Abstract
The current subgenus Drosophila (the traditional immigrans-tripunctata radiation) includes major elements of temperate drosophilid faunas in the northern hemisphere. Despite previous molecular phylogenetic analyses, the phylogeny of the subgenus Drosophila has not fully been resolved: the resulting trees have more or less varied in topology. One possible factor for such ambiguous results is taxon-sampling that has been biased towards New World species in previous studies. In this study, taxon sampling was balanced between Old and New World species, and phylogenetic relationships among 45 ingroup species selected from ten core species groups of the subgenus Drosophila were analyzed using nucleotide sequences of three nuclear and two mitochondrial genes. Based on the resulting phylogenetic tree, ancestral distributions and divergence times were estimated for each clade to test Throckmorton’s hypothesis that there was a primary, early-Oligocene disjunction of tropical faunas and a subsequent mid-Miocene disjunction of temperate faunas between the Old and the New Worlds that occurred in parallel in separate lineages of the Drosophilidae. Our results substantially support Throckmorton’s hypothesis of ancestral migrations via the Bering Land Bridge mainly from the Old to the New World, and subsequent vicariant divergence of descendants between the two Worlds occurred in parallel among different lineages of the subgenus Drosophila. However, our results also indicate that these events took place multiple times over a wider time range than Throckmorton proposed, from the late Oligocene to the Pliocene.
Introduction
The genus Drosophila is the most speciose and intensively studied species assemblage in the family Drosophilidae, yet the genus has long been recognized as paraphyletic since the 1975 report of Throckmorton [1], and this characterization has been confirmed in all subsequent, family-wide phylogenetic studies [2–6]. In the most recent molecular phylogenetic analysis [6], the genus Drosophila was considered paraphyletic with respect to 15 different genera. Together, these lineages constitute the tribe Drosophilini and are divided into two major clades: one comprises the genus Lordiphosa and the subgenus Sophophora of Drosophila, and the other includes 14 genera and four subgenera of Drosophila, including the subgenus Drosophila. The subgenus Drosophila is comprised of two main clades [4–14] which are interspersed with eight genera and two other subgenera of Drosophila [6]. One clade of the subgenus Drosophila had been referred to as the virilis section [15] or the virilis-repleta radiation [1,16], and the other as the quinaria section [15] or the immigrans-tripunctata radiation [1,16]. To resolve this paraphyly, Yassin [6] revised the phylogenetic classification of the subgenus Drosophila: the virilis-repleta clade (the virilis section) was revised as the subgenus Siphlodora, while the immigrans-tripunctata clade (the quinaria section) was left in the subgenus Drosophila. The revised subgenus Drosophila includes 21 species groups [6], of which 15 have been regarded as members of the immigrans-tripunctata clade [4]. They are widely distributed mainly in the northern hemisphere, with four species groups endemic to the Old World (the ancora, bizonata, histrio and immigrans groups), 14 to the New World (the appendiculata, calloptera, cardini, ecuadoriensis, guarani, guttifera, lutzii, macroptera, pallidipennis, peruensis, rubrifrons, sticta, tripunctata and xanthopallescens groups) and three distributed in both (the funebris, quinaria and testacea groups). Some of them, such as the funebris, guttifera, quinaria and testacea groups, represent major elements of temperate drosophilid faunas in the northern hemisphere.
The revised subgenera Siphlodora and Drosophila show parallel biogeographic patterns, i.e., the disjunctions between the Old and the New Worlds in tropical elements (species groups) and in species of temperate species groups. Throckmorton [1] hypothesized that this overall pattern of faunal disjunction between the Old and the New Worlds had emerged in parallel in five lineages (radiations in his descriptions) of Drosophilidae, i.e., in the Scaptodrosophila radiation, the sophophoran radiation, the virilis-repleta radiation (the revised Siphlodora), the immigrans-tripunctata radiation (the revised Drosophila) and the Hirtodrosophila radiation. However, this hypothesis was not presented in a testable way based on solid evidence of phylogenetic relationships. To remedy this, multiple DNA sequence-based studies have provided estimates of divergence times and biogeographic analyses of Old World and New World groups in some lineages of the Drosophilidae [17–20].
Phylogenetic relationships within the revised subgenus Drosophila or the immigrans-tripunctata radiation have been analyzed in a number of studies based on molecular data [2,3,5,6,12–14,19–24]. However, the resulting trees have varied in topology, especially with respect to the basal branches, and have not supported the monophyly of some species groups. Two recent molecular phylogeographic analyses failed to fully resolve the relationships in the immigrans-tripunctata radiation with high confidence [19,20]. With less resolved phylogenetic hypotheses, estimates of divergence times and ancestral distributions can become equivocal, and inferences for evolutionary history are controversial. Limited taxon sampling can contribute to these ambiguous results, and in many cases such sampling has been biased to New World taxa [18].
We gathered species widely from both the New World and the Old World, selecting 22 species from the Old World, 21 from the New World, one from the Australasian region and one cosmopolitan species in an attempt to reduce the taxon sampling bias. We estimated phylogenetic relationships by analyzing the nucleotide sequences of three nuclear and two mitochondrial genes and estimated ancestral distributions and divergence times in order to test Throckmorton’s [1] hypothesis about parallel patterns of Old World-New World divergence in the evolution of Drosophilidae.
Materials and Methods
Taxon sampling
A total of 45 species were selected as focal ingroup taxa from ten core species groups (bizonata, cardini, funebris, guarani, guttifera, histrio, immigrans, quinaria, testacea and tripunctata) of the revised subgenus Drosophila (the immigrans-tripunctata radiation). As outgroup taxa, D. virilis, D. hydei and Idiomyia grimshawi were selected from the sister clade of the subgenus Drosophila [6,19,20]. DNA was extracted from either live or alcohol-preserved specimens. The specimens of D. innubila, D. recens, D. tenebrosa and D. transversa were kindly provided by Prof. John Jaenike (University of Rochester). The remaining 41 specimens were obtained from the UC San Diego Drosophila Stock Center (DSSC), the Tokyo Metropolitan University (TMU) or field collections (Table 1). We determined the DNA sequences of alcohol dehydrogenase (Adh) for 35 species, glycerol-3-phosphate dehydrogenase (Gpdh) for 36 species, mitochondrial 28S ribosomal RNA (28S) for 45 species, cytochrome c oxidase subunit I (COI) for 45 species, and cytochrome c oxidase subunit II (COII) for 45 species. GenBank accession numbers of these sequences, along with those for known sequences used in the analyses, and taxonomic and biogeographic information are given in Table 1.
Table 1. List of studied species.
