Abstract
Charting differences between tumors and normal tissue is a mainstay of cancer research. However, clonal tumor expansion from complex normal tissue architectures potentially obscures cancer-specific events, including divergent epigenetic patterns. Using whole-genome bisulfite sequencing of normal B cell subsets, we observed broad epigenetic programming of selective transcription factor binding sites coincident with the degree of B cell maturation. By comparing normal B cells to malignant B cells from 268 patients with chronic lymphocytic leukemia (CLL), we showed that tumors derive largely from a continuum of maturation states reflected in normal developmental stages. Epigenetic maturation in CLL was associated with an indolent gene expression pattern and increasingly favorable clinical outcomes. We further uncovered that most previously reported tumor-specific methylation events are normally present in non-malignant B cells. Instead, we identified a potential pathogenic role for transcription factor dysregulation in CLL, where excess programming by EGR and NFAT with reduced EBF and AP-1 programming imbalances the normal B cell epigenetic program.
Identification of the cell of origin is essential to fully appreciate a tumor’s abnormal biology and the events that may give rise to the disease. Healthy tissue is usually composed of different normal cell types that retain distinct epigenomes1–3, which are important to establish and stabilize cellular phenotypes in mature cells4. A comparison of clonally expanded tumor cells to healthy tissue may identify cancer-specific genetic events; however, epigenetic alterations may merely reflect the highly specialized features of distinct cellular subtypes. Furthermore, epigenomic complexity is increased by differentiation pathways from progenitor (stem) cells within tissues. Variation among individuals is also observed5. As ongoing efforts uncover an expanding repertoire of tumor subtypes, a paradigm for comprehending the true uniqueness of a tumor sample in the context of normal cell complexity is lacking.
Epigenetic specialization is well described in the hematopoietic system6 and results from dynamic modifications occurring during lineage development7. The establishment of normal DNA methylation patterning is in part due to the activities of specific chromatin-interacting proteins and transcription factors8. Diseased tissues regularly exhibit degradation of DNA methylation patterns9. In CLL, genome-wide DNA methylation studies uncovered distinct methylation subtypes10,11, exhibiting remarkable longitudinal stability11–13. In addition, despite local pattern disorder14, the clonality of DNA methylation patterns is maintained to a higher degree in most CLLs than in other cancer types13. Clonal methylation likely reflects the methylation state present in very early disease stages and may, in part, derive from the founder cell. As broad epigenetic programming has recently been described to occur during B cell development15, here we address the complex relationship between individual CLLs and the variation in DNA methylation programming in normal cells.
RESULTS
DNA methylation programming during B cell maturation
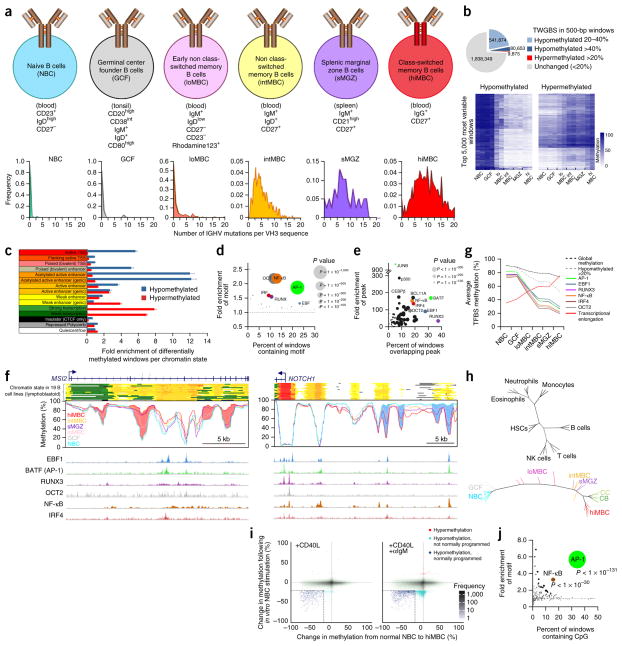
To capture dynamic DNA methylation programming during B cell maturation, we obtained discrete B cell subpopulations ranging in maturity from naive B cells to memory B cells, referred to as low-, intermediate- and high-maturity memory B cells; germinal center founder (GCF) cells, the subpopulation of B cells formed following antigen exposure16; and splenic marginal zone B cells (Fig. 1a). The maturity of the subpopulations was determined by examining the mutation status of IGHV3 gene rearrangements (Fig. 1a, bottom). To assess the DNA methylome of these populations, we performed tagmentation-based whole-genome bisulfite sequencing (TWGBS)17 on two donors for each subpopulation. Methylation levels were assessed by binning the genome into 5,009,715 windows of 500 bp in length. Only windows that contained ≥4 CpG sites (2,442,234) were considered (Supplementary Fig. 1a). Methylation differences were progressive (unidirectional) from naive B cells to high-maturity memory B cells (Fig. 1b, Supplementary Fig. 1b and Supplementary Table 1a,b). We observed prominent loss of methylation with increasing maturity, as previously reported10,15,18,19, shown here for 622,527 windows with a >20% decrease in methylation relative to naive B cells, representing 25.9% of the windows analyzed. Hypermethylation (an increase of >20% relative to naive B cells) occurred in 9,875 windows. A paucity of the total differences observed between naive and high-maturity memory B cells were unique to each of the intermediate subpopulations (<1% per subpopulation), indicating that these B cell subpopulations occupy a singular developmental trajectory. Next, we related the methylation changes that were acquired by the high-maturity memory B cell stage with chromatin states in a collection of 19 lymphoblastoid B cell lines5,20. Of note, lymphoblastoid B cells have an epigenetic signature similar to that of high-maturity memory B cells, making them suitable to assess the chromatin state acquired upon programming (Supplementary Fig. 1b). Hypomethylation was highly enriched in enhancer and promoter regions (Fig. 1c and Supplementary Fig. 1c), as observed previously10,15,19. Hypermethylation was enriched in regions of transcriptional elongation. Differential methylation was significantly enriched in genes involved in B cell– and lymphocyte-related processes and pathways, including B cell receptor (BCR) activation (Supplementary Fig. 1d–f). These findings suggest an important role for methylation programming in B cell maturation and function.
Figure 1.
Epigenetic programming during B cell maturation. (a) Top, FACS sorting markers used to isolate the analyzed B cell subsets after selection of CD19+ cells. Bottom, the frequency of IGHV3 mutations in each subpopulation. (b) Top, TWGBS summary comparing naive B cells and high-maturity memory B cells. Bottom, methylation heat maps for the top 5,000 most variable windows. (c) Enrichment of differentially methylated windows among chromatin states, defined using the 15-state ChromHMM model20 (hypermethylated, >20% change; hypomethylated, >40% change), in the comparison of naive B cells and high-maturity memory B cells. Fold enrichment was calculated independently in 19 lymphoblastoid B cell lines (error bars, s.e.m.). TSS, transcriptional start site. (d) Bubble scatterplot of 223 transcription factor motifs displaying the prevalence and fold enrichment of each motif in hypomethylated windows (>40% change). Bubble size corresponds to the P value. (e) Bubble scatterplot of 78 transcription factor ChIP-seq peaks (determined in GM12878 cells) in hypomethylated windows (>40% change). Bubbles are colored according to the cognate motif in d. (f) Examples of loci differentially methylated in normal B cells at single-CpG resolution. Regions of hypermethylation (pink) within the gene body of MSI2 (left) and hypomethylation (blue) upstream of NOTCH1 (right) are shown. Chromatin states (colors correspond to those used in c) and transcription factor binding site peaks from GM12878 cells are indicated. (g) Average methylation level of hypomethylated windows (>20% change) containing high-confidence transcription factor binding sites (TFBSs) and hypermethylated windows (>20% change) overlapping regions of transcriptional elongation in each B cell subtype. (h) Left, DNA methylation phylogenetic tree diagram of blood cell development of common hematopoietic cell types. Each branch tip represents a single sample. Right, diagram of B cell maturation constructed using high-confidence methylation (450K) at transcription factor binding sites. HSCs, hematopoietic stem cells; NK cells, natural killer cells; CC, centrocytes; CB, centroblasts. (i) Scatterplots displaying the change in methylation of 450K probes in naive B cells after in vitro stimulation and 5 d in culture (y axis) versus the difference in methylation between naive B cells and high-maturity memory B cells (x axis). Naive B cells were stimulated with CD40L (left) or with CD40L and antibody to IgM (αIgM; right). (j) Bubble scatterplot of transcription factor motifs overlapping hypomethylated CpGs following CD40L stimulation and 5 d in culture.
