SUMMARY
RAS binding is a critical step in the activation of BRAF protein serine/threonine kinase and stimulation of the mitogen-activated protein kinase signaling pathway. Mutations in both RAS and BRAF are associated with many human cancers. Here, we report the solution nuclear magnetic resonance (NMR) and X-ray crystal structures of the RAS-binding domain (RBD) from human BRAF. We further studied the complex between BRAF RBD and the GppNHp bound form of HRAS in solution. Backbone, side-chain, and 19F NMR chemical shift perturbations reveal unexpected changes distal to the RAS-binding face that extend through the core of the RBD structure. Moreover, backbone amide hydrogen/deuterium exchange NMR data demonstrate conformational ensemble changes in the RBD core structure upon complex formation. These changes in BRAF RBD reveal a basis for allosteric regulation of BRAF structure and function, and suggest a mechanism by which RAS binding can signal the drastic domain rearrangements required for activation of BRAF kinase.
Graphical abstract
INTRODUCTION
Intracellular signal transduction protein kinases play important roles in transferring signals from the cell surface to the nucleus, and are intimately involved in controlling numerous cellular processes including cell growth, proliferation, and death. In humans, the protein kinase family is encoded by more than 500 genes, which can be subdivided, based on enzyme specificity, into protein serine/threonine kinases, tyrosine kinases, and tyrosine kinase-like proteins (Manning et al., 2002). Serine/threonine kinases are intracellular enzymes that include a small family of protein kinases related to retroviral oncogenes discovered in 1983, named RAF (rapidly accelerated fibrosarcoma) (Lavoie and Therrien, 2015; Matallanas et al., 2011; Roskoski, 2010). The RAF kinase family comprises three known isoforms in humans: ARAF, BRAF, and CRAF (also called RAF-1). RAF kinases participate in the RAS-RAF-MEK-ERK signal transduction pathway, also called the mitogen-activated protein kinase (MAPK) cascade. All three intracellular RAF kinases share highly conserved regions (CR): CR1, CR2, and CR3. CR1 is composed of a RAS-binding domain (RBD) immediately followed by a cysteine-rich domain, which can bind two zinc ions. CR1 interacts with RAS and membrane phospholipids. CR2 is a serine/threonine-rich domain containing a 14-3-3 binding site, and CR3 features the kinase domain.
RAF kinases exhibit high substrate selectivity, exclusively targeting the dual-specificity (Tyr/Thr) kinases MEK1/2. Regulation of RAF kinases is extremely complex and strictly controlled by several factors and events, including protein-protein interactions, phosphorylation/dephosphorylation at numerous sites, and oligomerization state (Lavoie and Therrien, 2015; Matallanas et al., 2011; Roskoski, 2010). For example, RAF kinases are inhibited by binding of 14-3-3 and autoinhibitory domains, which precludes dimerization of the kinase domain and renders the enzyme inactive. Activation of RAF kinases is triggered by RAS-GTP binding, phosphorylation of the activation segment within the kinase, and conformational rearrangements involving both the αC helix and activation segment that lead to kinase domain dimerization required for enzymatic activity (Lavoie et al., 2014; Thevakumaran et al., 2015).
Mutations in the MAPK signaling pathway are associated with numerous human cancers. In particular, RAS mutations occur in ≈15% of all cancers, and mutations in BRAF, but not its isoforms, are prevalent in melanomas (≈66%), thyroid cancer (up to 70%), ovarian cancer (≈30%), colorectal cancer (up to 20%), and liver cancer (≈14%) (El-Nassan, 2014). By far the most common oncogenic mutation occurs in the activation segment of BRAF, V600E, and is found in ≈90% of cancers linked to this kinase. As a result, the kinase domain of BRAF has been the focal point of extensive structural studies and inhibitor design, which has led to the development of two anticancer drugs approved by the US Food and Drug Administration to date, sorafenib and vemurafenib.
Although not sufficient on its own, the interaction of the two oncoproteins RAS and BRAF is absolutely required for activation of the MAPK pathway (Matallanas et al., 2011; Roskoski, 2010). Consequently, understanding the nature of this interaction and the mechanism by which RAS binding activates BRAF is biologically important. Several nuclear magnetic resonance (NMR) and X-ray structures of RBD from CRAF, both free and in complex with HRAS and its homologs, and an NMR structure of RBD from ARAF have been reported. Initial structural and binding studies of human (Emerson et al., 1995) and rat (Terada et al., 1999) CRAF RBD by NMR established the interaction surface for HRAS on the RBD, which was subsequently confirmed by crystal structures of CRAF in complex with the RAS-like protein, Rap1A (Nassar et al., 1995) and more recently with HRAS (Fetics et al., 2015; Filchtinski et al., 2010). The reciprocal NMR studies of effector binding to RAS and its oncogenic mutants, where RAS is isotopically enriched, established a hierarchy of effector binding to HRAS in which BRAF RBD displayed the highest-affinity binding, and provide an elegant approach for directly monitoring the complex RAS signaling network (Smith and Ikura, 2014; Smith et al., 2013).
The RAS subfamily of guanine nucleotide-binding proteins (G proteins), comprising three major isoforms in humans (HRAS, KRAS, and NRAS), are small monomeric GTPases that play a critical role in numerous signal transduction pathways associated with cell growth and differentiation, and many human cancers (Stephen et al., 2014; Takai et al., 2001). RAS proteins function as molecular switches, oscillating between inactive GDP-bound and active GTP-bound states; the latter can bind and activate a variety of effector proteins and signaling pathways. The populations of active and inactive forms of RAS are tightly regulated by guanine nucleotide exchange factors (GEFs), which catalyze GDP-to-GTP exchange, and GTPase-activating proteins (GAPs), which lead to fast GTP hydrolysis and regenerate the inactive GDP form of RAS. Since the initial structural studies on RAS in the early 1990s, it has been well established that RAS proteins adopt multiple conformational states, and that such dynamics are largely associated with two flexible surface loops intimately involved with nucleotide binding, named switch I (residues 30–40) and switch II (residues 60–76) (Kraulis et al., 1994; Milburn et al., 1990).
Although BRAF RBD shares 55%–60% sequence identity with the corresponding domains in the ARAF and CRAF isoforms (Figure 1A), no structure for BRAF RBD has been reported to date. Here, we report the first solution NMR and X-ray crystal structures of RBD from the human protein serine/threonine kinase BRAF (UniProtKB/Swiss-Prot: P15056; NESG: HR4694F). BRAF RBD was selected for three-dimensional (3D) structure determination by the Northeast Structural Genomics Consortium (NESG; http://www.nesg.org) as part of the NIH Protein Structure Initiative program on the Human Cancer Protein Interaction Network (HCPIN) (Huang et al., 2008). We further prepared and characterized the complex of BRAF RBD and HRAS loaded with a non-hydrolyzable analog of GTP, guanosine 5′-[β,γ-imido]triphosphate (GppNHp). Backbone chemical shift perturbations (CSPs) establish the RAS-binding epitope on BRAF RBD. Moreover, backbone and side-chain CSPs, 19F NMR, and hydrogen/deuterium exchange NMR data reveal allosteric conformational changes upon RAS binding, propagated through the β sheet and α-helical core of the protein domain. These changes in BRAF RBD that accompany RAS binding provide a basis for allosteric regulation of BRAF structure and function, and suggest a mechanism by which RAS binding can signal domain rearrangements required for activation of BRAF kinase. Furthermore, our results are consistent with mounting evidence in the literature supporting the critical role of conformational dynamics in macromolecular recognition and allosteric regulation in biology.