| Genus | Subgenus | Species group | Species | Distribution | Source (Stock No.) | Adh | Gpdh | COI | COII | 28S |
|---|---|---|---|---|---|---|---|---|---|---|
| Drosophila | Drosophila | bizonata | bizonata | Palearctic, Oriental, Australasian | Kyoto, Japan | AB932648 | AB932684 | AB932720 | AB932765 | AB932810 |
| cardini | acutilabella | Nearctic, Neotropical | DSSC (15181–2171.10) | AB932645 | AB932681 | AB932716 | AB932761 | AB932806 | ||
| arawakana | Neotropic | DSSC (15182–2261.03) | AB932647 | AB932683 | AB932719 | AB932764 | AB932809 | |||
| cardini | Nearctic, Neotropical | DSSC (15181–2181.03) | AB932650 | AB932686 | AB932722 | AB932767 | AB932812 | |||
| cardinoides | Neotropic | DSSC (15181–2291.00) | AB932651 | AB932687 | AB932723 | AB932768 | AB932813 | |||
| dunni | Neotropic | DSSC (15182–2291.00) | AB932654 | AB932690 | AB932726 | AB932771 | AB932816 | |||
| nigrodunni | Neotropic | DSSC (15182–2311.00) | AB932663 | AB932698 | AB932741 | AB932786 | AB932831 | |||
| parthenogenetica | Neotropic | DSSC (15181–2221.00) | AB932666 | AB932701 | AB932745 | AB932790 | AB932835 | |||
| polymorpha | Neotropic | DSSC (15181–2231.00) | AB932668 | AB932703 | AB932747 | AB932792 | AB932837 | |||
| similis | Neotropic | DSSC (15182–2321.00) | AB932673 | AB932708 | AB932753 | AB932798 | AB932843 | |||
| funebris | funebris | Palearctic | TMU | AB932656 | AB932692 | AB932729 | AB932774 | AB932819 | ||
| macrospina | Nearctic | TMU | AB932661 | AB932696 | AB932736 | AB932781 | AB932826 | |||
| multispina | Palearctic, Oriental | TMU | AB932662 | AB932697 | AB932737 | AB932782 | AB932827 | |||
| guarani | ornatifrons | Neotropic | DSSC (15172–2151.00) | AB932657 | AB932693 | AB932730 | AB932775 | AB932820 | ||
| guttifera | guttifera | Nearctic | TMU | AB932658 | AB261155 | AB932731 | AB932776 | AB932821 | ||
| histrio | histrio | Palearctic, Oriental | Sapporo, Japan | AB932659 | AB932694 | AB932732 | AB932777 | AB932822 | ||
| sternopleuralis | Palearctic | Wakayama, Japan | AB932674 | AB932709 | AB932754 | AB932799 | AB932844 | |||
| immigrans | albomicans | Palearctic, Oriental, Australasian | TMU | AB033642 | AB261148 | AB932717 | AB932762 | AB932807 | ||
| formosana | Oriental | TMU | AB261131 | AB261143 | AB932728 | AB932773 | AB932818 | |||
| hypocausta | Oriental, Australasia | TMU | AB261133 | AB261151 | AB932733 | AB932778 | AB932823 | |||
| immigrans | Cosmopolitan | TMU | M97638 | AB261142 | AB932734 | AB932779 | AB932824 | |||
| nasuta | Oriental, Afrotropic, Neotropic | TMU | AB261132 | AB261144 | AB932738 | AB932783 | AB932828 | |||
| neohypocausta | Oriental, Australasia | TMU | AB261134 | AB261146 | AB932739 | AB932784 | AB932829 | |||
| neonasuta | Oriental | TMU | AB261138 | AB261151 | AB932740 | AB932785 | AB932830 | |||
| pallidifrons | Australasia | TMU | AB261136 | AB261150 | AB932744 | AB932789 | AB932834 | |||
| ruberrima | Oriental | TMU | AB932672 | AB932707 | AB932751 | AB932796 | AB932841 | |||
| siamana | Oriental | TMU | AB261135 | AB261147 | AB932752 | AB932797 | AB932842 | |||
| quinaria | angularis | Palearctic, Oriental | TMU | AB932646 | AB932682 | AB932718 | AB932763 | AB932808 | ||
| brachynephros | Palearctic, Oriental | Sapporo, Japan | AB932649 | AB932685 | AB932721 | AB932766 | AB932811 | |||
| curvispina | Palearctic | Sapporo, Japan | AB932652 | AB932688 | AB932724 | AB932769 | AB932814 | |||
| deflecta | Nearctic | TMU | AB932653 | AB932689 | AB932725 | AB932770 | AB932815 | |||
| falleni | Nearctic | TMU | AB932655 | AB932691 | AB932727 | AB932772 | AB932817 | |||
| innubila | Nearctic | University of Rochester | AB932660 | AB932695 | AB932735 | AB932780 | AB932825 | |||
| nigromaculata | Palearctic | Sapporo, Japan | AB932664 | AB932699 | AB932742 | AB932787 | AB932832 | |||
| phalerata | Palearctic | TMU | AB932667 | AB932702 | AB932746 | AB932791 | AB932836 | |||
| quinaria | Nearctic | DSSC (15130–2011.00) | AB932670 | AB932705 | AB932749 | AB932794 | AB932839 | |||
| recens | Nearctic | University of Rochester | AB932671 | AB932706 | AB932750 | AB932795 | AB932840 | |||
| subpalustris | Nearctic | TMU | AB932675 | AB932710 | AB932755 | AB932800 | AB932845 | |||
| tenebrosa | Nearctic | University of Rochester | AB932676 | AB932711 | AB932756 | AB932801 | AB932846 | |||
| transversa | Palearctic, Nearctic | University of Rochester | AB932678 | AB932713 | AB932758 | AB932803 | AB932848 | |||
| unispina | Palearctic, Oriental | Sapporo, Japan | AB932680 | AB932715 | AB932760 | AB932805 | AB932850 | |||
| testacea | orientacea | Palearctic | Sapporo, Japan | AB932665 | AB932700 | AB932743 | AB932788 | AB932833 | ||
| putrida | Nearctic | TMU | AB932669 | AB932704 | AB932748 | AB932793 | AB932838 | |||
| testacea | Palearctic, Oriental | Sapporo, Japan | AB932677 | AB932712 | AB932757 | AB932802 | AB932847 | |||
| tripunctata | tripunctata | Nearctic, Neotropical | TMU | AB932679 | AB932714 | AB932759 | AB932804 | AB932849 | ||
| Siphiodora | virilis | virilis | Cosmopolitan | GenBank | AB033640 | D10697 | JQ679111 | EU493791 | AF184014 | |
| repleta | hydei | Cosmopolitan | GenBank | X58694 | L41650 | JQ679112 | EU493736 | HQ110530 | ||
| Idiomyia | grimshawi | Australasian (Hawaii) | GenBank | CH916372 | CH916368 | CH916668 | CH916668 | CH930377 |
Classification to subgenus and species group, distribution range, source and Genbank accession numbers of mitochondrial and nuclear sequences (underlined for newly determined ones) are given for each species.
DNA extraction, PCR, cloning and sequencing
Genomic DNA was extracted from individual flies by the method of Stellaer [25] or Boom et al. [26] with some modifications [27]. PCR amplifications were carried out in 10-μl reaction volumes, each containing 1X Ex Taq buffer (Takara Bio), 200 μM dNTP, 0.5–1.0 μM of each primer, 0.25 U Ex Taq (Takara Bio), and approximately 10 ng of genomic DNA.
PCR thermal cycling conditions were 1 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at annealing temperature and 90 s at 72°C; and 7 min at 72°C. Annealing temperatures were 45°C (COI), 49°C (Adh), 51°C (COII), 55°C (28S), or 60°C (Gpdh). The primers used for the amplifications are listed in Table 2. Almost all of the PCR products were sequenced directly. However, 27 Adh and nine Gpdh sequences were sequenced after cloning into pGEM-T Easy vector (Promega) using E. coli DH5α as the host. Sequences were determined in both directions using a BigDye Terminator Sequencing Kit (Life Technologies) and an ABI 3130 and 3730 Genetic Analyzer, according to the manufacturer’s protocols.
Table 2. List of primers for the target genes.
| Locus | Length (bp) | Primers |
|---|---|---|
| Adh | 711 | Adh-F: 5’-ATGGCAATCGCTAAGAA-3’ |
| Adh-R:5’-TTAGATGCCAGAGTCCCAGT-3’ | ||
| Gpdh | 699 | Gpdh-F:5’-GTTTCTAGATCTGGTTGAGGCTGCCAAGAA-3’ |
| Gpdh-R:5’-ACATATGCTCTAGATGATTGCGTATGCA-3’ | ||
| 28S | 668 | 28S-F:5’-GACTACCCCCTGAATTTAAGGAT-3’ |
| 28S-R:5’-CTCCTTGGTGCGTGTTTC-3’ | ||
| COI | 699 | COI-F:5’-CAACATTTATTTTGATTTTTTGG-3’ |
| COI-R:5’-TYCATTGCACTAATCTGCCATATTAG-3’ | ||
| COII | 666 | COII-F:5’-ATGGCAGATTAGTGCAATGG-3’ |
| COII-R:5’-GTTTAAGAGACCACTTG-3’ |
Phylogenetic analyses
Nucleotide sequences were aligned with the MUSCLE [28] algorithm implemented in MEGA 5.05 [29] with the default settings: gap opening = -400, gap extend = 0, max iterations = 8, clustering method = UPGMB and minimum diagonal length = 24. The intron sequences of Adh and Gpdh were completely removed. Gaps were produced in the alignment procedure. The aligned sequence of 28S only included 15 gap sites. The resulting alignments were checked by eye. The aligned sequences of the five genes were concatenated by using FASconCAT [30] and the total length of the concatenated alignment was 3,443 bp (S1 Dataset).
