured by average radiance) for all infected mice depicted for Day 3, 10 and 14 (Day 7 is shown in Figure 4d). c, Bioluminescence in all wild-type (AAVR+/+), AAVR+/− and AAVR−/− FVB mice 7 days post AAV9-luciferase infection (does not include those displayed in Figure 4b). Radiance range of 2×105 – 1×107 p/s/cm2/sr. The P value was determined using an unpaired, two-sided Mann-Whitney t-test where ** - P <0.01, NS – not significant.
Extended Data Table S1.
Indel mutations in genes of isogenic knock-outs created by CRISPR/Cas9 or TALEN genome engineering.
| Gene | CRISPR/TALEN region (bold) | Cell line | Indel mutation in allele | Mutation |
|---|---|---|---|---|
| KIAA0319L (AAVR) | CCAGTGACGTAGTTACACCTATAGTG | HEK293 | CC--------AGTTACACCTATAGTGACAC | 8 bp del |
| CCAG--------------CTATAGTGACAC | 14bpdel | |||
| CCAGTG--GTAGTTACACCTATAGTGACAC | 2 bp del | |||
| A549 | CCAGT-ACGTAGTTACACCTATAGTGACAC | 1 bp del | ||
| CCAGTGAACGTAGTTACACCTATAGTGACAC | 1 bp ins | |||
| U20S | C-----------------CCTATAGTGACA | 17 bp del | ||
| CCAGTG-----173i-----ACGTAGTTAC | 173 bp ins | |||
| HuH7 | CCAGTG------271d------------ | 271 bp del | ||
| CCAGT-ACGTAGTTACACCTATAGTGACAC | 1 bp del | |||
| CCAGTG--GTAGTTACACCTATAGTGACAC | 2 bp del | |||
| K562 | CC--------AGTTACACCTATAGTGACAC | 8 bp del | ||
| CCAGTGAC---------69d---------- | 69 bp del | |||
| CTTGCTTTTGCTTCAGCGTTCTGTGG | HAP1 | CTTGCTTTTGCTTCAGCGTTTCTGTGGTTG | 1 bp ins | |
| HeLa | CTTGCTTTTGCTT-----147d-------- | 147 bp del | ||
| CTTGCTTTTGCTTCAGCG--TCTGTGGTTG | 2 bp del | |||
|
| ||||
| AU040320 (mouse AAVR) | GACTCTGCCTGCCACGCTCTATGGTG | MEF | GACTCTGCCTGCCACGCTCTA-GGTGGCTG | 1 bp del |
| GACTCTGCCTGCCACGCT----GGTGGCTG | 4 bp del | |||
|
| ||||
| FGFR1 | ACCGCTAGGCCGTCCCCGACCTTGCC | HAP1 | CACCGCTAGGGCCGTCCCCGACCTTGCCTG | 1 bp ins |
| HEK293 | CACC-------------CGACCTTGCCTG | 13 bp del | ||
| A549 | CACCGCTAGGGCCGTCCCCGACCTTGCCTG | 1 bp ins | ||
| K562 | CACCGCTAG-CCGTCCCCGACCTTGCCTG | 1 bp del | ||
| HeLa | CACCGCTA-----------------CCTG | 17 bp del | ||
|
| ||||
| cMET | CATTAGCTGTGGCAGCGTCAACAGAG | HAP1 | TCATTAGCTGTGGCAGCGTCA-CAGAGGGA | 1 bp del |
| HEK293 | TCATTAGCTGTGGCAGCGTCAAACAGAGGG | 1 bp ins | ||
| A549 | TCATTAGCTGTGGCA--27d & 5i--CCT | 27 bp del & 5 bp ins | ||
| K562 | TCATTAGCTGTGGCAGCGTCA-CAGAGGGA | 1 bp del | ||
| TCATTAGCTGTGGCAGCGTCAAACAGAGGG | 1 bp ins | |||
| HeLa | TCATTAGCTGTGGCAGCGTCA--AGAGGGA | 2 bp del | ||
| TCATTAGCTGTGGCAGCGTCAAACAGAGGG | 1 bp ins | |||
| B3GALT6 |
TGGCCATGCTGGCCTGGCTGGACGAGCAC GTGGCCTTCGAGTTCGTGCTCAAGGCGGA |
HAP1 | GCTGGACGAGCACGT--188i---------GGCCTTCGAG | 188 bp ins |
del = deletion; ins = insertion
Supplementary Material
Acknowledgments
The authors would like to thank: Karla Kirkegaard, Mark Kay and Thijn Brummelkamp for critical reading of the manuscript and valuable advice; Tom Lerch, Jeffrey Tyner, David Kabat and Hiroyuki Nakai for assistance with preliminary experiments; Hans Peter Bächinger for advice and assistance with surface plasmon resonance experiments, Stanford Shared FACS facility and its staff; Xuhuai Ji (Stanford Functional Genomics Facility); Stanford mouse facility; Tim Doyle for small animal imaging training; Lauren Popov for guidance in generating immunofluorescent images, Gabrielle Fuchs for technical assistance, and members of the Carette and Chapman labs for intellectual discussions and support. The work was funded in part by NIH R01 GM066875 (MSC), DP2 AI104557 (JEC) and U19 AI109662 (JEC). JEC is a David and Lucile Packard Foundation fellow.
