Abstract
Atopic dermatitis (AD) is characterized by reduced barrier function, reduced innate immune activation, and susceptibility to Staphylococcus aureus. Host susceptibility factors are suggested by monogenic disorders associated with AD-like phenotypes and can be medically modulated. S. aureus contributes to AD pathogenesis and can be mitigated by antibiotics and bleach baths. Recent work has revealed that the skin microbiome differs significantly between healthy controls and patients with AD, including decreased Gram-negative bacteria in AD. However, little is known about the potential therapeutic benefit of microbiome modulation. To evaluate whether parameters of AD pathogenesis are altered after exposure to different culturable Gram-negative bacteria (CGN) collected from human skin, CGN were collected from healthy controls and patients with AD. Then, effects on cellular and culture-based models of immune, epithelial, and bacterial function were evaluated. Representative strains were evaluated in the MC903 mouse model of AD. We found that CGN taken from healthy volunteers but not from patients with AD were associated with enhanced barrier function, innate immunity activation, and control of S. aureus. Treatment with CGN from healthy controls improved outcomes in a mouse model of AD. These findings suggest that a live-biotherapeutic approach may hold promise for treatment of patients with AD.
Gram-negative bacteria taken from the skin of healthy controls, but not patients with atopic dermatitis, improve atopic dermatitis parameters and models.
Introduction
The term “eczema,” often used to describe atopic dermatitis (AD), was coined in ancient Greece and roughly translates to “to boil out.” Modern science, however, recognizes the contribution of both host and environmental factors to this disease. Hallmarks of the disease include reduced barrier function, reduced innate immune activation, and susceptibility to infections with Staphylococcus aureus. Predisposing host factors are suggested by monogenic mutations in STAT3, filaggrin, and other genes associated with AD-like phenotypes (1). Host genetic influences can be therapeutically modulated through topical steroids or calcineurin inhibitors (2). S. aureus contributes to AD pathogenesis and can be mitigated by antibiotics (2, 3). Recent work has revealed that the skin microbiome is significantly different between healthy controls and patients with AD and that symptoms are associated with a loss of commensal diversity (4). However, it is unclear whether this dysbiosis is causal or could be therapeutically targeted. We found that culturable Gram-negative bacteria (CGN) from healthy controls were associated with activation of innate immunity, enhanced barrier function, and control of S. aureus. These bacteria provided benefit in a mouse model of AD and may hold therapeutic promise for human AD.
Results
Skin culturable Gram-negative microbiota differ between healthy controls and patients with AD.
Genetic-based microbiome identification has revealed that Gram-negative bacteria may represent 10%–50% of the skin microbiome and that significant differences in Gram-negative presence exist between patients with AD and healthy controls (4). Our recently described method for culturing Gram-negative skin flora through appropriate selection of media, antibiotics, and temperature (5) has allowed for functional characterization and comparison of CGN species. We found marked differences in the culturable bacteria present on the skin of 17 patients with the diagnosis of AD as compared with 26 healthy volunteers (HV) (Figure 1A and Supplemental Table 1; supplemental material available online with this article; doi:10.1172/jci.insight.86955DS1). These findings were consistent with published comparisons of the microbiome in HV and patients with AD (4). The predominant Gram-negative species identified by our culture methods in HV was Roseomonas mucosa. This and other Gram-negative bacteria grown by our methods were consistent with what has been identified by prior metagenomic analysis (6). However, incomplete genome characterization of organisms such as R. mucosa in current databases, and pooled analysis across individuals, limited species-level identification and determination of species diversity in a given individual in those metagenomic studies. Full genome sequencing of our cultured isolates will enable more detailed comparison of culturable and metagenomically identified microbiota in future studies. Roughly half of AD patients did not have any CGN, consistent with 16S rRNA data showing diminished abundance of Gram-negative bacteria and reduced bacterial diversity associated with flares of AD (4). We can not rule out that significant age differences in our two groups (32.2 years for HV, 18.5 for AD; Supplemental Table 2) may have contributed to the variation in microbiota, as has been found when contrasting the geriatric population with younger adults (7). However, although our numbers limit statistically valid subgroup analysis, there were no apparent correlations between CGN yield and age, sex, or AD disease severity (SCORAD), indicating that clinical control of disease may not impact presence of CGN (Supplemental Table 1).
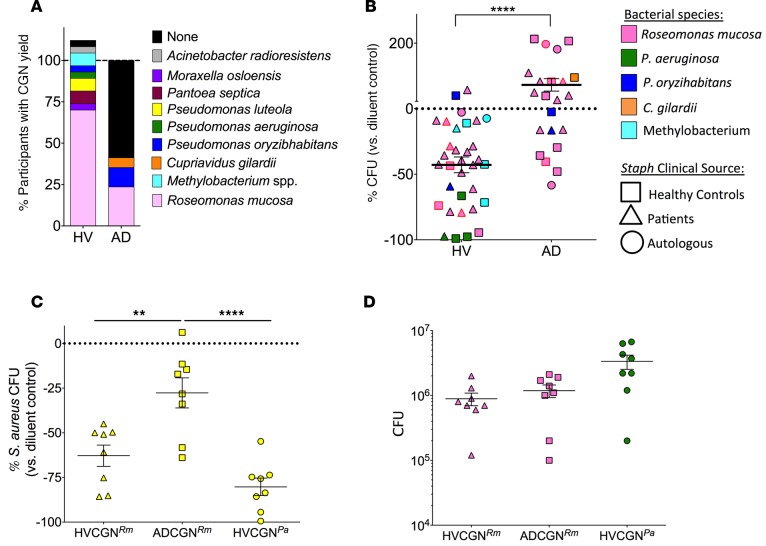
Figure 1. CGN isolates differ in presence and S.