We used the maximum-likelihood (ML) and Bayesian inference (BI) methods to construct phylogenetic trees. Partitioning concatenated data into more homogeneous subsets improves parameter estimation, reducing small data subset effects that can reduce signal-to-variance ratios [31]. Therefore, the coding sequences of each gene were partitioned into the 1st+2nd and 3rd codon positions, but the 28S sequences were not partitioned. The optimal substitution models for ML and BI were selected by using jModelTest 2.2 [32,33] on the basis of the corrected Akaike information criterion (AICc) [34]. Partitions and their associated substitution models are shown in Table 3.
Table 3. Partitions and their associated substitution models.
| Partition | Model | Base frequencies | Rate matrix | I | G |
|---|---|---|---|---|---|
| Adh-12 | GTR+G+I | A = 0.3111 | A-C = 1.4666 | 0.4520 | 0.6850 |
| C = 0.2243 | A-G = 2.7495 | ||||
| G = 0.2425 | A-T = 0.8934 | ||||
| T = 0.2222 | C-G = 1.4040 | ||||
| C-T = 4.1527 | |||||
| G-T = 1.0000 | |||||
| Adh-3 | GTR+G | A = 0.1335 | A-C = 2.5655 | - | 3.3950 |
| C = 0.3391 | A-G = 6.5409 | ||||
| G = 0.2597 | A-T = 3.4327 | ||||
| T = 0.2678 | C-G = 0.7696 | ||||
| C-T = 5.2957 | |||||
| G-T = 1.0000 | |||||
| Gpdh-12 | TrN+I | A = 0.2943 | A-C = 1.0000 | 0.8820 | - |
| C = 0.1853 | A-G = 1.3680 | ||||
| G = 0.2758 | A-T = 1.0000 | ||||
| T = 0.2447 | C-G = 1.0000 | ||||
| C-T = 15.8711 | |||||
| G-T = 1.0000 | |||||
| Gpdh-3 | GTR+G | A = 0.1502 | A-C = 1.2973 | - | 1.6310 |
| C = 0.3400 | A-G = 8.8768 | ||||
| G = 0.2256 | A-T = 2.1824 | ||||
| T = 0.2843 | C-G = 0.5332 | ||||
| C-T = 5.2132 | |||||
| G-T = 1.0000 | |||||
| 28S | HKY+I+G | A = 0.3705 | kappa = 4.1913 | 0.6400 | 0.3840 |
| C = 0.1220 | |||||
| G = 0.1746 | |||||
| T = 0.3329 | |||||
| COI-12 | GTR+I | A = 0.2382 | A-C = 4.8670 | 0.8840 | - |
| C = 0.2145 | A-G = 58.8736 | ||||
| G = 0.1955 | A-T = 10.2897 | ||||
| T = 0.3518 | C-G = 5.7327 | ||||
| C-T = 141.3394 | |||||
| G-T = 1.0000 | |||||
| COI-3 | TrN+I+G | A = 0.3972 | A-C = 1.0000 | 0.0310 | 0.6690 |
| C = 0.0688 | A-G = 149.3167 | ||||
| G = 0.0232 | A-T = 1.0000 | ||||
| T = 0.5108 | C-G = 1.0000 | ||||
| C-T = 62.3276 | |||||
| G-T = 1.0000 | |||||
| COII-12 | GTR+I | A = 0.2732 | A-C = 9.1372 | 0.8740 | - |
| C = 0.1767 | A-G = 30.9179 | ||||
| G = 0.1910 | A-T = 4.7341 | ||||
| T = 0.3591 | C-G = 0.0016 | ||||
| C-T = 102.0646 | |||||
| G-T = 1.0000 | |||||
| COII-3 | HKY+I | A = 0.4436 | kappa = 67.2853 | - | 0.3700 |
| C = 0.0652 | |||||
| G = 0.0239 | |||||
| T = 0.4673 |
Models listed here were used in Bayesian analyses.
The ML analysis was performed using RAxML 8.0.26 with RAxMLGUI 1.3.1 [35,36]. To obtain the ML tree, we used the rapid hill-climbing algorithm [37] with the 1,000 slow bootstrap (BP) option. The evolutionary model used for each data partition was GTR+I+G.
MrBayes 3.2.3 [38,39] was used to obtain the BI tree. For BI analyses, a starting tree was randomly selected and four chains were run. In each BI analysis, two independent runs were implemented in parallel, with trees sampled every 1,000 cycles. Runs were stopped after 5,000,000 Markov Chain Monte Carlo (MCMC) cycles. For each run, 5,001 samples were obtained, among which 2,500 early-phase trees were discarded as burn-in. The trace files generated by the MCMC runs were inspected in TRACER 1.6.0 [40] to check whether the number of sampling generations and effective sample sizes (ESS) were large enough for reliable parameter estimates. A consensus of sampled trees was computed using the“sumt” command, and the posterior probability (PP) for each interior branch was obtained to assess the robustness of the inferred relationships.
Estimation of divergence times
Divergence times were estimated using the relaxed molecular clock method implemented in BEAST 2.1.3 [41]. This method permits evolutionary rate variation among lineages. An uncorrelated lognormal rate evolutionary model was assumed for descendant lineages, and MCMC was used to estimate the parameters. The alignment and evolutionary models of nucleotide substitutions were the same as those used in the phylogenetic tree construction. A Yule prior was applied for the speciation process. Four MCMC chains of 50 million generations each were sampled every 1,000th generation. We collected 75% post burn-in samples and used TRACER to check whether the number of generations and ESS were large enough for reliable parameter estimation. No fossil records that can be used as calibration points exist in the subgenus Drosophila, hence we adopted the following estimates as calibration points from Russo et al. [20]: the estimated crown ages for the revised subgenera Drosophila (34.5 ± SD 12.3 Mya) and Siphlodora (27.1± 4.6 Mya) were introduced as priors with normal distributions in the analysis.
Biogeographical analysis
Distribution ranges of extant species were characterized by six unit biogeographic areas (OR: Oriental; PA: Palearctic; NA: Nearctic; NT: Neotropical; AU: Australasian; and AF: Afrotropical) and their combinations, referring to DrosWLD [42]. In order to infer possible ranges of ancestral species, the Bayesian Binary MCMC (BBM) method implemented in RASP 3.0 [43] was used. This method is primarily designed for reconstructing the ancestral state of a given node. In the BBM analysis, the probabilities of ancestral ranges at a node were estimated in terms of the MrBayes-generated probabilities of the unit biogeographic areas or their combinations. For estimating the possible ranges of ancestral species, 50,001 binary trees generated by BEAST 2.1.3 were used as input trees, with 12,501 discarded as burn-in. The maximum number of possible ancestral ranges at each node was set to six and the model of BBM analysis was as follows: the state frequency was fixed (Jukes-Canter) and the among-site rate variation was equal. For inferring possible ranges of ancestral species, 10 chains were run simultaneously for 50,000 generations of MCMC. The state was sampled every 100 trees and 100 early-phase trees were discarded as burn-in.