DNA methylation of transcription factor binding sites
Regions targeted for hypomethylation during B cell maturation showed highly significant enrichment of sequence motifs for six transcription factor families: AP-1, EBF, RUNX, OCT, IRF and NF-κB (Fig. 1d). Only a low level of enrichment was found for hypermethylated regions. We next compared hypomethylated regions with transcription factor chromatin immunoprecipitation and sequencing (ChIP-seq) data available for GM12878 lymphoblastoid cells. Among many transcription factors previously known to have enriched binding at hypomethylated sites in B cells10,19, transcription factors known to bind to the six motifs for these families were among the most significantly enriched of the 78 transcription factors with data available (Fig. 1e). Integration of chromatin state and transcription factor binding site information showed that hypomethylation programming is generally targeted to regions marked as enhancers as well as regions with transcription factor binding, whereas hypermethylation occurs in transcribed regions (Fig. 1f). We then compiled a set of hypomethylated windows representing high-confidence transcription factor binding sites by filtering for windows that contained both a predicted sequence motif and binding of the corresponding transcription factor. We found that these high-confidence transcription factor binding sites were hypomethylated to a greater degree than other hypomethylated sites: specifically, NF-κB, OCT2 and IRF4 binding sites were programmed earlier and showed a greater degree of hypomethylation than AP-1, EBF1 and RUNX3 binding sites (Fig. 1g). DNA methylation analysis of 3–6 independent healthy donors per subtype using Illumina Infinium HumanMethylation450 (450K) arrays confirmed the TWGBS findings (Supplementary Fig. 1b,g,h). A phylogenetic analysis using high-confidence transcription factor binding sites for the six transcription factors of interest, together with CpG sites in hypermethylated transcriptional elongation domains, demonstrated that all samples individually stemmed from a single trunk (Fig. 1h, right). This observation contrasts with the highly diverse and branched developmental architecture observed in the hematopoietic system at large (Fig. 1h, left).
To directly evaluate DNA methylation programming in B cells, we stimulated naive B cells under controlled conditions in vitro. Purified naive B cells were cultured either in the absence or presence of CD40 ligand (CD40L) and/or antibody against IgM. Treatment with CD40L selectively induced the hypomethylation of a subset of CpG sites that become hypomethylated during B cell maturation (Fig. 1i). Additional treatment with antibody against IgM caused further programming but also induced hypomethylation that was not observed in vivo. Separation of cells with high and low rates of proliferation showed that, although hypomethylation increased upon proliferation, selective programming was independent of proliferation in this setting (Supplementary Fig. 1i). Primarily AP-1 and NF-κB binding sites were targeted under these conditions, accounting for approximately 10% of the programmed sites in vivo (Fig. 1j and Supplementary Fig. 1j). Transcription factor binding site analysis confirmed that AP-1 and NF-κB family transcription factors occupied a highly significant proportion of these sites in GM12878 cells and in all other cell lines and were enriched at enhancers (Supplementary Fig. 1k). These results indicate that naive B cells possess an innate ability to program their epigenome in a highly selective and stimulus-dependent manner.
Relationship between normal B cells and CLL
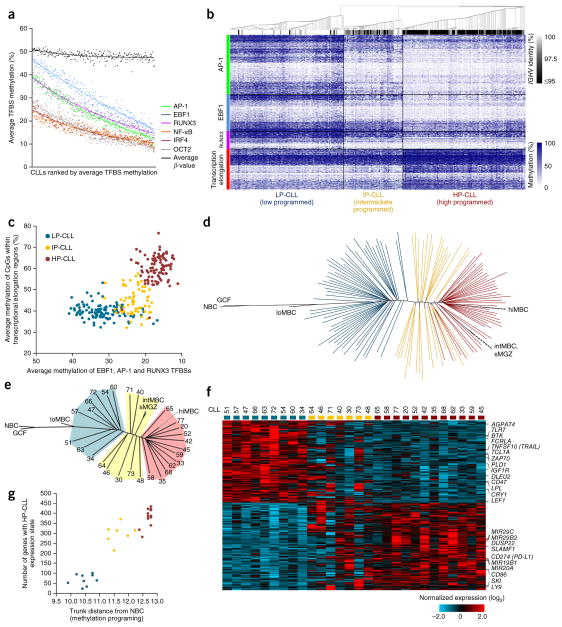
To assess the relationship between CLL and normal B cells, we used 450K methylation data from 68 CLL cases13 in combination with data for 60 new cases (DKFZ sample cohort, n = 128) and further included 139 cases published previously10 (International Cancer Genome Consortium (ICGC) sample cohort). To assess the degree of methylation programming achieved in each CLL, we analyzed high-confidence binding sites for the six transcription factors found to be most important for normal B cell methylation programming. By averaging methylation levels across transcription factor binding sites, we found that programming at NF-κB, OCT2 and IRF4 binding sites was largely complete in CLL; however, AP-1, EBF1 and RUNX3 binding sites demonstrated a greater variability in programming states among samples (Fig. 2a). Unsupervised clustering of all 267 CLL cases using these transcription factor binding sites together with hypermethylated CpG sites in transcriptional elongation domains identified three CLL subtypes (Fig. 2b), as found previously10. On the basis of the overall level of programming per cluster, we term these subtypes low-programmed (LP-CLL), intermediate-programmed (IP-CLL) and high-programmed (HP-CLL) CLL. This sample classification is highly concordant with classification using the most variable CpG sites on the 450K array or a published method based on a reduced number of CpG sites10 (with 95.1% and 92.7% concordance, respectively; Supplementary Table 2). Hypomethylation of AP-1, EBF1 and RUNX3 binding sites mostly discriminated IP-CLLs from LP-CLLs, whereas hypermethylation in transcriptional elongation regions was effective in discriminating HP-CLLs from IP-CLLs (Fig. 2c), reminiscent of the features distinguishing the normal B cell subtypes (Supplementary Fig. 1g). To illustrate the degree of programming achieved by individual CLL cases, we generated a phylogenetic tree incorporating normal B cell and CLL samples (Fig. 2d). As only minimal methylation differences exist between CD5+ and CD5− normal B cells10, we performed comparisons of CLLs to our normal B cells despite these not being sorted for CD5+ cells. Overall, CLLs demonstrated methylation states that were comparable with those of low-maturity to high-maturity memory B cells. In comparison to naive B cells, CLL samples had achieved at least ~70% of normal B cell maturation programming (Fig. 2d). Other than a gap between LP-CLL and IP-CLL cases, CLLs occupied virtually the entire remaining window of maturation states. Notably, all CLLs diverged off the common maturation axis (trunk), indicating that all CLLs derive from a continuum of potential maturation states and are not restricted to discrete maturation stages. Performing the same analysis using the ICGC sample cohort produced a similar result (Supplementary Fig. 2a).
Figure 2.
Evaluation of the maturation state of CLL using DNA methylation. (a) The average level of methylation at transcription factor binding sites for all six significantly programmed transcription factors in 267 CLL samples assessed by 450K array. Samples are ranked according to the average methylation level for all transcription factor binding sites. The average β value (global methylation) is also shown for each sample. (b) Heat map showing the methylation at all high-confidence AP-1, EBF1 and RUNX3 binding sites and in hypermethylated transcriptional elongation regions in 267 CLL samples. Consensus clustering of CLL samples identifies three methylation subtypes (LP-CLL, IP-CLL and HP-CLL). The level of IGHV homology (identity) for each CLL is indicated. (c) Scatterplot displaying the average methylation of AP-1, EBF1 and RUNX3 binding sites versus hypermethylated transcriptional elongation regions. (d) DNA methylation phylogenetic tree diagram of the DKFZ CLL sample cohort (n = 128) and normal B cell subtypes generated using AP-1, EBF1 and RUNX3 binding sites and hypermethylated transcriptional elongation regions. Each line represents a CLL sample; CpG methylation values for normal samples were averaged according to subtype. Colors indicate CLL subtypes as in c. (e) DNA methylation phylogenetic tree diagram of the CLL samples selected for RNA-seq analysis. Patient numbers are indicated for each sample; shaded areas represent the CLL subtypes. (f) Heat map of 459 genes differentially expressed in the LP-CLL and HP-CLL subtypes (FDR q < 0.05). Samples are ordered according to their position within the phylogenetic analysis in e. Genes that have been previously reported to be differentially expressed in the IGHV subtypes or to have a role in CLL pathogenesis are also indicated. (g) Scatterplot showing the correlation between the degree of methylation maturation (as assessed by the trunk distance from naive B cells in the phylogenetic analysis) and the acquisition of the HP-CLL expression state.
To investigate whether the degree of maturation achieved by individual CLLs influences their phenotype, we analyzed global gene expression using RNA sequencing (RNA-seq) of 28 CLL cases selected to represent a full range of maturation states (Fig. 2e). To prevent distortion of expression profiles due to artificial activation and cell stress artifacts resulting from technical handling21, CLL cells were isolated and purified from fresh blood and then allowed to recover during overnight culture. A comparison of the LP-CLL and HP-CLL subtypes identified 459 differentially expressed genes (false discovery rate (FDR) q < 0.05; Fig. 2f). Many of these genes have been previously described to be of prognostic and/or functional relevance in CLL, including TCL1A, ZAP70, BTK, MIR29B2-MIR29C, CD274, etc. The expression level of genes that are known to signify or contribute to a more indolent disease phenotype was found in the HP-CLL subtype. With the exception of one case (CLL30), IP-CLLs exhibited an expression pattern that combined the LP-CLL and HP-CLL expression states. The number of genes that acquired an HP-CLL–like expression state was significantly correlated with the degree of methylation maturation (P < 0.0001), with IP-CLL cases exhibiting an expression pattern closer to that of the HP-CLL than the LP-CLL subtype (Fig. 2g). Repeating this analysis using the ICGC sample cohort showed a similar relationship (Supplementary Fig. 2b–d). The association between global methylation programming and overall expression state occurred despite differentially expressed genes (versus consistently expressed genes) being only weakly enriched in harboring differentially methylated CpG sites (20.2% versus 16.9%; P = 0.035, χ2 test), consistent with previous observations10. These results suggest that CLL represents a continuum of phenotypes, observed in IP-CLL to HP-CLL.