Figure 1. Structure of BRAF RBD.
(A) Sequence alignment of the RAS-binding domains from the three human RAF kinase isoforms. Sequences, obtained from the RAF-like RBD protein domain family PF02196 (Finn et al., 2014), were aligned using Clustal Omega (Sievers et al., 2011) and the sequence alignment was rendered using ESPript 3.0 (Robert and Gouet, 2014). Boxed residues represent identical (white font, highlighted in red) and similar (red font) amino acid conservation. Residue numbering for BRAF RBD and secondary structural elements from its solution NMR structure (PDB: 2L05) are drawn above the alignment.
(B) Final ensemble of 20 conformers comprising the solution NMR structure of BRAF RBD (PDB: 2L05). Residues 149–232 are shown.
(C) X-Ray crystal structure of BRAF RBD (PDB: 3NY5). Residues 151–233 are shown; dotted lines represent missing electron density in the loop (L4) between strands β3 and β4.
(D) ConSurf conserved residue analysis for the entire PF02196 protein domain family (Pfam 27.0; 710 sequences) rendered on the solution NMR structure of BRAF RBD (residues 155–227). Residue coloring, reflecting the degree of residue conservation across the family, ranges from magenta (highly conserved) to cyan (variable).
(E) Superposition of BRAF RBD crystal structure (blue; residues 153–228) and the crystal structure of CRAF RBD (residues 54–131) in complex with HRAS (orange; PDB: 4G0N) (Fetics et al., 2015). The structure of HRAS has been omitted. All structures were rendered using PyMOL (PyMOL Molecular Graphics System, Version 1.4; Schrödinger LLC).
RESULTS
Structure of BRAF RBD
The solution NMR structure ensemble and 2.0-Å resolution crystal structure of BRAF RBD and structural statistics are presented in Figures 1B and 1C and Tables S1 and S2, respectively. In solution, BRAF RBDis a monomer under the conditions used in the NMR structure determination (pH 4.5 buffer), based on both analytical gel filtration with static light-scattering detection (Figure S1) and measurements of its rotational correlation time determined from 15N T1 and T2 relaxation data (Figure S2). Essentially complete backbone and side-chain resonance assignments (Table S1) were obtained using conventional triple-resonance NMR methods. These are reflected in its assigned 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum (Figure S3), and summary of sequential backbone triple-resonance connectivity and nuclear Overhauser effect spectroscopy (NOESY) data (Figure S4). For the crystal structure, electron density was interpretable for 323 of 384 residues (including the 11 amino acid residues of the N-terminal affinity tag) from the four protomers in the asymmetric unit, corresponding to residues 153–237 of BRAF. The first eight residues of the N-terminal tag and final four C-terminal residues are flexible and not visible on the electron density map. The final R factor of the atomic model is 21.3%, and Rfree is 27.1% (Table S2).
The structure BRAF RBD adopts a classic ubiquitin α/β roll composed of two α helices and a five-stranded mixed β sheet (Figures 1B and 1C). Strands β1-β2 and β3-β4-β5 are antiparallel to each other, while strands β1 and β5 are parallel. The two helices, α1 helix after β2 strand and α2 310 helix after β4 strand, pack against one face of the β sheet. The protein core, consisting of residues 154–228, is well ordered except for residues 202–204 (in loop L4 between β3 and β4 strands) and the disordered N and C termini. In general, core side-chain conformations are quite well defined in the NMR ensemble (Figure S5). The average backbone root-mean-square deviation (RMSD) between the medoid conformer of the NMR ensemble (the model with the lowest average RMSD to all the other models in the ensemble) and each conformer of NMR ensemble (0.75 ± 0.12 Å, over residues 154–228) is comparable with the RMSD between the medoid conformer and X-ray crystal structure (0.99 Å). The BRAF RBD solution and crystal structures are highly superimposable, with minor deviations confined to the termini and loop L4, which lacks electron density for three residues (Figure S6). ConSurf (Ashkenazy et al., 2010; Glaser et al., 2003) analysis across the entire Pfam RAF-like RAS-binding protein domain family, PF02196 (Finn et al., 2014), reveals that amino acid conservation is primarily clustered on the region of the domain encompassing β1 and β2 strands and the C terminus of α1 helix (Figure 1D).
A Dali search (Holm and Rosenström, 2010) for structurally similar proteins showed significant (high Z score) hits with the RBD of CRAF in complex with HRAS (Dali Z score 13.9; PDB: 4G0N-B, Cα RMSD 0.8 Å) (Figure 1E) and ubiquitin (Dali Z score 9.6, PDB: 3HM3-D, Cα RMSD 2.1A Å). X-Ray crystal structures of CRAF RBD bound to the RAS homolog Rap1A (Nassar et al., 1995) and, very recently, HRAS itself (Fetics et al., 2015) reveal that complex formation is mediated by an intermolecular interaction between two antiparallel β strands: the β2 strand of the RBD and β2 of HRAS or Rap1A. Interestingly, the RBDs from BRAF and CRAF share 100% sequence identity in the functionally important β1 and β2 strands (157RVFLPNKQRTVV169). Such sequence conservation suggests a highly similar HRAS binding mode for the BRAF and CRAF RBDs, which we have verified (see below).
BRAF RBD Forms a Ternary Complex with HRAS and GppNHp
To facilitate tight and persistent complex formation with BRAF RBD, bacterially expressed HRAS GTPase isolated in the GDP form must be bound to a non-hydrolyzable GTP analog, such as GppNHp. Using 31P NMR, we confirmed the complete enzymatic conversion of HRAS-GDP to HRAS-GppNHp and subsequent complex formation with BRAF RBD, which results in a small upfield shift of the γ-phosphate 31P resonance typically observed upon effector binding (Spoerner et al., 2004) (Figure 2A). Both analytical gel filtration with static light-scattering detection (Figure 2B) and rotational correlation times determined from 15N relaxation data (Figure 2C) reveal that the BRAF RBD and HRAS domains are monomers in solution, and that the resulting complex, RBD-HRAS-GppNHp, exhibits the molecular weight and τc changes expected for this ternary complex. As a consequence, the 15N T1 and T2 values for BRAF RBD, HRAS, and the complex follow the expected trend of increasing T1 and decreasing T2 with increasing macromolecular weight, in the slow motion limit (ω0τc >> 1) (Figure 2D).
Figure 2. Assessment of Phosphorylation and Oligomerization States of Free and Complexed BRAF RBD and HRAS.
(A) 1D 31P NMR spectra at 298 K of 0.6 mM HRAS-GDP (prior to alkaline phosphatase treatment), 0.6 mM HRAS-GppNHp after alkaline phosphatase treatment and reconstitution with GTP analog, and 0.3 mM [13C,15N]-BRAF RBD-HRAS-GppNHp. The 31P resonances for HRAS-bound GppNHp are labeled following the latest literature assignments (Spoerner et al., 2005).
(B) Analytical gel filtration/static light-scattering data for 0.6 mM [13C,15N]-BRAF RBD-HRAS-GppNHp at pH 7.5. Plots of relative differential refractive index (dn/dc) and experimental molecular weight (34.9 kDa) are shown in blue and red, respectively. The expected molecular weight of the complex, including isotope enrichment, nonnative residues, and the nucleotide, is 31.8 kDa.