Results
Phylogeny
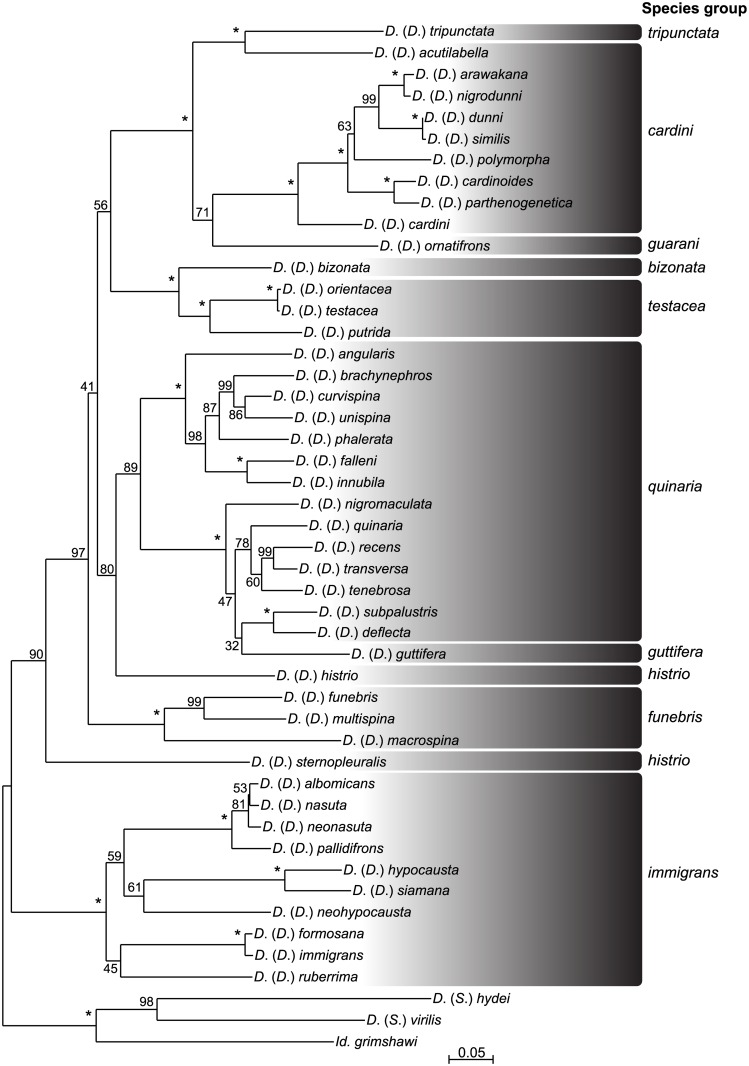
Fig 1 shows the best-scoring maximum likelihood (ML) tree obtained from the RaxmlGUI analysis using the concatenated dataset of five genes (Adh, Gpdh, 28S, COI, and COII). BEAST and MrBayes generated similar tree topologies, with only minor differences (Fig 2 and S1 Fig) that did not affect our conclusions.
Fig 1. Phylogenetic tree constructed by the Maximm likelihood (ML) analysis of the concatenated dataset.
Bootstrap values (BP) inferred by RAxML analysis are shown along interior branches. Symbol “*” indicates 100% BP.
Fig 2. Maximum clade credibility tree showing divergence time and reconstructed ancestral geographical distribution for each extant and ancestral species.
Blue bars show 95% time range intervals. Pie charts along nodes indicate probability of ancestral distribution estimated by BBM analysis. Ancestral distribution colored black means any ancestral distributions with relative probabilities <10%. Posterior probability (PP) inferred by BEAST analysis are shown along interior branch. Symbol “*” indicates 100% support value. Extant geographical distributions were retrieved from DrosWLD, shown in Table 1. The red typeface numbers indicate node numbers corresponding to those in Table 4.
In Figs 1 and 2, the ingroup species of the subgenus Drosophila were divided into two major clades, the immigrans species group and a clade that included all the remaining species groups. Within the immigrans group, the basal topology was not highly resolved except for the strongly supported monophyly of the nasuta species subgroup including D. albomicans, D. nasuta, D. neonasuta and D. pallidifrons. Although the immigrans subgroup (D. immigrans, D. formosana and D. ruberrima) and the hypocausta subgroup (D. hypocausta, D. neohypocausta and D. siamana) also formed monophyletic groups, the support for these clades was not as high, especially in the ML tree (Fig 1).
The other clade comprised 35 species belonging to nine species groups (bizonata, cardini, funebris, guarani, guttifera, histrio, quinaria, testacea and tripunctata). Among the other species groups that included multiple representatives, only two, the funebris group and the testacea group, were recovered as highly supported clades. The quinaria group was paraphyletic, but formed a highly supported clade along with the monotypic guttifera group. The clade including the quinaria + guttifera groups was split into two subclades with high confidence; one was comprised of D. angularis, D. brachynephros, D. curvispina, D. unispina, D. phalerata, D. falleni and D. innubila, and the other of D. quinaria, D. tenebrosa, D. deflecta, D. subpalustris, D. recens, D. transversa, D. nigromaculata and D. guttifera. In the latter subclade, the phylogenetic positions of D. guttifera, D. nigromaculata, D. quinaria and D. tenebrosa were largely different among the analyses (Figs 1 and 2 and S1 Fig). The two species of the histrio group were placed distantly: D. sternopleuralis was placed as the most basal branch to the clade comprising all the remaining 34 species, but D. histrio was assigned as the sister branch to the quinaria + guttifera clade. D. bizonata (the bizonata group) formed a highly supported clade with the testacea group. The three species groups (tripunctata, cardini and guarani) endemic to the New World formed another highly supported clade. Within this clade, however, the cardini group was not monophyletic. One of its members, D. acutilabella, formed a clade with D. tripunctata (the tripunctata group). However, the confidence for the basal relationships among four lineages (nodes 9, 12, 17 and 20 in Fig 2) was not very high.
Biogeographic history
Possible ancestral distributions inferred by the BBM analysis and divergence time estimates inferred by the BEAST analysis are shown in Fig 2. The Most Recent Common Ancestor (MRCA) of the subgenus Drosophila was estimated to have been distributed in the Oriental (OR) and/or Palearctic (PA) regions (OR: 39%; PA: 27%; PA+OR: 12%; Australasia: 11%) in the late Eocene to early Oligocene around 34.8 Mya (node 2, Table 4). From this ancestor, two lineages diverged in different geographic regions: the MRCA of the immigrans group was most likely distributed in OR (76%) at about 20.7 Mya (node 10), while the MRCA of the other lineage including the bizonata, cardini, funebris, guarani, guttifera, histrio, quinaria, testacea and tripunctata groups in PA (78%) was likely distributed at about 31.0 Mya (node 4). The most recent range expansion was seen in D. nasuta (node 43), which very recently invaded the Neotropical (NT) region from its original range of AF and OR [44].
Table 4. Divergence time among nodes, and ancestral distribution at each node estimated by the BBM analysis.
| Clade No. | Divergence time (Mya) | 95% range | Ancestral distribution (relative probability) |
|---|---|---|---|
| 1 | 40.39 | 47.63–33.44 | AU (0.37), OR (0.27), PA (0.19), * (0.17) |
| 2 | 34.78 | 41.24–28.56 | OR (0.39), PA (0.27), PA+OR (0.12), AU (0.11), * (0.11) |
| 3 | 32.79 | 39.68–26.68 | AU (0.49), * (0.51) |
| 4 | 31.02 | 37.17–25.11 | PA (0.78), PA+OR (0.17), * (0.05) |
| 5 | 26.74 | 32.13–21.50 | PA (0.67), PA+OR (0.29), * (0.05) |
| 6 | 26.11 | 30.45–21.87 | Cosmopolitan (0.88), * (0.11) |
| 7 | 25.71 | 30.93–20.62 | PA (0.48), PA+OR (0.48), * (0.05) |
| 8 | 24.51 | 29.71–19.69 | PA (0.45), PA+OR (0.38), * (0.07) |
| 9 | 22.70 | 27.85–17.84 | PA+OR (0.68), PA (0.30), * (0.02) |
| 10 | 20.74 | 25.97–15.70 | OR (0.76), * (0.12) |
| 11 | 19.50 | 24.28–15.16 | PA+OR (0.56), PA (0.35), * (0.09) |
| 12 | 19.24 | 23.69–15.14 | NA+NT (0.39), NT (0.38), NA (0.11), * (0.12) |
| 13 | 18.44 | 23.34–13.77 | OR+AU (0.55), OR (0.37), * (0.06) |
| 14 | 17.76 | 23.3–13.02 | OR (0.90), * (0.10) |
| 15 | 17.24 | 21.36–13.42 | NT (0.78), NA+NT (0.21), * (0.01) |
| 16 | 16.06 | 20.69–11.74 | OR+AU (0.83), OR (0.16), * (0.01) |
| 17 | 15.52 | 20.5–11.04 | NA (0.71), PA+OR (0.20), * (0.09) |
| 18 | 13.14 | 17.14–9.34 | NA+NT (0.98), * (0.02) |
| 19 | 12.34 | 15.85–9.25 | PA+OR (0.61), PA (0.36), * (0.02) |
| 20 | 12.02 | 16.26–8.40 | PA (0.44), PA+OR (0.43), * (0.13) |
| 21 | 10.95 | 13.88–8.25 | NT (0.51), NA+NT (0.48), * (0.01) |
| 22 | 9.58 | 12.37–7.02 | NA (0.34), PA (0.31), PA+NA (0.26), * (0.10) |
| 23 | 9.51 | 12.33–7.15 | PA (0.53), PA+OR (0.40), * (0.07) |
| 24 | 8.99 | 12.71–5.73 | PA (0.84), * (0.16) |
| 25 | 8.02 | 11.41–5.17 | PA (0.57), PA+OR (0.32), * (0.10) |
| 26 | 7.77 | 9.95–5.78 | NA (0.46), PA (0.33), * (0.19) |
| 27 | 7.25 | 9.55–5.29 | PA (0.74), * (0.26) |
| 28 | 7.25 | 9.33–5.38 | NA (0.98), * (0.02) |
| 29 | 7.21 | 9.31–5.32 | NT (0.93), * (0.07) |
| 30 | 6.34 | 8.21–4.55 | NT (1.00) |
| 31 | 5.36 | 7.11–3.79 | NA (1.00) |
| 32 | 5.27 | 7.59–3.39 | OR+AU (0.63), OR (0.36), * (0.01) |
| 33 | 5.12 | 6.91–3.53 | PA+OR (0.65), PA (0.34) |
| 34 | 4.28 | 6.22–2.69 | NA (0.93), * (0.07) |
| 35 | 4.26 | 5.80–2.93 | NA (0.99), * (0.01) |
| 36 | 3.87 | 5.50–2.46 | PA (0.55), PA+OR (0.45) |
| 37 | 3.77 | 5.23–2.47 | NT (1.00) |
| 38 | 3.69 | 5.28–2.35 | NA (1.00) |
| 39 | 3.06 | 4.49–1.91 | OR+AU (0.39), OR (0.38), AU (0.19), * (0.04) |
| 40 | 2.95 | 4.27–1.84 | NA (0.95), * (0.05) |
| 41 | 2.41 | 3.57–1.43 | NT (0.99), * (0.01) |
| 42 | 0.99 | 1.50–0.60 | OR (0.60), OR+AU (0.25), * (0.15) |
| 43 | 0.74 | 1.17–0.41 | OR (0.38), PA+OR (0.23), OR+AU (0.14), * (0.25) |