DNA methylation and clinical outcome in CLL
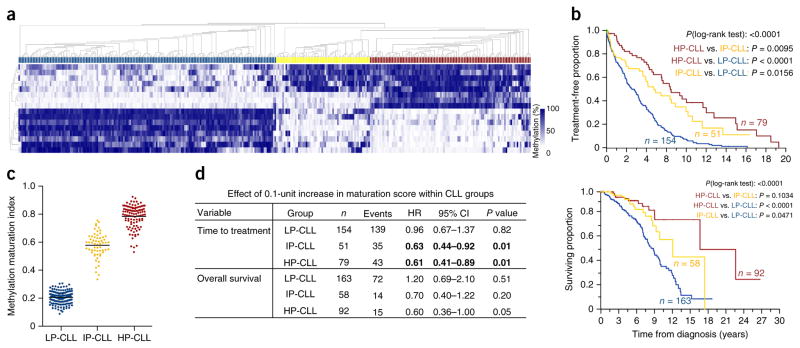
An independent sample set of 327 CLL cases collected as part of a prospective natural history study by the CLL Research Consortium (CRC) was used to compare methylation with clinical data. We selected 18 genomic regions that exhibited highly variable methylation across the CLL subtypes in the 450K methylation data for MassARRAY analysis. Consensus clustering identified three primary methylation subtypes (Fig. 3a). The subtypes displayed distinct distributions of IGHV homology (Supplementary Fig. 3), consistent with previous observations11. Using the time from diagnosis to treatment as an endpoint, we observed significant differences in pairwise comparisons of all subtypes (P < 0.05); notably, the IP-CLL subtype showed a distinct, intermediate outcome (Fig. 3b), supporting previous observations11. Analysis of data for overall survival highlighted a significant difference between the LP-CLL and IP-CLL subtypes. Next, we estimated the impact of DNA methylation levels within the subtypes by calculating a single methylation value that quantitatively scores the overall degree of methylation maturation for each case (Fig. 3c and Online Methods). By analyzing the methylation maturation score as a continuous variable, we observed that increasing maturity was significantly associated with an increase in the time to treatment within the IP-CLL and HP-CLL subtypes (Fig. 3d). Similar trends were observed for overall survival. These data demonstrate that, although separating cases into subtypes is informative (especially for LP-CLL cases), the quantitative degree of methylation maturation appears to provide further prognostic information for the HP-CLL and IP-CLL subtypes. Taken together with the finding of progressive acquisition of gene expression signatures with increasing methylation maturation, these data suggest that the continuum of methylation maturation observed for cases corresponds to a spectrum of phenotypes among patients.
Figure 3.
The impact of DNA methylation programming in patients with CLL. (a) DNA methylation heat map of 18 loci in an independent, clinically annotated set of 327 patients with CLL assessed by MassARRAY. Consensus clustering identifies three clusters of CLL cases (blue, LP-CLL; yellow, IP-CLL; brown, HP-CLL). (b) Kaplan-Meier plots showing time from diagnosis to treatment (top) and overall survival (bottom) for each CLL subtype (P < 0.0001, log-rank test). (c) The methylation maturation score of CLL samples separated by methylation subtype. This value depicts the degree of maturity by combining all available MassARRAY methylation data, taking into account the direction of programming (hyper- or hypomethylation) (LP-CLL, n = 163; IP-CLL, n = 61; HP-CLL, n = 103). Horizontal bars represent mean values. (d) Statistical summary using the methylation maturation score as a continuous variable within the CLL clusters. The hazard ratio (HR) and confidence interval (CI) of the effect of the score within each subtype per outcome variable were estimated from proportional hazards models. Significant associations (P < 0.05) are highlighted in bold.
Normal B cell methylation confounds CLL-specific findings
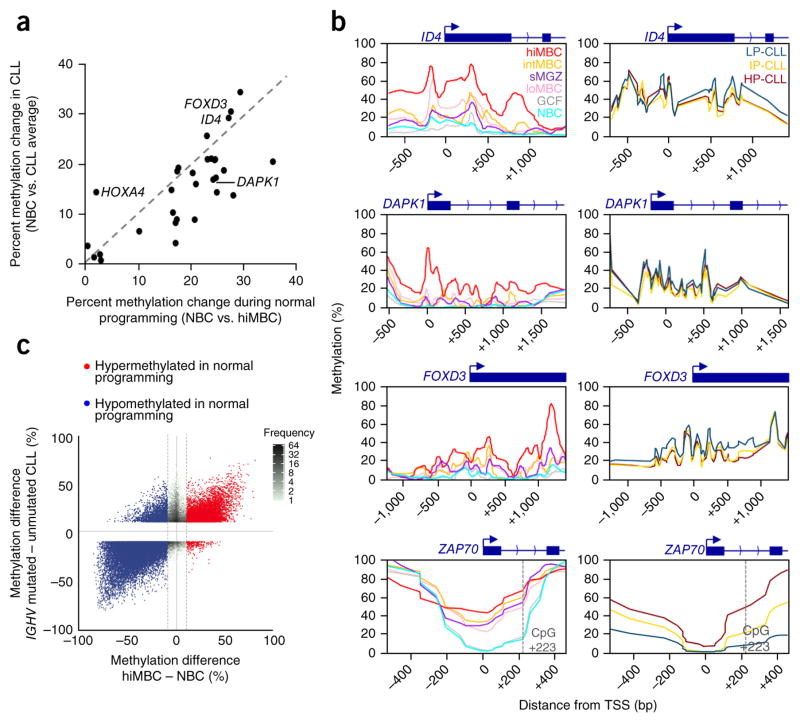
We next investigated CLL DNA methylation taking into consideration changes that occur during normal B cell maturation. We first obtained a list of genes that have been reported to be hypermethylated in CLL from a recent review22 and interrogated these loci again using 450K methylation data for normal B cells and 267 CLLs. This analysis showed that, for all genes except one (HOXA4), the degree of hypermethylation in CLL was equal to or exceeded by the extent of hypermethylation that normally occurs during normal B cell programming (Fig. 4a). A closer inspection of the reported hypermethylated regions using TWGBS showed a broad, progressive acquisition of methylation coincident with B cell maturation (Fig. 4b). We then measured the methylation levels in a subset of 12 CLL samples from each subtype using MassARRAY. The degree of hypermethylation in CLLs (consistent across subtypes) was generally similar to that in high-maturity memory B cells (Supplementary Table 3). Although the MassARRAY analysis covered fewer CpG sites than TWGBS, resulting in a slight difference in curve appearance, a CpG-wise comparison of the methylation gain using 450K array data in normal B cells and CLL samples indicated that the vast majority of individual CpG sites are equally hypermethylated in CLL and normal B cells at these loci (Supplementary Fig. 4a). These genes exhibited either very low or no expression in CLL and all normal B cell populations, including naive B cells, despite there being low levels of methylation in this subpopulation (Supplementary Fig. 4b and Supplementary Table 4). These findings are consistent with previous observations showing that hypermethylation in cancer is associated with gene suppression at earlier developmental stages23 and recurrent, slight hypermethylation during CLL progression24.
Figure 4.
Previously identified aberrant methylation in CLL is found in comparison of normal B cell subtypes. (a) Comparison of the methylation changes in CLL with those in high-maturity memory B cells using naive B cells as a reference. Data for 450K probes located within regions analyzed in previous studies were averaged for each cell type; CLL data from all three subtypes were combined. The gene list was obtained from Florean et al.22. (b) DNA methylation profiles of representative promoters that were falsely identified as aberrantly methylated regions shown for six normal B cell subsets (left) and the CLL subtypes (right). The promoter region of ZAP70, a region reported to be differentially methylated between IGHV subtypes, is also shown. TWGBS data were used to generate the profiles for the normal B cell subtypes, and the CLLs (n = 12 samples averaged per subtype) were analyzed by MassARRAY. The position of the highly prognostic CpG at +223 in the ZAP70 locus is indicated. (c) Comparison of the difference in methylation for CLLs with wild-type IGHV and CLLs with mutated IGHV versus the difference in methylation from naive B cells to high-maturity memory B cells, displaying all 450K probes that differ by >10% between the IGHV subtypes. Probes that are hyper- and hypomethylated between naive B cells and high-maturity memory B cells are highlighted.
Subclassification by IGHV status is commonly employed in CLL, and methylation differences specific to the IGHV subtypes have been reported22. We found that 82% of the CpG sites differentially methylated between the IGHV subtypes were also differentially methylated between naive B cells and low-maturity memory B cells (Fig. 4c). For instance, the region immediately flanking the ZAP70 promoter was strongly hypomethylated in most CLLs with wild-type IGHV relative to those with mutated IGHV and was a strong predictor of poor outcome25. We found that this difference in methylation arises from normally occurring low levels of methylation in naive B cells and subsequent hypermethylation during B cell maturation (Fig. 4b). Additional hypomethylation surrounding the ZAP70 promoter could be observed, most pronouncedly in LP-CLLs. We further investigated whether gene expression differences among the CLL subtypes corresponded to those observed among the normal B cell subpopulations. We found that some genes that were differentially expressed in the LP-CLL and HP-CLL subtypes also displayed parallel differential expression in the naive B cell and high-maturity memory B cell normal subpopulations, including ZAP70, TCL1A, BTK, CD274 and others, although this trend was observed for a minority of genes overall (Supplementary Fig. 4c). These results indicate that a large proportion of observations previously reported to be disease or IGHV subtype specific reflect differences among normal B cells.