aureusinhibition between healthy volunteers and patients with AD. (A) Percentage of individuals with CGN isolate yield from HV (n = 26) and AD (n = 17) subjects. Individuals with multiple CGN isolates were counted per isolate for >100% total in HV; see Supplemental Table 1 for details. (B) Eight strains of S. aureus isolated from HV and AD patients were grown in the presence of either CGN supernatant or control media. Each data point represents the effect on S. aureus growth of supernatant from one CGN isolate compared with media control (HV isolates = 9, AD isolates = 7); shapes represent sourcing of S. aureus as either the participant’s autologous S. aureus or one S. aureus isolate from HV or AD patients. Data points with red outline represent one HV- and one AD-derived CGN isolate selected for subsequent human challenge and mouse model experiments. Data are alternatively presented by CGN species in Supplemental Figure 1. (C) Healthy mouse ears were coinoculated with S. aureus (strain SAAS9) and R. mucosa from HV or AD patients or P. aeruginosa from a HV for 10 days. Day 12 ears were homogenized and plated by serial dilution for CFU. Percentage change in growth versus diluent (no CGN added) control is shown. (D) CGN CFU yield taken from same mice as in C. Data shown are a combination of 3 or more independent experiments (B) or representative of two independent experiments (C and D) and displayed as mean + SEM. HV, healthy volunteer; AD, atopic dermatitis; CGN, culturable Gram-negative; Rm, R. mucosa; Pa, P. aeruginosa. Significance determined by Student’s t test (B) or ANOVA with Bonferroni’s correction (C and D). **P < 0.01, ****P < 0.0001.
CGN from HV inhibit the growth of S. aureus.
Overgrowth of and infection with S. aureus are both contributors to and consequences of the immune imbalance and poor barrier function characteristic of AD. S. aureus can directly activate allergic mast cells (8, 9) and T cells (10). Treatment with antibiotics can reduce S. aureus burdens and improve symptoms but does not normalize the underlying pathology (2). To evaluate the effect of our CGN strains on S. aureus growth, we cultured 8 different isolates of S. aureus in the presence of the supernatant from cultures of CGN. Depending on the S. aureus strain, our yield after 2.5 hours of culture in the presence or absence of CGN supernatant ranged from 1.6 × 105 to 9 × 107 CFU (data not shown). On average, supernatants from HV-CGN inhibited S. aureus by nearly 50% versus the media control (Figure 1B). However, each CGN isolate supernatant displayed a range of inhibitory effects depending upon the isolate of S. aureus selected for challenge, suggesting a potentially dynamic interaction between these bacterial isolates (Supplemental Figure 1). In contrast, most strains of AD-CGN failed to inhibit S. aureus growth (Figure 1B and Supplemental Figure 1). Reinoculation of S. aureus from the inhibitory CGN supernatants into fresh media allowed normal growth, suggesting bacteriostatic rather than bactericidal activity (data not shown). We next coinoculated mouse ears with S. aureus and one of 3 CGN isolates: an HV-derived R. mucosa, an AD-derived R. mucosa, or an HV-derived Pseudomonas aeruginosa (R. mucosa isolates are indicated by red outlined symbols in Figure 1B). Consistent with our in vitro analysis, coinoculation of CGN and S. aureus on mouse ears reduced S. aureus yields, and this was most pronounced for the HV-derived CGN (Figure 1C) despite a lack of significant differences in yields between the strains of CGN recovered from the ear (Figure 1D).
CGN from HV induce select markers of innate immunity in humans.
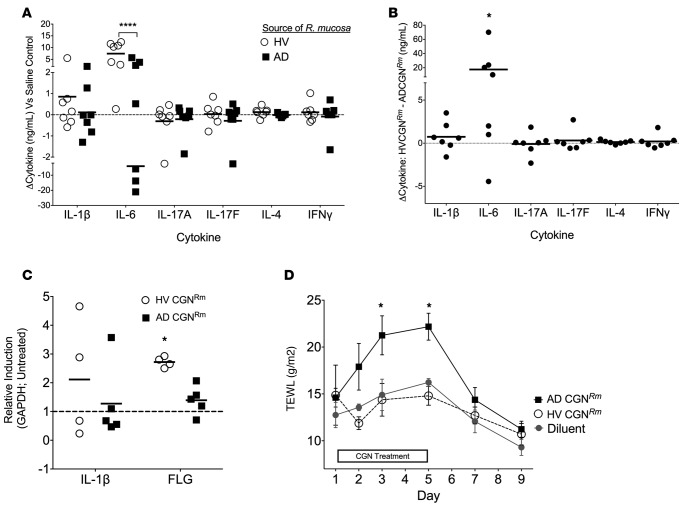
To measure in vivo human cutaneous immune reactivity to these bacteria, we induced suction blisters on the forearms of HV (Supplemental Figure 2A) and removed the epidermal blister roof (Supplemental Figure 2B) similarly to what has been previously described (11). Subjects undergoing blistering included HV1-2, HV19, and HV 21-24 (Supplemental Table 1). We then used challenge chambers (Supplemental Figure 2C) to expose the dermal blister base to lethally irradiated isolates of either an HV-derived or AD-derived R. mucosa, chosen based on their disparate inhibition of S. aureus (Figure 1B; red outline symbols). After 18 to 20 hours of stimulation, blister cytokines levels for IL-6 (Figure 2A) were significantly higher in response to the HV R. mucosa compared with that from a patient with AD (Figure 2A). Three subjects produced less IL-6 in response to the AD-sourced R. mucosa than their saline control (HV1, HV2, and HV22). There were no differences in levels of traditional Th cytokines, such as IL-17, IFN-γ, or IL-4 (Figure 2A), just as there were no significant differences in levels of many other cytokines (Supplemental Figure 3A) or antimicrobial peptides (Supplemental Figure 3B); however, later time points may be necessary to assess adaptive T cell cytokines. Paired comparison of responses to HV R. mucosa and AD R. mucosa within the same subject revealed that 6 of our 7 subjects (all but HV23) responded to the HV R. mucosa with significantly greater IL-6 production compared with that to the AD R. mucosa (Figure 2B). The effect of this differential inflammatory potential on AD pathogenesis remains to be elucidated. Of note, our blister results reflect the response of dermal exposure to the bacterial samples, since the epidermal roof was removed to allow application of the bacteria. Therefore, we next exposed human foreskin epidermis-derived primary keratinocytes (KC) to multiple isolates of live CGN in vitro. Compared with AD-CGN, HV-CGN enhanced mRNA abundance of defensin β4A (Supplemental Figure 4A), the upstream modulators (12, 13) CYP27b1 (a vitamin D converting enzyme; Supplemental Figure 4B), and the vitamin D receptor (Supplemental Figure 4C) as well as CAMP, the gene encoding cathelicidin (Supplemental Figure 4D) even when analysis was limited to R. mucosa strains (Supplemental Figure 4E). There were no differences in transcript levels for pro–IL-1β or IL-6 (Supplemental Figure 4, F and G). CD14, IL-8, TNF-α, TLR2, TLR3, TLR4, TLR9, or thymic stromal lymphopoietin (TSLP) mRNA abundance was also not different, and there was no apparent correlation between the ability of an isolate to inhibit S. aureus and activate KC (data not shown).
