Summary
This study revisited the Dohner prognostic hierarchy in a cohort of 1585 well‐documented patients with chronic lymphocytic leukaemia. The duration of both time to first treatment (TTFT) and overall survival (OS) were significantly longer than observed previously, and this is at least partly due to improved therapeutic options. Deletion 13q remains the most favourable prognostic group with median TTFT and OS from fluorescence in situ hybridization (FISH) testing of 72 months and >12 years, respectively. Deletion 11q had the poorest median TTFT (22 months) and 17p deletion the poorest median OS (5 years). The percentages of abnormal nuclei were significantly associated with differential TTFT for the trisomy 12, 13q and 17p deletion cohorts but not for the 11q deletion cohort. From the date of the first FISH study, patients with >85% 13q deletion nuclei had a notably shorter TTFT (24 months). Patients with ≤20% 17p deletion nuclei had longer median TTFT and OS from the date of the first FISH study (44 months and 11 years), and were more likely to be IGHV mutated.
Keywords: chronic lymphocytic leukaemia, cytogenetics, leukaemia, fluorescence in situ hybridization
For more than 15 years the Dohner hierarchical classification (Dohner et al, 2000), using fluorescence in situ hybridization (FISH) probes to identify trisomy 12 and deletions at 13q14.2‐q14.3 (including DLEU2, MIR15A/MIR16‐1), 11q22.3 (ATM) and 17p13.1 (TP53) has been the gold standard for the genetic characterization and assessment of prognosis for patients with chronic lymphocytic leukaemia (CLL). The Dohner classification has been validated by several groups (Dewald et al, 2003; Geisler et al, 2009). Given the clinical usefulness of the common recurring FISH detectable events found by the probes encompassing the Dohner hierarchy used to assess CLL patients, this genetic approach is used throughout the world. Its benefits are primarily to provide prognostic information for the clinician. Importantly, the FISH panel data can be used to counsel these patients, whether in early or more progressive disease status. In this era of utilization of highly effective chemoimmunotherapy, it is uncertain whether the original prognostic association is maintained between FISH patterns and clinical outcome variables. To investigate this, we took advantage of a robust clinical database that is part of the Chronic Lymphocytic Leukemia Research Consortium (CRC), a programme project dedicated to the conduct of basic and translational research in CLL. The CRC is composed of six active academic sites with significant numbers of CLL patients and each site is a source of CLL samples and highly detailed clinical information that are contributed to the CRC database. The CRC tissue bank and clinical database have been the source of multiple important published insights into the utility of prognostic markers in CLL (Rassenti et al, 2008; Visone et al, 2011; Claus et al, 2014).
We present here an assessment of the relationship between the common recurring FISH‐detectable genomic defects and CLL clinical outcome using the CRC multi‐institutional CLL cohort. This analysis included primarily a reassessment of the Dohner hierarchical classification (Dohner et al, 2000) and its associations with time to first treatment (TTFT) and overall survival (OS).
Methods
Clinical database
All the clinical data were captured via the CRC Tissue‐Core‐Management‐System (TCMS). We used closed data fields captured by the TCMS from all CRC investigators to assess the date of diagnosis, date of initial treatment and clinical status of each patient (Rassenti et al, 2008). A multiple closed unique patient identifier system was used that complied with the Health Insurance Portability and Accountability Act.
Patient cohort
We identified 1585 CLL patients in the CRC database for whom a complete FISH panel and relevant clinical data were available. Patients were diagnosed between 1972 and 2011; the most recent clinical follow‐up was in 2012. Blood samples were collected from consenting CLL patients who satisfied the diagnostic criteria for CLL and were enrolled in the CRC biorepository as per protocol approved by the Institutional Review Board (IRB 080918). FISH analyses were performed between 1998 and 2012. All of the CLL patients tested were negative for a CCND1/IGH fusion, thus ruling out the presence of mantle cell lymphoma.
FISH panel approach
All six participating CRC centres used standardized FISH panels as described previously (Smoley et al, 2010). For 59 of the 1585 patient samples, FISH analysis was done using cells harvested after cell culture stimulated with CpG oligonucleotide; the rest were harvested directly or after brief cell culture without mitogen. Thresholds used to designate 17p13.1 (17p) deletion, 11q22.3 (11q) deletion, trisomy 12, and 13q14.2‐q14.3 (13q) deletion were: ≥5%, ≥5%, ≥1·25% and ≥7%, respectively.
Statistical analysis
Demographic and clinical characteristics, including FISH abnormality, according to the Dohner hierarchical classification (Dohner et al, 2000) were summarized as number and percentage for categorical variable and median with interquartile ranges for continuous variables. Time to first treatment (TTFT) was measured either from the date of diagnosis (TTFT‐DX) or from the date of the initial FISH results (TTFT‐FISH) to initiation of first treatment. Patients who remained untreated at the time of analysis were censored on the date last confirmed to be without initiation of therapy. OS was calculated from either the date of the CLL diagnosis (OS‐DX) or the date of the initial FISH study (OS‐FISH) to the date of death or censored on the last known date alive. Kaplan–Meier estimates were used to summarize time to event data. The log rank test was used to assess the associations between FISH abnormality and TTFT and OS. P values < 0·05 were considered statistically significant.
To be consistent with the selection criteria in the original Dohner analysis (Dohner et al, 2000), we examined TTFT‐DX and OS‐DX in patients for whom FISH was done within 4 years of diagnosis. This subset of patients was, by design, the primary analysis cohort. We also examined the two clinical endpoints from the date of the initial FISH analysis for the entire cohort of 1585 patients (TTFT‐FISH and OS‐FISH), which permitted us to assess the performance of FISH testing completed at a time point not associated with diagnosis. To further clarify the prognostic value of FISH, we conducted additional analyses after excluding patients for whom treatment was initiated less than 14 d after the initial FISH analysis, reasoning that FISH results were not available at the time that the decision to treat was made.