(C) Plots of rotational correlation time (τc) determined from 15N T1 and T2 relaxation data as a function of protein molecular weight for 0.3–0.4 mM [U-13C,15N]-BRAF RBD (blue), [15N]-HRAS-GDP (magenta), and [13C,15N]-BRAF RBD-HRAS-GppNHp (green) at 298 K and pH 7.5; known monomeric proteins solved in the Northeast Structural Genomics project are shown in red.
(D) Plots of 60.82 MHz 15N T1 (left) and T2 (right) relaxation data for [U-13C,15N]-BRAF RBD (blue), [U-15N]-HRAS-GDP (magenta), and [U-13C,15N]-RBD-HRAS-GppNHp (green) at pH 7.5 and 298 K.
RAS Binding to BRAF RBD Results in Local and Long-Range CSPs
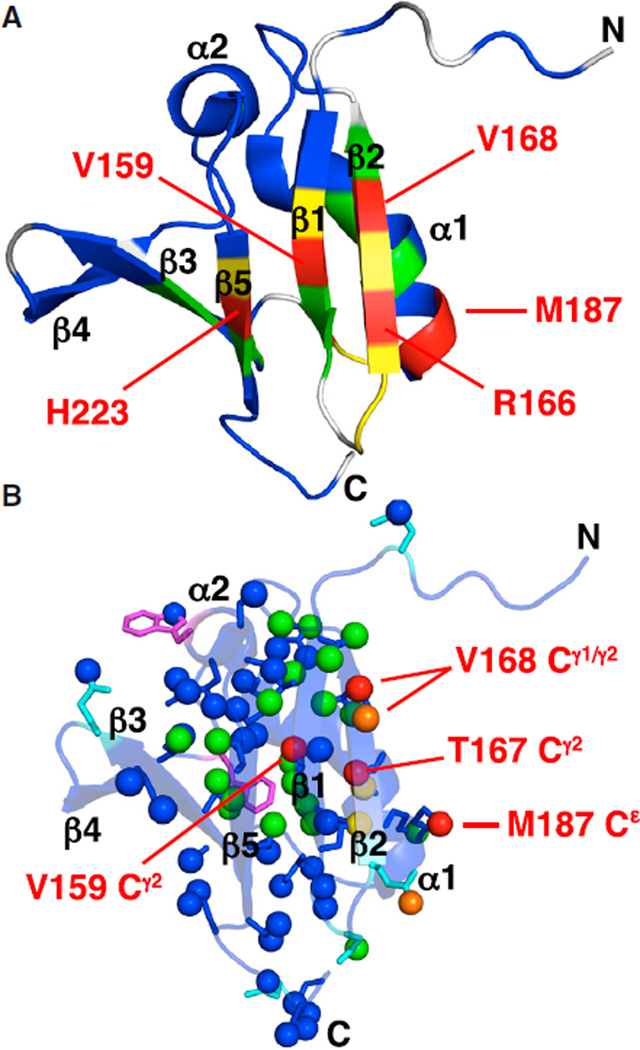
The effects of HRAS binding on BRAF RBD can be elucidated at an atomic level by comparing NMR spectral changes between apo and bound forms resulting from HRAS-GppNHp binding to [13C,15N]-BRAF RBD. Plots of HRAS-induced backbone amide, ΔδNH, and carbonyl, ΔδCO, CSPs along the sequence of BRAF RBD are shown in Figures 3A and 3B. The most dramatic amide and carbonyl CSPs correspond to key residues located in β2 strand (R166 and V168) and the end of α1 helix (M187). These trends correlate well with the HRAS-binding epitope of CRAF RBD, characterized by both X-ray crystallography (Fetics et al., 2015) and NMR CSPs (Emerson et al., 1995; Terada et al., 1999), in which the β2 strand of the RBD forms a network of intermolecular hydrogen bonds and the C-terminal end of α1 helix also mediates complex formation. For example, in the crystal structure of CRAF RBD-HRAS-GppNHp (PDB: 4G0N) (Fetics et al., 2015), the backbone NH of R67, CO of R67, and NH of V69 (corresponding to R166 and V168, respectively, in BRAF) interact with complementary moieties on β2 strand of HRAS, before this strand veers away from the RBD (Figure 4A). This pattern is recapitulated exactly in our backbone CSPs for HRAS binding to BRAF RBD; namely, very large ΔδNH values for R166 and V168, but only a large ΔδCO for R166 but not V168. The residue at the C-terminal end of α1 helix in CRAF RBD, V88, is completely buried in the protein-protein interface, including its backbone carbonyl oxygen (Figure 4B). The corresponding residue in BRAF RBD, M187, also exhibits sizable ΔδCO and ΔδNH CSPs consistent with a similar mode of binding for BRAF RBD. We observe no significant backbone amide CSPs for BRAF RBD in the presence of the GDP form of HRAS (data not shown).
Figure 3. NMR Chemical Shift Perturbations in BRAF RBD Resulting from HRAS Binding.
Plots of (A) backbone amide (ΔδNH) and (B) carbonyl (ΔδCO) CSPs versus residue number for BRAF RBD. Mean CSP values are indicated by blue lines, and residues exhibiting the largest CSPs are labeled. Secondary structural elements in BRAF RBD (PDB: 2L05) are shown above the plots.
Figure 4. Key Interfacial Residues in CRAF RBD-HRAS Structure.
(A) Conserved residues involved in intermolecular strand-strand hydrogen bonding in the crystal structure of CRAF RBD-HRAS (PDB: 4G0N) (Fetics et al., 2015). Residues R67 and V69 in CRAF correspond to R166 and V168 in BRAF.
(B) View from the same structure showing the environment around the C terminus of helix α1 from CRAF RBD and the buried carbonyl of V88 (corresponding to M187 in BRAF). Hydrogen atoms were added onto the structure using MolProbity 4.1 (Davis et al., 2007). Structures were rendered using PyMOL.
Mapping CSPs due to HRAS binding onto the structure of BRAF RBD yields further insights into the structural effects of complex formation. As expected, the largest backbone amide (Figure 5A) and side chain (Figure 5B) CSPs (in red) are generally clustered at the HRAS-binding interface. Even more significant are the numerous CSPs observed throughout the RBD structure, extending far beyond the RAS-binding epitope. Interestingly, these perturbations are propagated through much of the core of the domain, reflected by several backbone amide (including β1: R158, V159; β5: L222, H223), buried methyl (including β1: V157γ1, V159γ1/γ2; β2: T167γ2, V169γ2; α1: L181δ1, A184β, L185δ1/δ2; β3: V197γ1/γ2; β4: I208δ1; α2: I214δ1; β5: L222δ2, V224γ2), and indole NH (W210ε1) CSPs (Figures 5A and 5B). These results indicate allosteric conformational changes in BRAF RBD due to HRAS binding.
Figure 5. HRAS-Induced CSPs Mapped onto the Structure of BRAF RBD.
(A) Backbone amide CSPs (ΔδNH) mapped onto the solution NMR structure of BRAF RBD. Residues in the structure are colored according to the magnitude of their CSP compared with the mean, as follows: blue, CSP < mean; green, mean < CSP < mean + 1σ yellow, mean + 1σ < CSP < mean + 2σ; red, CSP > mean + 2σ (mean CSP = 0.1 ppm; σ = 0.1 ppm, excluding major outliers).