Non-normal methylation events
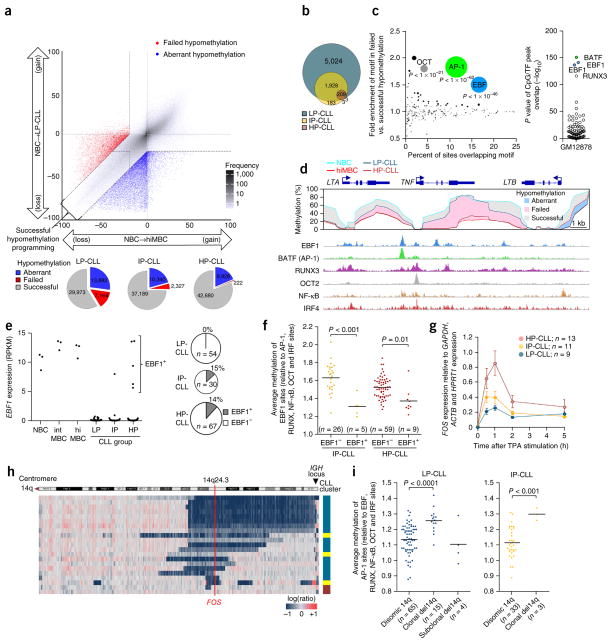
To compare CLL with normal development, we visualized methylation changes in the progression of naive B cells to high-maturity memory B cells versus the development of each CLL subtype individually using naive B cells as the reference (Fig. 5a). The CpG sites that changed in parallel with normal B cell subsets were termed ‘successful methylation programming’. Divergent methylation states in CLL can be a result of either a change that does not occur normally (termed ‘aberrant methylation programming’) or the failure to recapitulate a change that occurs during normal maturation (termed ‘failed methylation’). As the majority of differences between CLL and the normal subtypes involved hypomethylation, we chose to focus on these sites for further analyses. Failed hypomethylation events occurred primarily in LP-CLLs and were nearly absent in HP-CLLs (Fig. 5a,b). Failed hypomethylation sites were highly enriched in pathways relevant for the more aggressive phenotype of LP-CLLs (for example, apoptosis, leukocyte adhesion, response to antigenic stimulus, etc.; Supplementary Fig. 5a). These CpG sites were further enriched for AP-1, EBF1 and RUNX3 binding sites as compared to successfully programmed CpG sites (Fig. 5c,d), indicating that the development of LP-CLL involves a failure to properly program a significant proportion of these sites.
Figure 5.
Deficiency in DNA methylation programming in LP-CLLs results from loss of expression of the EBF1 and FOS transcription factors. (a) Differences in methylation from naive B cells to high-maturity memory B cells and to LP-CLL using the 450K array. Data for CpGs were averaged for each subtype (naive B cells, n = 5; high-maturity memory B cells, n = 5; LP-CLLs, n = 107). CpGs were categorized as having failed (red) or aberrant (blue) hypomethylation versus successful hypomethylation (gray). Bottom, the total number of CpGs per category per CLL subtype. (b) Proportional Venn diagram showing the number of CpGs that exhibit failed hypomethylation in the CLL subtypes. (c) Left, bubble scatterplot of transcription factor motif enrichment in failed (versus successful) hypomethylation. Bubble size corresponds to the P value. Right, enrichment P values of 78 transcription factor ChIP-seq peaks in GM12878 cells in failed hypomethylation. (d) DNA methylation in normal B cells and CLLs covering the TNF locus. Mean DNA methylation (450K) levels for the naive B cell, high-maturity memory B cell, LP-CLL and HP-CLL subtypes are designated by colored lines; shaded areas indicate differences in methylation (relative to naive B cells) that exhibit successful (gray), failed (pink) or aberrant (blue) programming. Bottom, transcription factor binding in GM12878 cells. (e) EBF1 expression from RNA-seq data highlighting EBF1+ CLL cases. Right, frequency of EBF1+ cells in the CLL subtypes, combining RNA-seq, qPCR and microarray data. RPKM, reads per kilobase of transcript per million mapped reads. (f) Average methylation levels of EBF1 binding sites (versus other programmed transcription factor binding sites) in EBF1+ and EBF1− CLLs. Differences were assessed by t test. (g) FOS expression time course after TPA induction as assessed by qPCR (error bars, s.e.m.). (h) Heat map showing genomic copy number profiling of chromosome 14q in 21 CLLs. CLL subtype is indicated. (i) Average methylation of AP-1 binding sites (versus other programmed transcription factor binding sites) in del14q versus disomic 14q CLLs. Cases with a subclonal deletion were considered separately. P values were assessed by t test.
Failed hypomethylation programming of EBF and AP-1 binding sites
To explore the potential mechanisms behind the blockage in AP-1 and EBF1 programming, we investigated the expression of EBF1 and AP-1 family members. EBF1 is essential for normal B cell maturation, and genetic disruption of EBF1 contributes to leukemogenesis26,27. A previous study found consistent loss of EBF1 expression in a small CLL cohort28. Investigation of EBF1 expression in our RNA-seq data combined with publically available data10,29 demonstrated that EBF1 expression was completely lost in the LP-CLL subtype (EBF1-); however, ~15% of IP-CLL and HP-CLL cases retained normal expression levels (Fig. 5e and Supplementary Fig. 5b). Although nonsynonymous mutations and deletions of EBF1 are exceedingly rare in CLL30,31, an increase in EBF1 promoter methylation was found in some EBF1− cases relative to EBF1+ CLLs (Supplementary Fig. 5c). Gene set enrichment analysis showed an enrichment of EBF1 target genes and genes involved in normal B cell maturation in EBF1+ CLL cases (Supplementary Fig. 5d). In addition, EBF1+ CLL cases lacked expression of genes previously found to be CLL specific, indicating that EBF1 loss may cause major transcriptional dysfunction in CLL. Notably, we found that EBF1+ CLLs exhibited lower methylation of EBF1 binding sites, suggesting that loss of EBF1 expression is associated with a failure to sufficiently program EBF binding sites (Fig. 5f).
Members of the canonical AP-1 family (JUN and FOS subfamilies) were silent or weakly expressed in unstimulated CLL cells from blood (Supplementary Fig. 6a) but were highly induced in CLL cells from lymph nodes32. Thus, to investigate functional levels of AP-1 expression, CLL cells were activated in vitro. To avoid the heterogeneity in BCR signaling capacity generally associated with the different IGHV subtypes33, we chose to activate cells downstream of the BCR signalo-some complex by using 12-O-tetradecanoylphorbol-13-acetate (TPA). Stimulation of cells with TPA uniformly induced gene activation across the CLL subtypes (Supplementary Fig. 6b). RNA-seq analysis after 1 h showed that, of the AP-1 family members, only FOS showed difference in expression between the subtypes (Supplementary Fig. 6c,d,f). Validation using quantitative PCR (qPCR) demonstrated impairment of FOS induction in LP-CLLs across a 5-h window (Fig. 5g). We next sequenced 192 CLL cases and failed to find non-synonymous protein-coding mutations; however, the FOS locus (at 14q24.3) has previously been shown to be recurrently deleted in approximately 2% of CLL cases34. To investigate whether reduced FOS levels lead to failed hypomethylation at AP-1 binding sites, we identified 21 CLL cases with 14q deletions from existing high-density SNP array data35 and by detecting copy number alterations from 450K data36. We found that 18 of the 21 deletions of 14q included FOS, and one case exhibited a proximal breakpoint (350 kb upstream) (Fig. 5h). The majority (16/19) of these cases belonged to the LP-CLL subtype, consistent with reduced maturation being associated with FOS loss. Indeed, two cases that showed deletions sparing the FOS locus were HP-CLLs. Notably, we observed that cases that exhibited clonal deletions of FOS retained higher levels of AP-1 binding site methylation, indicating that FOS has a role in the programming of these sites and that reduction of its expression may contribute to failed hypomethylation (Fig. 5i).