We also evaluated the association between the percentage of abnormal nuclei and both TTFT and OS for each FISH category using the log rank test. Using 15% increments for each FISH abnormality where feasible, we compared the outcomes of patients with increasing specific FISH defect burdens with those with a normal FISH result. Dohner FISH hierarchical order (Dohner et al, 2000) was further used to categorize for 11q deletion, trisomy 12 and 13q deletion in patients with 17p deletion. Fisher's exact test was used to assess the association of percentage of 17p deletion with other FISH anomaly by hierarchical order and IGHV mutation status.
Results
Distribution of FISH categories
Within the cohort of 1585 patients, 1048 had FISH testing performed within 4 years of diagnosis. The distribution of patients by FISH category did not differ between those diagnosed more than or less than 4 years prior to FISH testing (P = 0·14; Table 1). Although we preserved the structure of the Dohner study (FISH within 4 years of diagnosis, inclusion of previously treated patients), the distribution of FISH results within the CRC differed from those reported in the 325 patients studied by Dohner (P < 0·001). The CRC cohort includes 12% with 17p deletion, 11% with 11q deletion and 24% with trisomy 12, compared to the 7%, 17%, and 18% reported by Dohner et al (2000).
Table 1.
The distribution of patients by FISH category using the Dohner classification, for the entire cohort of 1585 CLL patients and for those with FISH testing within 4 years of diagnosis (N = 1048)
| Entire cohort | FISH within 4 years of diagnosis | |||
|---|---|---|---|---|
| N = 1585 | N = 1048 | |||
| N | % | N | % | |
| 17p deletion | 193 | 12 | 122 | 12 |
| 11q deletion | 187 | 12 | 114 | 11 |
| Trisomy 12 | 205 | 13 | 149 | 14 |
| Normal | 376 | 24 | 252 | 24 |
| 13q deletion | 624 | 39 | 411 | 39 |
FISH, fluorescence in situ hybridization.
TTFT for patients from the date of their diagnosis (TTFT‐DX)
The TTFT from diagnosis was examined in the cohort of 1048 patients whose FISH analyses were performed within 4 years of diagnosis. Treatment status was unavailable for three patients. As anticipated, median time to first treatment was most favourable for those with sole 13q deletion (72 months), followed by normal FISH (35 months) and trisomy 12 (30 months). The median TTFT of 22 months was similar for those with 11q deletion and 17p deletion (Table 2 and Fig 1).
Table 2.
Association of FISH category and time to first treatment from diagnosis for patients who had FISH testing done less than 4 years after diagnosis of chronic lymphocytic leukaemia (N = 1045)
| N | Treated (n) | Median TTFT–DX (months) | P‐value | |
|---|---|---|---|---|
| FISH category | ||||
| 17p deletion | 122 | 93 | 22 | <0·0001 |
| 11q deletion | 113 | 92 | 22 | |
| Trisomy 12 | 148 | 113 | 30 | |
| Normal | 252 | 164 | 35 | |
| 13q deletion | 411 | 183 | 72 | |
FISH, fluorescence in situ hybridization; TTFT‐DX, time to first treatment measured from the date of diagnosis.
Figure 1.
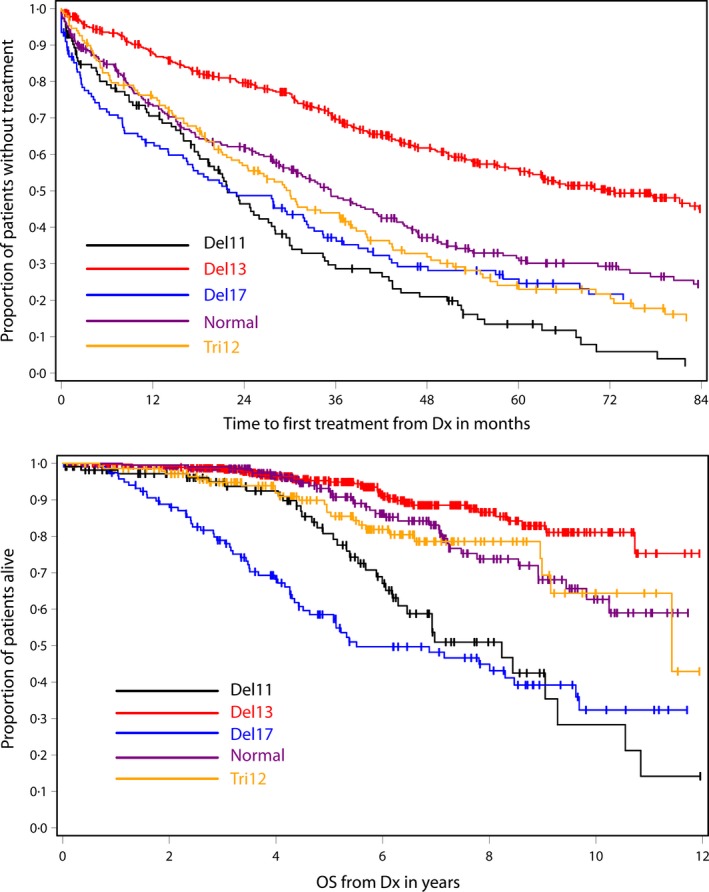
Time to first treatment and overall survival by FISH category for patients who had FISH analysis less than 4 years after diagnosis of chronic lymphocytic leukaemia. FISH, fluorescence in situ hybridization; Dx, diagnosis.