Footnotes
Author contributions: SP, MSC and JEC were responsible for overall design of the study. SP performed the haploid genetic screen, generated isogenic knock-out and AAVR complement cell lines, and performed antibody inhibition and AAVR tracking studies. NM designed soluble AAVR construct and performed binding and soluble AAVR inhibition studies. JEC designed AAVR-generated deletion mutant constructs. ASP performed the wild-type AAV2 infection assay and all in vivo studies, under the technical expertise of CMN. OD was responsible for heterologous overexpression and purification of soluble AAVR, JD assisted in the production of FGFR1KO and c-METKO cell lines, YI performed surface plasmon resonance measurements, LTJ generated the B3GALT6KO cell line, and JEW created the ΔC-tail construct. SP, MSC and JEC wrote the manuscript.
DNA sequencing data have been deposited in the NCBI sequencing read archive under NCBI Bioproject PRJNA284536 with BioSample SAMN03703230 (Gene trap control data set) and SAMN04244346 (AAV screen).
The authors declare competing financial interests: Stanford university is in the process of filing a patent claim regarding AAVR applications in AAV vector technology (Inventors: S.P., M.S.C., J.E.C., A.S.P., N.L.M. and O.D.).
References
- 1.Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nature reviews. Genetics. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathwani AC, et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. The New England journal of medicine. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaudet D, Methot J, Kastelein J. Gene therapy for lipoprotein lipase deficiency. Current opinion in lipidology. 2012;23:310–320. doi: 10.1097/MOL.0b013e3283555a7e. [DOI] [PubMed] [Google Scholar]
- 4.Lisowski L, et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. Journal of virology. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kashiwakura Y, et al. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. Journal of virology. 2005;79:609–614. doi: 10.1128/JVI.79.1.609-614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qing K, et al. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nature medicine. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 8.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonifacino JS, Hierro A. Transport according to GARP: receiving retrograde cargo at the trans-Golgi network. Trends in cell biology. 2011;21:159–167. doi: 10.1016/j.tcb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGough IJ, Cullen PJ. Recent advances in retromer biology. Traffic. 2011;12:963–971. doi: 10.1111/j.1600-0854.2011.01201.x. [DOI] [PubMed] [Google Scholar]
- 11.Poelmans G, Buitelaar JK, Pauls DL, Franke B. A theoretical molecular network for dyslexia: integrating available genetic findings. Molecular psychiatry. 2011;16:365–382. doi: 10.1038/mp.2010.105. [DOI] [PubMed] [Google Scholar]
- 12.Ellis BL, et al. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virology journal. 2013;10:74. doi: 10.1186/1743-422X-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen J, Qing K, Kwon HJ, Mah C, Srivastava A. Impaired intracellular trafficking of adeno-associated virus type 2 vectors limits efficient transduction of murine fibroblasts. Journal of virology. 2000;74:992–996. doi: 10.1128/jvi.74.2.992-996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibraghimov-Beskrovnaya O, et al. Strong homophilic interactions of the Ig-like domains of polycystin-1, the protein product of an autosomal dominant polycystic kidney disease gene, PKD1. Human molecular genetics. 2000;9:1641–1649. doi: 10.1093/hmg/9.11.1641. [DOI] [PubMed] [Google Scholar]
- 15.Bhella D. The role of cellular adhesion molecules in virus attachment and entry. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2015;370:20140035. doi: 10.1098/rstb.2014.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxfield FR, McGraw TE. Endocytic recycling. Nature reviews. Molecular cell biology. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 17.Kelly BT, Owen DJ. Endocytic sorting of transmembrane protein cargo. Current opinion in cell biology. 2011;23:404–412. doi: 10.1016/j.ceb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Nonnenmacher M, Weber T. Adeno-associated virus 2 infection requires endocytosis through the CLIC/GEEC pathway. Cell host & microbe. 2011;10:563–576. doi: 10.1016/j.chom.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nature reviews. Molecular cell biology. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 20.Beglova N, Blacklow SC. The LDL receptor: how acid pulls the trigger. Trends in biochemical sciences. 2005;30:309–317. doi: 10.1016/j.tibs.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Ohka S, et al. Receptor (CD155)-dependent endocytosis of poliovirus and retrograde axonal transport of the endosome. Journal of virology. 2004;78:7186–7198. doi: 10.1128/JVI.78.13.7186-7198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Molecular therapy : the journal of the American Society of Gene Therapy. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 23.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balazs AB, et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nonnenmacher M, Weber T. Intracellular transport of recombinant adeno-associated virus vectors. Gene therapy. 2012;19:649–658. doi: 10.1038/gt.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 27.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanjana NE, et al. A transcription activator-like effector toolbox for genome engineering. Nature protocols. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campeau E, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PloS one. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holster S, et al. Expression of the dyslexia candidate gene Kiaa0319-like in insect cells. Journal of Biochemistry and Molecular Biology in the Post Genomic Era. 2013;2:ePub. [Google Scholar]
- 31.Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. The Journal of cell biology. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jae LT, et al. Virus entry. Lassa virus entry requires a trigger-induced receptor switch. Science. 2014;344:1506–1510. doi: 10.1126/science.1252480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.