(B) Methyl (ΔδMe) and side-chain NH (ΔδNH) CSPs mapped onto the solution NMR structure of BRAF RBD. Methyl carbon and side-chain nitrogen atoms are represented as spheres, and colored using the same scheme as in (A) (mean CSP = 0.06 ppm; σ = 0.07 ppm, excluding M187). Side-chain resonances present in the apoprotein spectra but not assigned in the complex are shown in orange. Structures were rendered using PyMOL.
19F NMR to Probe for RAS Binding
To further probe the allosteric changes induced by HRAS binding, we incorporated 5-fluorotryptophan (5-F-Trp) into BRAF RBD and monitored complex formation using 19F NMR. BRAF RBD features a conserved tryptophan, W210, whose side chain is buried in the core of the α/β structure, and a second solvent-exposed tryptophan, W216, jutting out from helix α2 (Figure 6A). The ζ3 position in the analogous conserved buried tryptophan (W114) in CRAF RBD is more than 15 Å from the nearest HRAS backbone atom at the intermolecular strand-strand interface of the complex. Due to the numerous highly favorable properties of the 19F nucleus, including its high resonance frequency and the extraordinary sensitivity of its chemical shift to its local environment (Kitevski-LeBlanc and Prosser, 2012; Yu et al., 2013), we reasoned that 19F NMR combined with 5-F-Trp incorporation could serve as a sensitive monitor of HRAS conformational changes upon binding to BRAF RBD. The 19F NMR spectrum of 5-F-Trp-labeled BRAF RBD exhibits two 19F resonances in the expected chemical shift window (Figure 6B), which can be assigned on the basis of the solvent-induced isotope shift effect (Kitevski-LeBlanc and Prosser, 2012). Specifically, fully exposed fluorinated residues exhibit a Δδ ≈ −0.2 ppm upfield shift in 2H2O. In the case of 5-F-Trp-labeled BRAF RBD, the upfield 19F signal experiences a marked −0.16 ppm shift when the solvent is changed from 10% to 90% 2H2O, and is thus assigned to the exposed W216, whereas the downfield 19F resonance corresponding to the buried W210 experiences little dependence on solvent deuteration (Figure 6B). The titration of 5-F-Trp-labeled BRAF RBD with unlabeled HRAS-GppNHp is shown in Figure 6C. We observe classic slow exchange binding behavior, in which separate 19F signals for W210 in the apo (a) and bound (b) states decrease and increase, respectively, during the course of the titration. Such slow exchange behavior is expected for the reported high-affinity, low-nanomolar binding of HRAS-GTP to BRAF RBD (KD = 11.2 nM) (Fischer et al., 2007), and is also observed in the backbone 1H-15N chemical shift timescale (Terada et al., 1999). Moreover, the 19F signal due to W210 exhibits a significant CSP (Δδ = −0.16 ppm) induced by RAS binding, which is approximately twice as large as the side chain NH CSP observed for this residue (Figure 5B). The 19F signal due to W216 in helix α2 also exhibits a very small CSP (Δδ = −0.04 ppm). These results confirm that HRAS binding induces conformational changes that are sensed in the core of the RBD structure, distant from the HRAS-binding interface, by 19F NMR.
Figure 6. 19F NMR as a Probe for HRAS Binding to BRAF RBD.
(A) Locations of the two 5-F-Trp residues in the NMR structure of BRAF RBD. The fluorine atoms are shown as magenta spheres. The secondary structure elements were rendered (PyMOL) with some transparency to completely show the buried W210 side chain.
(B) Assignment of the 19F signals from 0.45 mM 5-F-Trp-labeled BRAF RBD using the solvent-induced isotope shift effect. Dotted lines indicate the shift of the 19F signal for W216 with increasing percentage of 2H2O in the buffer.
(C) Titration of 0.2 mM 5-F-Trp-labeled BRAF RBD with HRAS-GppNHp monitored by 19F NMR at 293 K. Resonances for W210 in the apo and bound states are labeled with a and b, respectively.
RAS Binding Changes the Free-Energy Landscape of BRAF RBD
To further characterize the conformational changes resulting from the binding of HRAS-GppNHp to BRAF RBD, we measured backbone amide hydrogen/deuterium exchange rates on both unbound and complexed RBD by NMR. In the limit of EX2 amide exchange conditions (Krishna et al., 2004), the experimentally determined residue-specific amide exchange rates, kex, for the protected amides can be converted to stabilization free energies, ΔGHX. When plotted across the RBD sequence, we observe a marked increase in ΔGHX (decrease in kex) across the sequence for the complex (in blue) compared with the apoprotein (in red) (Figure 7A). These include several additionally protected backbone amides in the complexed RBD, which are not protected in the free RBD. When mapped onto the 3D structure of BRAF RBD, we observe that these increases in the free energy of local and/or global conformational unfolding are transmitted throughout several secondary structure elements, including much of the β sheet and helices α1 and α2 (Figure 7B). A similar picture is obtained when the changes in amide exchange rates upon HRAS binding, expressed as ln(kexapo/kexcomplex), are represented on the structure of BRAF RBD (Figure 7C). Here, positive values reflect a decrease in kex, increased protection from exchange with solvent deuterons, and reduced protein flexibility upon complex formation, whereas negative values indicate increased kex and enhanced backbone motion induced by HRAS. The slowed amide exchange is not simply due to stabilizing a single conformation of RBD upon complex formation, as not all buried amide protons exhibit the same degree of reduced exchange rates in the complex. In fact, residue K206 in β4 strand, which exhibits a fast but measurable kex in the apoprotein, exchanges more rapidly in the complex. Taken together, these results demonstrate that RAS binding significantly alters the energy landscape of BRAF RBD, suggesting a shift from a structured yet somewhat dynamic, “breathing” apoprotein to a less conformationally dynamic bound domain structure.
Figure 7. Backbone Amide Hydrogen/Deuterium Exchange NMR Reveals Changes in the Free Energy Landscape of BRAF RBD upon HRAS Binding.
(A) Stacked bar graphs of stabilization free energies (ΔGHX) computed from backbone amide exchange rates (kex) versus sequence for apo (red, top) and complexed (blue, bottom) BRAF RBD. Secondary structural elements in BRAF RBD are shown above the plots.
(B) Free energies of amide exchange (ΔGHX) mapped onto the solution NMR structure of apo (left) and complexed (right) BRAF RBD. Residues in each structure are colored as a function of ΔGHX (kcal/mol) as follows: red, <5; yellow, 5 < ΔGHX < 6; green, 6 < ΔGHX < 7; cyan, 7 < ΔGHX < 8; blue, >8. Increasing ΔGHX correlates with increasing amide protection from exchange.
(C) Ratios of backbone amide exchange rates for apo (kexapo) and HRAS-bound (kexcomplex) BRAF RBD mapped onto its solution NMR structure (residues 154–228 shown). In cases where the exchange rate was too fast to be accurately measured, an upper limit of kex = 0.001 s−1 was assumed. Residues are colored as a function of ln(kexapo/kexcomplex) as follows: red, <0; yellow, 0–1; green, 1–2; cyan, 2–3; blue, 3–4; purple, >4; white, not measureable in both states. The approximate position of HRAS encompassing the binding interface, based on the crystal structure of its complex to CRAF RBD (PDB: 4G0N) (Fetics et al., 2015), is shown in orange. Structures were rendered using PyMOL.