OS for patients from the date of their diagnosis (OS‐DX)
For the patients with FISH analysis done within 4 years of diagnosis, OS was most favourable for the 13q deletion category, followed by normal FISH, trisomy 12, 11q deletion and then 17p deletion (Table 3 and Fig 1). The median survival was not reached for the 13q deletion and normal FISH cohorts, whereas median OS was 8 years for the 11q deletion cohort and 6 years for the 17p deletion group.
Table 3.
Association of FISH category and overall survival (OS) from diagnosis for patients who had FISH testing done less than 4 years after diagnosis of chronic lymphocytic leukaemia (N = 1048)
| N | Death (n) | Median OS‐DX (years)* | P‐value | |
|---|---|---|---|---|
| FISH category | ||||
| 17p deletion | 122 | 60 | 5 | <0·0001 |
| 11q deletion | 114 | 36 | 7 | |
| Trisomy 12 | 149 | 25 | 11 | |
| Normal | 252 | 35 | Not reached | |
| 13q deletion | 411 | 35 | Not reached | |
FISH, fluorescence in situ hybridization; OS‐DX, overall survival from the date of diagnosis. *[Correction added on 29 February 2016, after online publication: this has been corrected to years].
TTFT for patients from the date at which FISH was performed (TTFT‐FISH)
To evaluate the prognostic value of FISH from the date of the FISH study, even when the analysis was not done within 4 years of diagnosis, those patients (N = 404) whose FISH analysis was obtained after the initiation of first treatment was excluded from the entire cohort of 1585 patients (N studied = 1181). For this group, the median time to treatment was more favourable for those with sole 13q deletion (64 months) followed by normal FISH (30 months), trisomy 12 (14 months) and 17p deletion (10 months), and was significantly worse for those with 11q deletion (5 months) (Table 4 and Fig 2). For some patients, the first FISH analysis was performed at the same time when they received treatment, and thus the treatment decision was based on concurrent clinical evidence of worsening of disease rather than the prognostic influence of the FISH results. For this reason, we also excluded 220 patients who were treated within 14 d of the FISH analysis, and examined TTFT from the date of when the FISH analysis was performed for the remaining 961 patients. The results suggested similar findings to those that included all patients with FISH analysis done prior to initiation of first treatment (data not shown).
Table 4.
Association of FISH category and time to first treatment from the time of FISH testing (N = 1181)
| N | Treated (n) | Median TTFT–FISH (months) | P‐value | |
|---|---|---|---|---|
| FISH category | ||||
| 17p deletion | 120 | 80 | 10 | <0·0001 |
| 11q deletion | 124 | 102 | 3 | |
| Trisomy 12 | 163 | 123 | 14 | |
| Normal | 266 | 154 | 30 | |
| 13q deletion | 508 | 224 | 64 | |
FISH, fluorescence in situ hybridization; TTFT‐FISH, time to first treatment from the date of the initial FISH results.
Figure 2.
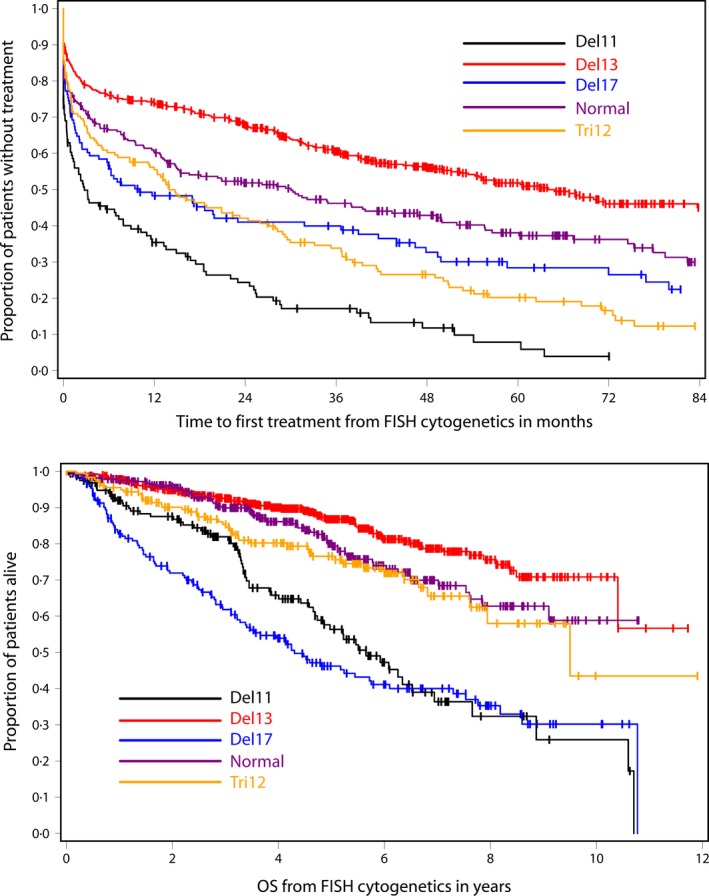
Time to first treatment and overall survival by FISH category from the date when FISH analysis was done. FISH, fluorescence in situ hybridization.
OS for patients from the date at which FISH is performed (OS‐FISH)
We also evaluated survival from time of FISH analysis for the entire cohort of patients with available FISH data (N = 1585). Patients with 13q deletion had the most favourable OS, and the median OS was not reached. The median OS for patients with normal cytogenetics was also not reached. The patients with trisomy 12 had a median OS of 11 years. Patients with 11q deletion had a median survival of 6 years and those with 17p deletion had the poorest median survival (4 years) (Table 5 and Fig 2).
Table 5.