Figure 2. CGN affects select innate immune markers and TEWL.
(A) Cytokine analysis for in vivo human blister challenge on healthy controls showing cytokine responses to a representative strain of R. mucosa from a healthy control versus one isolated from a patient with AD as compared with the saline control well (dotted line) (n = 7). (B) Paired analysis of data presented in A showing the cytokine production in the blister well exposed to the HV-sourced R. mucosa minus the paired cytokine production in the blister well exposed to the AD-sourced R. mucosa within in the same human subject. (C) Mouse ears were inoculated daily for 3 days with 1 × 107 CFU R. mucosa from either a HV or a patient with AD. Day 5 mRNA abundance for IL-1β and filaggrin standardized to GAPDH and compared with untreated mice is shown (n = 4–5 mice per group). (D) Mouse backs were shaved and hair was chemically removed on day 0. TEWL was then measured after daily application of 1 × 107 CFU R. mucosa from either a HV or a patient with AD (n = 4–5 mice per group). Data shown are a combination of 7 independent experiments (A and B) or representative of 2 or more independent experiments (C and D) and displayed as mean + SEM (C and D) or mean and individual participants (A and B). HV, healthy volunteer; AD, atopic dermatitis; CGN, culturable Gram-negative; Rm, R. mucosa; FLG, filaggrin; TEWL, transepidermal water loss. Significance from diluent control (or as indicated) determined by ANOVA with Bonferroni’s correction. *P < 0.05, ****P < 0.0001.
Lipid analysis of R. mucosa supernatants suggests possible mediators for S. aureus inhibition.
To begin dissection of the mechanistic basis for the differences in S. aureus inhibition by supernatants from different CGN strains, we incubated the supernatants with 80% ammonium sulfate, which will precipitate most proteins out of solution. The precipitated fraction of supernatants from R. mucosa isolates (HV, n = 4 and AD, n = 4) failed to inhibit S. aureus (strain USA300), whereas the unprecipitated fraction from HV–R. mucosa retained inhibitory effects (Supplemental Figure 5A). This suggests that inhibitory effects on S. aureus were due to a nonprotein product elaborated by R. mucosa. In contrast, inhibitory activity against S. aureus was retained in both the precipitated and unprecipitated fractions after ammonium sulfate treatment of the supernatant from our isolate of P. aeruginosa, suggesting multiple mechanisms of inhibition for this isolate (Supplemental Figure 5A). Extensive genomic and metabolomic studies will be needed to extend these findings and fully characterize the basis of our observed strain differences. However, some possible leads were provided by untargeted liquid chromatography–mass spectrometry that was performed on the supernatant from one inhibitory HV-derived and one noninhibitory AD-derived R. mucosa isolate (isolates identified in Figure 1B with red outline) to screen for relative concentrations of >3,000 lipids. After adjustment for multiple comparisons, this analysis revealed marked differences between these strains for only 3 lipid molecules (Supplemental Table 3). Two cardiolipin family lipids were elevated in the AD–R. mucosa supernatant (78:12, FA 16:0 at 12.1-fold and 78:12, FA 19:1 at 17.6-fold). Only lysophosphatidylcholine (LPC) (number of carbons:number of double bonds;double bond position, 34:5;3) was elevated (7.1-fold) in the HV–R. mucosa supernatant. While little is known about the effect of these classes of lipids on the immune system, some studies suggest that LPC modulates TLR2 and TLR4 (14) and T cell and macrophage activation (15) and is elevated in the lesional skin of patients with psoriasis (16). Cardiolipin has been implicated in inflammasome activation (17) and may impact leukotrienes and T cell adhesion molecules (18). The precise bacterially derived lipids detected are not commercially available; however, LPC isolated from chicken eggs was able to inhibit 3 different S. aureus strains in a dose-dependent manner at picomolar concentrations (Supplemental Figure 5B). In contrast, cardiolipin isolated from bovine heart failed to inhibit S. aureus. Similar concentrations of LPC failed to activate human KC cultures (Supplemental Figure 5C), indicating that LPC may contribute to S. aureus inhibition but does not account for the range of effects we observed with live R. mucosa. Further studies will be needed to confirm the role of LPC and identify other potential factors involved in the anti–S. aureus properties of CGN.
CGN from HV reduces transepidermal water loss in mice.
Transepidermal water loss (TEWL) is a surrogate for measuring barrier function in both mice and humans (19). Increase in TEWL is associated with the dry, itchy skin in AD (20) and increased cutaneous sensitization to antigens (21). For a subset of patients, this barrier defect is associated with dysfunction in the tight-junction protein filaggrin, loss of which has been directly associated with increased TEWL (22–24). Topical application to healthy mouse ears of the representative isolate of HV-CGN chosen based on S. aureus inhibitory effects and used in our human blister studies enhanced transcript levels of filaggrin compared with the AD-CGN isolate (Figure 2C) without any noticeable change in ear erythema or thickness (data not shown). Although further work will be needed to elucidate which of the numerous additional proteins and lipids contributing to skin permeability (25) are affected by our bacterial strains, application of AD-CGN increased TEWL, whereas HV-CGN had no effect (Figure 2D). Taken together, these data suggest that strains of CGN associated with a healthy status induce potentially beneficial immune outcomes related to activation of select vitamin D pathway and innate immunity markers as well as potential enhancement of barrier function.