DISCUSSION
In this study we present the solution and crystal structures of the RBD from BRAF kinase and characterize its complex with HRAS bound to a non-hydrolyzable analog of GTP, GppNHp, in solution. The solution and crystal structures of BRAF RBD confirm the classic ubiquitin α/β roll expected for RAS-binding domains. NMR CSPs for BRAF RBD resulting from HRAS-GppNHp binding corroborate the conserved RAS-binding epitope, but also demonstrate unexpected allosteric conformational changes that propagate from the binding epitope through the core of the RBD to distal elements in the domain. Moreover, backbone amide hydrogen/deuterium exchange NMR results reveal a general yet varied decrease in amide exchange rates across the structure of the domain upon HRAS binding. These changes in amide exchange rates distant from the RAS-binding epitope reveal a distribution of conformations in apo-RBD that are altered in the complex. This shift in conformational distribution is also indicated by CSPs in sites distant from the RAS-binding site upon complex formation. These data support an allosteric model for the RAS/RAF interaction, and suggest a mechanism by which RAS binding can signal the drastic domain rearrangements required for activation of BRAF kinase.
Two main conclusions can be drawn from our NMR studies of the BRAF RBD-HRAS-GppNHp complex. First, our CSP results on BRAF RBD demonstrate that the mode of RAS binding previously established for the CRAF isoform (Emerson et al., 1995; Fetics et al., 2015; Terada et al., 1999) is preserved in BRAF. Namely, specific determinants at the binding interface comprise conserved residues in the C-terminal region of helix α1 and intermolecular strand-strand hydrogen bonding mediated by the β2 strand of the RBD. Moreover, the expected interfacial methyl and amide side chains in BRAF RBD exhibit the largest CSPs upon HRAS binding, namely the methyls of T167, V169, and M187, and the NH2 of Q165 (Figure 5A). Taken together, these results indicate that the isoforms of RAF interact with RAS in a highly analogous fashion.
Second, the backbone amide and side-chain 1H, 13C, and 15N CSPs, and buried Trp 19F CSPs reported here clearly demonstrate that RAS binding to BRAF RBD results in a network of perturbations that extend into the RBD far beyond the immediate protein-protein interface. In addition, backbone amide hydrogen/deuterium exchange studies reveal that RAS binding results in dramatic changes in amide proton exchange rates permeating much of the secondary structure elements in the domain. The observation of both (predominantly) slower and (infrequent) faster backbone amide exchange rates in BRAF RBD induced by HRAS binding is consistent with a shift in the conformational distributions of the BRAF RBD structure upon complex formation. Taken together, these data demonstrate an allosteric change in the RBD structure due to RAS binding, which could provide the basis for its interactions with other domains in BRAF and/or other proteins.
The concept of allostery in this system was previously proposed on the basis of molecular dynamics (MD) simulations on the CRAF RBD-HRAS complex, where complex formation results in dynamic changes in the RBD emanating from the binding interface and “percolating” through the core of the structure via a network of interacting residues (Gohlke et al., 2003, 2004). Specifically, the MD calculations on apo and bound CRAF RBD predicted an increase in the rigidity of the β sheet of the RBD and increased flexibility of loop L4 in CRAF RBD upon complex formation. More recent MD studies confirmed the same principal network of coupled interactions in which the cluster R67, F61, and Q127 (R166, F160, and H223 in BRAF) plays a key role (Fetics et al., 2015). Moreover, both structural and MD results for the oncogenic Q61L mutant of HRAS indicate an altered allosteric effect on CRAF RBD, resulting in greatly reduced flexibility of loop L4 upon complex formation (Fetics et al., 2015). The results presented here on BRAF, which like ARAF features a much shorter loop L4 (Figure 1A), extend this idea of dynamic allostery to BRAF RBD and provide direct spectroscopic evidence that the RAS-binding signal is transmitted via both backbone and side-chain conformational changes in BRAF RBD. Specifically, our solution NMR data reveal that the RAS-binding signal is also transmitted through several buried side chains in the core of the RBD, in addition to the in silico predicted network of residues on the surface of the aforementioned β sheet.
In recent years numerous structural, spectroscopic, and MD studies have established that the structure of the RAS protein is also highly dynamic, featuring several conformational substates whose populations can be modulated by the phosphorylation state and nature of the bound guanosine nucleotide, interactions with effector proteins, and mutations in RAS itself (Fetics et al., 2015; Rosnizeck et al., 2014; Smith and Ikura, 2014; Spoerner et al., 2010). In the GTP-bound state, for example, RAS adopts two major conformational states, termed state 1(T) and state 2(T), which have disparate functions; state 1(T) exhibits a lower affinity for effector proteins and is recognized by GEFs, whereas binding of effector proteins strongly shifts the equilibrium toward state 2(T) (Spoerner et al., 2010). Manipulation of the conformational equilibrium between states 1(T) and 2(T) by RAS mutations, particularly in switch I, and inhibitor binding can abrogate or alter binding to effector proteins (Rosnizeck et al., 2014; Smith and Ikura, 2014; Spoerner et al., 2004). In light of the results presented here on BRAF RBD and recent studies on CRAF RBD (Fetics et al., 2015), it is interesting to note that the allosteric changes that originate from the binding interface observed in RAF-RAS complexes result from a high-affinity interaction between two proteins which each have their own conformational plasticity.
Finally, the interplay between conformational dynamics, molecular plasticity, and allostery is emerging as a central paradigm for understanding the basis of molecular recognition, regulation, and signaling in nature (Motlagh et al., 2014). The historic perception of protein structure as being static has been supplanted in recent years by a more dynamic view centered on the concept of conformational selection (Boehr et al., 2009), whereby proteins fluctuate between a range of structures and are thereby predisposed for interaction with binding partners (Huang and Montelione, 2005). To this end, NMR relaxation techniques have taken center stage in establishing the link between the intrinsic internal conformational dynamics of proteins, even for buried methyl-bearing residues, and macromolecular recognition and allosteric regulation (Aramini et al., 2014; Sekhar and Kay, 2013; Tzeng and Kalodimos, 2011, 2012; Wand, 2013). Moreover, β-sheet structures are particularly well suited for mediating allostery and signal transduction via correlated backbone motions causing global sheet bending and twisting, resulting in “channels of communication” running perpendicular to the strands (Fenwick et al., 2014). In accordance with these views, the results presented here provide empirical evidence that RAS binding is translated through the RBD structure via conformational perturbations. Since the exact structural basis for how RAS binding to RAF kinases unlocks the inactive multi-domain structure of RAF and how this event ultimately leads to a catalytically active kinase remains an open question, this work provides a starting point for further investigations into how the binding of RAS, as well as oncogenic RAS mutants, alters allosterically regulated domain-domain interactions in the full-length BRAF kinase.