Association of FISH category and overall survival (OS) from time of FISH testing (N = 1585)
| N | Death (n) | Median OS–FISH (years)* | P‐value | |
|---|---|---|---|---|
| FISH category | ||||
| 17p deletion | 193 | 93 | 4 | <0·0001 |
| 11q deletion | 187 | 65 | 6 | |
| Trisomy 12 | 205 | 45 | 10 | |
| Normal | 376 | 63 | Not reached | |
| 13q deletion | 624 | 81 | Not reached | |
FISH, fluorescence in situ hybridization; OS‐FISH, time to first treatment from the date of diagnosis. *[Correction added on 29 February 2016, after online publication: this has been corrected to years].
Association of the percentage of abnormal nuclei on TTFT and OS from when FISH analysis was performed
Here we studied if the extent of each FISH detectable defect is related to clinical outcome. The median and range of percentages of abnormal nuclei for each FISH abnormality are provided in Table 6. We found that the number of patients was not always distributed evenly by percentage of abnormal nuclei (Table 7). For example, 59 of 120 patients (49%) from the 17p deletion group exhibited ≤20% abnormal nuclei and only 14 (%) had between 20% and 50% abnormal nuclei (Table 7). Similarly, 42% of patients with sole 13q deletion had >70% abnormal nuclei.
Table 6.
Percentage of abnormal nuclei for each FISH category (N = 1585)
| N | Median (%) | Interquartile range | Range | |
|---|---|---|---|---|
| FISH anomaly | ||||
| 17p deletion | 193 | 38·5 | (10·5%, 76%) | (5%, 99·5%) |
| 11q deletion | 187 | 70 | (34·5%, 87%) | (6·5%, 100%) |
| Trisomy 12 | 205 | 55 | (26·5%, 72%) | (2·5%, 94%) |
| 13q deletion | 624 | 64·8 | (38%, 84%) | (7%, 100%) |
FISH, fluorescence in situ hybridization.
Table 7.
Association of percentage of abnormal nuclei and time to first treatment from the time of FISH testing
| N (%) | Treated | Median TTFT (months) | P‐value | |
|---|---|---|---|---|
| % of 17p – (Fig 4) | ||||
| 5–20% | 59 (49) | 33 | 44 | 0·002 |
| 20–50% | 14 (12) | 10 | 1 | |
| 50–65% | 15 (13) | 12 | 1 | |
| 65–80% | 11 (9) | 8 | 8 | |
| >80% | 21 (18) | 17 | 1 | |
| % of 11q – | ||||
| 5–20% | 13 (10) | 11 | 2 | 0·96 |
| 20–35% | 18 (15) | 15 | 3 | |
| 35–50% | 12 (10) | 8 | 3 | |
| 50–65% | 16 (13) | 15 | 2 | |
| 65–80% | 24 (19) | 21 | 3 | |
| >80% | 41 (33) | 32 | 3 | |
| % of trisomy 12 | ||||
| 1·5–15% | 23 (14) | 15 | 54 | 0·03 |
| 15–30% | 17 (10) | 10 | 30 | |
| 30–45% | 19 (12) | 14 | 15 | |
| 45–60% | 29 (18) | 22 | 29 | |
| 60–75% | 44 (27) | 34 | 7 | |
| >75% | 31 (19) | 28 | 3 | |
| % of 13q – (Fig 3) | ||||
| 7–25% | 78 (15) | 26 | Not reached | <0·0001 |
| 25–40% | 54 (11) | 17 | 85 | |
| 40–55% | 81 (16) | 31 | 89 | |
| 55–70% | 80 (16) | 32 | 71 | |
| 70–85% | 111 (22) | 53 | 52 | |
| >85% | 104 (20) | 65 | 24 | |
FISH, fluorescence in situ hybridization; TTFT, time to first treatment.
For the 11q deletion, there was no significant difference in median TTFT by the percentage of abnormal nuclei, whereas trends in TTFT were observed for trisomy 12, 13q deletion and 17p deletion. The percentage of 13q deletion nuclei was significantly associated with TTFT in the time from when FISH analysis was performed (P < 0·0001, Table 7 and Fig 3) For 13q deletion, patients who exhibited >85% abnormal nuclei had a shorter TTFT from time of FISH analysis (24 months), similar to those with normal FISH and in contrast to those patients who had <85% abnormal nuclei (52 months to median not reached) (P < 0·0001). There was no significant association between percentage of 13q deletion nuclei and IGHV mutation status (P = 0·1).
Figure 3.
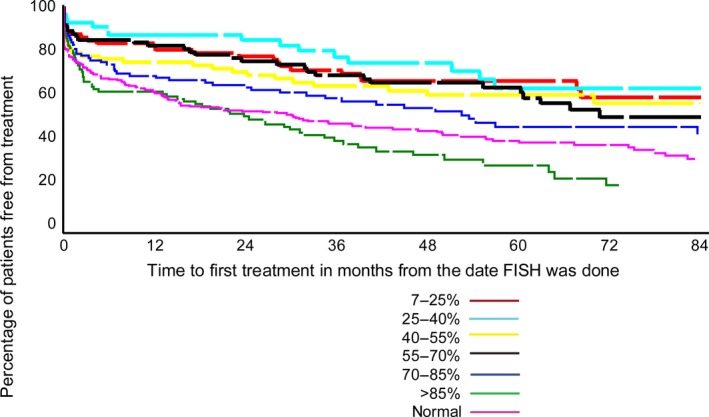
Association of percentage of 13q deletion nuclei and TTFT from when FISH analysis was done. FISH, fluorescence in situ hybridization; TTFT, time to first treatment.