| 44 | 0.66 | 1.10–0.32 | NT (1.00) |
| 45 | 0.49 | 0.83–0.24 | OR (0.74), PA+OR (0.12), * (0.14) |
| 46 | 0.17 | 0.35–0.06 | PA (0.74), PA+OR (0.25), * (0.01) |
| 47 | 0.15 | 0.30–0.04 | NT (1.00) |
Asterisks indicates the ancestral areas each of which with the relative probability of < 0.1.
Dispersals from the Old World to the New World were inferred to have occurred five times in the latter lineage. The first migratory event occurred from the PA (45%) or PA+OR (38%) distribution (node 8) to Nearctic (NA)+NT (39%) or NT (38%) (node 12) in the tripunctata, cardini and guarani group clade. The second event was detected in the ancestor of the funebris group, which migrated from PA (67%) or PA+OR (29%) to the NA (71%) during the period of 26.7 (node 5) to 15.5 (node 17) Mya. The third event was a migration or range expansion in the ancestor of the Nearctic quinaria-group clade including D. guttifera, which occurred at 19.5 (node 11) to 9.6 (node 22) Mya. The fourth was a migration in the ancestor of D. falleni and D. innubila from PA (53%) or PA+OR (40%) to NA (93%) 9.5 (node 23) to 4.3 (node 34) Mya. The fifth was a migration of D. putrida from PA (57%) or PA+OR (32%) to NA (current range) after 8.0 Mya (node 25).
In addition, dispersals in the reverse direction from the New World to the Old World were inferred in a few individual species and a small clade. From the above-mentioned data, Nearctic MRCA of the funebris group, the ancestor of the Old World species D. funebris and D. multispina, migrated back to PA (84%) or PA+OR (16%) 26.7 (node 17) to 15.5 (node 24) Mya. Within the Nearctic clade of the quinaria group, there are two species that are not endemic to NA: D. nigromaculata distributed in PA and D. transversa distributed in the Holarctic (PA+NA). The PA distribution of D. nigromaculata would have resulted from the backward migration of this species from the NA ancestor or vicariant speciation of the Holarctic ancestor into this PA species and the NA ancestor of the other species. However, this inference was not supported in the ML tree (Fig 1) or BI tree (S1 Fig): D. nigromaculata was placed basal to the clade containing D. guttifera and the other six species of the quinaria group. D. transversa expanded its range to PA after 3.0 Mya (node 40).
Discussion
Phylogeny
The phylogeny reconstructed by the present study was largely consistent with those in previous studies. The subgenus Drosophila (the traditional immigrans-tripunctata radiation) was consistently divided into two clades: one was the immigrans species group and the other comprised the remaining species groups [2,5,13,14,20,23,24]. Although the immigrans group has thus far been regarded as monophyletic, this may be due to insufficient taxon-sampling for this species group. Recent studies have revealed that D. quadrilineata, a member of the quadrilineata subgroup of the immigrans group, was placed outside the clade of the immigrans group proper [4,12,19], and that it is closely related to the genus Samoaia [6,45]. Moreover, Katoh et al. [12] suggested that D. annulipes, another species of the quadrilineata subgroup, is related to the genus Idiomyia (or Hawaiian Drosophila), along with D. maculinotata. Thus, the quadrilineata subgroup is still questionable in terms of its monophyly and phylogenetic position [12]. Another questionable species subgroup in the immigrans group is the curviceps subgroup. When Zhang and Toda [46] established this species subgroup, they suggested that it is closer to the quadrilineata subgroup than to the other subgroups of the immigrans group. Yassin [6] did not assign these two species subgroups to any subgenus in his revised classification of Drosophila. This taxonomic situation is inconvenient: some members (the immigrans, nasuta and hypocausta subgroups) of the immigrans group belong to the subgenus Drosophila, but the others (the quadrilineata and curviceps subgroups) are incertae sedis for the subgenus. To partly solve this problem, Pradhan et al. [47] have made the curviceps subgroup independent, as a species group, from the immigrans group, but left its subgeneric assignment undetermined, i.e., incertae sedis. Thus, more taxonomic revision is needed for the traditional immigrans group and its related taxa.
The clade including the species groups other than the immigrans group is here termed the quinaria-tripunctata radiation and is characterized by the less resolved, basal branching of major component clades. Within this radiation, two large, well-supported clades were recognized in agreement with previous studies. One is the so-called tripunctata radiation, the monophyly of which has been repeatedly inferred in the previous studies [13,14,22–24]. Currently, this radiation includes 123 species of six (calloptera, cardini, guarani, pallidipennis, sticta and tripunctata) species groups that diversified mostly in the Neotropical region [22,24,42]. Phylogenetic relationships among these species groups and the monophyly for some of them are still questionable [5,13,14,23,24], and the present study cannot address these issues because of poor taxon-sampling (only 11 species from three species groups). The other large, well-supported clade comprises the quinaria and guttifera species groups. It has often been indicated in previous analyses [4,5,13,19–21,48] that D. guttifera is closely related to the quinaria group. Accordingly, Markow and O'Grady [49] treated D. guttifera as a species of the quinaria group. The present study has confirmed the inference by Perlman et al. [21] that this “quinaria” clade is further divided into two subclades. These subclades can be more or less distinguished from each other by their choice of breeding niches: one consists of fungal breeders such as D. phalerata, D. brachynephros and D. falleni, whereas the other includes decaying plant breeders such as D. nigromaculata, D. deflecta, D. quinaria, D. limbata, D. subpalustris and D. palustris [21,50,51].
However, some phylogenetic inferences drawn from the present study are new or different from those of previous studies. The phylogeny of the histrio species group has been unclear for a long time, although either D. histrio or D. sternopleuralis has been included in some previous studies [3–6,20,21]. The present study included both species and revealed that the histrio group is not monophyletic. With high confidence, D. sternopleuralis represents the most basal lineage in the quinaria-tripunctata radiation, while D. histrio is the sister taxon to the “quinaria” clade. The phylogenetic position of D. sternopleuralis has been problematic in previous studies [3,4,20]. It should be noted, however, that Russo et al. [20] placed D. sternopleuralis along with D. pruinosa of the Afrotropical loiciana species complex [52] in the sister clade to the immigrans group proper, though not highly supported. Furthermore, although D. sternopleuralis is distributed in warm temperate regions of Japan and Korea, a number of its relatives have been recorded in the tropical Australasian to the subtropical Oriental regions, i.e., Papua New Guinea, Borneo, Sri Lanka, India, Nepal, Myanmar, southern China and Taiwan [43,53–56]. Although the phylogenetic position of D. histrio has also been controversial [4–6,21], our results strongly support that it is a sister relationship to the “quinaria” clade, which is consistent with the topology inferred by van der Linde and Houle [4].