Aberrant hypomethylation of NFAT and EGR binding sites
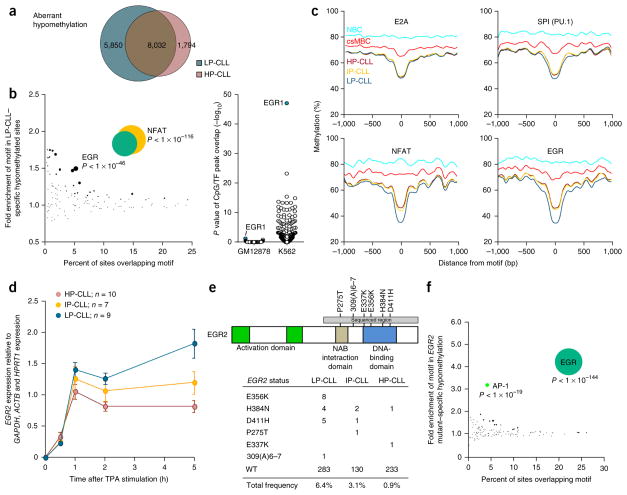
We next focused on aberrant (CLL-specific) hypomethylation (Fig. 5a, blue dots). We found 13,882 and 9,826 CpG sites aberrantly hypomethylated (subtype average >20% loss) in LP-CLL and HP-CLL, respectively, of which 8,032 overlapped (Fig. 6a). Aberrantly hypomethylated CpG sites common to both subtypes were highly enriched in sequence motifs for NFAT, EGR, E2A and SPI1 (PU.1) relative to successfully hypomethylated CpG sites (Supplementary Table 5). Comparison of the non-overlapping aberrantly hypomethylated CpG sites showed enrichment for NFAT and EGR motifs specific to the LP-CLL subtype (Fig. 6b). The HP-CLL subtype did not show any unique enrichment. We did not observe transcription factor binding site enrichment in these disease-specific regions (consistent with GM12878 cells being derived from normal B cells); however, consistent with the EGR family being important in myeloid cell development37, we found that EGR1 was the most enriched transcription factor at these sites in the K562 chronic myeloid leukemia cell line (Fig. 6b, right). A composite analysis of methylation levels proximal to motifs demonstrated that hypomethylation occurs locally (approximately ±100 bp with respect to the motif) in aberrantly hypomethylated sequences (Fig. 6c). Aberrantly methylated CpG sites are also programmed in normal B cells but to a much lower degree, implying that aberrant hypomethylation may result from excess activity by the transcription factors that perform normal programming. NFAT and EGR motifs exhibited excess programming in LP-CLL (Fig. 6c, bottom), despite an overall reduction in normal programming in this subtype (Fig. 2b–d).
Figure 6.
Transcription factor binding sites enriched in CLL-specific hypomethylation. (a) Proportional Venn diagram showing the number of aberrantly hypomethylated CpGs in LP-CLL and HP-CLL (illustrated in Fig. 5a). (b) Bubble scatterplot of transcription factor motifs displaying the prevalence and fold enrichment of each motif in aberrant hypomethylation. Bubble size corresponds to the P value of the association. Right, enrichment P value of 78 transcription factor ChIP-seq peaks in GM12878 and K562 cells in LP-CLL–specific hypomethylated regions. P values were assessed by Fisher’s exact test. (c) Composite CpG methylation levels surrounding transcription factor motifs (range of ±1 kb) in aberrantly hypomethylated regions from 450K data. E2A and SPI1 motifs exemplify motifs equally aberrantly hypomethylated across the CLL subtypes, whereas NFAT and EGR motifs display additional hypomethylation in LP-CLLs. (d) Expression of EGR2 in each CLL subtype as determined by qPCR during a 5-h time window after TPA induction (error bars, s.e.m.). (e) Summary of the nonsynonymous EGR2 DNA-binding domain mutations found in 670 CLL cases showing the amino acid changes for each CLL subtype. WT, wild type. (f) Bubble scatterplot of transcription factor motif enrichment in regions specifically hypomethylated in EGR2-mutated CLLs. Bubble size corresponds to the P value of the association.
Comparable to the AP-1 family, the EGR family is highly inducible and exhibited very low constitutive expression in resting normal B cells and CLL cells (Supplementary Fig. 6a). Indeed, EGR1, EGR2 and EGR3 were the most inducible genes in CLL after TPA stimulation (Supplementary Fig. 6f). However, only EGR2 exhibited a difference in induction between the LP-CLL and HP-CLL groups (Supplementary Fig. 6e). Interestingly, we observed higher EGR2 induction in LP-CLLs, the opposite of the pattern of induction observed with FOS (Fig. 5g and Supplementary Fig. 6c,f) and consistent with EGR2-dependent suppression of FOS38. Further evaluation showed higher sustained EGR2 expression in LP-CLL versus HP-CLL cells after exposure to TPA (Fig. 6d). Recurrent, gain-of-function EGR2 mutations mapping to the DNA-binding domain have recently been described in CLL38. To test whether EGR2 can influence DNA methylation patterns in CLL, we sequenced 670 CLL cases for mutations mapping to the DNA-binding domain. We uncovered 24 mutations in total, 21 of which comprised recurrent mutations encoding p.Glu356Lys, p.His384Asn and p.Asp411His substitutions (Fig. 6e). Consistent with the enrichment of EGR motifs in LP-CLL–specific hypomethylation regions, we observed an over-representation (17/21) of EGR2 mutations in LP-CLL. Analysis of 450K methylation data from EGR2-mutated samples showed a significant over-representation of EGR motifs in hypomethylated sequences as compared to cases with wild-type EGR2 (Fig. 6f), consistent with abnormal EGR2 activity having a role in establishing aberrant hypomethylation of transcription factor binding sites in CLL.
DISCUSSION
Here we find that knowledge of dynamic and subtype-specific differences in normal tissue is a prerequisite to accurately resolve events that are truly disease specific. We show that all CLLs by methylation status are like memory B cells (exhibiting ~70–100% of the programming for high-maturity memory B cells), consistent with previous findings39. Previous efforts to uncover CLL-specific methylation events were largely based on comparisons to CD19+ normal B cells, a mixture with a majority of naive B cells40. Virtually all reported ‘CLL-specific’ differences reevaluated here reflected normal B cell maturation. Although it is possible that parallel methylation events contribute to the disease phenotype, large numbers of memory B cells persist for long periods of time in the body and are clearly not malignant despite possessing these modifications. IGHV subtype–specific differences in methylation also occur during normal B cell maturation and thus are likely not causative. However, it is probable that these differences would modify the behavior of the disease if features linked to immaturity were maintained in a given CLL (failed programming). For example, ZAP70 is unmethylated and expressed in naive B cells, and the failure to hypermethylate and repress its expression (as occurs in normal cells) in LP-CLL may further enhance the aggressiveness of this subtype41. This paradigm of complexity within normal tissues obscuring the interpretation of cancer-specific events could be applied to other tissues where different cell types with distinct or developing methylation patterns are present.
DNA methylation data analysis classifies CLLs into three primary subtypes using either 450K data or a reduced panel of loci10,11. We confirmed the existence of these subtypes in independent samples, using 450K data or a reduced panel by MassARRAY analysis, and also confirmed previously determined relationships between the subtypes and clinical outcomes9,12. Notably, the degree of DNA methylation maturity within subtypes significantly associates with outcome as a continuous variable (IP-CLL and HP-CLL). These findings suggest that, although segregation of CLL cases into subtypes is a useful classification approach, a continuous spectrum of phenotypes exists among IP-CLL and HP-CLL cases. We observe a striking difference in the degree of DNA methylation maturity (and gene expression) between cases that have 100% IGHV identity and ones that have minimal IGHV mutation (generally representative of the LP-CLL and IP-CLL subtypes, respectively). As LP-CLLs display less variation among cases, the within-subtype relationship between maturity and phenotype may only be relevant for cases progressed beyond LP-CLL.
By focusing on disease-specific DNA methylation events, we uncover a previously unappreciated role for transcription factors in CLL (although described in other leukemias and lymphomas42,43) in potentially establishing specific aberrant methylation patterns (summarized in Fig. 7). Interestingly, our data suggest that the development of CLL, especially the more aggressive LP-CLL subtype, involves an imbalance in the normal configuration of transcription factor–dependent epigenetic programming. However, as the transcription factors implicated are known to have important roles in hematopoietic development44–47, we cannot exclude the possibility that a yet unanalyzed normal B cell subtype may possess these features. The reduced programming activity of EBF1 and FOS in the early developmental window establishes a cellular state of phenotypic immaturity coincident with a failure to upregulate the expression of many genes involved in differentiation and growth suppression. JUN-FOS heterodimers can normally recognize methylated DNA and reverse epigenetic silencing in B cells48. Conversely, the transcription factors that are implicated in aberrant hypomethylation of binding sites, including NFAT, EGR, SPI1 and E2A, are likely overactive. These transcription factors are known downstream targets of BCR signaling pathways, raising the possibility that increased transcription factor activity may be related to autoreactive BCRs49. Furthermore, as most of the transcription factor genes are weakly expressed or inactive in resting cells, the aberrant induction of these genes or function of the encoded proteins, like mutant EGR2, would be observed only after activation38. Although the mechanism of EBF1 and FOS downregulation in the vast majority of cases is currently unclear, the activity of JUN and FOS is inhibited by direct interaction with TCL1A50, whose overexpression causes a CLL-like disease in mice. Taking these results together, we propose that multiple genetic and epigenetic events converge in very early stages of B cell maturation to perturb normal programming networks and thereby promote the initiation of a premalignant clone.
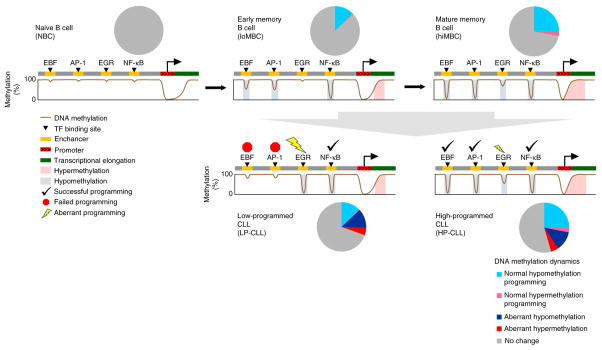
Figure 7.