The percentage of 17p deletion nuclei was significantly associated with TTFT from when FISH analysis was performed (P = 0·002, Table 7 and Fig 4). Patients with ≤20% abnormal nuclei had a significantly longer TTFT from diagnosis (44 months) than did those with >20% abnormal nuclei: 1 month for the 20–65% group, 8 months for the 65–80% group and 1 month for >85% (P < 0·0001). To further assess the TTFT for the cohort who had >20% abnormal we categorized them into 4 groups (described in methods) and examined the TTFT, but we were unable to find any further cut‐off point that was significantly different from the 20% cut‐off (data not shown). However, the percentage of 17p deletion using the >20% deletion nuclei was significantly associated with OS in time from initial FISH analysis (P < 0·0001). As with TTFT, patients with ≤20% 17p deletion nuclei had a longer OS of 11 years compared to median OS of 3 years for those with ≥20% 17p deletion nuclei.
Figure 4.

Association of percentage of 17p deletion nuclei and TTFT from when FISH analysis was done. FISH, fluorescence in situ hybridization; TTFT, time to first treatment.
The percentage of trisomy 12 nuclei was significantly associated with TTFT from when FISH analysis was performed (P = 0·03, Table 7). Patients with > 60% of abnormal trisomy 12 nuclei appeared to have shorter TTFT (P < 0·0001).
The Dohner hierarchical order for 11q deletion, trisomy 12 and 13q deletion (Dohner et al, 2000) was further applied to patients with 17p deletion. For instance, if a patient had 11q deletion along with 17p this patient was then categorized as having 11q deletion when assessing association of percentage of 17p deletion with other FISH anomalies by hierarchical order. There was no significant association observed between percentage of 17p deletion and other FISH anomalies summarized by hierarchical order. However there was a significant association between percentage of abnormal 17p nuclei and IGHV mutation status (Table 8); patients with ≤20% 17p deletion nuclei were more likely than those with ≥20% 17p deletion nuclei to have mutated IGHV status (51% vs. 25%, P = 0·005).
Table 8.
Other FISH anomalies and IGHV mutation status by percentage of 17p deletion in patients with 17p deletion
| % of 17p abnormal nuclei | P‐value | ||
|---|---|---|---|
| ≤20% | >20% | ||
| FISH hierarchical excluding 17p deletion | |||
| 11q deletion | 7 | 6 | 0·44 |
| Trisomy 12 | 9 | 7 | |
| Normal | 10 | 18 | |
| 13q deletion | 33 | 30 | |
| Other FISH anomalies | |||
| 11q deletion | 7 | 6 | |
| Trisomy 12 | 10 | 8 | |
| 13q deletion | 40 | 36 | |
| IGHV mutations status | |||
| Mutated | 30 | 15 | 0·005 |
| Unmutated | 29 | 46 | |
FISH, fluorescence in situ hybridization.
Discussion
We employed a cohort of 1585 patients diagnosed with CLL to revisit the FISH‐based prognostic hierarchy described by Dohner et al (2000) based on their population sample of 325 CLL patients. By comparison, although the FISH hierarchy remains similar when OS is considered, the TTFT and OS appears more favourable than originally reported. This clinical trend is, of course, compatible with the enhanced ability to treat CLL patients more effectively with chemoimmunotherapy over the time frame of our study.
The FISH hierarchy remained consistent for all patients in our CRC cohort, as well as when excluding patients whose FISH analysis was done at the time of treatment. Our major findings also include that, within the FISH hierarchy, 11q deletion patients had the shortest TTFT, and a shorter TTFT was observed with increasing numbers of abnormal interphase nuclei exhibiting either 13q or 17p deletion.
The patients with 13q deletion clearly comprised the most favourable prognostic group. TTFT for the normal FISH category fell consistently between the 13q deletion and the trisomy 12 and 11q deletion groups. Although the median TTFT from diagnosis was similar (Table 2 and Fig 1), the 11q deletion patients tended to fare more poorly than the 17p group after the median was reached (22 months). Thereafter, TTFT was similar for the 17p deletion and trisomy 12 groups. In a somewhat striking deviation from the Dohner hierarchy, when we excluded patients who received treatment shortly after the initial FISH analysis, TTFT was more favourable for those with a 17p deletion than for those with either an 11q deletion or trisomy 12. We suggest, along with others (Davids et al, 2015), that using the date of the first FISH study offers a more objective analysis of TTFT and OS than that obtained using the date of diagnosis of CLL.
For OS, the 17p deletion patients fared most poorly until approximately 7 years of follow‐up, whereupon the 11q deletion patients tended to exhibit a similar unfavourable survival, and these two groups exhibited dramatically poorer survival than did the other three hierarchical categories. Although the median OS for trisomy 12 was 10–11 years in comparison with the normal FISH group, their Kaplan–Meier survival curves followed similar trajectories (see Figs 1 and 2). As with TTFT, the 13q deletion group exhibited the most favourable prognosis, with roughly 75% of these patients alive at 10 years post‐diagnosis and post‐initial FISH testing.