Only two previous studies [5,6] examined the monophyly of the funebris species group, including D. funebris and D. macrospina, in molecular phylogenetic analyses. However, the results were controversial. The present study suggests that three species, D. multispina in addition to the two above, of the funebris group form a highly supported clade. Although its placement within the quinaria-tripunctata radiation is consistent with most previous studies [2,3–6,9,14,19–22], its phylogenetic position within the radiation has been uncertain in previous studies.
The testacea and bizonata species groups formed a clade in previous, phylogenetic studies [5,6], and our analyses confirm this, but differ from that of van der Linde et al. [5] with respect to the phylogenetic position of D. bizonata. We placed it as the sister to the testacea group, which is monophyletic. However, van der Linde et al. [5] placed D. bizonata within the testacea group, rendering the latter paraphyletic. The discrepancy may have been due to the difference in gene markers employed for the phylogenetic analyses. In addition, both studies included only one species (D. bizonata) of the bizonata group, of which six more species are known. This issue should be addressed using better taxon-sampling.
Evolutionary history
Throckmorton [1] hypothesized that two major biogeographic patterns resulted in the evolution of the Drosophilidae. The first is the primary disjunction of tropical faunas between the Old and the New Worlds in the early Oligocene, and the second is the disjunction of temperate faunas between the two Worlds in the mid-Miocene. A number of recent studies have tested this hypothesis by molecular phylogenetic analyses in specific lineages [17,18,24] or across large taxa such as the genus Drosophila [19], the subfamily Drosophilinae [23] and the entire family Drosophilidae [20]. However, the time estimates for these disjunctions varied among the studies because of differences in the timescale placed on the phylogenies that were used [57]. Dates from two sources have been used as calibration points to estimate divergence times within the Drosophilidae: one from fossils, and the other from biogeography. The former source is rare in Drosophilidae, with only few Baltic and Dominican amber fossils dated geologically [58,59]. The latter source comes from the phylogeography of Hawaiian drosophilids, and has more often been used to establish calibration points in molecular phylogenetic studies on Drosophilidae [60,61]. However, Obbard et al. [57] pointed out some biases of the Hawaiian-calibrated dates. Secondarily, taxon sampling affects the inferences on evolutionary history through estimations of ancestral distributions and divergence times. Gao et al. [18] pointed out the importance of recognizing genuine sister groups in obtaining reliable estimates for these attributes.
Since neither fossil-based nor phylogeography-based calibration points are available for the species employed in the present study, we applied the divergence time estimates at two nodes (node 2 and node 6) from Russo et al. [20] as calibration points. The study by Russo et al. employed the largest number of calibration points (two fossil-based and six Hawaiian-phylogeographic calibrations) for estimating divergence times, and was based on the largest taxon samplings (358 species) from the Drosophilidae, but it was somewhat biased to New World species for the quinaria-tripunctata radiation. The present study was conducted to compensate for this bias by including more Old World species of this radiation.
Russo et al. [20] inferred that the pattern of drosophilid faunal disjunctions between the Old and the New Worlds was not as simple as Throckmorton [1] depicted. Such disjunctions were regarded as having occurred multiple times during the long geological period from the late Eocene to the Holocene. However, the most important aspect of Throckmorton’s [1] hypothesis, i.e., the parallelism of disjunctions on separate lineages of Drosophilidae, was recovered, indicating that such disjunctions may have been caused by vicariant events. The present study, focusing on the revised subgenus Drosophila (the immigrans-tripunctata radiation), has corroborated the general pattern of disjunctions inferred by Russo et al. [20], but has shown that the evolutionary history of this lineage should largely be revised.
The first difference was seen in the estimation of ancestral distribution for the quinaria-tripunctata radiation. Russo et al. [20] estimated that the MRCA of this radiation was most likely distributed in the New World, while our results suggested that it was distributed in the Palearctic region. Certainly, this could be due to the difference in taxon sampling between the two studies. Accordingly, Russo et al. [20] inferred that the first Old World–New World disjunction within the subgenus Drosophila occurred in the late Eocene to produce two tropical lineages, the immigrans species group in the Oriental region and the tripunctata radiation in the Neotropical region. In parallel with this disjunction, another vicariant divergence of tropical/subtropical elements was estimated to have concurred in the late Eocene between the Oriental/Palearctic genus Lordiphosa and the Neotropical saltans and willistoni species groups of the subgenus Sophophora. However, our results suggest that the ancestor of the quinaria-tripunctata radiation had once entered the temperate Palearctic region in the early to mid-Oligocene. From this ancestor, then, two lineages, the funebris species group and the tripunctata radiation, arose by nearly concurrent migrations of their ancestors from the Palearctic region to the New World in the late Oligocene to the early Miocene. Gao et al. [18] estimated that the primary, tropical disjunctions such as that between Lordiphosa and Neotropical Sophophora occurred in the mid-Eocene when the North Atlantic Land Bridge (NALB) broke. Even if the NALB could serve as a migrating route for organisms such as drosophilid flies with larger dispersal abilities until later ages [20,62], our estimate of the ancestral age of the Neotropical tripunctata radiation was much younger than the time of the NALB break. In addition, this lineage was regarded as derived from an ancestor distributed in the Palearctic and/or the Oriental regions in the late Oligocene, when tropical forests continuously graded into temperate forests only in East Asia [62,63]. This suggests that the ancestor of the tripunctata radiation, along with the ancestor of the funebris group, migrated through the Bering Land Bridge into the Nearctic region. Robe et al. [24] inferred, based on molecular phylogenetic and biogeographic analyses, that after the colonization to the Nearctic region this lineage had expanded its range to the Neotropical region and much diversified there.
The second difference was seen in the estimated time and direction of ancestral migration between the Old and the New Worlds in the “quinaria” clade. Russo et al. [20] estimated that the ancestor of this clade had migrated from the New World to the Old World in the late Oligocene. Our divergence time estimates (Fig 2) suggest that it is likely that the ancestor was distributed in the Oriental and/or the Palearctic regions. From this Old World ancestor, one lineage was derived by migration or range expansion to the Nearctic region in the mid-Miocene. Another species concurrently migrated, but in the opposite direction, from the Nearctic to the Palearctic region and became the ancestor of Old World species in the funebris group. The Old World–New World temperate disjunctions dated to the mid-Miocene age were detected in the lineages of the genus Hirtodrosophila and the Drosophila (Siphlodora) melanica species group as well [20].
The third phase of Old World-New World disjunctions was estimated to have occurred on three lineages in the “quinaria” clade and the testacea species group, as well as in the Drosophila (Sophophora) obscura species group [20], in the late Miocene. In the fourth phase, the subarctic species D. transversa achieved its Holarctic distribution by expanding its range from the Nearctic to the Palearctic region in the Pliocene. In this phase, cool-temperate species of the Drosophila (Siphlodora) robusta and virilis species groups became disjunct between the Palearctic and the Nearctic regions [20].