Summary of global and transcription factor binding site DNA methylation programming in normal B cells and CLL. Pie charts display the proportion of CpGs across the genome that are modified relative to naive B cells during normal DNA methylation programming and in CLL development. The cartoons present a summary of the dynamic DNA methylation programming in normal and CLL samples. Selective transcription factor binding sites and regions of transcriptional elongation are significantly enriched for DNA methylation changes. Many of the regions targeted for hypomethylation are marked with a histone enhancer chromatin signature, implying that hypomethylation of these regions is involved in the establishment of enhancers during maturation. Some binding sites, such as those for NF-κB, IRF and OCT transcription factors, are programmed earlier than others, such as binding sites for EBF, AP-1 and RUNX transcription factors. Overall, hypermethylation occurs more rarely but is enriched in regions of transcriptional elongation. CLL samples mostly retain the state of methylation programming at transcription factor binding sites corresponding to the stage from which the founder cell arose, with from ~70–100% of the program in normal memory cells achieved overall. In the more aggressive LP-CLL subtype, EBF and AP-1 binding sites fail to achieve mature levels of programming relative to other successfully programmed transcription factor binding sites. EGR (and NFAT) sites are aberrantly hypomethylated relative to normal cells.
URLs
International Immunogenetics Information System (IMGT) analysis website, http://www.imgt.org/; International Cancer Genome Consortium (ICGC) Data Portal, https://dcc.icgc.org/; Gene Expression Omnibus (GEO) data sets, http://www.ncbi.nlm.nih.gov/gds; RnBeads analysis software, http://rnbeads.mpi-inf.mpg.de/; GREAT analysis, http://bejerano.stanford.edu/great/public/html; Gene Set Enrichment Analysis (GSEA), http://www.broadinstitute.org/gsea; HOMER analysis software, http://homer.salk.edu/homer/motif; ENCODE, https://www.encodeproject.org/; bsseq, http://bioconductor.org/packages/release/bioc/html/bsseq.html; CLL Research Consortium (CRC), http://cll.ucsd.edu/; chromatin variation across 19 human lymphoblastoid cell lines (Kundaje laboratory), https://sites.google.com/site/anshulkundaje/projects/chromatinvariationpilot#TOC-Chromatin-State-Maps.
ONLINE METHODS
Normal B cell isolation
Normal peripheral blood B cell subsets were isolated from full blood donated by healthy adults. Tonsillar tissue was derived from tonsillectomies, and splenic tissue was derived from surgery due to traumatic rupture or tumor removal without a direct effect from the tumor on spleen tissue. The study protocol was approved by the Internal Review Board of the Medical School in Essen, Germany. All subjects gave informed consent. Peripheral blood mononuclear cells were isolated by Ficoll-Paque density centrifugation (Amersham). CD19+ B cells were enriched to >98% of the population by magnetic cell separation using the MACS system (Miltenyi Biotec). Peripheral blood B cells were sorted as CD23+IgDhighCD27− (naive B cell), IgM+IgDlowCD27−CD23−Rhodamine123+ (low-maturity memory B cell), IgM+IgD+CD27+ (intermediate-maturity memory B cell) and IgG+CD27+ (high-maturity memory B cell) populations; tonsillar GCF B cells were sorted as CD20highCD38intIgM+IgD+CD80high, CD20highCD38intCD83+CD 184− (centrocyte) and CD20highCD38intCD83−CD184+ (centroblast) populations; and splenic marginal zone B cells were sorted as IgM+CD21highCD27+ cells. Cells were stained with APC-, PE- or FITC-conjugated antibody to human CD38 (HIT2), FITC-, APC- or PE-Cy7–conjugated antibody to CD27 (MT271), PE- or APC-conjugated antibody to CD23 (M-L233), FITC- or PerCP-Cy5.5–conjugated antibody to CD20 (2H7), PE-conjugated antibody to CD80 (L307.4), PerCP-Cy5.5–conjugated antibody to CD83 (HBI5e, BioLegend), PE-Cy7–conjugated antibody to CD184 (I2G5, BioLegend), FITC-conjugated antibody to IgG (G18-145), PE-conjugated antibody to CD21 (1048), FITC-conjugated antibody to IgM (G20-127), and PE-, PE-Cy7–, PerCP-Cy5.5– or V500-conjugated antibody to IgD (IA6-2) and with Rhodamine123 (Sigma-Aldrich) in appropriate combinations. If not stated otherwise, antibodies were purchased from BD Biosciences (Becton Dickinson). B cells were sorted with a FACSAria cell sorter (BD Biosciences), and purity was determined by reanalysis on a FACSCanto flow cytometer (BD Biosciences) with FACSDiva software.
IGHV mutation analysis
DNA was extracted from sorted B cells using Gentra PureGene Core kit A (Qiagen) according to the manufacturer’s instructions. IGHV gene PCR of IGHV3 family rearrangements was carried out with an annealing temperature of 60 °C for 40 cycles with Phusion High-Fidelity DNA polymerase (Finnzymes, Thermo Scientific). Primer sequences are listed in Supplementary Table 6. Amplified DNA was gel purified, cloned using the pGEM-T-Easy cloning kit (Promega) and then sequenced with an ABI 3130 Sequencer (Life Technologies). IGHV mutation analysis was performed using the International Immunogenetics Information System (IMGT). Naive B cells and GCF B cells showed virtually germline configurations and low-maturity memory B cells showed a mutation frequency of 2.0%, with increasing maturation rates for splenic marginal zone B cells (3.5%), intermediate-maturity memory B cells (3.5%) and high-maturity memory B cells (7.1%).
In vitro stimulation of naive B cells
Naive B cells were sorted as CD23+IgDhighCD27− cells and cultured in RPMI-1640 supplemented with 30% FCS and 1% penicillin-streptomycin (Pan Biotech) in the presence of 100 mg/l recombinant human interleukin (IL)-15 (Immunotools). After labeling with carboxyfluorescein succinimidyl ester (CFSE; 5 μM final concentration; BioLegend), cells were stimulated on day 0 with 10 mg/l goat antibody to human immunoglobulin (Dianova, 109-606-064) alone or in combination with 2 mg/l recombinant human CD40L with a histidine tag (2706-CL) plus 1 mg/l antibody to histidine tag (R&D Systems, clone MAB050) for up to 11 d. Proliferating and non-proliferating cells were collected on days 1, 5 and 11, according to CFSE content, and DNA methylation analysis was performed using Illumina 450K arrays. Methylation values were averaged for replicates. Stimulated samples were compared to unstimulated cells from the same day of culture.
CLL samples
For the DKFZ cohort, peripheral blood samples from patients with CLL were obtained from the University Hospital in Ulm, Germany, and the National Center for Tumor Diseases (NCT) in Heidelberg, Germany, after obtaining informed consent by a procedure approved by the Ethics Committee of Ulm University or the University Hospital of Heidelberg. Samples were obtained from fresh blood using Lymphoprep (Axis-Shield). For patients with CLL who retained less than 100,000 lymphocytes/μl in blood, CLL cells were purified using magnetic cell sorting with selection for CD19+ cells. Cell purity was >98% for CD5+CD19+ cells after selection. The clinically annotated cohort of 348 CLL cases was obtained from CRC following informed consent and approval by the Institutional Review Board of the University of California, San Diego; a table summarizing the available data on clinicobiological features for these cases separated by DNA methylation subtype is provided as Supplementary Table 7. The CLL cell purity of unsorted samples from this sample set was inferred from DNA methylation analysis, and 21 samples showing <80% purity were removed (final n = 327). On the basis of available data, 313 cases were followed for overall survival analysis and 287 cases were followed for analysis of the time to first treatment from diagnosis. DNA was extracted using the Qiagen DNeasy Blood and Tissue kit. Sequencing and analysis of the IGHV region was performed as previously described51. The primers used for sequencing of the EGR2 region encoding the DNA-binding domain are listed in Supplementary Table 6. CLL samples used for RNA extraction were cultured overnight (18–24 h) after purification in RPMI-1640 and 10% autoserum before RNA extraction. For in vitro activation of CLL cells, 10 ng/μl TPA in DMSO was used. RNA was extracted using the RNeasy kit from Qiagen.