CGN from HV improve outcomes in a mouse model of AD.
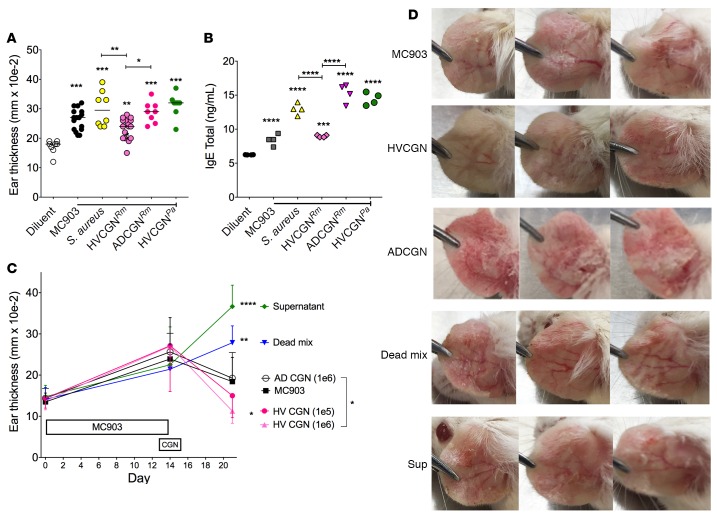
MC903, a vitamin D analog, induces an AD-like dermatitis when applied to mouse ears (26). Similar to human AD, the pathogenesis of the MC903 model depends on interactions between Langerhans and Th2 cells (27, 28) and involves TSLP (29), CCL5, and CCL8 (30). Parameters affected by the MC903 model include visual redness, ear thickness, and induction of serum IgE (31). To test the effect of CGN on development of AD in this model, we applied a HV-sourced R. mucosa, an AD-sourced R. mucosa, or an HV-sourced isolate of P. aeruginosa to mouse ears daily starting 2 days prior and continuing concurrently with MC903 application. Application of these bacteria did not alter MC903-induced ear redness and thickness, but HV-sourced R. mucosa elicited less thickening than AD-sourced R. mucosa (Figure 3A). Application of the AD-sourced R. mucosa also enhanced serum IgE induction, whereas the HV-sourced R. mucosa showed no significant impact (Figure 3B). Consistent with inoculation of healthy mouse ears, transcript levels of filaggrin were significantly lower in MC903-treated mice exposed to AD-sourced R. mucosa (Supplemental Figure 6A).
Figure 3. CGN from HV improves outcomes in the MC903 mouse model of AD-like dermatitis.
(A and B) Both ears of mice were inoculated daily with R. mucosa from a HV or an AD patient or P. aeruginosa from a HV or SAAS9 strain of S. aureus for 2 days prior to MC903 application. Then, bacteria were coapplied with MC903 daily for 13 days. Day 14 ear thickness (A) for each ear and serum total IgE levels (B) are shown; n = 4–8 mice per group. (C and D) Mice were treated with MC903 for 14 days to induce AD-like dermatitis. Starting on day 13, mice were treated daily for 3 days with 1 × 106 CFU of live R. mucosa from a HV or AD patient (HVCGN and ADCGN), 1 × 106 CFU of killed R. mucosa from the same HV resuspended in the supernatant from 3 × 106 CFU of autologous R. mucosa (Dead mix), or supernatant from 1 × 107 CFU of the HV R. mucosa. Day 21 ear thickness (C) and visual redness (D) are shown; n = 3–5 mice per group. Significance versus the diluent group is shown unless otherwise indicated. Data shown are representative of 3 independent experiments and displayed as mean + SEM. HV, healthy volunteer; AD, atopic dermatitis; CGN, culturable Gram-negative; Rm, R. mucosa; Pa, P. aeruginosa. Significance determined by ANOVA with Bonferroni’s correction. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To test the therapeutic potential of CGN, we induced AD-like dermatitis with MC903 and then inoculated the ears with CGN daily for 3 days. Treatment with our same representative isolate of HV–R. mucosa was associated with a reduction in ear thickness and visible redness, whereas AD–R. mucosa did not alter thickness but increased visual redness (Figure 3, C and D). To evaluate whether a live biotherapeutic was necessary or if surface and secreted factors alone could provide similar results, we applied an equivalent CFU concentration of lethally irradiated HV-CGN suspended in its supernatant. Neither this “dead mix” nor the supernatant alone provided any benefit, and each increased ear thickness (Figure 3, C and D). Of note, R. mucosa could be recovered from the mouse ear as far out as 3 weeks after inoculation (either in isolation or after MC903 treatments) (Supplemental Figure 6B), suggesting the potential to colonize mouse ears as well as human skin.
Discussion
Current therapeutic approaches can significantly improve AD symptoms, yet, even when successful, these treatments are not curative and extract a significant emotional toll on patients and their families (32). While numerous insights into the pathogenesis of AD have been discovered, many causal underpinnings remain enigmatic. Genetic host factors implicated in AD pathogenesis include the aforementioned filaggrin as well as polymorphisms in serine protease inhibitor Kazal-type 5 (SPINK5) (33), IL-4 (34), and IL-13 (35). However, the genetic determinants of AD have proven to be highly complex (36). The contribution of S. aureus to the disease process of AD is suggested by the near 100% rate of colonization for patients with AD as compared with only 5% to 20% of controls (37). Many S. aureus virulence factors have been suggested to contribute to AD pathogenesis and have been recently well reviewed (38). However, reduction of S. aureus burden through antibiotics and bleach baths fails to provide lasting relief from AD symptoms (39). More recently, understanding of the topographic distribution, temporal variations, and environmental modulators of the microbiome is rapidly expanding through 16S rRNA DNA and metagenomic approaches (40). The potential for Gram-positive commensal bacteria to influence AD has been suggested by recent findings that Staphylococcus epidermidis can activate TLR2, leading to increased production of antimicrobial peptides, tight-junction proteins, and vitamin D activation (13, 38). Furthermore, S. epidermidis has been shown to produce several factors that can inhibit S. aureus growth and/or biofilm formation (41–43), suggesting an additional protective role for this commensal against the development of AD symptoms.