Thus, ancestral migrations and subsequent vicariant divergences of descendants between the Old and the New Worlds occurred multiple times in parallel among different lineages in the quinaria-tripunctata radiation of the subgenus Drosophila. Such faunal disjunctions between the two Worlds concurred in other drosophilid lineages as well, and shaped a part of the overall phylogeographic pattern in the family Drosophilidae [20]. These faunal vicariant events correspond to the secondary disjunctions of temperate elements in Throckmorton’s [1] hypothesis, but would have occurred repeatedly in a wider time range, from the late Oligocene to the Pliocene, than Throckmorton [1] proposed. Such multiple migration episodes between the Old and the New World have also been well documented in terrestrial plants and mammals. Tiffney [64] proposed that floristic exchanges between the two Worlds most likely occurred in five major periods: (1) the pre-Tertiary, (2) the early Eocene, (3) the late Eocene–Oligocene, (4) the Miocene, and (5) the late Tertiary–Quaternary periods. The four phases of Old World-New World disjunctions detected for the quinaria-tripunctata radiation in the present study seem to correspond to the last two of the five flora-exchange periods. Zhanxiang [65] recognized three major waves of dispersals from Eurasia to North America in carnivoran mammals: the first in the early Miocene, the second in the late Miocene and the third in the Pliocene: corresponding to the first, third and fourth phases of faunal disjunctions in the quinaria-tripunctata radiation, respectively. These parallel, spatiotemporal patterns observed in terrestrial organisms of the northern hemisphere strongly suggest the contribution of the Bering Land Bridge as a common factor to exchanges and isolation of temperate elements between the Old and the New Worlds [20,62,65]. Paleo-climatic and -vegetational conditions around Beringia gradually changed with global cooling after the Early Eocene Climatic Optimum [66]. The vegetation around the Beringia was dominated by rich deciduous and some coniferous temperate forests in the late Oligocene to the early Miocene, but changed to the cool-temperate forest with decreased diversity of deciduous angiosperms and conifers thereafter from the late Miocene to the Pliocene and to the taiga/tundra in the Holocene [62,63]. The changing climatic and vegetational conditions around the Beringia should have filtered different elements of the temperate biota in the process of faunal/floral exchanges between the Old and the New Worlds after the early or mid-Miocene age: elements adapted to the warmer climate became disjunct earlier in geological time.
Our results clearly show the importance of wider and denser taxon sampling for the reconstruction of evolutionary histories, although there still remain a number of species groups of the subgenus Drosophila to be included in phylogenetic analyses. Further expansion of taxon sampling across the whole family Drosophilidae should provide more evidence for parallel biogeographic disjunctions in separate lineages, and will help to document the vicariant nature of regional faunal development.
Supporting Information
(NEX)
(PDF)
Acknowledgments
We thank M. Watada and K. Tamura for their valuable suggestions in the course of this study; J. Jaenike, T. Ide and the late T. Aotsuka for the material support; and H. Katakura, H. Kajihara, and M. H. Dick for valuable comments and helpful suggestions.
Data Availability
All relevant data are fully available without restriction. All sequences are available from GenBank (accession numbers are in supplementary table).
Funding Statement
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Japan (No. 24370033). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Throckmorton LH. The phylogeny, ecology and geography of Drosophila In: King RC editor, Handbook of Genetics. vol. 3 New York: Plenum Press; 1975. pp. 421–469. [Google Scholar]
- 2.Remsen J, O’Grady P. Phylogeny of Drosophilinae (Diptera: Drosophilidae), with comments on combined analysis and character support. Mol Phylogenet Evol. 2002;24: 249–264. [DOI] [PubMed] [Google Scholar]
- 3.Da Lage JL, Kergoat GJ, Maczkowiak F, Silvain JF, Cariou ML, Lachaise D. A phylogeny of Drosophilidae using the Amyrel gene: questioning the Drosophila melanogaster species group boundaries. J Zool Syst Evol Res. 2007;45: 47–63. [Google Scholar]
- 4.van der Linde K, Houle D. A supertree analysis and literature review of the genus Drosophila and closely related genera (Diptera, Drosophilidae). Insect Syst Evol. 2008;39: 241–297. [Google Scholar]
- 5.van der Linde K, Houle D, Spicer DG, Steppan SJ. A supermatrix-based molecular phylogeny of the family Drosophilidae. Genet Res (Camb). 2010;92: 25–38. [DOI] [PubMed] [Google Scholar]
- 6.Yassin A. Phylogenetic classification of the Drosophilidae Rondani (Diptera): the role of morphology in the postgenomic era. Syst Entomol. 2013;38: 349–364. [Google Scholar]
- 7.Pélandakis M, Solignac M. Molecular phylogeny of Drosophila based on ribosomal RNA sequences. J Mol Evol. 1993;37: 525–543. [DOI] [PubMed] [Google Scholar]
- 8.Tamura K, Toba G, Park J, Aotsuka T. Origin of Hawaiian drosophilids inferred from alcohol dehydrogenase gene sequences In: Nei M, Takahata N, editors. Current Topics on Molecular Evolution. University Park: Institute of Molecular Evolutionary Genetics, Pennsylvania State University; 1995. pp. 9–18. [Google Scholar]
- 9.Remsen J, DeSalle R. Character congruence of multiple data partitions and the origin of the Hawaiian Drosophilidae. Mol Phylogenet Evol. 1998;9: 225–235. [DOI] [PubMed] [Google Scholar]
- 10.Tatarenkov A, Ayala FJ. Phylogenetic relationships among species groups of the virilis–repleta radiation of Drosophila. Mol Phylogenet Evol. 2001;21: 327–331. [DOI] [PubMed] [Google Scholar]
- 11.Katoh T, Tamura K, Aotsuka T. Phylogenetic position of the subgenus Lordiphosa of the genus Drosophila (Diptera: Drosophilidae) inferred from alcohol Dehydrogenase (Adh) gene sequences. J Mol Evol. 2000;51: 122–130. [DOI] [PubMed] [Google Scholar]
- 12.Katoh T, Nakaya D, Tamura K, Aotsuka T. Phylogeny of the Drosophila immigrans species group (Diptera: Drosophilidae) based on Adh and Gpdh sequences. Zoolog Sci. 2007;24: 913–921. [DOI] [PubMed] [Google Scholar]
- 13.Yotoko KS, Medeiros HF, Solferini VN, Klaczko LB. A molecular study of the systematics of the Drosophila tripunctata group and the tripunctata radiation. Mol Phylogenet Evol. 2003;28: 614–619. [DOI] [PubMed] [Google Scholar]
- 14.Robe LJ, Valente VLS, Budnik M, Loreto EL. Molecular phylogeny of the subgenus Drosophila (Diptera, Drosophilidae) with an emphasis on Neotropical species and groups: a nuclear versus mitochondrial gene approach. Mol Phylogenet Evol. 2005;36: 623–640. [DOI] [PubMed] [Google Scholar]
- 15.Hsu TC. The external genital apparatus of male Drosophilidae in relation to systematics. Univ Texas Publ. 1949;4920: 80–142. [Google Scholar]
- 16.Malogolowkin C. Sobre a genitália dos drosofilídeos. IV. A genitália masculina no subgênero Drosophila (Diptera, Drosophilidae). Rev Bras Biol. 1953;13: 245–264. [Google Scholar]
- 17.Gao JJ, Watabe HA, Aotsuka T, Pang JF and Zhang YP. Molecular phylogeny of the Drosophila obscura species group, with emphasis on the Old World species. BMC Evol Biol. 2007;7: 87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao JJ, Hu YG, Toda MJ, Katoh T, Tamura K. Phylogenetic relationships between Sophophora and Lordiphosa, with proposition of a hypothesis on the vicariant divergences of tropical lineages between the Old and New Worlds in the family Drosophilidae. Mol Phylogenet Evol. 2011;60: 98–107. 10.1016/j.ympev.2011.04.012 [DOI] [PubMed] [Google Scholar]
- 19.Morales-Hojas R, Vieira J. Phylogenetic patterns of geographical and ecological diversification in the subgenus Drosophila. PLoS One. 2012;7: e49552 10.1371/journal.pone.0049552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russo CAM, Mello B, Frazão A, Voloch CM. Phylogenetic analysis and a time tree for a large drosophilid data set (Diptera: Drosophilidae). Zool J Linn Soc. 2013;119: 765–775. [Google Scholar]