We also examined TTFT for each FISH category by the percentage of abnormal nuclei. Here, we found that the prognosis was more favourable when the percentage of 17p deletion nuclei was low, consistent with the findings of Tam et al (2009). For deletion 13q, the prognosis was less favourable when the percentage of abnormal nuclei was high, and this held whether TTFT was observed from diagnosis or from the date of the initial FISH analysis. In another deviation from the Dohner hierarchy (Dohner et al, 2000), patients with 5–20% 17p deletion nuclei exhibited a very favourable 44 and 57 months of TTFT from time of FISH analysis and diagnosis, which was comparable to TTFT of the 13q deletion patients as a group. In contrast, patients with the highest percentage of 13q deletion nuclei (>85%) exhibited a much less favourable TTFT than those with fewer than 85% nuclei with 13q deletion. Those with >85% 13q deletion nuclei exhibited a TTFT more comparable to those with a normal FISH pattern or trisomy 12, and this is similar to earlier observations (Van Dyke et al, 2010; Dal Bo et al, 2011). In contrast to 17p and 13q deletion groups, patients with 11q deletion did not exhibit a statistically significant association between TTFT and percentage of abnormal nuclei. Thus, we were unable to confirm an association between high percentage 11q deletion and adverse outcomes reported by Marasca et al (2013) and Jain et al (2015). Similarly, we were unable to confirm an association between small clones with TP53 mutation and poor survival (Rossi et al, 2014), assuming that small clones with TP53 mutation are comparable to our patients with FISH detectable but low percentages of 17p deletion. It was not possible to carry out a similar analysis for an association between OS and percentage of abnormal nuclei, because the survival data are not sufficiently mature in our CRC database. It is our intent to conduct further analyses on the relationship of abnormal nuclei as the clinical data mature.
In terms of our study, the results are most applicable to patients with CLL whose FISH analysis is done within 4 years of diagnosis and prior to starting therapy. Sequential evaluation of genetic defects via FISH or with more detailed sequencing methods will be of great interest in monitoring clinical progression in CLL, and clonal genetic abnormalities in CLL have been shown to evolve and change over time (Shanafelt et al, 2006; Braggio et al, 2012; Chuang et al, 2012; Landau et al, 2013; Ouillette et al, 2013; Ojha et al, 2015). Changes in prognosis upon disease progression associated with cytogenetic evolution (Guieze & Wu, 2015) highlight the association of clinical features with specific genetic changes. Future studies of genetic changes over time using a large and well annotated clinical cohort of CLL patients will be required to understand the potential benefits of chromosomal microarray, selected gene sequencing platforms, or deep sequencing of whole genomes for a more complete understanding of prognosis and new therapeutic targets in CLL (Edelmann et al, 2012; Rossi et al, 2013; Baliakas et al, 2015; Puente et al, 2015).
Limitations of our study include that the cohort consisted of patients who were referred or self‐referred to these six academic institutions. However the large size of this cohort is likely to reflect the overall CLL patient base. We have assumed that the improvements in outcome for patients who are assessed via FISH testing are probably due to the dramatic impact of chemoimmunotherapy on patient outcome. However our database does not include the specific therapies that each patient received and thus we cannot know what percentages of patients received these treatments. It is also unlikely that there were significant percentages of CLL patients who received the more novel targeted signal inhibitors now being widely tested as this study analyses only used data acquired prior to 2011.
In summary our updated data on this large cohort of CLL cases validates the Dohner hierarchy (Dohner et al, 2000). The duration of both TTFT and survival are estimated to be considerably longer than during the years of the Dohner study, probably because of the much more effective combination therapies instituted over the last 20 years. We also observed that the percentage of abnormal nuclei is an important prognostic variable. Finally, we found that the FISH panel commonly used by CLL practitioners remains a valuable prognostic tool even as the therapeutic options change for our CLL patients.
Author contributions
DLVD, LZR, EG and NEK designed the study, provided patient data, verified and collated the data, contributed to the analysis of the results and wrote the manuscript; LW and DN carried out the statistical analyses, NAH, PDC, MDA and TK provided patient data, contributed to the analysis of the results and edited the manuscript.
Acknowledgements
‘This work was supported by a grant (PO1‐CA81534) from the National Institutes of Health to T. J. Kipps for the CLL Research Consortium’. We appreciate the technical support and cytogenetic data from C Sreekantaiah and SA Smoley.
References