DNA methylation analysis
For whole-genome bisulfite sequencing of normal B cell subsets, 30 ng of DNA was used to produce four independent barcoded sequencing libraries per DNA sample using the tagmentation (TWGBS) method17. Libraries were sequenced using paired-end 100-bp sequencing on an Illumina HiSeq machine, and reads were pooled after the removal of duplicate and low-quality reads. Overall genomic coverage ranged from 10- to 17-fold per sample; a summary of the sequencing data for each sample is provided in Supplementary Table 8. Bisulfite sequencing reads were processed using MethylCTools with default parameters (V. Hovestadt, S. Picelli, B. Radlwimmer, M.Z. and P.L., unpublished data), which uses the Burrows-Wheeler alignment algorithm52. CpG methylation was calculated using MethylCTools and bsseq in R/Bioconductor. CpG methylation was summarized by tiling the genome into 500-bp windows, and methylation was averaged for all windows containing >4 interrogated CpGs; only windows containing sufficient information in all B cell subtypes were used for analyses. The window size of 500 bp was selected in accordance with the size distribution of regions differentially methylated in normal B cells and CLL cells in published data10. To determine the optimal threshold number of CpGs to consider for each window, windows were grouped according to the number of CpGs they contained and were compared for total genomic coverage, variation between biological replicates of the same subtype and the distribution of methylation values (Supplementary Fig. 7). A threshold of four CpGs per window provided the best combination of coverage and reproducibility while including regions that displayed a broad range of methylation levels. Window methylation levels were averaged for biological replicates to produce lists of hyper- and hypomethylated windows for downstream analysis. The Infinium methylation assay was carried out as described previously53. 450K data for the ICGC cohort were obtained from the European Genome-phenome Archive under accession EGAS00001000272. Methylation data for the GM12878 cell line were obtained from ENCODE/HAIB data54. Raw 450K data for both the DKFZ and ICGC sample sets were normalized by the BMIQ method55 without background subtraction using RnBeads software56. After probes overlapping SNPs and the X and Y chromosomes were removed, 459,625 probes were considered for downstream analysis. To test the association between DNA methylation subtype, methylation programming status and clinical variables, targeted DNA methylation analysis was performed using the MassARRAY system (Agena Biosciences). To select a reduced panel of genomic regions that represent overall DNA methylation status, CpGs were selected from 450K analysis of the DKFZ sample cohort that possessed high statistical power to discriminate between subtypes. Briefly, CpGs were separated into groups by their methylation pattern across all three DNA methylation subtypes to ensure the selection of CpGs that represented both hyper- and hypomethylation programming directionality and early (change included in IP-CLL cells) and late (change excluded in IP-CLL cells) events. CpGs were ranked by P value and fold difference between groups using Qlucore Omics Explorer software. The discriminatory potential of the top candidate CpGs was then tested in the ICGC cohort. From this analysis, we chose the 18 CpGs that best recapitulated the three DNA methylation subtypes in both cohorts as defined by 450K data. MassARRAY analysis of 348 CLL samples was performed as previously described. Values for multiple CpGs analyzed within each region were averaged for each amplicon. Primer sequences and amplicon coordinates are shown in Supplementary Table 6. Information regarding chromatin state, transcription factor binding sites, and sequence motifs overlapping or contained within the MassARRAY amplicons is summarized in Supplementary Table 9. DNA methylation values for all amplicons in all samples are listed in Supplementary Table 10. Estimates of treatment-free and overall survival probabilities were obtained by the Kaplan-Meier method. The log-rank test compared differences between survival curves, and statistical significance was declared at P < 0.05. The reduction of amplicon methylation data to a single value representing the methylation maturation score for each sample was performed by subtracting from 100% the percent methylation values for amplicons associated with hypomethylation programming and then calculating the mean methylation values for all 18 amplicons. Higher maturation scores indicate a higher level of maturation. Hazard ratio estimates and 95% confidence intervals describing the association of 0.1-point increases in the methylation maturation score with time to treatment and overall survival from diagnosis were obtained from proportional hazards models that included the cluster and methylation maturation score and their interaction term. This analysis was purely exploratory and was performed to support laboratory data.
Gene expression analysis
RNA-seq libraries were generated using the TruSeq Stranded Total RNA kit (Illumina). Sequencing reads were aligned using the STAR algorithm, and RPKM values were calculated for each gene. Differential gene expression for the LP-CLL and HP-CLL subtypes was calculated using the Qlucore Omics Explorer and considered significant at FDR q < 0.05. The number of genes (of 459 genes differentially expressed in the LP-CLL and HP-CLL subtypes) that had acquired the HP-CLL expression state was calculated by totaling the number of genes per sample whose expression was closer to the levels in HP-CLL (versus LP-CLL) than the median expression value across all samples. CLL expression data used for validation were obtained from the ICGC Consortium. RNA profiles derived from frozen samples and samples that showed activation of cell stress pathways were excluded from the analysis. For qPCR analysis of gene expression, RNA was reverse transcribed into cDNA using Superscript II (Invitrogen) and was analyzed using the Universal Probe Library System (Roche). The LightCycler 480 Probes Master Mix kit was used for amplification in the LightCycler 480 Real-Time PCR instrument (Roche). Target gene expression is expressed relative to average expression for the housekeeping genes GAPDH, ACTB and HPRT1.
Data analysis
The search for transcription factor sequence recognition motifs was performed using HOMER software v4.5. To determine the relative enrichment of known motifs in TWGBS data, sequence contained in 500-bp windows was searched against a selected background of windows that were adjusted to have equal GC content and the same number of CpGs as well as a similar distribution of methylation levels in naive B cells. Windows showing a >40% loss of methylation between naive B cells and high-maturity memory B cells were used. Motifs displaying a high degree of similarity found among significant motifs (P < 1 × 10−10) were replaced with a single consensus motif determined by the HOMER de novo search algorithm (Supplementary Table 11).
Chromatin states were defined with the standard 15-state model, as described previously, using the ChromHMM algorithm20. Enrichment of DNA methylation differences in chromatin states was assessed using chromatin state data produced by Kasowski et al.5 with a minimum overlap of 1 bp with a given 500-bp window. Statistical analysis for enrichment of windows showing over 20% gain of methylation or over 40% loss of methylation between naive B cells and high-maturity memory B cells versus adjusted background windows (selected as for the motif search) was performed using Fisher’s exact test.
Association of DNA methylation differences with transcription factor binding was assessed by comparing window or CpG position with transcription factor ChIP-seq peak data available for cell lines from the ENCODE Project6. An association was considered if a transcription factor ChIP-seq peak overlapped the 500-bp window in TWGBS analyses or overlapped a region ±100 bp to a 450K probe. Fold enrichment was calculated using a background of windows or probes adjusted to have equal CpG content as well as a similar distribution of methylation levels in naive B cells. P values were calculated using Fisher’s exact test. A set of high-confidence transcription factor binding site–containing windows or 450K probes or CpGs was compiled by an intersection of transcription factor ChIP-seq peaks and corresponding motif determination using HOMER. The average methylation of binding sites for the six transcription factors of interest was calculated by first removing windows or 450K probes that were associated with >2 of the six transcription factors. To mitigate potential differences between normal high-maturity memory B cells and the GM12878 cell line, 450K probes that differed by >20% were removed from the probe set. The windows and probes used for transcription factor binding site methylation analysis and subsequent analyses are listed in Supplementary Tables 12 and 13.
Phylogenetic analysis of DNA methylation was performed as previously described57. Briefly, phylogenetic trees were inferred by the minimal evolution method58 using the fastme.bal function in the R package ape. Phylogenies were generated by applying the minimal evolution algorithm on Euclidean distance matrices based on continuous methylation values. The 450K probes used for phylogenetic analysis included high-confidence binding sites for the six transcription factors of interest plus CpGs in hypermethylated transcriptional elongation domains (totaling 2,184 CpGs) and are listed in Supplementary Table 13. 450K data for normal hematopoietic cell types were obtained from data sets GSE35069 and GSE49618 in the Gene Expression Omnibus. Quantitative genomic copy number profiles of CLL samples were ascertained from 450K data36. Gene set enrichment analysis59 of expression levels in EBF1+ CLL samples was carried out by comparing all annotated gene sets for enrichment in EBF1+ relative to EBF1− CLL cells. The EBF1 target gene set was obtained from previously published data60.
Principal-component analysis, consensus clustering and data visualization were performed using Qlucore. Hierarchical clustering was carried out using the Euclidean distance metric and average linkage criteria. Molecular pathway and Gene Ontology (GO) term enrichment for DNA methylation programming was assessed by analyzing the top 10,000 hypomethylated windows and the top 2,000 hypermethylated windows in comparison to matched sets of background windows adjusted to have equal GC content and the same number of CpG sites as well as a similar distribution of methylation levels in naive B cells. Association of windows with genes and downstream pathway and GO term enrichment analysis were carried out using GREAT61. Windows were assigned to genes by using the basal plus extension association rule setting with a maximum distance of 100 kb for distal associations.
For statistical analysis of DNA methylation and gene expression, the data were processed using SAS v9.3 and GraphPad Prism software. Data distributions were tested for normality, and comparisons of non-normal data were performed using non-parametric statistical tests as appropriate. To identify differential expression across DNA methylation subtypes in RNA-seq data, an FDR threshold of 0.05 was used. The sample size of patients with CLL required to detect a minimally significant difference of 20% in overall methylation for transcription factor binding sites between patients grouped by the presence or absence of a genetic mutation or difference in gene expression was estimated using a type I error of 0.05 with power of 0.95.
Code availability
An R script is provided to summarize DNA methylation into genomic tiling windows in the Supplementary Note. Links and references to previously developed algorithms used are provided in the URLs, “DNA methylation analysis,” “Gene expression analysis” and “Data analysis” sections.