EXPERIMENTAL PROCEDURES
Cloning, Expression, Purification, and Sample Preparation
RBD from human serine/threonine-protein kinase BRAF (UniProtKB/Swiss-Prot: P15056, BRAF_HUMAN; NESG: HR4694F) and the first 171 residues (out of 189 in the mature protein) of human HRAS (UniProtKB/Swiss-Prot: P01112, RASH_HUMAN; NESG: HR9664), referred to here as BRAF RBD and HRAS, respectively, were cloned, expressed, and purified based on the standard procedures of the NESG (Acton et al., 2011). Complete cloning, protein expression, purification, and sample preparation protocols are provided in Supplemental Experimental Procedures. In brief, isotopically enriched BRAF RBD for NMR spectroscopy and X-ray crystallography (BRAF[149–232] and BRAF[153–237], respectively), each containing an 11-residue N-terminal affinity tag (MGHHHHHHSHM), were expressed in Escherichia coli BL21(DE3)-Gold cells (Agilent) grown in MJ9 minimal medium (Jansson et al., 1996) containing U-(15NH4)2SO4 and [U-13C]-glucose as the sole nitrogen and carbon sources for NMR, or supplemented with selenomethionine for crystallography (Doublie et al., 1996). Incorporation of 5-F-Trp into BRAF RBD was performed as previously described (Aramini et al., 2014). Tagless and unlabeled HRAS used for binding studies with BRAF RBD was expressed in E. coli Tuner(DE3) cells (EMD Millipore) grown in LB medium and cleaved using N-terminal hexaHis-tagged TEV protease (Kapust et al., 2001). To activate HRAS for binding to BRAF RBD, the purified bacterially expressed GDP form of HRAS was treated with calf intestinal alkaline phosphatase (New England BioLabs) in the presence of a non-hydrolyzable analog of GTP, GppNHp (Sigma) (Smith and Rittinger, 2002). Samples of [U-13C,15N]- and [U-5%-13C,100%-15N]-BRAF RBD for NMR structure determination were concentrated by centrifugation to 0.7–0.9 mM in 20 mM ammonium acetate, 100 mM NaCl, 10 mM DTT, 5 mM CaCl2, 50 µM 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS), and 10% 2H2O (v/v) at pH 4.5. Samples of 0.3–0.6 mM [U-13C,15N]-BRAF RBD-HRAS-GppNHp complex for NMR spectroscopy were prepared in 20 mM Tris-HCl, 100 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine (TCEP), 5 mM MgCl2, 50 µM DSS, 10% (v/v) 2H2O at pH 7.5 by combining equimolar amounts of apo-[U-13C,15N]-BRAF RBD and unlabeled HRAS-GppNHp in the same buffer followed by concentration by centrifugation. Samples of 5-F-Trp BRAF RBD for 19F NMR spectroscopy were concentrated to 0.2–0.5 mM in pH 7.5 buffer (see above). All pET expression vectors used in this work have been deposited in the PSI Materials Repository (http://psimr.asu.edu/).
Static Light-Scattering Measurements
The oligomerization states of free BRAF RBD and HRAS, and the stoichiometry of their complex, were assessed by analytical gel filtration coupled with static light scattering and molecular rotational correlation times determined from 15N T1 and T2 relaxation measurements (see below). Analytical gel filtration with multi-angle static light-scattering detection data were collected at λ = 690 nm on a miniDAWN (TREOS) Light Scattering instrument (Wyatt Technology) coupled with an analytical gel filtration column. All measurements were performed at room temperature and with a flow rate of 0.5 ml/min. Data were analyzed using the ASTRA software package (Wyatt Technology).
NMR Spectroscopy
All 1H, 13C, and 15N NMR data were collected at 298 K on Bruker AVANCE 600- and 800-MHz spectrometers equipped with 1.7-mm TCI and 5-mm TXI cryoprobes, respectively, processed with NMRPipe (Delaglio et al., 1995), and visualized using SPARKY 3 (T.D. Goddard and D.G. Kneller, University of California San Francisco). All spectra were referenced to internal DSS. 1D 31P NMR spectra were acquired at 298 K on a Bruker AVANCE III 500-MHz spectrometer equipped with a 5-mm QCI-P cryoprobe at a resonance frequency of 202.65 MHz, using a 3.0-s repetition time, 20,000-Hz sweep width, and 1,000–1,600 scans. 1D 19F NMR spectra were acquired locked and at 20°C on a Varian INOVA 500-MHz spectrometer equipped with a room-temperature 5-mm 1H/19F switchable probe at a frequency of 470.18 MHz, using a 5.35-s repetition time, 20,000-Hz sweep width, and 1,000–4,000 scans, and referenced to external neat CFCl3 (Aramini et al., 2014). 31P and 19F spectra were processed with 5- and 10-Hz exponential multiplication, respectively, and analyzed using Mnova 9.0 (Mestrelab Research). Near complete 1H, 13C, and 15N resonance assignments for BRAF RBD at pH 4.5 were determined using conventional triple-resonance NMR methods (see Supplemental Experimental Procedures). In brief, automated backbone resonance assignments were made using PINE 1.0 (Bahrami et al., 2009), and the final connectivity information was visualized and confirmed using AutoAssign 2.4.0 (Moseley et al., 2001). Side-chain assignments were completed manually using 3D HBHA(CO)NH, HBHANH, HCCH-COSY, HCCH-TOCSY, and (H)CCH-TOCSY experiments. The final resonance assignments, NOESY spectral peak lists, and time domain data for BRAF RBD were deposited in the BioMagResDB (BMRB: 17030). Backbone resonance assignments of apo-[13C,15N]-BRAF RBD at pH 7.5 were confirmed using standard 3D triple-resonance NMR experiments. Backbone and side-chain methyl resonance assignments of [13C,15N]-BRAF RBD in complex with HRAS-GppNHp at pH 7.5 were deduced on the basis of TROSY-based 3D triple-resonance backbone experiments, and 3D (H) CCH-TOCSY and 13C- and 15N-edited NOESY spectra, respectively. Global protein rotational correlation times (τc) were computed from the ratio of 15N T1 and T2 relaxation times, derived from pseudo-2D 15N-edited T1 and T2 (CPMG) relaxation experiments (Farrow et al., 1994), and the nuclear frequency (νN) according to Equation 1 (Fushman et al., 1994; Kay et al., 1989).
| (Equation 1) |
Solution NMR Structure Refinement
The solution NMR structure of BRAF RBD was calculated using CYANA 3.0 (Güntert et al., 1997; Herrmann et al., 2002) supplied with peak intensities from 3D 15N-edited NOESY (τm = 100 ms), 3D 13C-edited aliphatic NOESY (τm = 100 ms), and 3D 13C-edited aromatic NOESY (τm = 120 ms) spectra, together with broad dihedral angle constraints derived by TALOS+ (Shen et al., 2009) (ϕ, ψ ± 30°) for ordered residues with confidence scores of 10. The 20 structures with lowest target function out of 100 calculated in the final cycle were further refined by restrained MD in explicit water using CNS 1.2 (Brünger et al., 1998; Linge et al., 2003) and the PARAM19 force field, supplied with the final NOE-derived distance and TALOS+ dihedral angle constraints. The final refined ensemble of 20 structures for BRAF RBD (excluding the first nine residues of the N-terminal 6xHis purification tag) was deposited in the PDB (PDB: 2L05). Structural statistics and global structure quality factors (Table S1) were computed using the PSVS 1.5 (Bhattacharya et al., 2007) and MolProbity (Davis et al., 2007) servers. The global goodness-of-fit of the final structure ensemble with the NOESY peak list data and resonance assignments was determined using the RPF server (Huang et al., 2005, 2012).
Crystallization, Data Collection, and Structure Refinement
Crystallization screening was performed using a microbatch-under-oil crystallization method at 18°C (Chayen et al., 1990). After optimization, BRAF RBD crystals useful for structure determination were grown in drops composed of 1.0 µl of protein and 1.0 µl of precipitant solution (100 mM Bis-Tris [pH 6.5], 28% [w/v] PEG 2000) under paraffin oil (Hampton Research). The crystals were cryoprotected with 15% ethylene glycol prior to flash-freezing in liquid nitrogen for data collection. A selenomethionyl single-wavelength anomalous diffraction (SAD) data set (Hendrickson, 1991) was collected at the peak wavelength of the selenium K edge using beamline X4A at the National Synchrotron Light Source (λ = 0.97903 Å). The diffraction data from a single crystal was processed with the HKL2000 package (Otwinowski and Minor, 1997).