- Baliakas, P. , Hadzidimitriou, A. , Sutton, L.A. , Rossi, D. , Minga, E. , Villamor, N. , Larrayoz, M. , Kminkova, J. , Agathangelidis, A. , Davis, Z. , Tausch, E. , Stalika, E. , Kantorova, B. , Mansouri, L. , Scarfo, L. , Cortese, D. , Navrkalova, V. , Rose‐Zerilli, M.J. , Smedby, K.E. , Juliusson, G. , Anagnostopoulos, A. , Makris, A.M. , Navarro, A. , Delgado, J. , Oscier, D. , Belessi, C. , Stilgenbauer, S. , Ghia, P. , Pospisilova, S. , Gaidano, G. , Campo, E. , Strefford, J.C. , Stamatopoulos, K. & Rosenquist, R. (2015) Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia, 29, 329–336. [DOI] [PubMed] [Google Scholar]
- Braggio, E. , Kay, N.E. , VanWier, S. , Tschumper, R.C. , Smoley, S. , Eckel‐Passow, J.E. , Sassoon, T. , Barrett, M. , Van Dyke, D.L. , Byrd, J.C. , Jelinek, D.F. , Shanafelt, T.D. & Fonseca, R. (2012) Longitudinal genome‐wide analysis of patients with chronic lymphocytic leukemia reveals complex evolution of clonal architecture at disease progression and at the time of relapse. Leukemia, 26, 1698–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, H.Y. , Rassenti, L. , Salcedo, M. , Licon, K. , Kohlmann, A. , Haferlach, T. , Foa, R. , Ideker, T. & Kipps, T.J. (2012) Subnetwork‐based analysis of chronic lymphocytic leukemia identifies pathways that associate with disease progression. Blood, 120, 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus, R. , Lucas, D.M. , Ruppert, A.S. , Williams, K.E. , Weng, D. , Patterson, K. , Zucknick, M. , Oakes, C.C. , Rassenti, L.Z. , Greaves, A.W. , Geyer, S. , Wierda, W.G. , Brown, J.R. , Gribben, J.G. , Barrientos, J.C. , Rai, K.R. , Kay, N.E. , Kipps, T.J. , Shields, P. , Zhao, W. , Grever, M.R. , Plass, C. & Byrd, J.C. (2014) Validation of ZAP‐70 methylation and its relative significance in predicting outcome in chronic lymphocytic leukemia. Blood, 124, 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Bo, M. , Rossi, F.M. , Rossi, D. , Deambrogi, C. , Bertoni, F. , Del Giudice, I. , Palumbo, G. , Nanni, M. , Rinaldi, A. , Kwee, I. , Tissino, E. , Corradini, G. , Gozzetti, A. , Cencini, E. , Ladetto, M. , Coletta, A.M. , Luciano, F. , Bulian, P. , Pozzato, G. , Laurenti, L. , Forconi, F. , Di Raimondo, F. , Marasca, R. , Del Poeta, G. , Gaidano, G. , Foa, R. , Guarini, A. & Gattei, V. (2011) 13q14 deletion size and number of deleted cells both influence prognosis in chronic lymphocytic leukemia. Genes, Chromosomes & Cancer, 50, 633–643. [DOI] [PubMed] [Google Scholar]
- Davids, M.S. , Vartanov, A. , Werner, L. , Neuberg, D. , Dal Cin, P. & Brown, J.R. (2015) Controversial fluorescence in situ hybridization cytogenetic abnormalities in chronic lymphocytic leukaemia: new insights from a large cohort. British Journal of Haematology, 170, 694–703. [DOI] [PubMed] [Google Scholar]
- Dewald, G.W. , Brockman, S.R. , Paternoster, S.F. , Bone, N.D. , O'Fallon, J.R. , Allmer, C. , James, C.D. , Jelinek, D.F. , Tschumper, R.C. , Hanson, C.A. , Pruthi, R.K. , Witzig, T.E. , Call, T.G. & Kay, N.E. (2003) Chromosome anomalies detected by interphase fluorescence in situ hybridization: correlation with significant biological features of B‐cell chronic lymphocytic leukaemia. British Journal of Haematology, 121, 287–295. [DOI] [PubMed] [Google Scholar]
- Dohner, H. , Stilgenbauer, S. , Benner, A. , Leupolt, E. , Krober, A. , Bullinger, L. , Dohner, K. , Bentz, M. & Lichter, P. (2000) Genomic aberrations and survival in chronic lymphocytic leukemia. The New England Journal of Medicine, 343, 1910–1916. [DOI] [PubMed] [Google Scholar]
- Edelmann, J. , Holzmann, K. , Miller, F. , Winkler, D. , Buhler, A. , Zenz, T. , Bullinger, L. , Kuhn, M.W. , Gerhardinger, A. , Bloehdorn, J. , Radtke, I. , Su, X. , Ma, J. , Pounds, S. , Hallek, M. , Lichter, P. , Korbel, J. , Busch, R. , Mertens, D. , Downing, J.R. , Stilgenbauer, S. & Dohner, H. (2012) High‐resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood, 120, 4783–4794. [DOI] [PubMed] [Google Scholar]
- Geisler, C. , Jurlander, J. , Bullinger, L. , Sander, S. , Brown, P. , Benner, A. , Philip, P. , Dohner, H. & Stilgenbauer, S. (2009) Danish CLL2‐Study revisited: FISH on a cohort with a 20‐yr follow‐up confirms the validity of the hierarchical model of genomic aberrations in chronic lymphocytic leukaemia. European Journal of Haematology, 83, 156–158. [DOI] [PubMed] [Google Scholar]
- Guieze, R. & Wu, C.J. (2015) Genomic and epigenomic heterogeneity in chronic lymphocytic leukemia. Blood, 126, 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, P. , Keating, M. , Thompson, P.A. , Trinh, L. , Wang, X. , Wierda, W. , Ferrajoli, A. , Burger, J. , Kantarjian, H. , Estrov, Z. , Abruzzo, L. & O'Brien, S. (2015) High fluorescence in situ hybridization percentage of deletion 11q in patients with chronic lymphocytic leukemia is an independent predictor of adverse outcome. American Journal of Hematology, 90, 471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau, D.A. , Carter, S.L. , Stojanov, P. , McKenna, A. , Stevenson, K. , Lawrence, M.S. , Sougnez, C. , Stewart, C. , Sivachenko, A. , Wang, L. , Wan, Y. , Zhang, W. , Shukla, S.A. , Vartanov, A. , Fernandes, S.M. , Saksena, G. , Cibulskis, K. , Tesar, B. , Gabriel, S. , Hacohen, N. , Meyerson, M. , Lander, E.S. , Neuberg, D. , Brown, J.R. , Getz, G. & Wu, C.J. (2013) Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell, 152, 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasca, R. , Maffei, R. , Martinelli, S. , Fiorcari, S. , Bulgarelli, J. , Debbia, G. , Rossi, D. , Rossi, F.M. , Rigolin, G.M. , Gattei, V. , Del Poeta, G. , Laurenti, L. , Forconi, F. , Montillo, M. , Gaidano, G. & Luppi, M. (2013) Clinical heterogeneity of de novo 11q deletion chronic lymphocytic leukaemia: prognostic relevance of extent of 11q deleted nuclei inside leukemic clone. Hematological Oncology, 31, 88–95. [DOI] [PubMed] [Google Scholar]