Supplementary Material
Acknowledgments
We would like to thank the Genomics and Proteomics Core Facility at the German Cancer Research Center, in particular R. Fisher and M. Schick for their excellent technical support and expertise. We thank Imaging Center Essen (IMCES) for support in B cell sorting. We are grateful to M. Bähr, O. Mücke, M. Helf and S. Ohl for technical support. We also thank J. Edelmann, M. Seiffert, L. Sellner, B. Wu, V. Hovestadt, A. Kundaje and J.I. Martín-Subero for providing samples, data and/or analytical tools. S.S. is supported by the Else Kröner Fresenius Stiftung (2012_A146), the Virtual Helmholtz Institute (VH-VI-404) and the Deutsche Forschungsgemeinschaft (SFB 1074 projects B1 and B2). This work was supported in part by the Helmholtz Association, from the DKFZ–Heidelberg Center for Personalized Oncology (DKFZ-HIPO), the German Cancer Consortium (DKTK), the CLL Research Consortium (CRC), the German Federal Ministry of Education and Research CancerEpiSys network (BMBF 031 6049C), the Virtual Helmholtz Institute (VH-VI-404), the Deutsche Forschungsgemeinschaft (GKR1431 and SE1885/2-1), the Leukemia and Lymphoma Society (P01 CA081534), the Four Winds Foundation, the European Union’s Seventh Framework Programme through the Blueprint Consortium, the German Ministry of Education and Research (BMBF) through the ICGC MMML-Seq Project (01KU1002A-J) and the US National Institutes of Health (PO1-CA81534).
Footnotes
Accession codes. Data are available at the European Genome-phenome Archive under accession EGAS00001000534.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
C.C.O., M.S., M.P., A.S., D.W. and C.P. designed and performed experimental work. C.C.O., Y.A., L.G., A.S.R., Q.W., C.D.I., S.D.K., D.B., D.B.L. and O.B. performed data analysis. M.S., L.R., T.J.K., H.D., R.K., T.Z., S.S. and J.C.B. provided clinical samples or data. C.C.O., M.S. and C.P. prepared the manuscript and figures. B.B., D.M., M.Z., P.L., T.Z., S.S., J.C.B. and C.P. provided project leadership. All authors contributed to the final manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 5.Kasowski M, et al. Extensive variation in chromatin states across humans. Science. 2013;342:750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lara-Astiaso D, et al. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabezas-Wallscheid N, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15:507–522. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 9.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulis M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 11.Queiros AC, et al. A B-cell epigenetic signature defines three biologic subgroups of chronic lymphocytic leukemia with clinical impact. Leukemia. 2015;29:598–605. doi: 10.1038/leu.2014.252. [DOI] [PubMed] [Google Scholar]
- 12.Cahill N, et al. 450K-array analysis of chronic lymphocytic leukemia cells reveals global DNA methylation to be relatively stable over time and similar in resting and proliferative compartments. Leukemia. 2013;27:150–158. doi: 10.1038/leu.2012.245. [DOI] [PubMed] [Google Scholar]
- 13.Oakes CC, et al. Evolution of DNA methylation is linked to genetic aberrations in chronic lymphocytic leukemia. Cancer Discov. 2014;4:348–361. doi: 10.1158/2159-8290.CD-13-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landau DA, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26:813–825. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulis M, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47:746–756. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebecque S, de Bouteiller O, Arpin C, Banchereau J, Liu YJ. Germinal center founder cells display propensity for apoptosis before onset of somatic mutation. J Exp Med. 1997;185:563–571. doi: 10.1084/jem.185.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, et al. Tagmentation-based whole-genome bisulfite sequencing. Nat Protoc. 2013;8:2022–2032. doi: 10.1038/nprot.2013.118. [DOI] [PubMed] [Google Scholar]
- 18.Shaknovich R, et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood. 2011;118:3559–3569. doi: 10.1182/blood-2011-06-357996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai AY, et al. DNA methylation profiling in human B cells reveals immune regulatory elements and epigenetic plasticity at Alu elements during B-cell activation. Genome Res. 2013;23:2030–2041. doi: 10.1101/gr.155473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dvinge H, et al. Sample processing obscures cancer-specific alterations in leukemic transcriptomes. Proc Natl Acad Sci USA. 2014;111:16802–16807. doi: 10.1073/pnas.1413374111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florean C, Schnekenburger M, Grandjenette C, Dicato M, Diederich M. Epigenomics of leukemia: from mechanisms to therapeutic applications. Epigenomics. 2011;3:581–609. doi: 10.2217/epi.11.73. [DOI] [PubMed] [Google Scholar]
- 23.Teschendorff AE, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith EN, et al. Genetic and epigenetic profiling of CLL disease progression reveals limited somatic evolution and suggests a relationship to memory-cell development. Blood Cancer J. 2015;5:e303. doi: 10.1038/bcj.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claus R, et al. Validation of ZAP-70 methylation and its relative significance in predicting outcome in chronic lymphocytic leukemia. Blood. 2014;124:42–48. doi: 10.1182/blood-2014-02-555722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zandi S, et al. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol. 2008;181:3364–3372. doi: 10.4049/jimmunol.181.5.3364. [DOI] [PubMed] [Google Scholar]
- 27.Heltemes-Harris LM, et al. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. J Exp Med. 2011;208:1135–1149. doi: 10.1084/jem.20101947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seifert M, et al. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J Exp Med. 2012;209:2183–2198. doi: 10.1084/jem.20120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira PG, et al. Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome Res. 2014;24:212–226. doi: 10.1101/gr.152132.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landau DA, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herishanu Y, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-κB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117:563–574. doi: 10.1182/blood-2010-05-284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mockridge CI, et al. Reversible anergy of sIgM-mediated signaling in the two subsets of CLL defined by VH-gene mutational status. Blood. 2007;109:4424–4431. doi: 10.1182/blood-2006-11-056648. [DOI] [PubMed] [Google Scholar]
- 34.Reindl L, et al. Biological and clinical characterization of recurrent 14q deletions in CLL and other mature B-cell neoplasms. Br J Haematol. 2010;151:25–36. doi: 10.1111/j.1365-2141.2010.08299.x. [DOI] [PubMed] [Google Scholar]
- 35.Edelmann J, et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood. 2012;120:4783–4794. doi: 10.1182/blood-2012-04-423517. [DOI] [PubMed] [Google Scholar]
- 36.Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 37.Pham TH, et al. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood. 2012;119:e161–e171. doi: 10.1182/blood-2012-01-402453. [DOI] [PubMed] [Google Scholar]
- 38.Damm F, et al. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov. 2014;4:1088–1101. doi: 10.1158/2159-8290.CD-14-0104. [DOI] [PubMed] [Google Scholar]
- 39.Klein U, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194:1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein U, Rajewsky K, Küppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998;188:1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, et al. ZAP-70 enhances IgM signaling independent of its kinase activity in chronic lymphocytic leukemia. Blood. 2008;111:2685–2692. doi: 10.1182/blood-2006-12-062265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prange KH, Singh AA, Martens JH. The genome-wide molecular signature of transcription factors in leukemia. Exp Hematol. 2014;42:637–650. doi: 10.1016/j.exphem.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 43.Kretzmer H, et al. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat Genet. 2015;47:1316–1325. doi: 10.1038/ng.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 45.Gururajan M, et al. Early growth response genes regulate B cell development, proliferation, and immune response. J Immunol. 2008;181:4590–4602. doi: 10.4049/jimmunol.181.7.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng SL, Gerth AJ, Ranger AM, Glimcher LH. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity. 2001;14:13–20. doi: 10.1016/s1074-7613(01)00085-1. [DOI] [PubMed] [Google Scholar]
- 47.Garrett-Sinha LA, et al. PU.1 and Spi-B are required for normal B cell receptor–mediated signal transduction. Immunity. 1999;10:399–408. doi: 10.1016/s1074-7613(00)80040-0. [DOI] [PubMed] [Google Scholar]
- 48.Gustems M, et al. c-Jun/c-Fos heterodimers regulate cellular genes via a newly identified class of methylated DNA sequence motifs. Nucleic Acids Res. 2014;42:3059–3072. doi: 10.1093/nar/gkt1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosén A, Murray F, Evaldsson C, Rosenquist R. Antigens in chronic lymphocytic leukemia—implications for cell origin and leukemogenesis. Semin Cancer Biol. 2010;20:400–409. doi: 10.1016/j.semcancer.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 50.Pekarsky Y, et al. Tcl1 functions as a transcriptional regulator and is directly involved in the pathogenesis of CLL. Proc Natl Acad Sci USA. 2008;105:19643–19648. doi: 10.1073/pnas.0810965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kröber A, et al. VH mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416. [PubMed] [Google Scholar]
- 52.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bibikova M, et al. Genome-wide DNA methylation profiling using Infinium® assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 54.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 55.Teschendorff AE, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Assenov Y, et al. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11:1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brocks D, et al. Epigenetic intratumor heterogeneity and clonal evolution in aggressive prostate cancer. Cell Rep. 2014;8:798–806. doi: 10.1016/j.celrep.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 58.Desper R, Gascuel O. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comput Biol. 2002;9:687–705. doi: 10.1089/106652702761034136. [DOI] [PubMed] [Google Scholar]
- 59.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bohle V, Döring C, Hansmann ML, Küppers R. Role of early B-cell factor 1 (EBF1) in Hodgkin lymphoma. Leukemia. 2013;27:671–679. doi: 10.1038/leu.2012.280. [DOI] [PubMed] [Google Scholar]
- 61.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.