The program SHELXE/D (Schneider and Sheldrick, 2002) was used to locate a selenium site and to calculate phases to 2.6 Å resolution. The model was completed using iterative cycles of manual rebuilding in Coot (Emsley and Cowtan, 2004), and was refined against 1.99 Å data with the program PHENIX (Adams et al., 2002). The data processing and refinement statistics for the crystal structure determination are summarized in Table S2. The quality of the final structure was assessed using PSVS (Bhattacharya et al., 2007) and PROCHECK (Laskowski et al., 1993). The atomic coordinates and structure factors are available in the PDB under accession code PDB: 3NY5.
NMR CSP Measurements
CSPs on BRAF RBD resulting from complex formation with HRAS were computed from 2D 1H-15N TROSY-HSQC, TROSY- and standard 3D HNCO, and 2D 1H-13C HSQC spectra of [13C,15N]-BRAF RBD, free and in complex with HRAS-GppNHp acquired at identical field strength, temperature, and buffer conditions (800 MHz, 298 K, pH 7.5). Composite 1H and 15N backbone amide chemical shift perturbations, ΔδNH, were calculated using Equation 2 with weighting factors, ωH and ωN, of 1.0 and 0.154, respectively; carbonyl CSPs were multiplied by a weighting factor, ωCO, of 0.341 (Evenäs et al., 2001).
| (Equation 2) |
Composite side-chain methyl CSPs were computed using Equation 3.
| (Equation 3) |
Hydrogen/Deuterium Exchange NMR Experiments
Exchange rates for slowly exchanging backbone amides were measured by NMR using the following procedure. Samples of 0.5–0.6 mM [13C,15N]-BRAF RBD free or in complex with HRAS-GppNHp in pH 7.5 buffer were lyophilized and redissolved in the same volume (50 µl) of 2H2O and immediately placed into a 1.7-mm microtube for NMR. The NH signal intensities for slowly exchanging backbone amides were then measured over time in a series of 600-MHz 2D 1H-15N SOFAST HMQC spectra (Schanda and Brutscher, 2005) at 298 K. Amide exchange rates, kex, were obtained by exponential fitting of the signal intensity decays using KaleidaGraph 4.0 (Synergy Software).
According to the classic Linderstrøm-Lang model, the exchange of structurally protected amide hydrogen atoms with solvent proceeds through a transiently exposed state according to Equation 4 (Krishna et al., 2004):
| (Equation 4) |
where kop and kcl are the rate constants of opening and closing, and kint is the intrinsic amide exchange rate, which depends on residue type, neighboring sequence, pH, and temperature. Under conditions of EX2 exchange (kcl ≫ kint), the free energy of amide exchange per residue can be computed using Equation 5 (Krishna et al., 2004).
| (Equation 5) |
Hence, knowledge of the empirical amide exchange rates, kex, and intrinsic exchange rates, kint, obtained for the BRAF RBD protein sequence using the Sphere server (www.fccc.edu/research/labs/roder/sphere/) (Bai et al., 1993), yields ΔGHX for slowly exchanging amides.
Supplementary Material
Highlights.
Solution NMR and X-ray crystal structures of BRAF RAS-binding domain
RAS-binding surface mapped onto the structure of BRAF RBD
RAS binding alters amide exchange rates throughout BRAF RBD
Evidence for RAS-induced allosteric changes in BRAF RBD
In Brief.
RAS binding is a critical step in the activation of BRAF kinase and the MAPK signaling pathway. Aramini et al. report the solution NMR and X-ray crystal structures of the RAS-binding domain (RBD) from human BRAF, and use NMR to reveal unexpected allosteric changes in BRAF RBD upon RAS binding.
Acknowledgments
We thank I. Pelczer and K. Conover for assistance with acquiring 31P NMR data at Princeton University, and S. Anderson, S. Arunkumar, D. Case, C. Ciccosanti, J. Everett, S. Krishna Murthy, D. Patel, R. Shastry, and R. Tejero for valuable discussions and technical support. This work was supported by a grant from the National Institute of General Medical Sciences Protein Structure Initiative U54-GM094597 (to G.T.M.).
Footnotes
ACCESSION NUMBERS
The PDB and BioMagRes DataBank accession numbers for the structures presented in this paper are PDB: 2L05 and BMRB: 17030 (NMR) and PDB: 3NY5 (X-ray).
Supplemental Information includes Supplemental Experimental conline at http://dx.doi.org/10.1016/j.str.2015.06.003.
AUTHOR CONTRIBUTIONS
Conceptualization: J.M.A., S.M.V., Y.J.H., L.T., G.T.M. Investigation: J.M.A., S.M.V., L.M.T., H.J., E.T.C., J.S., M.S., T.B.A., R.X. Writing—original draft: J.M.A., S.M.V., L.T., G.T.M. Writing—review and editing: J.M.A., S.M.V., L.T., G.T.M.
REFERENCES
- Acton TB, Xiao R, Anderson S, Aramini J, Buchwald WA, Ciccosanti C, Conover K, Everett J, Hamilton K, Huang YJ, et al. Preparation of protein samples for NMR structure, function, and small-molecule screening studies. Methods Enzymol. 2011;493:21–60. doi: 10.1016/B978-0-12-381274-2.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Aramini JM, Hamilton K, Ma LC, Swapna GV, Leonard PG, Ladbury JE, Krug RM, Montelione GT. 19F NMR reveals multiple conformations at the dimer interface of the nonstructural protein 1 effector domain from influenza A virus. Structure. 2014;22:515–525. doi: 10.1016/j.str.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami A, Assadi AH, Markley JL, Eghbalnia HR. Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput. Biol. 2009;5:e1000307. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chayen NE, Stewart PDS, Maeder DL, Blow DM. An automated-system for microbatch protein crystallization and screening. J. Appl. Crystallogr. 1990;23:297–302. [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Doublie S, Kapp U, Aberg A, Brown K, Strub K, Cusack S. Crystallization and preliminary X-ray analysis of the 9 kDa protein of the mouse signal recognition particle and the selenomethionyl-SRP9. FEBS Lett. 1996;384:219–221. doi: 10.1016/0014-5793(96)00316-x. [DOI] [PubMed] [Google Scholar]
- El-Nassan HB. Recent progress in the identification of BRAF inhibitors as anti-cancer agents. Eur. J. Med. Chem. 2014;72:170–205. doi: 10.1016/j.ejmech.2013.11.018. [DOI] [PubMed] [Google Scholar]
- Emerson SD, Madison VS, Palermo RE, Waugh DS, Scheffler JE, Tsao KL, Kiefer SE, Liu SP, Fry DC. Solution structure of the Ras-binding domain of c-Raf-1 and identification of its Ras interaction surface. Biochemistry. 1995;34:6911–6918. doi: 10.1021/bi00021a001. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evenäs J, Tugarinov V, Skrynnikov NR, Goto NK, Muhandiram R, Kay LE. Ligand-induced structural changes to maltodextrin-binding protein as studied by solution NMR spectroscopy. J. Mol. Biol. 2001;309:961–974. doi: 10.1006/jmbi.2001.4695. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Fenwick RB, Orellana L, Esteban-Martin S, Orozco M, Salvatella X. Correlated motions are a fundamental property of b-sheets. Nat. Commun. 2014;5:4070. doi: 10.1038/ncomms5070. [DOI] [PubMed] [Google Scholar]