Our work may further the emerging hypothesis that the dysbiosis seen in AD is not simply an associated finding, but may be a contributor to pathology. We have found that select strains of the Gram-negative bacteria R. mucosa from healthy controls were able to influence hallmarks of AD, including S. aureus growth, TEWL, and certain parameters of innate immune activation. Further work will be needed to generalize these findings to additional isolates of CGN, to elucidate the relative contribution of each of these effects to the observed therapeutic benefit, to investigate whether the clinical utility of R. mucosa involves interactions with other microbiome species, and to characterize how differences in R. mucosa strains differentially drive these mechanisms. Outcomes in the MC903 mouse model of AD differed after inoculation with a R. mucosa strain from a healthy control versus a patient with AD, suggesting therapeutic potential for human disease. However, our strain of P. aeruginosa failed to produce in vivo therapeutic benefit, despite similar effects on KC activation and S. aureus inhibition, suggesting that benefit is not a universal finding of CGN harvested from healthy controls. The therapeutic implications of our findings may be limited, given that our treatment did not affect many central immunologic markers in AD, such as TSLP or Th2 cytokines, despite affecting filaggrin transcript levels. Differential effects on additional induced and genetic models of AD (44) may provide further insight into the beneficial mechanisms of R. mucosa.
Prior reports have suggested that addition of Gram-negative Vitreoscilla filiformis lysates to skin creams may provide benefit in AD treatment (45). However, the loss of efficacy upon killing of our R. mucosa isolate suggests that use of isolated secreted and/or surface products without live commensals may not provide benefit for all bacteria. This dynamic host-commensal interaction may be required for clinical utility of R. mucosa, and the potential for colonization could offer the advantages of limited reapplications and more physiologic pharmacokinetics. Further human studies will be needed to assess the limitations, effects on other microbiota species, and therapeutic potential of R. mucosa and other bacterial strains to normalize skin homeostasis in AD.
Methods
Gram-negative bacterial collection and identification.
Two FloqSwabs (Copan) moistened in sterile PBS (Corning Cellgro) were rubbed on the subject’s skin at the antecubital fossa and volar forearm vigorously for 15 to 30 seconds. For patients with AD, sampling was done at these sites from affected lesional skin if present. One swab was placed into a 15-ml conical tube (Corning Life) with 2 ml sterile HBSS (Sigma-Aldrich) containing vancomycin (300 μg/ml) and amphotericin B (5 μg/ml; Sigma-Aldrich) to inhibit growth of Gram-positive bacteria and fungi. The remaining swab was placed into a 15-ml conical tube containing 2 ml Reasoner’s 2A (R2A) broth (Teknova) with similar concentrations of vancomycin and amphotericin B. The tubes, with swabs left in place, were then incubated at 32°C with constant shaking for 48 to 72 hours before plating 100 μl from each tube onto an R2A agar plate (Remel). Evaluation at earlier time points did not yield additional species (5). Colonies were then taken for species identification by mass spectrometry using MALDI-TOF analysis. Bacterial protein extraction for MALDI-TOF mass spectrometry using the BioTyper (v3.1, Bruker Daltonics Inc.) was performed by the NIH Clinical Center microbiology lab using previously described methods (46), instrument settings, and calibration (47, 48). BioTyper identification was supplemented by additional mass spectra profiles provided by several NIH-developed databases (46, 49, 50). Ten of the healthy control R. mucosa strains reported in Figure 1 were identified based solely on the unique colony morphology, UV light reactivity, and Gram stain characteristics that had been observed in the other R. mucosa isolates identified by MALDI-TOF analysis. Five of the HV–R. mucosa were previously reported (5). All R. mucosa isolates used for subsequent studies were verified by MALDI-TOF analysis. Enumeration of individuals with CGN yield was performed by dividing the number of isolates from the indicated species by the total population (27 for HV and 17 for AD). For individuals with more than one CGN isolate, each isolate was considered separately for a total of >100% in the HV group.
In vitro S.
aureus inhibition assay. Gram-negative isolates were cultured as above in 5 ml of R2A for 8 to 10 days. Bacteria were pelleted at 5,000 g for 12 minutes. Supernatants were harvested, frozen, and then lyophilized (Labconco FreeZone 2.5). The lyophilized product and a control of concurrently incubated and lyophilized R2A without bacteria were suspended in 1 ml of tryptic soy broth (TSB). An overnight culture of S. aureus strains grown in 5 ml TSB was diluted 1:100 into fresh media, and then 100 μl of the newly diluted culture was combined with 100 μl of the TSB containing the CGN supernatant or lyophilized R2A control. Samples were incubated for 3 hours at 37°C under constant agitation. Serial dilutions were performed and plated; the following day colonies were counted and averaged across all countable plates. Percent impact on S. aureus growth was calculated by dividing the number of CFU in the CGN supernatant–exposed culture by the number of CFU in the R2A-only exposed culture. Experiments using LPC (Sigma-Aldrich; item L4129) or cardiolipin (Sigma-Aldrich; item C0563) were performed by incubating S. aureus in lipids resuspended in TSB at indicated concentrations.
Ammonium sulfate precipitation.
R. mucosa isolates from 4 HV and 4 AD were grown for 10 days, along with our HV–P. aeruginosa isolate. Bacteria were pelleted and ammonium sulfate precipitation was performed on the supernatants as previously described (51).
Untargeted liquid chromatography–mass spectrometry.
The representative R. mucosa isolates from HV and AD were grown in quintuplet for 10 days. The bacteria were pelleted and supernatants were lyophilized. They were then sent for untargeted liquid chromatography–mass spectrometry, arranged through sourcing services provided by Sheena Sharma (Science Exchange), and performed by Dayanjan Wijesinghe (Virginia Commonwealth University, Richmond, Virginia, USA) using previously described methodologies (52).