- 21.Perlman SJ, Spicer GS, Shoemaker DD, Jaenike J. Associations between mycophagous Drosophila and their Howardula nematode parasites: a worldwide phylogenetic shuffle. Mol Ecol. 2003;12: 237–249. [DOI] [PubMed] [Google Scholar]
- 22.Hatadani LM, McInerney JO, de Medeiros HF, Junqueira ACM, de Azeredo-Espin AM, Klaczko LB. Molecular phylogeny of the Drosophila tripunctata and closely related species groups (Diptera: Drosophilidae). Mol Phylogenet Evol. 2009;51: 595–600. 10.1016/j.ympev.2009.02.022 [DOI] [PubMed] [Google Scholar]
- 23.Robe LJ, Loreto ELS, Valente VLS. Radiation of the “Drosophila” subgenus. (Drosophilidae, Diptera) in the Neotropics. J Zool Syst Evol Res. 2010;48: 310–321. [Google Scholar]
- 24.Robe LJ, Valente VLS, Loreto ELS. Phylogenetic relationships and macro-evolutionary patterns within the Drosophila tripunctata “radiation” (Diptera: Drosophilidae). Genetica. 2010;138: 725–735. 10.1007/s10709-010-9453-0 [DOI] [PubMed] [Google Scholar]
- 25.Steller HD. Rapid small-scale isolation of Drosophila DNA and RNA In: Rubin L, editor. Rubin Lab Method Book 2nd edition Rubin Lab:1990. pp. 185–186. [Google Scholar]
- 26.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi N, Ohta Y, Katoh T, Kahono S, Hartini S, Katakura H. Molecular phylogenetic analysis of three groups of Asian epilachnine ladybird beetles recognized by the female internal reproductive organs and modes of sperm transfer. J Nat Hist. 2009;43: 1637–1649. [Google Scholar]
- 28.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol. Evol. 2011;28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kück P, Meusemann K, FASconCAT: Convenient handling of data matrices. Mol Phylogenet Evol. 2010;56: 1115–1118. 10.1016/j.ympev.2010.04.024 [DOI] [PubMed] [Google Scholar]
- 31.Phillips MJ, Penny D. The root of the mammalian tree inferred from whole mitochondrial genomes. Mol Phylogenet Evol. 2003;28: 171–185. [DOI] [PubMed] [Google Scholar]
- 32.Guindon S, Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol. 2003;52: 696–704. [DOI] [PubMed] [Google Scholar]
- 33.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat methods. 2012;9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19: 716–723. [Google Scholar]
- 35.Stamatakis A. RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics. 2014;30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012;12: 335–337. [Google Scholar]
- 37.Stamatakis A, Blagojevic F, Nikolopoulos DS, Antonopoulos CD. Exploring new search algorithms and hardware for phylogenetics: RAxML meets the IBM cell. J VLSI Signal Process Syst Signal Image Video Technol. 2007;48: 271–286. [Google Scholar]
- 38.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17: 754–755. [DOI] [PubMed] [Google Scholar]
- 39.Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 40.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.6. 2013. Available: http://tree.bio.ed.ac.uk/software/tracer/
- 41.Bouckaert R, Heled J, Kühnert D, Vaughan TG, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. BEAST2: A software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10: e1003537 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toda MJ. DrosWLD-Species: taxonomic information database for world species of Drosophilidae [Internet]. c2006 [updated 2015 Sep 15; cited 2015 Dec 18]. Available: http://bioinfo.lowtem.hokudai.ac.jp/db/modules/stdb/
- 43.Yu Y, Harris AJ, Blair C, He XJ. RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol Phylogenet Evol. 2015;87: 46–49. 10.1016/j.ympev.2015.03.008 [DOI] [PubMed] [Google Scholar]
- 44.Vilela CR. Goñi B. Is Drosophila nasuta Lamb (Diptera, Drosophilidae) currently reaching the status of a cosmopolitan species? Rev Bras Entomol. 2015;59: 346–350. [Google Scholar]
- 45.Yassin A, Da Lage JL, David JR, Kondo M, Madi-Ravazzi L, Prigent SR, Toda MJ. Polyphyly of the Zaprionus genus group (Diptera: Drosophilidae). Mol Phylogenet Evol. 2010;55: 335–339. 10.1016/j.ympev.2009.09.013 [DOI] [PubMed] [Google Scholar]
- 46.Zhang W, Toda M. A new species-subgroup of the Drosophila immigrans species group (Diptera, Drosophilidae), with description of two new species from China and revision of taxonomic terminology. Jpn J Ent. 1992;60: 839–850. [Google Scholar]
- 47.Pradhan S, Sati PC, Fartyal RS, Chatterjee RN, Sarswat M, Kandpal MC, et al. Drosophila curviceps Species-Group (Diptera: Drosophilidae) from India, with Description of a New Species and Redescription of a Known Species. Proc Zool Soc. 2015;68: 178–183. [Google Scholar]
- 48.Spicer GS, Jaenike J. Phylogenetic analysis of breeding site use and α-amanitin tolerance within the Drosophila quinaria species group. Evolution. 1996: 2328–2337. [DOI] [PubMed] [Google Scholar]
- 49.Markow TA, O'Grady P. Drosophila: a guide to species identification and use. London: Academic Press; 2005. [Google Scholar]
- 50.Kimura M, Toda M, Beppu K, Watabe H. Breeding sites of drosophilid flies in and near Sapporo, northern Japan, with supplementary notes on adult feeding habits. Kontyû. 1977;45: 571–582. [Google Scholar]
- 51.Shorrocks B. The breeding sites of temperature woodland Drosophila In: Ashburner M, Carson HL, Thompson JN Jr editors. Genetics and biology of Drosophila. Vol. 3b London: Academic Press; 1982. pp. 385–428. [Google Scholar]
- 52.Tsacas L. The new African complex Drosophila loiciana and the allied species D. matileana n. sp. (Diptera: Drosophilidae). Ann Soc entomol Fr. 2002;38: 57–70. [Google Scholar]
- 53.Okada T. Diptera from Nepal, Cryptochaetidae, Diastatidae and Drosophilidae. Bull Br Mus. 1966;Suppl 6: S1–129. [Google Scholar]
- 54.Okada T, Carson HL. Drosophilidae Associated with Flowers in Papua New Guinea: IV. Araceae, Compositae, Convolvulaceae, Rubiaceae, Leguminosae, Malvaceae. Kontyû. 1982;50: 511–526. [Google Scholar]
- 55.Toda MJ. Drosophilidae (Diptera) in Burma. III. The subgenus Drosophila, excepting the D. immigrans species-group. Kontyû. 1988;56: 625–640. [Google Scholar]
- 56.Toda MJ, Peng TX. Eight species of the subgenus Drosophila (Diptera: Drosophilidae) from Guangdong Province, southern China. Zool Sci. 1989;6: 155–166. [Google Scholar]
- 57.Obbard DJ, Maclennan J, Kim KW, Rambaut A, O’Grady PM, Jiggins FM. Estimating divergence dates and substitution rates in the Drosophila phylogeny. Mol Biol Evol. 2012;29: 3459–3473. 10.1093/molbev/mss150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hennig W. Die Acalyptratae des Baltischen Bernsteins und ihre Bedeutung für die Erforschung der phylogeneticchen Entwicklung dieser Dipteren-Gruppe. Stuttgart Beitr Naturkd. 1965;145: 1–215. [Google Scholar]
- 59.Grimaldi DA. Amber fossil Drosophilidae (Diptera), with particular reference to the Hispaniolan taxa. Am Mus Novit. 1987;2880: 1–23. [Google Scholar]
- 60.Hardy DE. Introduction and background information In: White MJD editors. Genetic mechanisms of speciation in insects. Boston: Reidel;1974. pp. 71–80. [Google Scholar]
- 61.Clague DA, Dalrymple GB. Tectonics, geochronology and origin of the Hawaiian-Emperor volcanic chain In: Decker RW, Wright TL, Stauffer PH, editors. US Geological Survey Professional Paper 1350. Washington (DC): US Government Printing Office; 1987. pp. 1–55. [Google Scholar]
- 62.Tiffney BH, Steven RM. The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the Northern Hemisphere Tertiary. Int J Plant Sci. 2001;162(suppl 6): S3–17. [Google Scholar]
- 63.Wilis KJ, McElwain JC. The Evolution of Plants. 2nd ed Oxford: Oxford University Press; 2014. [Google Scholar]
- 64.Tiffney BH. The Eocene North Atlantic land bridge: its importance in Tertiary and modern phytogeography of the Northern Hemisphere. J Arnold Arbor. 1985;66: 243–273. [Google Scholar]
- 65.Zhanxiang Q. Dispersals of Neogene Carnivorans between Asia and North America. Bull Am Mus Nat Hist. 2003;279: 18–31. [Google Scholar]
- 66.Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292: 686–693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(NEX)
(PDF)
Data Availability Statement
All relevant data are fully available without restriction. All sequences are available from GenBank (accession numbers are in supplementary table).