- Ojha, J. , Ayres, J. , Secreto, C. , Tschumper, R. , Rabe, K. , Van Dyke, D. , Slager, S. , Shanafelt, T. , Fonseca, R. , Kay, N.E. & Braggio, E. (2015) Deep sequencing identifies genetic heterogeneity and recurrent convergent evolution in chronic lymphocytic leukemia. Blood, 125, 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouillette, P. , Saiya‐Cork, K. , Seymour, E. , Li, C. , Shedden, K. & Malek, S.N. (2013) Clonal evolution, genomic drivers, and effects of therapy in chronic lymphocytic leukemia. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 19, 2893–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente, X.S. , Bea, S. , Valdes‐Mas, R. , Villamor, N. , Gutierrez‐Abril, J. , Martin‐Subero, J.I. , Munar, M. , Rubio‐Perez, C. , Jares, P. , Aymerich, M. , Baumann, T. , Beekman, R. , Belver, L. , Carrio, A. , Castellano, G. , Clot, G. , Colado, E. , Colomer, D. , Costa, D. , Delgado, J. , Enjuanes, A. , Estivill, X. , Ferrando, A.A. , Gelpi, J.L. , Gonzalez, B. , Gonzalez, S. , Gonzalez, M. , Gut, M. , Hernandez‐Rivas, J.M. , Lopez‐Guerra, M. , Martin‐Garcia, D. , Navarro, A. , Nicolas, P. , Orozco, M. , Payer, A.R. , Pinyol, M. , Pisano, D.G. , Puente, D.A. , Queiros, A.C. , Quesada, V. , Romeo‐Casabona, C.M. , Royo, C. , Royo, R. , Rozman, M. , Russinol, N. , Salaverria, I. , Stamatopoulos, K. , Stunnenberg, H.G. , Tamborero, D. , Terol, M.J. , Valencia, A. , Lopez‐Bigas, N. , Torrents, D. , Gut, I. , Lopez‐Guillermo, A. , Lopez‐Otin, C. & Campo, E. (2015) Non‐coding recurrent mutations in chronic lymphocytic leukaemia. Nature, 526, 519–524. [DOI] [PubMed] [Google Scholar]
- Rassenti, L.Z. , Jain, S. , Keating, M.J. , Wierda, W.G. , Grever, M.R. , Byrd, J.C. , Kay, N.E. , Brown, J.R. , Gribben, J.G. , Neuberg, D.S. , He, F. , Greaves, A.W. , Rai, K.R. & Kipps, T.J. (2008) Relative value of ZAP‐70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood, 112, 1923–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, D. , Rasi, S. , Spina, V. , Bruscaggin, A. , Monti, S. , Ciardullo, C. , Deambrogi, C. , Khiabanian, H. , Serra, R. , Bertoni, F. , Forconi, F. , Laurenti, L. , Marasca, R. , Dal‐Bo, M. , Rossi, F.M. , Bulian, P. , Nomdedeu, J. , Del Poeta, G. , Gattei, V. , Pasqualucci, L. , Rabadan, R. , Foa, R. , Dalla‐Favera, R. & Gaidano, G. (2013) Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood, 121, 1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, D. , Khiabanian, H. , Spina, V. , Ciardullo, C. , Bruscaggin, A. , Fama, R. , Rasi, S. , Monti, S. , Deambrogi, C. , De Paoli, L. , Wang, J. , Gattei, V. , Guarini, A. , Foa, R. , Rabadan, R. & Gaidano, G. (2014) Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood, 123, 2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanafelt, T.D. , Witzig, T.E. , Fink, S.R. , Jenkins, R.B. , Paternoster, S.F. , Smoley, S.A. , Stockero, K.J. , Nast, D.M. , Flynn, H.C. , Tschumper, R.C. , Geyer, S. , Zent, C.S. , Call, T.G. , Jelinek, D.F. , Kay, N.E. & Dewald, G.W. (2006) Prospective evaluation of clonal evolution during long‐term follow‐up of patients with untreated early‐stage chronic lymphocytic leukemia. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology, 24, 4634–4641. [DOI] [PubMed] [Google Scholar]
- Smoley, S.A. , Van Dyke, D.L. , Kay, N.E. , Heerema, N.A. , Dell' Aquila, M.L. , Dal Cin, P. , Koduru, P. , Aviram, A. , Rassenti, L. , Byrd, J.C. , Rai, K.R. , Brown, J.R. , Greaves, A.W. , Eckel‐Passow, J. , Neuberg, D. , Kipps, T.J. & Dewald, G.W. (2010) Standardization of fluorescence in situ hybridization studies on chronic lymphocytic leukemia (CLL) blood and marrow cells by the CLL Research Consortium. Cancer Genetics and Cytogenetics, 203, 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam, C.S. , Shanafelt, T.D. , Wierda, W.G. , Abruzzo, L.V. , Van Dyke, D.L. , O'Brien, S. , Ferrajoli, A. , Lerner, S.A. , Lynn, A. , Kay, N.E. & Keating, M.J. (2009) De novo deletion 17p13.1 chronic lymphocytic leukemia shows significant clinical heterogeneity: the M. D. Anderson and Mayo Clinic experience. Blood, 114, 957–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke, D.L. , Shanafelt, T.D. , Call, T.G. , Zent, C.S. , Smoley, S.A. , Rabe, K.G. , Schwager, S.M. , Sonbert, J.C. , Slager, S.L. & Kay, N.E. (2010) A comprehensive evaluation of the prognostic significance of 13q deletions in patients with B‐chronic lymphocytic leukaemia. British Journal of Haematology, 148, 544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone, R. , Veronese, A. , Rassenti, L.Z. , Balatti, V. , Pearl, D.K. , Acunzo, M. , Volinia, S. , Taccioli, C. , Kipps, T.J. & Croce, C.M. (2011) miR‐181b is a biomarker of disease progression in chronic lymphocytic leukemia. Blood, 118, 3072–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]