Blister induction and bacterial challenge.
An 8-well suction chamber was designed (Ammnra Creations) and 3D printed in bronze (Shapeways) (Supplemental Figure 1A). This was placed onto the participants’ ethanol-sterilized volar forearms and placed under 30 mm Hg of suction for 2 hours using a microderm abrasion device (Kendal Diamond HB-SF02). After device removal, the resultant epidermal blister roofs were surgically removed. Eight challenge chambers were designed (Ammnra Creations) and 3D printed in bronze (Shapeways) (Supplemental Figure 1C). Using a matching 8-well template (Ammnra Creations), 10-mm punch biopsies (Acu-Punch, Acu-derm) were cut into one 4-inch × 4-inch Duoderm dressing (ConvaTec). The rims of the challenge chambers were brushed with Dermabond adhesive (Ethicon) before being placed over the denuded blisters. Each well was filled with 1 ml of either sterile saline (Corning Life) or 2 × 107 CFU of irradiated bacteria suspended in sterile saline. Blister fluid was removed via pipette (Eppendorf) the following morning. Collected samples were centrifuged at 350 g for 7 minutes, and supernatants were removed and frozen for batch analysis.
Cytokine and antimicrobial peptide detection.
Standards of human reg3gamma (Sino Biologicals) or blister fluid samples (100%–12.5% diluted in 0.9% NaCl) were coated on Nunc Maxisorp plates overnight. The next day, wells were washed 5 times with PBS and blocked in 3% BSA in PBS for 1 hour. Wells were washed once with PBS. Polyclonal, protein A–purified rabbit anti-mouse reg3gamma (13.2 mg/ml; gift from J. Kolls, University of Pittsburgh, Pittsburgh, Pennsylvania, USA) was added at a 1:1,000 dilution in 1% BSA/PBS and incubated 90 minutes at room temperature. Anti-rabbit HRP (Santa Cruz) and TMB substrate (Ebioscience) were added, and the plate was incubated at room temperature for 5 to 10 minutes. 2 N H2SO4 was added to stop the reaction; plates were read at 450 nm. LL-37 (Hycult; HK321-02) and human β-defensin 3 (PeproTech; 900-K2) were measured by commercial ELISA kits. Cytokine levels were determined by Bio-Plex (BIO-Rad) per the manufacturer’s instructions.
KC cultures.
Primary foreskin KC cultures were collected and stimulated as previously described (53). 1 × 107 CFU of live CGN were added to KC culture media. mRNA was extracted after 24 hours for PCR analysis by the ΔΔCT method. Experiments using LPC or cardiolipin were performed by adding the lipids resuspended in media at indicated concentrations and assessed in an identical manner.
Mice.
BALB/c mice were purchased from Taconic Farms. Mice were used between 8 and 14 weeks of age. Experiments were performed in both male and female mice, but age and sex were matched within each experiment. All experiments were done in compliance with the guidelines of the NIAID Institutional Animal Care and Use Committee.
TEWL measurements.
Mice were shaved and hair chemically removed (Nair). Starting the following day, approximately 1 × 107 CFU of live bacteria were placed on the nude areas daily. Immediately prior to inoculation, TEWL was measured daily by VapoMeter (Delfin), per the manufacturer’s instructions.
MC903 and ear inoculations.
Experiments in the MC903 mouse model of AD were performed as previously described (28). For prevention studies, 1 × 107 CFU of Gram-negative bacteria were suspended in sterile PBS and dripped onto the mouse ears in 10 μl of volume. Inoculations were initiated 2 days prior to treatment with MC903 and continued throughout the MC903 exposure. MC903 was placed first, and the ethanol was allowed to evaporate for 2 to 5 minutes prior to placement of bacterial isolates. Ear thickness was measured on day 14. mRNA isolation and PCR were performed as previously described (53). For experiments using coinoculation of S. aureus, 1 × 106 CFU of the SAAS9 strain of S. aureus (gift from C. Nagao, NCI, Bethesda, Maryland, USA) was dripped onto the ear immediately prior to inoculation with the Gram-negative isolate. Treatment studies were performed by exposing mice to MC903 daily for 14 days and inoculating with 1 × 107 CFU of Gram-negative bacteria on days 13 to 15. Ear thickness was measured and photos were taken on day 21.
Serum total IgE analysis.
Serum was collected on day 14 of MC903 treatment. Total IgE was determined as previously described (53) using a commercial kit (Bethyl Laboratories; E90-115).
Statistics.
Means were compared using 2-tailed unpaired t test, or ANOVA for comparison of multiple samples, with Prism software (GraphPad). P values less than 0.05 were considered significant
Study approval.
All animal experiments were conducted under approved National Institute of Allergy and Infectious Disease Office of Animal Care and Use procedures. All human sample collection and processing were performed with approval of the National Institute of Allergy and Infectious Disease IRB, which approved the associated clinical trial (NCT02262819). All subjects gave full consent to sample collection, and all participants provided their written consent to the research protocol.
Author contributions
IAM designed the studies, performed and analyzed experiments, and wrote the manuscript. MLJ performed ammonium sulfate precipitation and assisted on bacterial collection and sample analysis. KWW, NBP, JDR, and DS performed or assisted on bacterial collections and sample analysis. IS performed ELISA for antimicrobial peptides. KDS provided clinical supervision for all patients. SKD oversaw the project and wrote the manuscript. All authors critically reviewed the manuscript.
Supplementary Material
Acknowledgments
We would like to acknowledge the participants for their assistance in this project. We thank Karen Frank, Frida Stock, Julia Shah, Elim Cho, Bevin Feutrier, Pam Welch, and Dirk Darnell for technical and administrative assistance. This work was supported by the Intramural Research Program of the NIH and the National Institute of Allergy and Infectious Diseases.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2016;1(10):e86955. doi:10.1172/jci.insight.86955.
Contributor Information
Inka Sastalla, Email: sastallai@niaid.nih.gov.