- Fetics SK, Guterres H, Kearney BM, Buhrman G, Ma B, Nussinov R, Mattos C. Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure. 2015;23:505–516. doi: 10.1016/j.str.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filchtinski D, Sharabi O, Rüppel A, Vetter IR, Herrmann C, Shifman JM. What makes Ras an efficient molecular switch: a computational, biophysical, and structural study of Ras-GDP interactions with mutants of Raf. J. Mol. Biol. 2010;399:422–435. doi: 10.1016/j.jmb.2010.03.046. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Hekman M, Kuhlmann J, Rubio I, Wiese S, Rapp UR. B- and C-RAF display essential differences in their binding to Ras: the isotype-specific N terminus of B-RAF facilitates Ras binding. J. Biol. Chem. 2007;282:26503–26516. doi: 10.1074/jbc.M607458200. [DOI] [PubMed] [Google Scholar]
- Fushman D, Weisemann R, Thuring H, Ruterjans H. Backbone dynamics of ribonuclease T1 and its complex with 2’GMP studied by two-dimensional heteronuclear NMR spectroscopy. J. Biomol. NMR. 1994;4:61–78. doi: 10.1007/BF00178336. [DOI] [PubMed] [Google Scholar]
- Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- Gohlke H, Kiel C, Case DA. Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J. Mol. Biol. 2003;330:891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- Gohlke H, Kuhn LA, Case DA. Change in protein flexibility upon complex formation: analysis of Ras-Raf using molecular dynamics and a molecular framework approach. Proteins. 2004;56:322–337. doi: 10.1002/prot.20116. [DOI] [PubMed] [Google Scholar]
- Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Montelione GT. Structural biology: proteins flex to function. Nature. 2005;438:36–37. doi: 10.1038/438036a. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J. Am. Chem. Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Hang D, Lu LJ, Tong L, Gerstein MB, Montelione GT. Targeting the human cancer pathway protein interaction network by structural genomics. Mol. Cell Proteomics. 2008;7:2048–2060. doi: 10.1074/mcp.M700550-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Rosato A, Singh G, Montelione GT. RPF: a quality assessment tool for protein NMR structures. Nucleic Acids Res. 2012;40:W542–W546. doi: 10.1093/nar/gks373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson M, Li YC, Jendeberg L, Anderson S, Montelione GT, Nilsson B. High-level production of uniformly 15N- and 13C-enriched fusion proteins in Escherichia coli. J. Biomol. NMR. 1996;7:131–141. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- Kapust RB, Tözsér J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- Kitevski-LeBlanc JL, Prosser RS. Current applications of 19F NMR to studies of protein structure and dynamics. Prog. Nucl. Magn. Reson. Spectrosc. 2012;62:1–33. doi: 10.1016/j.pnmrs.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ, Domaille PJ, Campbell-Burk SL, Van Aken T, Laue ED. Solution structure and dynamics of ras p21 • GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 1994;33:3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- Krishna MM, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK - a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lavoie H, Therrien M. Regulation of RAF protein kinases in ERK signalling. Nat. Rev. Mol. Cell Biol. 2015;16:281–298. doi: 10.1038/nrm3979. [DOI] [PubMed] [Google Scholar]
- Lavoie H, Li JJ, Thevakumaran N, Therrien M, Sicheri F. Dimerization-induced allostery in protein kinase regulation. Trends Biochem. Sci. 2014;39:475–486. doi: 10.1016/j.tibs.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A, Kolch W. Raf family kinases: old dogs have learned new tricks. Genes Cancer. 2011;2:232–260. doi: 10.1177/1947601911407323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn MV, Tong L, deVos AM, Brünger A, Yamaizumi Z, Nishimura S, Kim SH. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- Moseley HN, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. The 2.2 Å crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature. 1995;375:554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R., Jr RAF protein-serine/threonine kinases: structure and regulation. Biochem. Biophys. Res. Commun. 2010;399:313–317. doi: 10.1016/j.bbrc.2010.07.092. [DOI] [PubMed] [Google Scholar]
- Rosnizeck IC, Filchtinski D, Lopes RP, Kieninger B, Herrmann C, Kalbitzer HR, Spoerner M. Elucidating the mode of action of a typical Ras state 1(T) inhibitor. Biochemistry. 2014;53:3867–3878. doi: 10.1021/bi401689w. [DOI] [PubMed] [Google Scholar]
- Schanda P, Brutscher B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Sekhar A, Kay LE. NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc. Natl. Acad. Sci. USA. 2013;110:12867–12874. doi: 10.1073/pnas.1305688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SJ, Rittinger K. Preparation of GTPases for structural and biophysical analysis. Methods Mol. Biol. 2002;189:13–24. doi: 10.1385/1-59259-281-3:013. [DOI] [PubMed] [Google Scholar]
- Smith MJ, Ikura M. Integrated RAS signaling defined by parallel NMR detection of effectors and regulators. Nat. Chem. Biol. 2014;10:223–230. doi: 10.1038/nchembio.1435. [DOI] [PubMed] [Google Scholar]
- Smith MJ, Neel BG, Ikura M. NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proc. Natl. Acad. Sci. USA. 2013;110:4574–4579. doi: 10.1073/pnas.1218173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoerner M, Wittinghofer A, Kalbitzer HR. Perturbation of the conformational equilibria in Ras by selective mutations as studied by 31P NMR spectroscopy. FEBS Lett. 2004;578:305–310. doi: 10.1016/j.febslet.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Spoerner M, Nuehs A, Ganser P, Herrmann C, Wittinghofer A, Kalbitzer HR. Conformational states of Ras complexed with the GTP analogue GppNHp or GppCH2p: implications for the interaction with effector proteins. Biochemistry. 2005;44:2225–2236. doi: 10.1021/bi0488000. [DOI] [PubMed] [Google Scholar]
- Spoerner M, Hozsa C, Poetzl JA, Reiss K, Ganser P, Geyer M, Kalbitzer HR. Conformational states of human rat sarcoma (Ras) protein complexed with its natural ligand GTP and their role for effector interaction and GTP hydrolysis. J. Biol. Chem. 2010;285:39768–39778. doi: 10.1074/jbc.M110.145235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging Ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol. Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Terada T, Ito Y, Shirouzu M, Tateno M, Hashimoto K, Kigawa T, Ebisuzaki T, Takio K, Shibata T, Yokoyama S, et al. Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins. J. Mol. Biol. 1999;286:219–232. doi: 10.1006/jmbi.1998.2472. [DOI] [PubMed] [Google Scholar]
- Thevakumaran N, Lavoie H, Critton DA, Tebben A, Marinier A, Sicheri F, Therrien M. Crystal structure of a BRAF kinase domain monomer explains basis for allosteric regulation. Nat. Struct. Mol. Biol. 2015;22:37–43. doi: 10.1038/nsmb.2924. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Protein dynamics and allostery: an NMR view. Curr. Opin. Struct. Biol. 2011;21:62–67. doi: 10.1016/j.sbi.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- Wand AJ. The dark energy of proteins comes to light: conformational entropy and its role in protein function revealed by NMR relaxation. Curr. Opin. Struct. Biol. 2013;23:75–81. doi: 10.1016/j.sbi.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JX, Hallac RR, Chiguru S, Mason RP. New frontiers and developing applications in 19F NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2013;70:25–49. doi: 10.1016/j.pnmrs.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.