Danial Saleem, Email: danial.saleem@nih.gov.
References
- 1.Lyons JJ, Milner JD, Stone KD. Atopic dermatitis in children: clinical features, pathophysiology, and treatment. Immunol Allergy Clin North Am. 2015;35(1):161–183. doi: 10.1016/j.iac.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boguniewicz M, Leung DY. The abc’s of managing patients with severe atopic dermatitis. J Allergy Clin Immunol. 2013;132(2):511–2.e5. doi: 10.1016/j.jaci.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi T, et al. Dysbiosis and staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity. 2015;42(4):756–766. doi: 10.1016/j.immuni.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong HH, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myles IA, Reckhow JD, Williams KW, Sastalla I, Frank KM, Datta SK. A method for culturing Gram-negative skin microbiota. BMC Microbiol. 2016;16(1):e86955. doi: 10.1186/s12866-016-0684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh J, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514(7520):59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlievert PM, Strandberg KL, Lin YC, Peterson ML, Leung DY. Secreted virulence factor comparison between methicillin-resistant and methicillin-sensitive Staphylococcus aureus, and its relevance to atopic dermatitis. J Allergy Clin Immunol. 2010;125(1):39–49. doi: 10.1016/j.jaci.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura Y, et al. Staphylococcus -toxin induces allergic skin disease by activating mast cells. Nature. 2013;503(7476):397–401. doi: 10.1038/nature12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brauweiler AM, Goleva E, Leung DY. Th2 cytokines increase Staphylococcus aureus alpha toxin-induced keratinocyte death through the signal transducer and activator of transcription 6 (STAT6) J Invest Dermatol. 2014;134(8):2114–2121. doi: 10.1038/jid.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Follin P, Dahlgren C. A skin chamber technique as a human model for studies of aseptic inflammatory reactions. Methods Mol Biol. 2007;412:333–346. doi: 10.1007/978-1-59745-467-4_22. [DOI] [PubMed] [Google Scholar]
- 12.Miller J, Gallo RL. Vitamin D and innate immunity. Dermatol Ther. 2010;23(1):13–22. doi: 10.1111/j.1529-8019.2009.01287.x. [DOI] [PubMed] [Google Scholar]
- 13.Schauber J, Gallo RL. The vitamin D pathway: a new target for control of the skin’s immune response? Exp Dermatol. 2008;17(8):633–639. doi: 10.1111/j.1600-0625.2008.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carneiro AB, et al. Lysophosphatidylcholine triggers TLR2- and TLR4-mediated signaling pathways but counteracts LPS-induced NO synthesis in peritoneal macrophages by inhibiting NF-κB translocation and MAPK/ERK phosphorylation. PLoS One. 2013;8(9):e86955. doi: 10.1371/journal.pone.0076233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabarowski JH, Xu Y, Witte ON. Lysophosphatidylcholine as a ligand for immunoregulation. Biochem Pharmacol. 2002;64(2):161–167. doi: 10.1016/S0006-2952(02)01179-6. [DOI] [PubMed] [Google Scholar]
- 16.Ryborg AK, Grøn B, Kragballe K. Increased lysophosphatidylcholine content in lesional psoriatic skin. Br J Dermatol. 1995;133(3):398–402. doi: 10.1111/j.1365-2133.1995.tb02667.x. [DOI] [PubMed] [Google Scholar]
- 17.Iyer SS, et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39(2):311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan M, et al. Oxidized but not native cardiolipin has pro-inflammatory effects, which are inhibited by Annexin A5. Atherosclerosis. 2014;235(2):592–598. doi: 10.1016/j.atherosclerosis.2014.05.913. [DOI] [PubMed] [Google Scholar]
- 19.Fluhr JW, Feingold KR, Elias PM. Transepidermal water loss reflects permeability barrier status: validation in human and rodent in vivo and ex vivo models. Exp Dermatol. 2006;15(7):483–492. doi: 10.1111/j.1600-0625.2006.00437.x. [DOI] [PubMed] [Google Scholar]
- 20.Boguniewicz M, Leung DY. Recent insights into atopic dermatitis and implications for management of infectious complications. J Allergy Clin Immunol. 2010;125(1):4–13. doi: 10.1016/j.jaci.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pyun BY. Natural history and risk factors of atopic dermatitis in children. Allergy Asthma Immunol Res. 2015;7(2):101–105. doi: 10.4168/aair.2015.7.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelleher M, et al. Skin barrier dysfunction measured by transepidermal water loss at 2 days and 2 months predates and predicts atopic dermatitis at 1 year. J Allergy Clin Immunol. 2015;135(4):930–5.e1. doi: 10.1016/j.jaci.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Bantz SK, Zhu Z, Zheng T. The atopic march: progression from atopic dermatitis to allergic rhinitis and asthma. J Clin Cell Immunol. 2014;5:e86955. doi: 10.4172/2155-9899.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hon KL, Leung AK, Barankin B. Barrier repair therapy in atopic dermatitis: an overview. Am J Clin Dermatol. 2013;14(5):389–399. doi: 10.1007/s40257-013-0033-9. [DOI] [PubMed] [Google Scholar]
- 25.Jensen JM, Proksch E. The skin’s barrier. G Ital Dermatol Venereol. 2009;144(6):689–700. [PubMed] [Google Scholar]
- 26.Leyva-Castillo JM, Hener P, Jiang H, Li M. TSLP produced by keratinocytes promotes allergen sensitization through skin and thereby triggers atopic march in mice. J Invest Dermatol. 2013;133(1):154–163. doi: 10.1038/jid.2012.239. [DOI] [PubMed] [Google Scholar]
- 27.Elentner A, et al. Langerhans cells are critical in the development of atopic dermatitis-like inflammation and symptoms in mice. J Cell Mol Med. 2009;13(8B):2658–2672. doi: 10.1111/j.1582-4934.2009.00797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Du J, Zhu J, Yang X, Zhou B. Thymic stromal lymphopoietin signaling in CD4(+) T cells is required for TH2 memory. J Allergy Clin Immunol. 2015;135(3):781–91.e3. doi: 10.1016/j.jaci.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M, Hener P, Zhang Z, Ganti KP, Metzger D, Chambon P. Induction of thymic stromal lymphopoietin expression in keratinocytes is necessary for generating an atopic dermatitis upon application of the active vitamin D3 analogue MC903 on mouse skin. J Invest Dermatol. 2009;129(2):498–502. doi: 10.1038/jid.2008.232. [DOI] [PubMed] [Google Scholar]
- 30.Rebane A, et al. MicroRNA-146a alleviates chronic skin inflammation in atopic dermatitis through suppression of innate immune responses in keratinocytes. J Allergy Clin Immunol. 2014;134(4):836–847.e11. doi: 10.1016/j.jaci.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 31.Li M, Hener P, Zhang Z, Kato S, Metzger D, Chambon P. Topical vitamin D3 and low-calcemic analogs induce thymic stromal lymphopoietin in mouse keratinocytes and trigger an atopic dermatitis. Proc Natl Acad Sci U S A. 2006;103(31):11736–11741. doi: 10.1073/pnas.0604575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis-Jones S. Quality of life and childhood atopic dermatitis: the misery of living with childhood eczema. Int J Clin Pract. 2006;60(8):984–992. doi: 10.1111/j.1742-1241.2006.01047.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhao LP, et al. Association of SPINK5 gene polymorphisms with atopic dermatitis in Northeast China. J Eur Acad Dermatol Venereol. 2012;26(5):572–577. doi: 10.1111/j.1468-3083.2011.04120.x. [DOI] [PubMed] [Google Scholar]
- 34.Kawashima T, et al. Linkage and association of an interleukin 4 gene polymorphism with atopic dermatitis in Japanese families. J Med Genet. 1998;35(6):502–504. doi: 10.1136/jmg.35.6.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsunemi Y, et al. Interleukin-13 gene polymorphism G4257A is associated with atopic dermatitis in Japanese patients. J Dermatol Sci. 2002;30(2):100–107. doi: 10.1016/S0923-1811(02)00065-8. [DOI] [PubMed] [Google Scholar]
- 36.Bieber T. Atopic dermatitis. N Engl J Med. 2008;358(14):1483–1494. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 37.Gong JQ, et al. Skin colonization by Staphylococcus aureus in patients with eczema and atopic dermatitis and relevant combined topical therapy: a double-blind multicentre randomized controlled trial. Br J Dermatol. 2006;155(4):680–687. doi: 10.1111/j.1365-2133.2006.07410.x. [DOI] [PubMed] [Google Scholar]
- 38.Williams MR, Gallo RL. The role of the skin microbiome in atopic dermatitis. Curr Allergy Asthma Rep. 2015;15(11):e86955. doi: 10.1007/s11882-015-0567-4. [DOI] [PubMed] [Google Scholar]
- 39.Gilani SJ, Gonzalez M, Hussain I, Finlay AY, Patel GK. Staphylococcus aureus re-colonization in atopic dermatitis: beyond the skin. Clin Exp Dermatol. 2005;30(1):10–13. doi: 10.1111/j.1365-2230.2004.01679.x. [DOI] [PubMed] [Google Scholar]
- 40.Tsai YC, et al. Resolving the complexity of human skin metagenomes using single-molecule sequencing. MBio. 2016;7(1):e01948–e01915. doi: 10.1128/mBio.01948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otto M. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol. 2010;5(2):183–195. doi: 10.1586/edm.10.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugimoto S, et al. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J Bacteriol. 2013;195(8):1645–1655. doi: 10.1128/JB.01672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cogen AL, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J Invest Dermatol. 2010;130(1):192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin H, He R, Oyoshi M, Geha RS. Animal models of atopic dermatitis. J Invest Dermatol. 2009;129(1):31–40. doi: 10.1038/jid.2008.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gueniche A, et al. Effects of nonpathogenic gram-negative bacterium Vitreoscilla filiformis lysate on atopic dermatitis: a prospective, randomized, double-blind, placebo-controlled clinical study. Br J Dermatol. 2008;159(6):1357–1363. doi: 10.1111/j.1365-2133.2008.08836.x. [DOI] [PubMed] [Google Scholar]
- 46.Lau AF, et al. A rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 2014;52(8):2804–2812. doi: 10.1128/JCM.00694-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lau AF, Drake SK, Calhoun LB, Henderson CM, Zelazny AM. Development of a clinically comprehensive database and a simple procedure for identification of molds from solid media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51(3):828–834. doi: 10.1128/JCM.02852-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youn JH, Drake SK, Weingarten RA, Frank KM, Dekker JP, Lau AF. Clinical performance of a matrix-assisted laser desorption ionization-time of flight mass spectrometry method for detection of certain blakpc-containing plasmids. J Clin Microbiol. 2016;54(1):35–42. doi: 10.1128/JCM.01643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevenson LG, Drake SK, Shea YR, Zelazny AM, Murray PR. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically important yeast species. J Clin Microbiol. 2010;48(10):3482–3486. doi: 10.1128/JCM.00687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saleeb PG, Drake SK, Murray PR, Zelazny AM. Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011;49(5):1790–1794. doi: 10.1128/JCM.02135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.[No authors listed] Protein precipitation using ammonium sulfate. Curr Protoc Protein Sci. 2016;84:e86955. doi: 10.1002/0471140864.psa03fs84. [DOI] [PubMed] [Google Scholar]
- 52.Wijesinghe DS, Chalfant CE. Systems-level lipid analysis methodologies for qualitative and quantitative investigation of lipid signaling events during wound healing. Adv Wound Care (New Rochelle) 2013;2(9):538–548. doi: 10.1089/wound.2012.0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myles IA, et al. Signaling via the IL-20 receptor inhibits cutaneous production of IL-1 and IL-17A to promote infection with methicillin-resistant Staphylococcus aureus. Nat Immunol. 2013;14(8):804–811. doi: 10.1038/ni.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.